Abstract
Over the past years, numerous methods have emerged to automate the quantification of animal behavior at a resolution not previously imaginable. This has opened up a new field of computational ethology and will, in the near future, make it possible to quantify in near completeness what an animal is doing as it navigates its environment. The importance of improving the techniques with which we characterize behavior is reflected in the emerging recognition that understanding behavior is an essential (or even prerequisite) step to pursuing neuroscience questions. The use of these methods, however, is not limited to studying behavior in the wild or in strictly ethological settings. Modern tools for behavioral quantification can be applied to the full gamut of approaches that have historically been used to link brain to behavior, from psychophysics to cognitive tasks, augmenting those measurements with rich descriptions of how animals navigate those tasks. Here we review recent technical advances in quantifying behavior, particularly in methods for tracking animal motion and characterizing the structure of those dynamics. We discuss open challenges that remain for behavioral quantification and highlight promising future directions, with a strong emphasis on emerging approaches in deep learning, the core technology that has enabled the markedly rapid pace of progress of this field. We then discuss how quantitative descriptions of behavior can be leveraged to connect brain activity with animal movements, with the ultimate goal of resolving the relationship between neural circuits, cognitive processes and behavior.
Tracking, from coarse to fine
Quantitative descriptions of behavior begin by tracking movements. In this section we describe the computational tools for extracting measurements of animal motion from video recordings and the challenges associated with capturing progressively more detailed descriptions such as pose (Fig. 1a).
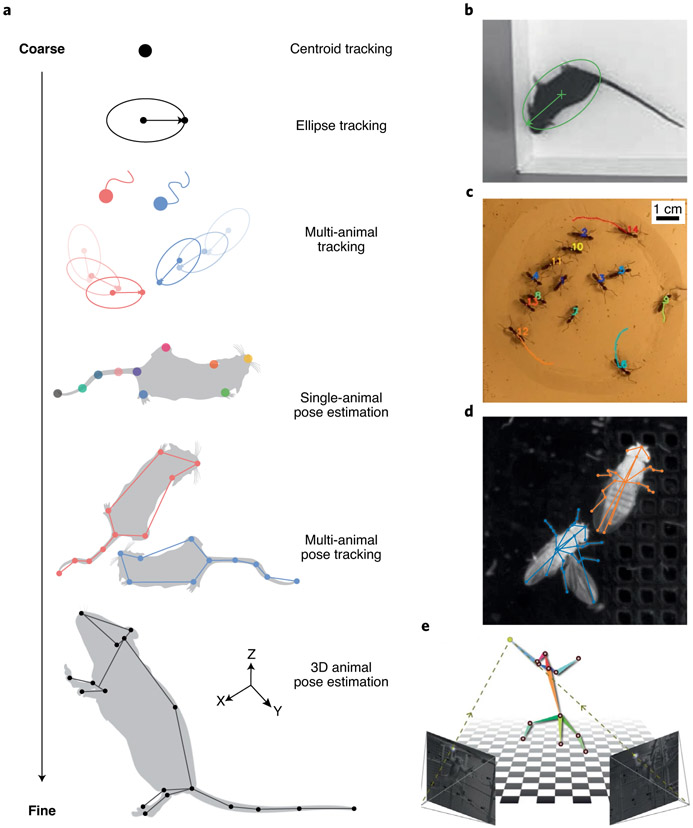
Fig. 1 ∣. Tracking, from coarse to fine.
a, Representations extracted by different forms of tracking, ranging from a single point to full 3D pose. b, Single mouse tracked with ellipse and orientation2. c, Multi-animal tracking of ants with reliable identity assignment14. d, Multi-animal pose tracking of a pair of socially interacting fruit flies31. e, 3D pose estimation of a monkey from multiple camera views46. Images in a adapted with permission from SciDraw.io119 or created with BioRender.com120; in b from ref. 2, Nature Publishing Group; in c from ref. 14, Nature Publishing Group; in e from ref. 46, Nature Publishing Group.
Animal centroids, ellipses and identities.
At its coarsest, animal behavior can be quantified by estimating the position of its centroid (i.e., the midpoint or the center of mass) over time. These centroid trajectories, quantified as sequences of image coordinates, reflect the motion of an animal within its environment and can be used to measure spatial navigation or locomotion behavior. The centroid treats the animal as a single point, which fails to capture its heading, but this description can be augmented by finding the major and minor axes of an ellipse encircling the animal (Fig. 1b). This is a conveniently universal description, as most animals with a CNS share a similar body plan, in which a spinal or ventral nerve cord forms a line at the center of an elongated body.
Classical approaches to estimating centroids and ellipses primarily relied on background subtraction, an algorithm that identifies the image pixels belonging to the animal (i.e., the foreground) from which the centroid can be computed by finding the midpoint of their coordinates. When the background contrasts with the animal, such as in backlit arenas, background subtraction can be performed through simple thresholding of the image intensity. If the background is static, it can be modeled by finding the median image frame; however, this fails often if the animal does not move for prolonged periods of time. Classical approaches employ robust algorithms to model the background1, but newer methods have begun to use deep learning to better deal with more complex backgrounds, affording the ability to track animals in more naturalistic conditions2.
Extending ellipse tracking to multiple animals adds even more richness to behavioral descriptions, where quantities such as relative distances and orientations can be used to infer complex social interactions. For example, close interactions that occur during aggression or courtship may be detectable using the distance between centroids, while the relative angle between animals can indicate the directedness of the behavior, such as chasing.
Assuming centroids or ellipses can be detected reliably within individual images, the multi-animal setting introduces the particularly challenging problem of estimating identity, i.e., the task of associating animal detections correctly over time (Fig. 1c). In the broader domain of multi-object tracking, the most common approach to the identity assignment problem is tracking by detection, in which objects (for example, animals) are detected within single images and then subsequently linked together across frames. These algorithms must contend with challenging cases such as objects occluding one another, disappearing for periods of time or failing to be detected in the first stage. We refer the reader to previous reviews for more in-depth analyses of classical multi-animal tracking methodology3-5.
Modern approaches to multi-animal tracking typically address the association problem by modeling the appearance and/or motion of the animals. This allows for identity association by matching new detections to previous tracks based on their similarity to the modeled features. Motion models will typically make constant velocity assumptions to enable extrapolation from past trajectories6. These types of models will often fail when animals are closely (socially) interacting with each other, as is common in multi-animal experiments. Techniques that leverage machine learning to model appearance may rely on artificial distinguishing visual features, such as painting the animals’ fur with unique patterns7 or using a different colored tag for each animal8. Some approaches may rely on combining videography with implanted radio-frequency identification (RFID) tags, enabling reliable and wireless identification for highly robust tracking, making them particularly well-suited for monitoring behavior over longer timescales9. These potentially invasive manipulations, however, may be prohibitively laborious when using many animals and may hinder the ethological validity of experiments intended to measure natural behavior10.
Although identity association in multi-object tracking remains an open problem, the state-of-the-art techniques now rely on deep learning for learning distinguishable appearance features without artificial markers11. A common approach is to employ a technique known as contrastive learning: the objective is to find a mapping in which images with the same identity are closer to each other than to images with different identities; this is the basis of modern facial recognition systems12 and has also been applied to animal facial recognition13. In the domain of animal tracking, this approach has been demonstrated to be highly effective with socially behaving animals in complex environments14,15. Despite their impressive performance, the downside to these methods is that they typically require more training data to adapt to new animals, new imaging conditions and new experimental settings. This requirement is particularly burdensome, because manual annotation of animal identities over video frames can be prohibitively laborious as it may require annotators to step through the video frame-by-frame to ensure they do not mislabel animals when they are closely interacting. Some approaches to ameliorate this bootstrap the labeling using classical tracking for self-supervised learning14, and advances in unsupervised learning may soon enable fully automated deep-learning-based tracking without previous annotations16.
Animal pose estimation.
Centroid and ellipse tracking, though highly descriptive, fail to capture the movements of limbs and appendages and consequently cannot be used to detect behaviors such as grooming, rearing, tapping and locomotor coordination. Animal pose, on the other hand, is represented by the location of all of its body-part landmarks (typically at the skeleton joints). Pose estimation is able to capture nearly all of the degrees of freedom of body motion—and by extension, the degrees of freedom that the brain can actuate via its motor system.
Human pose estimation has long been studied, both from the perspective of biological motion perception17,18, i.e., how humans and animals perceive the motion of other organisms, and from the engineering point of view, i.e., the design of algorithms to retrieve pose19. While the former provides a theoretical grounding for the use of pose as a biologically relevant representation of behavior—particularly in social contexts—the latter has enabled accurate and automated pose estimation from conventional videography.
State-of-the-art deep-learning-based approaches to human pose estimation have drastically improved accuracy over classical methods, owing to the effectiveness of neural networks for computer vision tasks in general, but more specifically due to the development of the heatmap (also referred to as confidence map; Fig. 2a) representation of landmark locations20-22. This representation encodes the location of each landmark as the density function of a two-dimensional (2D) Gaussian distribution centered on the ground-truth image coordinates of each landmark, i.e., a heatmap image in which the brightest pixel is at the location of the landmark within the image. This representation is particularly well-suited for convolutional neural networks (CNNs), which excel at learning complex transformations of image patches. Pose estimation CNNs are trained to predict heatmaps from input images by learning from labeled examples where the ground truth landmark coordinates are known, enabling the correct heatmaps to be computed for comparison with the CNN’s prediction. Once trained, the landmark coordinates on unlabeled images can be decoded from the predicted heatmaps via peak detection.
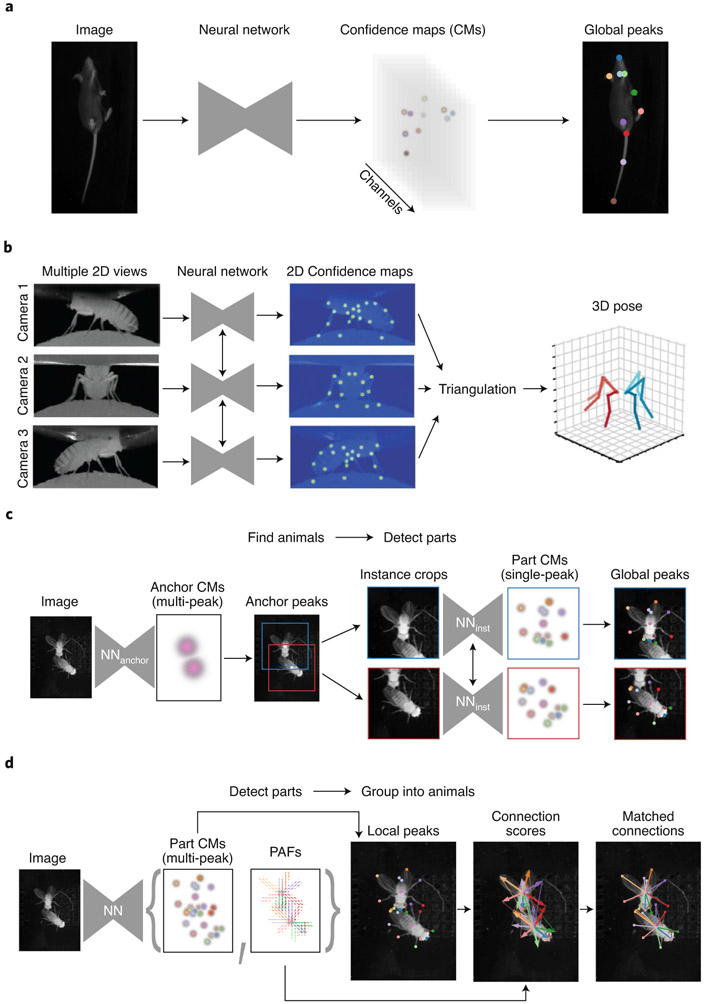
Fig. 2 ∣. Anatomy of pose estimation systems.
a, In single-instance pose estimation32, each body-part type is encoded as a confidence map that is predicted by a convolutional neural network given an image as input (left). The network is trained to predict confidence maps (CMs) with only a single peak per channel (middle), enabling the coordinates to be decoded by finding the global peak in each channel of the confidence maps (right). b, The 3D pose estimation system employed in DeepFly3D (ref. 45). These systems may use a single neural network (left) to predict 2D confidence maps (middle) for each independent view. These landmarks are then triangulated based on the geometry of the cameras and the consistency of the 2D predictions (right). c, A top-down multi-animal pose estimation system employed in SLEAP31. All instances of an ‘anchor part’ are first located by a CNN trained to predict anchor part confidence maps (left). The image is cropped around each anchor (middle) and a CNN trained to predict all part confidence maps is applied to each crop (right). d, A bottom-up multi-animal pose estimation system employed in SLEAP31. A single neural network detects all instances of all body parts and simultaneously predicts part affinity fields (PAFs)58, a representation of the connectivity between body parts (left). The grouping of body parts to the appropriate animals via a matching procedure uses the PAFs to score candidate connections (right). Images in b adapted with permission from ref. 45, eLife.
Although conceptually there is no difference between human and animal heatmaps, the biggest challenge to adapting the deep-learning-based approach to animal pose estimation is the need for labeled training data. To enable human pose estimation ‘in the wild’ that generalizes to arbitrary viewpoints, illumination, body sizes, clothing and other variability in image features, the computer vision community has generated datasets with sizes ranging from tens of thousands to millions of labeled images23-25. Efforts to generate these employed crowdsourcing and required thousands of hours of manual labor, but these costs are amortized over time, as human anatomy is static. In contrast, a CNN that is trained to locate human hands would not be able to generalize to predict insect leg tips.
To address the labeling problem, three main approaches have been employed to enable pose estimation for new animals when no training data is available: transfer learning and efficient neural network design. The first of these, transfer learning, formed the basis and main contribution of the widely used animal pose estimation software DeepLabCut26. Transfer learning is a widely used method for reducing the need for large datasets; it works by reusing the parameters (and therefore visual feature detectors) learned in CNNs trained on a broader set of natural images (typically ImageNet27). This relies on the assumption that reducing the need for learning general-purpose visual features, such as oriented edges and textured patches, will facilitate fine-tuning the parameters of the network with less training data. This approach is a topic of active research in computer vision, and recent empirical studies have reported conflicting results on its advantages for general computer vision tasks28,29 and for animal pose estimation30,31.
In contrast, the second approach formed the primary contributions of the LEAP software framework32 and was improved upon in DeepPoseKit33. Efficient neural network design, in which the CNN architecture is kept small, has fewer parameters to tune than the general-purpose architectures normally used in transfer learning34. This reduction is justified by the assumption that variability of imaging conditions in animal behavioral data is relatively low—a foundational feature of reproducible laboratory experimental design—and therefore requires lower representational capacity. The added benefit of designing neural networks to the needs of the data is that it is considerably faster to train and predict, making human-in-the-loop training more efficient, but this approach may not be as well suited to the prediction of animal pose in less constrained, non-laboratory settings.
Active learning, closely related to human-in-the-loop training35, is a technique that can drastically reduce the time required for generating large datasets by proposing images to label that are representative of diverse data features; these can then be used to train CNNs with a small number of very distinct images. After training with few labels and generating predictions on unlabeled data, new labels can be generated by simply correcting the predictions, iteratively reducing the time required to label with every training loop. This approach has been adopted by all major animal pose estimation frameworks to reduce the effort required to label new datasets.
The success and accessibility of these methods have spurred a revolution in fields dealing with animal movement, from neuroscience36 to ecology37. Future research in animal pose estimation will continue to reduce the need for labeled data through the use of techniques such as self-supervised learning and domain adaptation38-41 and to improve the precision of landmark localization by incorporating temporal information without new labels42,43.
Animal pose estimation in three dimensions.
For many animals, 2D pose estimation will be insufficient to capture all body landmarks, as some will typically be occluded in any single fixed viewpoint. This issue is especially compounded in highly deformable animals such as mice whose skeleton landmarks may be difficult to localize through fur44 and during out-of-plane behaviors such as rearing. The standard approach to three-dimensional (3D) pose estimation, then, is to record behavior using multiple cameras arranged such that they collectively capture all of the landmarks of interest across viewpoints (Fig. 1e).
The standard approach to 3D pose estimation consists of three steps: 2D pose estimation, triangulation and refinement (Fig. 2b). The 2D pose estimation step is typically performed as described for the monocular case, but 3D animal pose estimation frameworks will often leverage the ability to map points from one view to another to reduce the labeling effort45,46. The triangulation step is preceded by a one-time calibration procedure, in which an object with distinct features (typically a checkerboard-like pattern47) is used to compute the camera calibration matrices that enable projection of 2D points in the image plane to consistent 3D world coordinates. In practice, triangulation is noisy, so some form of refinement is typically employed to eliminate false detections and resolve inconsistencies in projections of the same point from different views. Tools for 3D animal pose estimation have employed a variety of approaches to refinement, such as incorporating constraints on the geometry of the animal’s body (for example, limb lengths)45,46, temporal smoothing48 or parametric shape modeling49,50. The rapid pace of progress in 3D animal pose estimation is likely to yield a series of advances in the coming years; the 3D human pose estimation field provides an outlook on what’s to come, in particular, lifting from monocular 2D51, which will reduce the technical challenges associated with multicamera animal behavioral setups52.
While 3D landmark localization helps to improve the completeness of behavioral representations, it may still fail to capture movement of non-rigid parts of the body, particularly in animals with amorphous body shapes, such as hydra53. One approach to capturing the full shape information of animals is to fit articulated 3D models of animals to new images39,40,54. This model-based technique can be robust and may require relatively little training data, but comes at the cost of professional 3D computer-aided design (CAD) expertise to design and articulate (‘rig’) models for new animal body types, and it would not be robust to large deformations such as in experiments that employ amputations of body parts. A more general approach is to explicitly fit a deformable surface to capture the detailed shape of animals within images; although this is an active area of research in computer vision, early results hold promise55-57 for development of general-purpose tools to enable the routine use of animal shape estimation for behavioral quantification.
Multi-animal pose tracking.
The level of description afforded by pose estimation is uniquely advantageous when quantifying social behaviors, as relative features such as inter-body-part distances and orientations can be used to detect directed interactions between animals (Fig. 1d). Just as in multi-animal centroid and ellipse tracking, however, extending pose estimation to multiple animals introduces new technical challenges. First, part association: dealing with multiple animals with the same body plan means that sets of detected landmarks must be correctly assigned to each animal. Second, identity assignment: detections in one frame must be correctly associated with detections belonging to the same animal in subsequent frames. Approaches to multi-instance pose estimation (in both humans and animals) can generally be categorized as either ‘top-down’, in which the animals are first detected (for example, by finding their centroids) and then their body parts are located within a cropped image of the animal, or ‘bottom-up’, in which all body parts first located and then grouped by animal.
Top-down pose estimation systems (Fig. 2c) solve the part-association problem implicitly by using the location of image features relative to the center of the crop. In this approach individual animals are first detected within the frame, such as by standard centroid detection methods (see “Animals, centroids, ellipses and identities” section above) or by a neural network trained to generate region proposals. These regions are cropped such that the animal is centered within the image. Given these crops, the neural network responsible for part localization is trained to predict confidence maps with a single peak, just as in the single-instance case, even if body parts of other animals are present within the crop.
In contrast, bottom-up pose estimation systems (Fig. 2d) encode all instances of each body part in the same set of confidence maps and encode their connectivity or grouping separately. A commonly used representation of the connectivity between body parts are part affinity fields58, which are composed of vectors whose orientation follow the direction of the animal’s skeleton within the image. A grouping procedure uses the similarity between the line integral of part affinity field vectors between body parts and the line segment between the two points as a scoring function for grouping pairs of body part detections.
While there is a variety of theoretical trade-offs between the two approaches, empirical studies indicate that selecting the appropriate one may depend on features specific to the dataset, such as the morphology of the animals and the relative scale of their image features31. See Box 1 for more details on these and other considerations for practitioners.
Box 1 ∣. Choosing a tracking system.
Selecting the appropriate behavioral quantification tool is an important decision that should be made at the experimental design stage. Some types of experiments may preclude some forms of tracking altogether, for example, multi-camera setups. Constraints on the data acquisition, such as resolution, illumination and arena size should be balanced with the desire to get as much information out of the behavioral recordings as possible.
The primary consideration is the level of description necessary for capturing the behavioral signals that best test a given hypothesis. Centroid and orientation tracking may suffice, for example, to obtain coarse locomotor statistics, information about navigational strategies, place preference or characteristics of some social interactions. For single animal tracking, a classical tracker such as Ctrax1 or ToxTrac6 may be a practical solution because additional labeling is not required. If imaging conditions are not optimal, for example, due to low illumination or complex backgrounds, it may be preferable to use a deep-learning-based solution even for centroid tracking2. When tracking multiple animals, addressing the identity assignment problem will be the primary challenge in ensuring the quality of the tracking. If the animals frequently occlude one another or disappear from the field of view, it may be necessary to employ a deep-learning-based multi-animal tracker such as idtracker.ai (https://idtrackerai.readthedocs.io/en/latest/) to minimize identity switches14.
If a pose estimation system is optimal, it is important to consider the resolution required to capture the smallest feature of an animal. For insects with thin appendages, high spatial resolution is required, which must be balanced with the size of the field of view; for rodents, a lower resolution may suffice, but note that some body parts like the tail may require additional considerations if imaging against a low-contrast background. Next, consider the viewpoint of the camera. For ideal 2D pose estimation, movements out of the image plane should be minimized, which is most commonly achieved by carefully aligned cameras either overhead or from below a transparent floor of an elevated arena. For immobilized animals, higher resolution may be more easily achieved, but it may be difficult to get a viewpoint where movements are within the image plane. Finally, ensure that the signal-to-noise ratio of the images is maximized by providing sufficient, constant and uniform illumination (potentially in infrared to reduce corruption by room light), focused camera optics to reduce spatial blur and low camera exposure time to reduce temporal blur.
In the majority of cases, any of the mature animal-pose-estimation frameworks (for example,DeepLabCut26, SLEAP31 and DeepPoseKit33) should produce satisfactory results and can be configured to achieve similar speed and accuracy. All of these frameworks provide a shared base set of features: graphical user interfaces (GUI) for labeling and inspecting results, usage documentation with example data, active learning, multiple neural network architectures with optional pretraining for transfer learning, and Python-based implementations. Their differences derive primarily from their capacity for customization and extension for specialized applications. DeepLabCut has a large user base that has adapted it to a diverse range of specialized applications; DeepPoseKit provides a minimalist implementation of the core functionality required to build a pose estimation system; and SLEAP provides a flexible codebase, developed with a standardized code style and documentation formats, continuous integration, and native multiplatform support and modular application programming interfaces to facilitate extensibility and customization for specialized applications.
For multi-animal tracking, all three frameworks support top-down approaches; a recent update to DeepLabCut provides experimental support for bottom-up, while SLEAP offers native support for both. MARS121 provides an alternative solution for tracking rodents with different fur colors and is integrated with tools for advanced downstream analyses. The SLEAP GUI offers functionality for proofreading tracking, as it was designed with special emphasis on multi-animal pose tracking.
When extending to 3D, it is important to first consider how many cameras may be necessary and to ensure that they are synchronized and calibrated. For larger animals, such as non-human primates that can occupy large spaces, OpenMonkeyStudio46 describes a state-of-the-art system using an array of 62 cameras. For smaller animals, DeepFly3D45 and Anipose48 provide flexible general-purpose toolkits to deal with calibration and triangulation of 2D landmarks. At the cutting edge, the 3D-from-2D pose estimation framework LiftPose3D52 may be suitable for settings in which multicamera triangulation is not possible.
In the multi-animal context, the problem of pose estimation is naturally promoted to that of pose tracking due to the second problem: identity assignment. The primary challenge in identity assignment with multi-animal pose tracking, as opposed to multi-animal centroid tracking, is the considerably increased manual annotation requirements of labeling consecutive frames with both pose and identity. In lieu of labor-intensive labeling, the same solutions employed for multi-object centroid tracking can address this problem through conventional motion models (see “Animal centroids, ellipses and identities” above) to track the centroid59 or collections of keypoints60,61. More sophisticated approaches that rely on identity and pose annotation in consecutive frames may yield considerable improvements for animal pose tracking by leveraging temporal information to perform top-down detection within clips rather than single images62. In the bottom-up approach, however, the representations used for part association must be explicitly extended to the temporal dimension, such as in temporal associative embeddings63 or spatiotemporal affinity fields64, but they provide the same benefits as the single-image bottom-up approaches.
Quantifying the dynamics of behavior
Behavior is a dynamic phenomenon that involves changes to an animal’s pose over time. Unlike the tracking of body parts, quantification of this temporal structure is a fundamentally difficult problem without a clear ground truth. It is often assumed that behavior can be described as a sequence of discrete behavioral states, such as ‘walking’ or ‘grooming’. The techniques discussed below use advances in machine learning to classify these states from video data or features derived from tracking (Box 2). This type of behavioral quantification can facilitate comparison between instances of individual behaviors (for example, in response to specific sensory inputs or across experimental conditions) and generate hypotheses about the neural circuitry that gives rise to them (for example, by demarcating event boundaries or timescales of computation).
Box 2 ∣. Choosing a behavioral feature representation.
Tracking data, whether at the resolution of centroids or pose, is ultimately represented as a set of coordinates in space, typically in units of pixels in the reference frame of the camera. Converting these into a meaningful representation amenable to interpretation and analysis requires the choice of a feature representation that can be derived from the raw coordinates. This step precedes the analysis of dynamics, and most methods for quantifying dynamical structure are agnostic to the specific behavioral feature (Fig. 3a) on which they operate.
A simple and effective approach when dealing with pose is to adjust the coordinates to an egocentric representation, which can be achieved by centering the coordinates such that the origin is at a fixed anatomical position (e.g., thorax in insects, spine base in rodents or centroid more generally). With a second reference point, such as the head or neck, the remaining points can then be rotated such that the animal is always facing in a consistent orientation, enabling a fully egocentric reference frame for the pose coordinates32. In 3D, a third point of reference is required to ensure this rotational invariance, and its selection depends on the available landmarks and the animal’s anatomy46,50,80. Once transformed to egocentric coordinates, displacements can be interpreted in relation to the body, and their occupancy reflects body configuration independent of global position or orientation.
From coordinates, a number of hand-crafted features can easily be derived, such as velocities, distances and orientations, both between body parts and in relation to other animals or objects in the environment. A number of these have been previously described for various species and experimental contexts1,9,50. Some frameworks facilitate the computation of exhaustive sets of combinations of pairwise features, such as all-to-all distances, angles and velocities66,68,79; these can provide a superset of behavioral features agnostic to their semantic interpretation, but much less precise than deliberately designed features. For behaviors involving highly periodic movements, such as locomotion or grooming, spectral features can provide an effective representation by expressing the behavioral feature in time–frequency space32,75,122.
An alternative approach that may be better suited to capturing the correlation structure between postural coordinates (or even between pairwise features) is to employ dimensionality-reduction techniques. Principal component analysis (PCA), for example, is often used to describe body shape in few dimensions, as it naturally takes advantage of the high correlation between articulated body segments in the kinematic tree of animal skeletons. This is particularly useful for animals such as worms89 and fish78 that have many degrees of articulation along a centerline. More complex correlation structure may be analogously identified using nonlinear dimensionality reduction on the coordinates or pairwise features, such as variational auto-encoders95.
Some dynamics may not be effectively captured by tracking, particularly for non-rigid and ‘blobby’ body parts, such as subtle facial movements44. For these, traditional methods for extracting behavioral features operate directly on the images, typically by performing dimensionality reduction on egocentrically aligned crops of the animals75,81 or on regions of interest in immobilized animals44,123,124. In one notable application, a facial emotion recognition technique was applied to mouse faces to extract estimates of their emotional state from facial image features directly125. A recently described database of animal faces may be of particular interest for future work on general purpose animal face features13.
Finally, while many approaches to quantifying dynamics may be effective at capturing the structure and describing the correlation between semantically meaningful behavioral features (body kinematics), for the task of directly classifying behaviors from video frames, methods that capture general image motion may improve accuracy over pose-based features. In the human action recognition field, while some methods do operate on pose126, many opt instead to use deep neural networks to learn to extract motion features by training them to classify actions directly from raw video frames127. In animals, ABRS128 and DeepEthogram129 describe systems for classifying behaviors directly from video by using optical flow features and other learned image features. These techniques alone, however, do not lend themselves directly to understanding the neural control of behavior, which necessarily involves motion and actuation of specific body parts, but can be useful tools to predict the occurrence of well-defined behaviors.
Animal behavior, as defined by humans.
The simplest way to define a behavior is by defining a fixed set of rules that describe the criteria that must be met to determine its occurrence at a given instant. These can be as simple as classifying instances when the animal’s centroid is moving at a speed greater than a minimum threshold as ‘locomotion’, but can quickly become complex when establishing detailed inclusion and exclusion criteria based on fine descriptions of postural features9. Although easy to evaluate and interpret, fixed rules may fail to capture the full variability of behaviors that can be flexibly expressed, particularly when subject to experimental manipulations that may alter the statistics of the features used in the classification criteria65.
A common approach that strikes a balance between human definitions and computer-aided classification is to leverage supervised machine learning. Given user-provided examples of times when particular behaviors are (or are not) occurring, these methods derive classification criteria using specific features (for example, body-part positions or speeds) extracted from the raw data (Fig. 3b). Popular toolkits use decision trees (or random forest ensembles)66-68 to learn potentially complex or abstract classifiers from animal tracking features. These methods leverage data to avoid the tedious and potentially flawed manual design of classification criteria, in addition to providing measures of robustness to overfitting through standard statistical techniques such as cross-validation.
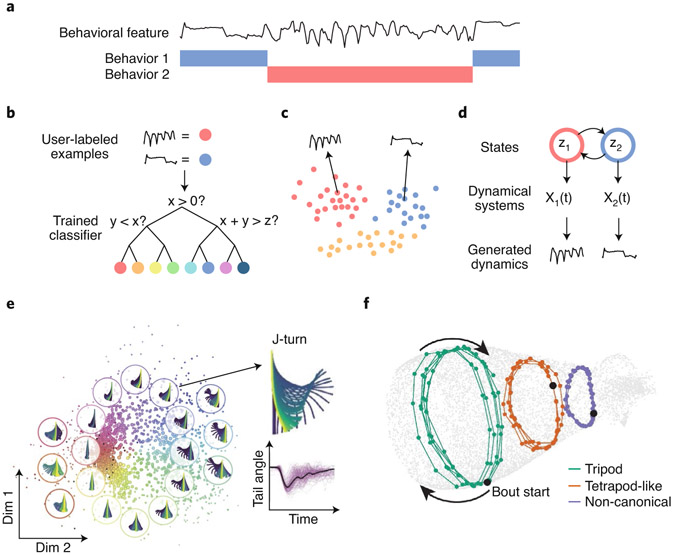
Fig. 3 ∣. Quantifying behavioral dynamics.
a, A snippet of behavioral dynamics during which two types of behavior occur. Behavior 1 (blue) is characterized by slow, step-like dynamics, whereas behavior 2 (red) is characterized by fast oscillations with sharp peaks. b, In supervised classification, a human first annotates examples of each type of behavior (top). A classifier such as a decision tree will learn criteria to classify new data based on the examples provided (bottom). c, In clustering, examples are grouped by their similarity rather than human annotations. The resulting clusters correspond to distinct behaviors. Points represent short windows of time reduced to two dimensions for visualization. d, In dynamical models, behaviors are represented by states that the model is permitted to transition between (top). These states parametrize the models that generate the state-specific dynamics (middle). The observed dynamics are assumed to come from the model that is most likely to generate similar dynamics (bottom). e, Clusters of zebrafish hunting behaviors based on the similarity of their postural trajectories (depicted within the bubbles)90. Points correspond to individual bouts after applying nonlinear dimensionality reduction to the zebrafish pose trajectories as a preprocessing step. f, Manifold embedding of fruit fly gait with the cyclical continuous structure of different gait modes highlighted91. Note that although this representation does not capture cluster-like structure, it does identify both the phase of gait strides (circles) and a continuous axis of variation that transitions smoothly from slow (non-canonical) to fast (tripod) locomotion. Images in e adapted with permission from ref. 90, Cell Press; in f from ref. 91, eLife.
Though these methods can achieve high accuracy, they often rely on user-defined features derived from the input data, such as numerical derivatives66, generic dimensionality reduction67, or exhaustive combinations of relative features68. These may fail to capture more complex relationships in higher-dimensional descriptions (for example, multi-animal poses) or higher-order temporal patterns. A promising direction for more robust general-purpose supervised behavioral classification in animals is to adopt state-of-the-art techniques from human action recognition (i.e., human ‘behaviors’), in which deep-learning-based systems have excelled at data-driven feature extraction from kinematic features69 (Box 2).
Animal behavior, as defined by the data.
Supervised behavior classification, though more robust than classification using hand-crafted criteria, still suffers from the bias of subjective human annotation. Studies demonstrating the large degree of disagreement even among experts with clear guidelines for when to annotate a behavior reveal the shortcomings of depending on human definitions of a behavior70-72. An alternative approach is to ask a computer to learn patterns from the data alone using unsupervised classification methods. In these techniques, the statistics of the behavioral time series themselves are used to determine the criteria used to classify a given time point into a distinct ‘cluster’ or ‘state’ The common assumption across these methods is that data belonging to the same state exhibit similar, stereotyped dynamics given some measure of similarity73,74.
The simplest form of unsupervised classification involves clustering (Fig. 3c). In the typical clustering workflow, dimensionality reduction and feature extraction (for example, using principal component analysis, spectral estimation and manifold embedding techniques such as t-stochastic neighbor embedding (t-SNE)75) are applied to the raw data (which can be videos or pose data derived from videos). The similarity between behaviors at two points in time is then quantified using a similarity metric between the two feature vectors (for example, using the Euclidean distance or a probabilistic measure of similarity such as the Kullback–Leibler divergence). Given a set of time points and the similarity between them, clustering algorithms attempt to group the points into discrete sets in which each point is more similar to the other members of its set than it is to points outside the set. The simplest algorithms, such as k-means and Gaussian mixture models76, have been used successfully to categorize behaviors, but they require the researcher to specify the number of clusters a priori (though this can be estimated using statistical testing). Density-based clustering, using algorithms such as the watershed transform75, avoid having the user specify the exact number of behaviors to be found by identifying peaks in the estimated distribution of time points in the space of extracted features. This approach, first introduced in MotionMapper75, is widely used and has been effective at clustering behavioral dynamics across species and input representation types (Fig. 3e)32,45,46,75,77-80.
Interpretation of the behavioral clusters will depend on the application. These clusters describe groupings of points that are self-similar, but do not directly describe what precisely distinguishes one cluster from another. The first pass approach to interpreting behavioral clusters relies on qualitative observation of raw data exemplars from each group, such as video clips75. Visual inspection may reveal that one cluster corresponds to locomotor behavior and another to grooming, but may fail to provide an explanation for why one cluster of locomotor behavior differs from a separate locomotor cluster. The next step in interpreting these clusters is to compute the empirical feature distributions of the data that falls within the clusters. These may reveal that one form of locomotion is distinct from another based on the peak frequency of limb oscillations32, but may be more difficult to interpret when differences are small or the input data is too high-dimensional to easily interpret differences.
Although clustering is an effective method when little is known about the structure of the behavioral dynamics, more sophisticated approaches afford the ability to explicitly model the characteristics that define the representation of the behavioral dynamics (Fig. 3d). These have the advantage of being potentially more interpretable through direct examination of the model parameters, and some have the added benefit of being able to generate new examples sampled from the model rather than relying on exemplars in the data. One such class of methods that build on probabilistic graphical models are termed state-space models. Rather than attempting to divide the data based on its similarity, these models assume that there exist unobservable (hidden) discrete ‘states’ that parametrize the processes underlying the data. MoSeq81 uses autoregressive generative processes with a sticky hidden Markov model to identify the hidden states and the transition structure between states. This form of modeling can also be combined with simpler generative processes like a multivariate Gaussian distribution49. Other approaches leverage non-parametric Bayesian statistics to model recurrent dependencies in the transitions between different linear dynamical systems, combining the expressivity of graphical models for describing sequence structure with the algebraic interpretability of a linear dynamical system82. An alternative approach to leveraging linear dynamical systems uses an adaptive segmentation algorithm based on statistical model testing rather than an explicit model of transitions between states83.
Behavioral programs operate at many timescales and can be described at different levels of abstraction, all of which could potentially be useful representations of underlying neural computations. For example, it may be desirable to describe the fast timescale kinematics of locomotion while simultaneously representing the longer timescale motivational state of the animal that sets up its navigational goal. Hierarchical clustering can capture structure in behavioral dynamics in a similar way to simple clustering, with the added benefit of decomposing higher-order behavioral clusters into progressively finer-grained subtypes84. Information theoretic techniques for grouping behaviors based on the structure of state transitions have shown a link between a hierarchical temporal organization of the behaviors and the similarity between behaviors85. Explicitly hierarchical dynamical models such as the hierarchical hidden Markov model86 or adaptive segmentation algorithm83 organize behavioral states in a hierarchy based on the structure of their generating processes, affording additional interpretability to the multiscale representations. Assuming a flat discretization of behavioral states, higher-order sequence models such as those used in bioinformatic algorithms to discover motifs in genetic data87 or formal grammars used to model natural language88 can be co-opted to describe behavioral state sequences.
Though these methods address the problems of representing behavior across multiple timescales, they do not effectively provide a solution to the problem of simultaneously occurring behaviors such as walking and sniffing. Though recognizing parallel behaviors is possible through supervised classification by simply using multiple independent classifiers, an unsupervised solution has not yet been proposed. Future work in this domain may be able to achieve this by taking into account that parallel dynamical processes involve different appendages, for example.
Adding continuous structure to discrete representations.
Although the typical behavior map presumes strictly discrete boundaries between distinct behaviors, the execution of motor commands is ultimately expressed through continuous motion73. For example, while walking and jogging may require distinct motor programs, slow walking and fast walking may differ only in the frequency of the stride cycle, smoothly transitioning through ‘moderate walking’. Ultimately, a complete representation of behavior would include both discrete boundaries between behaviors and the continuous variation within them.
Dimensionality reduction methods such as principal component analysis are an effective means of discovering continuous patterning within a behavior, as they compress behavioral dynamics into fewer dimensions, along which the dynamics smoothly vary. This approach has been used to describe worm postural dynamics, revealing an oscillator structure during locomotion89, as well as to describe continuous dynamics of postural trajectories within zebrafish locomotion78 and hunting90 behaviors. More complex kinematics that are not as easily reduced can be captured through nonlinear manifold embedding algorithms and have been employed, for example, to reveal complex periodic structure in fruit fly locomotion (Fig. 3f)91 (akin to the worm oscillator).
Recent methods have begun to employ neural networks as a means of extracting continuous dynamic representations from behavioral time series. These methods afford greater flexibility by enabling robust feature extraction while simultaneously inferring discrete clusters in tandem with continuous representations92,93 or by imposing variational constraints on the distribution of the representations, thereby encouraging more interpretable quantities to be captured in the manifold of dynamics94,95.
Linking brain activity and behavior
A core application of the methods described above is to use descriptions of behavior to understand the neural activity that generates it. As we have discussed above, tools for tracking movement quantify the motor output of the brain, from coarse centroid tracking that describes spatial navigation to fine-grained pose estimation that captures the dynamics of muscle control (Fig. 1). Since these tools can locate the sensory organs of the animal, they also make it possible to reconstruct the sensory inputs animals receive. For example, the visual field of an animal can be estimated from the position of objects in its environment relative to its eyes (Fig. 4a). This is particularly advantageous in freely-moving behavioral setups, which sacrifice the ability to control (and therefore precisely know) the visual stimuli in a given psychophysics or virtual reality experiment96-99. Stimuli of other sensory modalities, such as mechanosensation, can also be estimated from videography, while others such as acoustic stimuli will require different instrumentation and methods for feature extraction (Box 3).
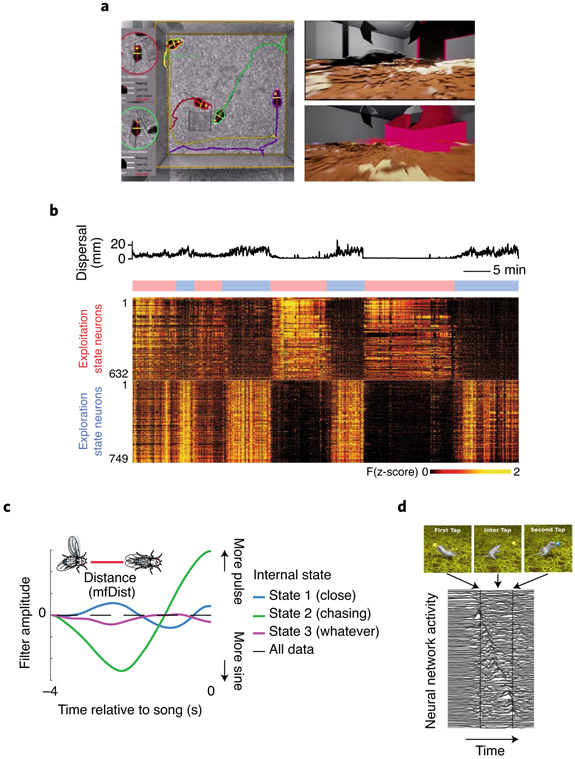
Fig. 4 ∣. Approaches to linking brain to behavior.
a, Tracking centroids and orientations of animals enables the reconstruction of their sensory inputs by simulating a first-person view of their environment9. b, Zebrafish tracking and whole-brain imaging during hunting behavior shows how representations of internal states (exploration vs exploitation) are revealed when aligning to behavioral data104. c, Model of courting flies captures the shape and timescale of visual sensory inputs (mfDist; distance between animals) that predicts behavioral output (courtship song) modulated by internal state110. d, An ANN can learn to control a simulated rat via motor commands to perform a tapping task. Top: rendering of the simulated rat performing the task. Bottom: latent representations learned by the ANN that is used to drive the behavior113. Images in a adapted with permission from ref. 9, Nature Publishing Group; in b from ref. 104, Nature Publishing Group; in c from ref. 110, Nature Publishing Group; in d from ref. 113, arXiv.
Box 3 ∣. Quantifying acoustic behaviors.
Not all motor output can be quantified using conventional videography. In particular, vocalizations and other forms of acoustic communication are either not clearly visible or are produced at frequencies much higher than the frame rates of standard video cameras. Acoustic behaviors, however, can be easily measured using microphones placed in proximity of interacting animals. Similar to the challenges described for tracking and segmenting behaviors from video, the primary challenge in quantifying acoustic behaviors is detecting and classifying individual acoustic events, such as courtship song syllables.
Classical approaches rely on signal processing techniques to filter and extract acoustic events based on spectral and temporal features of animal songs. These have been used with much success to detect, for example, Drosophila courtship song130, zebra finch song131 and mouse vocalizations132,133. The automated detection of fly song has facilitated analysis of large behavioral datasets linking visual feedback cues (the moving female fly) to the dynamic patterning of male song, which would not have been possible otherwise134. Newer approaches have been developed to ease the difficulty of hand-crafting specialized signal-processing pipelines by leveraging deep learning to learn from user annotations135-137. These tools are crucial in improving the robustness of acoustic behavior detection to new experimental conditions with different noise properties.
However, these methods are limited by a priori knowledge of the syllables or modes that comprise song. A parallel approach to the supervised techniques leverages unsupervised learning to discover new acoustic behavioral subtypes, analogous to the unsupervised approaches for tracking. Manifold embedding has enabled the discovery of previously undiscovered fruit fly song types138 and immature songbird sequences139, as well as mapping of less-stereotyped mouse vocalizations140-142.
As tools to measure behavioral features improve, so does the resolution of the estimated representations of the inputs and outputs to the nervous system. Here we highlight the major classes of approaches that have successfully leveraged behavioral quantification to dissect brain function.
Peri-behavior time histograms.
Analogously to how peristimulus time histograms enable circuit interrogation by aligning the activity of neurons relative to onset or offset of stimuli, a major approach to linking neural function to behavior is to describe the distribution of behavioral quantities surrounding neural events. For example, manipulation of neural activity through genetic tools has enabled brain-wide screens that associate activation of precise subsets of neurons with the animal’s entire behavioral repertoire84,100,101.
While neuromodulation experiments provide precise temporal control over stimulation, techniques for awake and freely-moving in vivo recording can be used to align neural activity to more naturalistic and spontaneous behaviors. Combined with tracking and quantification of dynamics, this approach has been successfully used to discover neural correlates of a number of behavioral features described in this review. Instantaneous animal pose has been found to be represented across cortical regions in freely-moving rodents102. Discrete behavioral motifs identified using state-space models such as MoSeq81, as well as their sequences, have been associated with neural activity in the striatum, revealing a code for action selection103. Higher-order behavioral states (hierarchies), such as the exploration-versus-exploitation dichotomy in zebrafish hunting behavior, have been associated with internal neural states through the use of whole-brain imaging and pose estimation (Fig. 4b)104. Multi-timescale behavioral structures have been found to correlate with a hierarchy of neural dynamics in freely moving worms, connecting population-level codes with fast timescale motor control105.
Finally, simultaneous recording of neural activity across multiple animals has recently been used to identify neural correlates of social behavioral features in both bats and mice106,107. By aligning neural activity to social behavior quantified from multi-animal tracking, neural representations were discovered that encode both fast timescale features, such as the current and future behavior of the animal’s social partner, as well as higher-order cognitive features, such as their social hierarchy. These codes appear to be synchronized across animals, revealing a potential mechanistic basis for coordination of social behaviors, reflecting previous reports of synchronization identified from behavioral data alone108.
Models of sensorimotor transformations.
Given representations of sensory inputs and motor outputs, another way to link behavior to neural activity is through explicit modeling of this sensorimotor transformation. Modeling the transformation from sensory input to motor output can recover stimulus filters109 and even infer internal states110. These models attempt to fit simple but easily interpretable transformations between sensory input and behavioral output (Fig. 4c). This level of modeling has the benefit of being a general-purpose approach to discovering the relative importance of different sensory features and their timescales.
Approaches that more comprehensively model the internal computations of the sensorimotor transformation afford the ability to incorporate knowledge about the underlying biological structure of the computations at the cost of increased model complexity. By simulating known neural connectivity and their biophysics, these forms of models enable in silico experimentation111,112. For example, performing ablations of specific model neurons and observing the changes in behavioral responses can provide insights into the computations being performed, which can be validated with analogous experiments in vivo.
Designing network models becomes increasingly difficult as the behaviors they attempt to predict become more detailed and less constrained, as is the case in freely moving and naturally behaving animals. An emerging approach to address this is to use artificial neural networks (ANNs) that can learn to perform the sensorimotor transformations while abstracting away details about the underlying biological neural networks. This form of modeling naturally leads to agent-based models, i.e., models that can perceive and respond to their environment. If the environment can be fully simulated, these agents are able to be trained without any data by providing them with a behavioral task and constraining their kinematics to realistic biomechanics. Recently, it was demonstrated that an agent-based model of rodents trained to perform classical cognitive–behavioral tasks, such as the two-tap task or navigating a Y-maze, are not only able to attain comparable performance to real animals, but also learn to compute internal representations of motor planning and control from sensory inputs (Fig. 4d)113,114.
Finally, an approach called imitation learning combines the ability of ANNs to efficiently learn complex transformations with the ability to impose biological fidelity derived from empirical data. These are constructed as agent-based models, but rather than being trained in simulation, they instead learn from behavioral data to predict motor outputs (for example, changes in pose) from reconstructed sensory inputs (for example, visual field-of-view). These have been applied to fruit fly data and shown to be capable of learning high-level representations of the computations underlying unconstrained behavior115,116. ANNs can also be constructed with architectures based on biological neuroanatomy to further constrain the types of representations it learns (a feature that is particularly useful in light of recent advances in connectomics117). Recent work has shown that the computations learned by these models exhibit a remarkable degree of similarity to physiology, even when trained on behavioral data alone118.
Conclusion
In this Review we have detailed the existing and emerging methods in quantifying animal behavior to better understand the brain. From tracking to dynamics, it is clear that advances in deep learning and computer vision have revolutionized our ability to extract increasingly detailed descriptions of behavior. Further, as modeling frameworks continue to evolve, so will our ability to use behavior as a means of comparing neural dynamics and structure across diverse animals and experimental paradigms, toward developing holistic theories of brain function. We believe that these advances position behavioral quantification as a core instrument in the neuroscience toolbox, essential to the quest of understanding the brain.
Acknowledgements
T.D.P. is supported by NSF GRFP (DGE-1148900) and the Princeton Porter Ogden Jacobus Fellowship. M.M. and J.W.S. are supported by an NIH BRAIN Initiative R01 (R01 NS104899) and an NSF Physics Frontier Center grant (NSF PHY-1734030). M.M. is also supported by an HHMI Faculty Scholar award and an NIH NINDS R35 research program award. We thank B. Cowley and J. Pillow for very helpful comments and suggestions.
Footnotes
Competing interests
The authors declare no competing interests.
Peer review information Nature Neuroscience thanks Ann Kennedy, Pavan Ramdya, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Branson K, Robie AA, Bender J, Perona P & Dickinson MH High-throughput ethomics in large groups of Drosophila. Nat. Methods 6, 451–457 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geuther BQ et al. Robust mouse tracking in complex environments using neural networks. Commun Biol 2, 124 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson DJ & Perona P Toward a science of computational ethology. Neuron 84, 18–31 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Robie AA, Seagraves KM, Egnor SER & Branson K Machine vision methods for analyzing social interactions. J. Exp. Biol 220, 25–34 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Sridhar VH, Roche DG & Gingins S Tracktor: image-based automated tracking of animal movement and behaviour. Methods Ecol. Evol 10, 815–820 (2019). [Google Scholar]
- 6.Rodriguez A et al. ToxTrac: a fast and robust software for tracking organisms. Methods Ecol. Evol 9, 460–464 (2018). [Google Scholar]
- 7.Ohayon S, Avni O, Taylor AL, Perona P & Roian Egnor SE Automated multi-day tracking of marked mice for the analysis of social behaviour. J. Neurosci. Methods 219, 10–19 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gal A, Saragosti J & Kronauer DJC anTraX: high throughput video tracking of color-tagged insects. Preprint at bioRxiv 10.1101/2020.04.29.068478 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Chaumont F et al. Real-time analysis of the behaviour of groups of mice via a depth-sensing camera and machine learning. Nat. Biomed. Eng 3, 930–942 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Krakauer JW, Ghazanfar AA, Gomez-Marin A, MacIver MA & Poeppel D Neuroscience Needs Behavior: Correcting a Reductionist Bias. Neuron 93, 480–490 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Ciaparrone G et al. Deep learning in video multi-object tracking: a survey. Preprint at arXiv https://arxiv.org/abs/1907.12740 (2019). [Google Scholar]
- 12.Schroff F, Kalenichenko D & Philbin J FaceNet: a unified embedding for face recognition and clustering. in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 815–823 (2015). [Google Scholar]
- 13.Khan MH et al. AnimalWeb: a large-scale hierarchical dataset of annotated animal faces. in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 6939–6948 (2020). [Google Scholar]
- 14.Romero-Ferrero F, Bergomi MG, Hinz RC, Heras FJH & de Polavieja GG idtracker.ai: tracking all individuals in small or large collectives of unmarked animals. Nat. Methods 16, 179–182 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Bozek K, Hebert L, Portugal Y & Stephens GJ Markerless tracking of an entire insect colony. Preprint at bioRxiv 10.1101/2020.03.26.007302 (2020). [DOI] [Google Scholar]
- 16.Karthik S, Prabhu A & Gandhi V Simple unsupervised multi-object tracking. Preprint at arXiv https://arxiv.org/abs/2006.02609 (2020). [Google Scholar]
- 17.Johansson G Visual perception of biological motion and a model for its analysis. Percept. Psychophys 14, 201–211 (1973). [Google Scholar]
- 18.Marr D & Vaina L Representation and recognition of the movements of shapes. Proc. R. Soc. Lond. B Biol. Sci 214, 501–524 (1982). [DOI] [PubMed] [Google Scholar]
- 19.O’Rourke J & Badler NI Model-based image analysis of human motion using constraint propagation. in IEEE Transactions on Pattern Analysis and Machine Intelligence (PAMI-2), 522–536 (1980). [Google Scholar]
- 20.Carreira J, Agrawal P, Fragkiadaki K & Malik J Human pose estimation with iterative error feedback. in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 4733–4742 (2016). [Google Scholar]
- 21.Wei S-E, Ramakrishna V, Kanade T & Sheikh Y Convolutional pose machines. in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 4724–4732 (2016). [Google Scholar]
- 22.Newell A, Yang K & Deng J Stacked hourglass networks for human pose estimation in Computer Vision – ECCV 2016 483–499 (Springer, 2016). [Google Scholar]
- 23.Lin T-Y et al. Microsoft COCO: common objects in context in Computer Vision – ECCV 2014 740–755 (Springer, 2014). [Google Scholar]
- 24.Andriluka M, Pishchulin L, Gehler P & Schiele B 2D human pose estimation: new benchmark and state of the art analysis. in Proc. IEEE Conf. Computer Vision and Pattern Recognition 3686–3693 (2014). [Google Scholar]
- 25.Ionescu C, Papava D, Olaru V & Sminchisescu C Human3.6M: large scale datasets and predictive methods for 3D human sensing in natural environments. IEEE Trans. Pattern Anal. Mach. Intel 36, 1325–1339 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Mathis A et al. DeepLabCut: markerless pose estimation of user-defined body parts with deep learning. Nat. Neurosci. 21, 1281–1289 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Russakovsky O et al. ImageNet large scale visual recognition challenge. Int. J. Comput. Vis 115, 211–252 (2015). [Google Scholar]
- 28.Kornblith S, Shlens J & Le QV in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 2661–2671 (2019). [Google Scholar]
- 29.He K, Girshick R & Dollár P Rethinking ImageNet pre-training. in Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV) 4918–4927 (2019). [Google Scholar]
- 30.Mathis A, Yüksekgönül M, Rogers B, Bethge M & Mathis MW Pretraining boosts out-of-domain robustness for pose estimation. Preprint at arXiv https://arxiv.org/abs/1909.11229 (2019). [Google Scholar]
- 31.Pereira TD et al. SLEAP: multi-animal pose tracking. Preprint at bioRxiv 10.1101/2020.08.31.276246 (2020). [DOI] [Google Scholar]
- 32.Pereira TD et al. Fast animal pose estimation using deep neural networks. Nat. Methods 16, 117–125 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Graving JM et al. DeepPoseKit, a software toolkit for fast and robust animal pose estimation using deep learning. eLife 8, e47994 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He K, Zhang X, Ren S & Sun J Deep residual learning for image recognition. in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 770–778 (2016). [Google Scholar]
- 35.Yu F et al. LSUN: construction of a large-scale image dataset using deep learning with humans in the loop. Preprint at arXiv https://arxiv.org/abs/1506.03365 (2015). [Google Scholar]
- 36.Mathis MW & Mathis A Deep learning tools for the measurement of animal behavior in neuroscience. Curr. Opin. Neurobiol 60, 1–11 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Christin S, Hervet É & Lecomte N Applications for deep learning in ecology. Methods Ecol. Evol 10, 1632–1644 (2019). [Google Scholar]
- 38.Suwajanakorn S, Snavely N, Tompson JJ & Norouzi M Discovery of latent 3D keypoints via end-to-end geometric reasoning. Adv. Neural Inf. Process. Syst 31 2059–2070 (2018). [Google Scholar]
- 39.Li S et al. Deformation-aware unpaired image translation for pose estimation on laboratory animals. in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 13158–13168 (2020). [Google Scholar]
- 40.Mu J, Qiu W, Hager G & Yuille A Learning from synthetic animals. arXiv https://arxiv.org/abs/1912.08265 (2019). [Google Scholar]
- 41.Cao J et al. Cross-domain adaptation for animal pose estimation. arXiv https://arxiv.org/abs/1908.05806 (2019). [Google Scholar]
- 42.Liu X et al. OptiFlex: video-based animal pose estimation using deep learning enhanced by optical flow. Preprint at bioRxiv 10.1101/2020.04.04.025494 (2020). [DOI] [Google Scholar]
- 43.Wu A, Buchanan EK, Whiteway M & Schartner M Deep Graph Pose: a semi-supervised deep graphical model for improved animal pose tracking. Preprint at bioRxiv 10.1101/2020.08.20.259705 (2020). [DOI] [Google Scholar]
- 44.Musall S, Kaufman MT, Juavinett AL, Gluf S & Churchland AK Single-trial neural dynamics are dominated by richly varied movements. Nat. Neurosci 22, 1677–1686 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Günel S et al. DeepFly3D, a deep learning-based approach for 3D limb and appendage tracking in tethered, adult Drosophila. eLife 8, 640375 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bala PC et al. Automated markerless pose estimation in freely moving macaques with OpenMonkeyStudio. Nat. Commun 11, 4560 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garrido-Jurado S, Muñoz-Salinas R, Madrid-Cuevas FJ & Marín-Jiménez MJ Automatic generation and detection of highly reliable fiducial markers under occlusion. Pattern Recognit. 47, 2280–2292 (2014). [Google Scholar]
- 48.Karashchuk P et al. Anipose: a toolkit for robust markerless 3D pose estimation. Preprint at bioRxiv 10.1101/2020.05.26.117325 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ebbesen CL & Froemke RC Automatic tracking of mouse social posture dynamics by 3D videography, deep learning and GPU-accelerated robust optimization. Preprint at bioRxiv 10.1101/2020.05.21.109629 (2020). [DOI] [Google Scholar]
- 50.Storchi R et al. A high-dimensional quantification of mouse defensive behaviors reveals enhanced diversity and stimulus specificity. Curr. Biol 10.1016/j.cub.2020.09.007 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen Y, Tian Y & He M Monocular human pose estimation: a survey of deep learning-based methods. Comput. Vis. Image Underst 192, 102897 (2020). [Google Scholar]
- 52.Gosztolai A et al. LiftPose3D, a deep learning-based approach for transforming 2D to 3D pose in laboratory animals. Preprint at bioRxiv 10.1101/2020.09.18.292680 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tzouanas CN, Kim S, Badhiwala KN, Avants BW & Robinson JT Stable behavioral and neural responses to thermal stimulation despite large changes in the Hydra vulgaris nervous system. Preprint at bioRxiv 10.1101/787648 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kearney S, Li W, Parsons M, Kim KI & Cosker D RGBD-Dog: predicting canine pose from RGBD sensors. in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 8336–8345 (2020). [Google Scholar]
- 55.Zuffi S, Kanazawa A & Black MJ Lions and tigers and bears: Capturing non-rigid, 3D, articulated shape from images. in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 3955–3963 (2018). [Google Scholar]
- 56.Kulkarni N, Gupta A, Fouhey DF & Tulsiani S Articulation-aware canonical surface mapping. in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 452–461 (2020). [Google Scholar]
- 57.Badger M et al. 3D bird reconstruction: a dataset, model, and shape recovery from a single view. Preprint at arXiv https://arxiv.org/abs/2008.06133 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cao Z, Simon T, Wei S-E & Sheikh Y Realtime multi-person 2D pose estimation using part affinity fields. in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 7291–7299 (2017). [Google Scholar]
- 59.Francisco FA, Nührenberg P & Jordan A High-resolution animal tracking with integration of environmental information in aquatic systems. Preprint at bioRxiv 10.1101/2020.02.25.963926 (2020). [DOI] [Google Scholar]
- 60.Xiao B, Wu H & Wei Y Simple baselines for human pose estimation and tracking. in Proceedings of the European Conference on Computer Vision (ECCV) 466–481 (2018). [Google Scholar]
- 61.Jiang Z et al. Detection and tracking of multiple mice using part proposal networks. Preprint at arXiv https://arxiv.org/abs/1906.02831 (2019). [Google Scholar]
- 62.Wang M, Tighe J & Modolo D Combining detection and tracking for human pose estimation in videos. in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 11088–11096 (2020). [Google Scholar]
- 63.Jin S, Liu W, Ouyang W & Qian C Multi-person articulated tracking with spatial and temporal embeddings. in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 5664–5673 (2019). [Google Scholar]
- 64.Raaj Y, Idrees H, Hidalgo G & Sheikh Y Efficient online multi-person 2d pose tracking with recurrent spatio-temporal affinity fields. in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 4620–4628 (2019). [Google Scholar]
- 65.Datta SR, Anderson DJ, Branson K, Perona P & Leifer A Computational neuroethology: a call to action. Neuron 104, 11–24 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kabra M, Robie AA, Rivera-Alba M, Branson S & Branson K JAABA: interactive machine learning for automatic annotation of animal behavior. Nat. Methods 10, 64–67 (2013). [DOI] [PubMed] [Google Scholar]
- 67.Hong W et al. Automated measurement of mouse social behaviors using depth sensing, video tracking, and machine learning. Proc. Natl Acad. Sci. USA 112, E5351–E5360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nilsson SRO et al. Simple Behavioral Analysis (SimBA) – an open source toolkit for computer classification of complex social behaviors in experimental animals. Preprint at bioRxiv 10.1101/2020.04.19.049452 (2020). [DOI] [Google Scholar]
- 69.Ren B, Liu M, Ding R & Liu H A survey on 3D skeleton-based action recognition using learning method. Preprint at arXiv https://arxiv.org/abs/2002.05907 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Levitis DA, Lidicker WZ & Freund G Behavioural biologists don’t agree on what constitutes behaviour. Anim. Behav. 78, 103–110 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Szigeti B, Stone T & Webb B Inconsistencies in C. elegans behavioural annotation. Preprint at bioRxiv 10.1101/066787 (2016). [DOI] [Google Scholar]
- 72.Leng X, Wohl M, Ishii K, Nayak P & Asahina K Quantitative comparison of Drosophila behavior annotations by human observers and a machine learning algorithm. Preprint at bioRxiv 10.1101/2020.06.16.153130 (2020). [DOI] [Google Scholar]
- 73.Brown AEX & de Bivort B Ethology as a physical science. Nat. Phys 14, 653–657 (2018). [Google Scholar]
- 74.Berman GJ Measuring behavior across scales. BMC Biol. 16, 23 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Berman GJ, Choi DM, Bialek W & Shaevitz JW Mapping the stereotyped behaviour of freely moving fruit flies. J. R. Soc. Interface 11, 20140672 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Todd JG, Kain JS & de Bivort BL Systematic exploration of unsupervised methods for mapping behavior. Phys. Biol 14, 015002 (2017). [DOI] [PubMed] [Google Scholar]
- 77.Klaus A et al. The spatiotemporal organization of the striatum encodes action space. Neuron 95, 1171–1180.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marques JC, Lackner S, Felix R & Orger MB Structure of the zebrafish locomotor repertoire revealed with unsupervised behavioral clustering. Curr. Bid 28, 181–195.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 79.Hsu AI & Yttri EA B-SOiD: an open source unsupervised algorithm for discovery of spontaneous behaviors. Preprint at bioRxiv 10.1101/770271 (2020). [DOI] [Google Scholar]
- 80.Zimmermann C, Schneider A, Alyahyay M, Brox T & Diester I FreiPose: a deep learning framework for precise animal motion capture in 3D spaces. Preprint at bioRxiv 10.1101/2020.02.27.967620 (2020). [DOI] [Google Scholar]
- 81.Wiltschko AB et al. Mapping sub-second structure in mouse behavior. Neuron 88, 1121–1135 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Linderman S et al. Bayesian learning and inference in recurrent switching linear dynamical systems. in Proceedings of the 20th International Conference on Artificial Intelligence and Statistics (eds. Singh A & Zhu J) 54, 914–922 (PMLR, 2017). [Google Scholar]
- 83.Costa AC, Ahamed T & Stephens GJ Adaptive, locally linear models of complex dynamics. Proc. Natl Acad. Sci. USA 116, 1501–1510 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vogelstein JT et al. Discovery of brainwide neural-behavioral maps via multiscale unsupervised structure learning. Science 344, 386–392 (2014). [DOI] [PubMed] [Google Scholar]
- 85.Berman GJ, Bialek W & Shaevitz JW Predictability and hierarchy in Drosophila behavior. Proc. Natl Acad. Sci. USA 113, 11943–11948 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tao L, Ozarkar S, Beck JM & Bhandawat V Statistical structure of locomotion and its modulation by odors. eLife 8, e41235 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ligon RA, Scholes E & Sheehan MJ RAD-Behavior (Recombining Atomized, Discretized, Behavior): a new framework for the quantitative analysis of behavioral execution. Preprint at bioRxiv 10.1101/739151 (2019). [DOI] [Google Scholar]
- 88.Gupta S & Gomez-Marin A A context-free grammar for Caenorhabditis elegans behavior. Preprint at bioRxiv 10.1101/708891 (2019). [DOI] [Google Scholar]
- 89.Stephens GJ, Johnson-Kerner B, Bialek W & Ryu WS Dimensionality and dynamics in the behavior of C. elegans. PLOS Comput. Biol 4, e1000028 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mearns DS, Donovan JC, Fernandes AM, Semmelhack JL & Baier H Deconstructing hunting behavior reveals a tightly coupled stimulus-response loop. Curr. Biol 30, 54–69.e9 (2020). [DOI] [PubMed] [Google Scholar]
- 91.DeAngelis BD, Zavatone-Veth JA & Clark DA The manifold structure of limb coordination in walking Drosophila. eLife 8, e46409 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Su K, Liu X & Shlizerman E PREDICT & CLUSTER: unsupervised skeleton based action recognition. in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 9631–9640 (2020). [Google Scholar]
- 93.Graving JM & Couzin ID VAE-SNE: a deep generative model for simultaneous dimensionality reduction and clustering. Preprint at bioRxiv 10.1101/2020.07.17.207993 (2020). [DOI] [Google Scholar]
- 94.Johnson MJ, Duvenaud DK, Wiltschko A, Adams RP & Datta SR Composing graphical models with neural networks for structured representations and fast inference. Adv. Neural Inf. Process. Syst 29, 2946–2954 (2016). [Google Scholar]
- 95.Luxem K, Fuhrmann F, Kürsch J, Remy S & Bauer P Identifying behavioral structure from deep variational embeddings of animal motion. Preprint at bioRxiv 10.1101/2020.05.14.095430 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Harvey CD, Collman F, Dombeck DA & Tank DW Intracellular dynamics of hippocampal place cells during virtual navigation. Nature 461, 941–946 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stowers JR et al. Virtual reality for freely moving animals. Nat. Methods 14, 995–1002 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Haberkern H et al. Visually guided behavior and optogenetically induced learning in head-fixed flies exploring a virtual landscape. Curr. Biol 29, 1647–1659.e8 (2019). [DOI] [PubMed] [Google Scholar]
- 99.Naik H, Bastien R, Navab N & Couzin ID Animals in virtual environments. IEEE Trans. Vis. Comput. Graph 26, 2073–2083 (2020). [DOI] [PubMed] [Google Scholar]
- 100.Robie AA et al. Mapping the neural substrates of behavior. Cell 170, 393–406.e28 (2017). [DOI] [PubMed] [Google Scholar]
- 101.Cande J et al. Optogenetic dissection of descending behavioral control in Drosophila. eLife 7, e34275 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mimica B, Dunn BA, Tombaz T, Bojja VPTNCS & Whitlock JR Efficient cortical coding of 3D posture in freely behaving rats. Science 362, 584–589 (2018). [DOI] [PubMed] [Google Scholar]
- 103.Markowitz JE et al. The striatum organizes 3D behavior via moment-to-moment action selection. Cell 174, 44–58.e17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Marques JC, Li M, Schaak D, Robson DN & Li JM Internal state dynamics shape brainwide activity and foraging behaviour. Nature 577, 239–243 (2020). [DOI] [PubMed] [Google Scholar]
- 105.Kaplan HS, Salazar Thula O, Khoss N & Zimmer M Nested neuronal dynamics orchestrate a behavioral hierarchy across timescaleS. Neuron 105, 562–576.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang W & Yartsev MM Correlated neural activity across the brains of socially interacting bats. Cell 178, 413–428.e22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kingsbury L et al. Correlated neural activity and encoding of behavior across brains of socially interacting animals. Cell 178, 429–446.e16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Klibaite U & Shaevitz JW Paired fruit flies synchronize behavior: uncovering social interactions in Drosophila melanogaster. PLoS Comput. Biol 16, e1008230 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gepner R, Mihovilovic Skanata M, Bernat NM, Kaplow M & Gershow M Computations underlying Drosophila photo-taxis, odor-taxis, and multi-sensory integration. eLife 4, e06229 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Calhoun AJ, Pillow JW & Murthy M Unsupervised identification of the internal states that shape natural behavior. Nat. Neurosci 22, 2040–2049 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Maesani A et al. Fluctuation-driven neural dynamics reproduce Drosophila locomotor patterns. PLOS Comput. Biol 11, e1004577 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kim J, Santos JA, Alkema MJ & Shlizerman E Whole integration of neural connectomics, dynamics and bio-mechanics for identification of behavioral sensorimotor pathways in Caenorhabditis elegans. Preprint at bioRxiv 10.1101/724328 (2019). [DOI] [Google Scholar]
- 113.Merel J et al. Deep neuroethology of a virtual rodent. in International Conference on Learning Representations https://openreview.net/forum?id=SyxrxR4KPS (2020). [Google Scholar]
- 114.Crosby M, Beyret B & Halina M The Animal-AI Olympics. Nature Machine Intelligence 1, 257 (2019). [Google Scholar]
- 115.Eyjolfsdottir E, Branson K, Yue Y & Perona P Learning recurrent representations for hierarchical behavior modeling. in International Conference on Learning Representations https://openreview.net/forum?id=BkLhzHtlg (2017). [Google Scholar]
- 116.Teng M, Le TA, Scibior A & Wood F Imitation learning of factored multi-agent reactive models. Preprint at arXiv https://arxiv.org/abs/1903.04714 (2019). [Google Scholar]
- 117.Dorkenwald S et al. FlyWire: online community for whole-brain connectomics. Preprint at bioRxiv 10.1101/2020.08.30.274225 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Michaels JA, Schaffelhofer S, Agudelo-Toro A & Scherberger H A modular neural network model of grasp movement generation. Preprint at bioRxiv 10.1101/742189 (2020). [DOI] [Google Scholar]
- 119.Sci draw. Mouse top 10.5281/zenodo.3925916 (2020). [DOI]
- 120.BioRender. https://biorender.com (2020).
- 121.Segalin C et al. The Mouse Action Recognition System (MARS): a software pipeline for automated analysis of social behaviors in mice. Preprint at bioRxiv 10.1101/2020.07.26.222299 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Klibaite U, Berman GJ, Cande J, Stern DL & Shaevitz JW An unsupervised method for quantifying the behavior of paired animals. Phys. Biol 14, 015006 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Stringer C et al. Spontaneous behaviors drive multidimensional, brainwide activity. Science 364, 255 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Batty E et al. BehaveNet: nonlinear embedding and Bayesian neural decoding of behavioral videos. Adv. Neural Inform. Process. Syst 32, 15706–15717 (2019). [Google Scholar]
- 125.Dolensek N, Gehrlach DA, Klein AS & Gogolla N Facial expressions of emotion states and their neuronal correlates in mice. Science 368, 89–94 (2020). [DOI] [PubMed] [Google Scholar]
- 126.Gupta P et al. Quo vadis, skeleton action recognition? Preprint at arXiv https://arxiv.org/abs/2007.02072 (2020). [Google Scholar]
- 127.Carreira J & Zisserman A Quo vadis, action recognition? A new model and the kinetics dataset. in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 6299–6308 (2017). [Google Scholar]
- 128.Ravbar P, Branson K & Simpson JH An automatic behavior recognition system classifies animal behaviors using movements and their temporal context. J. Neurosci. Methods 326, 108352 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bohnslav JP et al. DeepEthogram: a machine learning pipeline for supervised behavior classification from raw pixels. Preprint at bioRxiv 10.1101/2020.09.24.312504 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Arthur BJ, Sunayama-Morita T, Coen P, Murthy M & Stern DL Multi-channel acoustic recording and automated analysis of Drosophila courtship songs. BMC Biol. 11, 11 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pearre B, Perkins LN, Markowitz JE & Gardner TJ A fast and accurate zebra finch syllable detector. PLoS One 12, e0181992 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Van Segbroeck M, Knoll AT, Levitt P & Narayanan S MUPET-Mouse Ultrasonic Profile ExTraction: a signal processing tool for rapid and unsupervised analysis of ultrasonic vocalizations. Neuron 94, 465–485.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sangiamo DT, Warren MR & Neunuebel JP Ultrasonic signals associated with different types of social behavior of mice. Nat. Neurosci 23, 411–422 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Coen P et al. Dynamic sensory cues shape song structure in Drosophila. Nature 507, 233–237 (2014). [DOI] [PubMed] [Google Scholar]
- 135.Coffey KR, Marx RG & Neumaier JF DeepSqueak: a deep learning-based system for detection and analysis of ultrasonic vocalizations. Neuropsychopharmacology 44, 859–868 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fonseca AHO, Santana GM, Bampi S & Dietrich MO Analysis of ultrasonic vocalizations from mice using computer vision and machine learning. Preprint at bioRxiv 10.1101/2020.05.20.105023 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Cohen Y, Nicholson DA & Gardner TJ TweetyNet: a neural network that enables high-throughput, automated annotation of birdsong. Preprint at bioRxiv 10.1101/2020.08.28.272088 (2020). [DOI] [Google Scholar]
- 138.Clemens J et al. Discovery of a new song mode in Drosophila reveals hidden structure in the sensory and neural drivers of behavior. Curr. Biol 28, 2400–2412.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Mackevicius EL et al. Unsupervised discovery of temporal sequences in high-dimensional datasets, with applications to neuroscience. eLife 8, e38471 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Tabler JM et al. Cilia-mediated Hedgehog signaling controls form and function in the mammalian larynx. eLife 6, e19153 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sainburg T, Thielk M & Gentner TQ Latent space visualization, characterization, and generation of diverse vocal communication signals. Preprint at bioRxiv 10.1101/870311 (2019). [DOI] [Google Scholar]
- 142.Goffinet J, Mooney R & Pearson J Inferring low-dimensional latent descriptions of animal vocalizations. Preprint at bioRxiv 10.1101/811661 (2019). [DOI] [Google Scholar]