Abstract
Objective:
This study aims to construct a systematic mRNA-miRNA-lncRNA network to identify novel lncRNAs and miRNAs biomarkers for laryngeal squamous cell carcinoma (LSCC).
Methods:
The mRNA, miRNA and lncRNA expression profiles of LSCC were obtained from Gene Expression Omnibus (GEO) database. The differentially expressed mRNAs, miRNAs and lncRNAs (DEmRNAs, DEmiRNAs and DElncRNAs) were screened between LSCC tissues and controls. Functional analysis of DEmRNAs, DEmRNAs targeted by DEmiRNAs and DEmRNAs targeted by DElncRNAs were respectively performed. The miRWalk, starbase and DIANA-LncBase were respectively used to predict DEmiRNAs-DEmRNAs, DElncRNAs-DEmRNAs and DElncRNAs-DEmiRNAs pairs. ceRNA network was built by DEmiRNAs-DEmRNAs and DElncRNAs-DEmiRNAs pairs. LncRNA subcellular localization was predicted using lncLocator. Using published The Cancer Genome Atlas (TCGA) and external datasets (GSE127165 and GSE133632), we also validated the expression of key DElncRNAs and DEmiRNAs in ceRNA network. The diagnostic and prognostic value of candidate genes was evaluated by ROC curve analysis and survival analysis, respectively.
Results:
There were 5 mRNA datasets, 3 miRNA datasets and 2 lncRNA datasets in this study. Totally, 2957 DEmRNAs, 61 DElncRNAs and 23 DEmiRNAs were identified. Functional analysis of DEmRNAs shows that they were significantly enriched in cancer-related pathways, such as DNA replication and extracellular matrix organization. There were 11 DEmiRNAs, 17 DElncRNAs and 967 DEmRNAs in the ceRNA network. Notably, up-regulated lncRNA DGCR5-down-regulated has-miR-338-3p/has-miR-139-5p pairs in this network were experimentally validated. Moreover, down-regulated AL121839.2, down-regulated LINC02147, up-regulated AC079328.2, up-regulated AC004943.2 and up-regulated HMGA2-AS1 were located in the cytoplasm. AL121839.2 and LINC02147 interacted with has-miR-1246. AC004943.2, AC079328.2 and HMGA2-AS1 targeted has-miR-3185, has-miR-3137 and has-miR-582-5p, respectively. Based on the TCGA and external datasets (GSE127165 and GSE133632), DGCR5 and AC004943.2 were significantly up-regulated while AL121839.2 and LINC02147, has-miR-338-3p, has-miR-139-5p and has-miR-582-5p were significantly down-regulated, which were consistent with our integration analysis. DGCR5, AL121839.2, LINC02147, AC004943.2, has-miR-338-3p, has-miR-139-5p and has-miR-582-5p could predict the occurrence of LSCC. Survival analysis suggested that only, AL121839.2 has potential prognostic value for LSCC.
Conclusion:
This study provided novel insights into the ceRNA network and uncovered novel lncRNAs and miRNAs with diagnostic value in LSCC.
Keywords: laryngeal squamous cell carcinoma, long non-coding lncRNAs, competing endogenous RNA network, cytoplasm
Introduction
Laryngeal squamous cell carcinoma (LSCC) is a common malignant tumor in head and neck and occupies approximately 90% of the larynx cancers.1 Studies show that the incidence and mortality rate of LSCC have been on the rise in the past few years.2,3 Although diagnostic strategies have been improved, nearly 60% of LSCC patients are diagnosed at the advanced stages and have poor clinical outcomes.4,5 Therefore, it is an urgent need to investigate the underlying mechanism of LSCC initiation and progression, which will help determine effective therapeutic targets and improve overall prognosis. Currently, existing research has identified a wide variety of dysregulated RNA transcripts, and bioinformatics analysis have suggested that they may be associated with the development of LSCC.6,7 However, a major limitation of these studies is that the sample size is too small.
Non-coding RNAs, such as long non-coding lncRNA (lncRNA) and microRNA miRNAs) are discovered to involve in the pathogenesis of LSCC.8,9 More importantly, previous studies present a competing endogenous RNA (ceRNA) theory that lncRNAs act as miRNAs sponges and compete with messenger RNAs (mRNAs).10 Accordingly, numerous researchers have indicated that lncRNA-miRNA-mRNA network play essential roles in progression of cancers, including LSCC. Liu and Yu analyze the expression profiles of lncRNAs, miRNAs and mRNAs to construct a ceRNA network, and identified 5 lncRNAs biomarkers for LSCC, including TSPEAR-AS, CASK-AS1, MIR137HG, PART1 and LSAMP-AS1.11 Kong et al point out that 2 lncRNAs (AC016773.1 and C00299) may be involved in the progression of LSCC based on a lncRNA-miRNA-mRNA network analysis.12 Notably, those lncRNAs in the cytoplasm function as ceRNAs to regulate the expression of target mRNAs.13 Thus, the subcellular localization of lncRNAs can greatly improve understandings of the pathogenesis of LSCC via ceRNA network.
In this study, the 5 mRNA expression profiles, 3 miRNA expression profiles and 2 lncRNA expression profiles were obtained from Gene Expression Omnibus (GEO) database. The differentially expressed miRNAs, lncRNAs, and mRNAs (DEmiRNAs, DElncRNAs, and DEmRNAs) were identified between LSCC and adjacent tissues. Functional analysis was performed to explore the potential biological roles of DEmRNAs. Moreover, DElncRNAs-DEmRNAs-DEmiRNAs network was constructed, and finally, the subcellular localization of lncRNAs was carried out to predict key lncRNAs as ceRNAs in LSCC. The expression level and diagnostic and prognostic values of key DElncRNAs and DEmiRNAs were evaluated in external published datasets. This study will provide deeper insights of underlying molecular mechanism of LSCC by a cytoplasmic lncRNAs-miRNAs-mRNAs regulatory network.
Materials and Methods
Data Collection
The mRNA/miRNA/lncRNA expression profiles were firstly retrieved from GEO database (https://www.ncbi.nlm.nih.gov/) using keywords of (“larynx”[MeSH Terms] OR laryngeal[All Fields]) AND (“carcinoma, squamous cell”[MeSH Terms] OR squamous cell carcinoma[All Fields]). Then, the datasets were further selected based on the inclusion and exclusion criteria as follows: 1) the data types were expression profiling by array or non-coding RNA profiling by array; 2) included datasets should involve whole-genome expression data; and 3) the data was from LSCC and adjacent tissues (controls). Finally, a total of 5 mRNA datasets (GSE143224, GSE84957, GSE59652, GSE59102, and GSE51985), including 69 LSCC tissues and 50 adjacent tissues samples, were included this study. There were 3 miRNA datasets (GSE124678, GSE70289, and GSE62819), which consisted of 14 LSCC tissues and 49 adjacent tissues samples. And, 2 lncRNA datasets (GSE84957 and GSE59652) consisting 16 LSCC tissues and 16 adjacent tissues samples were obtained (Table 1).
Table 1.
The List of mRNA, miRNA and lncRNA Datasets Included in This Study.
| Datasets | GEO ID | Platform | Samples(N:P) |
|---|---|---|---|
| mRNA datasets | GSE143224 | GPL5175 [HuEx-1_0-st] Affymetrix Human Exon 1.0 ST Array [transcript (gene) version] | 11:14 |
| GSE84957 | GPL17843 Agilent-042818 Human lncRNA Micorarray 8_24_v2 [Probe Name version] | 9:9 | |
| GSE59652 | GPL13825 Arraystar Human LncRNA microarray V2.0 (Agilent-033010 Feature Number version) | 7:7 | |
| GSE59102 | GPL6480 Agilent-014850 Whole Human Genome Microarray 4x44K G4112F (Probe Name version) | 13:29 | |
| GSE51985 | GPL10558 Illumina HumanHT-12 V4.0 expression beadchip | 10:10 | |
| miRNA datasets | GSE124678 | GPL16770 Agilent-031181 Unrestricted_Human_miRNA_V16.0_Microarray (miRBase release 16.0 miRNA ID version) | 32:5 |
| GSE70289 |
GPL15018 Agilent-031181 Unrestricted_Human_miRNA_V16.0_Microarray 030840 (Feature Number version) GPL20228 Agilent-053955 Human miRBase 16.0 Human_v19 [Feature Number Version] |
12:4 | |
| GSE62819 | GPL16384 [miRNA-3] Affymetrix Multispecies miRNA-3 Array | 5:5 | |
| lncRNA datasets | GSE84957 | GPL17843 Agilent-042818 Human lncRNA Micorarray 8_24_v2 [Probe Name version] | 9:9 |
| GSE59652 | GPL13825 Arraystar Human LncRNA microarray V2.0 (Agilent-033010 Feature Number version) | 7:7 |
GEO: Gene Expression Omnibus; N: the number of control tissues; P: the number of laryngeal squamous cell carcinoma tissues.
Identification of Differentially Expressed RNAs
We used the metaMA package in R language to combine data from different datasets of each type of RNA and calculate P-value and false discovery rate (FDR).14 For mRNA/lncRNA data, the DEmRNAs/DElncRNAs were extracted between LSCC and controls with the cutoff of FDR < 0.05. Similarly, the DEmiRNAs were obtained between LSCC and adjacent tissues using the selection criterion of P < 0.05. Besides, the hierarchical clustering analyses of these DEmRNAs/DElncRNAs/DEmRNAs) were carried out using R pheatmap package (https://cran.r-project.org/package=pheatmap).
Functional Analyses of DEmRNAs
In order to understand a deep understanding of potential biological functions of DEmRNAs in LSCC, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were performed by the R clusterprofiler package, which can automate the process of biological-term classification and the enrichment analysis of gene clusters.15 There were 3 categories in GO analysis: molecular function (MF), biological process (BP), and cellular component (CC). FDR < 0.05 was set as the screening threshold for significant enrichment.
Prediction of Target mRNAs of DEmiRNAs and Functional Analyses
The target mRNAs regulated by DEmiRNAs were predicted using miRWalk (http://mirwalk.umm.uni-heidelberg.de/), which provides the largest available collection of predicted and experimentally verified miRNAs-targets pairs.16 Herein, the DEmiRNAs-mRNA network was constructed by cytoscape (http://www.cytoscape.org/). Furthermore, GO and KEGG analyses of the target mRNAs were performed with the R clusterprofiler package. The significantly enriched GO terms and KEGG pathways were screened with FDR < 0.05.
Prediction of DElncRNAs-DEmRNAs Interactions and Functional Analyses
Firstly, we utilized starbase (http://starbase.sysu.edu.cn/index.php) database to predict the DElncRNA-mRNAs pairs. Then, the cytoscape (http://www.cytoscape.org/) was used to visualize the DElncRNA-mRNAs regulatory network. To further explore the underlying biological functions of DElncRNAs, the GO and KEGG analyses of the mRNAs regulated by DElncRNAs were undertaken by the R clusterprofiler package. FDR < 0.05 was set as the cutoff criterion for the significant enrichment.
Prediction of DEmiRNAs-DElncRNAs Interactions
DIANA-LncBase provides an extensive compendium of miRNA: experimentally supported and in silico predicted miRNA recognition elements on lncRNAs.17 Herein, the DIANA-LncBase was used to predict lncRNAs targeted by DEmiRNAs, and subsequently, the DEmiRNAs-lncRNAs network was established using the cytoscape (http://www.cytoscape.org/). Herein, the relationships of DEmiRNAs and lncRNAs with the opposite expression pattern were finally retained.
CeRNA Network Construction
Increasing studies have reported that lncRNA-miRNA-mRNA ceRNA network may be involved in the pathogenesis of LSCC.11,18 In this study, the reversely regulated DElncRNAs-DEmiRNAs pairs and DEmiRNAs-DEmRNAs pairs were extracted and used for construction of DElncRNA-DEmiRNA-DEmRNA regulatory network. Notably, the abundance and subcellular localization of ceRNA components are key factors for ceRNA activity.19 LncRNAs in cytoplasm participate in ceRNA network to exert molecular regulatory functions. Therefore, lncRNA subcellular localization is essential for understanding their biological roles. LncLocator, an ensemble classifier-based predictor, has been used for the lncRNA subcellular localization.20,21 Herein, the lncRNA subcellular localization was predicted using lncLocator based on deep learning. Furthermore, those DElncRNAs in cytoplasm were screened, and accordingly, the DElncRNAs-associated ceRNA network was built.
Validation in The Cancer Genome Atlas (TCGA)
The lncRNA and miRNA gene expression profiles and clinical data of head and neck squamous cell carcinoma were downloaded by the Cancer Genome Atlas (TCGA) (http://tcga data.nci.nih.gov/). Among which, patients with LSCC who has no history of malignancy and neoadjuvant treatment were included in our study. For lncRNA expression analysis, 92 LSCC tissues and 12 normal adjacent samples from patients with LSCC were included. For miRNA expression analysis, 116 LSCC tissues and 12 normal adjacent samples from patients with LSCC were enrolled. The above data was used to verify the expression of key DElncRNAs and DEmiRNAs, respectively. In order to evaluate the diagnostic value of key DElncRNAs and DEmiRNAs in LSCC, the “pROC” package was performed to generate ROC, and the area under the ROC curve (AUC) represents the diagnostic value. When AUC value was greater than 0.7, the DElncRNAs and DEmiRNAs was thought to be able to distinguish between LSCC and normal adjacent with good specificity and sensitivity. Finally, we also evaluated the prognostic of candidate DElncRNAs and DEmiRNAs in ceRNA network, survival analysis was generated using clinical data from TCGA. Kaplan–Meier curve was plotted using the survival (https://cran.r-project.org/web/packages/survival/index.html) in R.
Evaluation the Expression Level and Diagnostic Values of Key DElncRNAs and DEmiRNAs in External Datasets (GSE127165 and GSE133632)
GSE127165 dataset was obtained from the GEO (https://www.ncbi.nlm.nih.gov/geo/), which consisted of LSCC tissues (57 samples) and paired adjacent normal mucosa tissues (57 samples). GSE133632 dataset was downloaded from the GEO, which consisted of LSCC tissues (57 samples) and paired adjacent normal mucosa tissues (57 samples). The GEO dataset GSE127165 and GSE133632 were used to confirm the expression of key DElncRNAs and DEmiRNAs, respectively. The diagnostic values of key DElncRNAs and DEmiRNAs were performed by “pROC” package.
Results
Identification of Differentially Expressed RNAs
Our differential expression analysis showed that there were 2957 DEmRNAs (1722 up-regulated mRNAs and 1235 down-regulated mRNAs) between LSCC and adjacent normal. The top 50 DEmRNAs were listed in Table S1. In addition, 23 DEmiRNAs (4 up-regulated miRNAs and 19 down-regulated miRNAs) and 61 DElncRNAs (34 up-regulated lncRNAs and 27 down-regulated lncRNAs) were also identified between LSCC and adjacent normal. These DEmiRNAs and DElncRNAs were exhibited in Tables 2 and 3. Moreover, we found that the differentially expressed RNAs can significantly distinguished LSCC and control samples (Figure S1-3).
Table 2.
The List of Differentially Expressed miRNAs in This Study.
| MiRNAs | P value | Regulation | MiRNAs | P value | Regulation |
|---|---|---|---|---|---|
| hsa-miR-4286 | 0.0038 | Up-regulation | hsa-miR-375 | 0.0001 | Down-regulation |
| hsa-miR-658 | 0.0047 | Up-regulation | hsa-miR-193a-5p | 0.0011 | Down-regulation |
| hsa-miR-1246 | 0.0063 | Up-regulation | hsa-miR-3185 | 0.0013 | Down-regulation |
| hsa-miR-554 | 0.0088 | Up-regulation | hsa-miR-1244 | 0.0015 | Down-regulation |
| hsa-miR-3654 | 1.24E-08 | Down-regulation | hsa-miR-936 | 0.0032 | Down-regulation |
| hsa-miR-582-5p | 1.05E-06 | Down-regulation | hsa-miR-532-5p | 0.0038 | Down-regulation |
| hsa-miR-3182 | 1.76E-06 | Down-regulation | hsa-miR-3154 | 0.0044 | Down-regulation |
| hsa-miR-362-3p | 2.46E-06 | Down-regulation | hsa-miR-3137 | 0.0068 | Down-regulation |
| hsa-miR-338-3p | 1.88E-05 | Down-regulation | hsa-miR-590-5p | 0.0076 | Down-regulation |
| hsa-miR-4324 | 4.08E-05 | Down-regulation | hsa-miR-3663-5p | 0.0080 | Down-regulation |
| hsa-miR-3919 | 4.51E-05 | Down-regulation | hsa-miR-1273d | 0.0081 | Down-regulation |
| hsa-miR-139-5p | 0.0001 | Down-regulation |
Table 3.
The List of Differentially Expressed lncRNAs in This Study.
| LncRNAs | FDR | Regulation | LncRNAs | FDR | Regulation |
|---|---|---|---|---|---|
| AC007620.2 | 0.0002 | Up-regulation | AL160236.2 | 0.0406 | Up-regulation |
| HMGA2-AS1 | 0.0018 | Up-regulation | AC034223.1 | 0.0440 | Up-regulation |
| AC123768.5 | 0.0029 | Up-regulation | AC108477.2 | 0.0473 | Up-regulation |
| AC023593.1 | 0.0029 | Up-regulation | AL355773.1 | 0.0029 | Down-regulation |
| DGCR9 | 0.0060 | Up-regulation | LINC02256 | 0.0095 | Down-regulation |
| DGCR5 | 0.0060 | Up-regulation | AL121872.1 | 0.0112 | Down-regulation |
| AC060234.1 | 0.0100 | Up-regulation | AC104825.1 | 0.0112 | Down-regulation |
| AL669830.1 | 0.0112 | Up-regulation | AC010636.2 | 0.0141 | Down-regulation |
| AP000487.1 | 0.0125 | Up-regulation | LINC02147 | 0.0165 | Down-regulation |
| AC024267.6 | 0.0137 | Up-regulation | LINC01484 | 0.0185 | Down-regulation |
| AC006064.2 | 0.0162 | Up-regulation | GHRLOS | 0.0225 | Down-regulation |
| MELTF-AS1 | 0.0182 | Up-regulation | AL121839.2 | 0.0229 | Down-regulation |
| AP000695.2 | 0.0210 | Up-regulation | PRKAG2-AS1 | 0.0248 | Down-regulation |
| AP000695.1 | 0.0210 | Up-regulation | ZNF503-AS1 | 0.0291 | Down-regulation |
| AC106820.5 | 0.0210 | Up-regulation | AL356259.1 | 0.0291 | Down-regulation |
| LINC01234 | 0.0245 | Up-regulation | AC005920.3 | 0.0298 | Down-regulation |
| AC092115.3 | 0.0252 | Up-regulation | HORMAD2-AS1 | 0.0311 | Down-regulation |
| AP001439.1 | 0.0267 | Up-regulation | AL713852.1 | 0.0311 | Down-regulation |
| AP000331.1 | 0.0271 | Up-regulation | AC005618.2 | 0.0311 | Down-regulation |
| AC139100.1 | 0.0272 | Up-regulation | AL359715.1 | 0.0320 | Down-regulation |
| LINC01614 | 0.0291 | Up-regulation | LINC01364 | 0.0349 | Down-regulation |
| AC005606.1 | 0.0291 | Up-regulation | AL513008.1 | 0.0361 | Down-regulation |
| AC004943.2 | 0.0300 | Up-regulation | AC129507.1 | 0.0376 | Down-regulation |
| AL354956.1 | 0.0365 | Up-regulation | AL390726.2 | 0.0396 | Down-regulation |
| AC078778.1 | 0.0376 | Up-regulation | AL353742.1 | 0.0403 | Down-regulation |
| AL445183.2 | 0.0394 | Up-regulation | BX284668.5 | 0.0406 | Down-regulation |
| AC093001.1 | 0.0401 | Up-regulation | AL049543.1 | 0.0406 | Down-regulation |
| AL049569.1 | 0.0402 | Up-regulation | LINC00278 | 0.0423 | Down-regulation |
| AC084024.3 | 0.0402 | Up-regulation | AC110619.1 | 0.0430 | Down-regulation |
| LINC02005 | 0.0403 | Up-regulation | AC022007.1 | 0.0434 | Down-regulation |
| AC079328.2 | 0.0403 | Up-regulation |
FDR: false discovery rate.
Functional Analyses of DEmRNAs
GO and KEGG analysis of DEmRNAs were performed to uncover their underlying biological roles. The results showed that these DEmRNAs were significantly enriched in 521 GO-BP terms, such as extracellular matrix/structure organization, DNA-dependent DNA replication, and neutrophil activation involved in immune response; 123 GO-CC terms, such as collagen-containing extracellular matrix, basolateral plasma membrane, extracellular matrix component and chromosomal region; and 30 GO-MF terms, such as extracellular matrix structural constituent, cell adhesion molecule binding, and cadherin binding. The top 15 GO-BP/CC/MF terms were displayed in Table 4 and Figure S4. For KEGG analysis, we found 12 significantly enriched KEGG pathways, including DNA replication, cell cycle, proteasome, fanconi anemia pathway, homologous recombination, ECM-receptor interaction, mismatch repair, IL-17 signaling pathway, nucleotide excision repair, fatty acid degradation, small cell lung cancer, and tryptophan metabolism (Table 4; Figure S4).
Table 4.
The Top 15 Significantly Enriched GO-BP/MF/CC Terms and All Significantly Enriched KEGG Pathways.
| Terms/Pathways | GO-ID | Description | No. | FDR |
|---|---|---|---|---|
| GO-BP terms | GO:0030198 | extracellular matrix organization | 110 | 9.19E-12 |
| GO:0043062 | extracellular structure organization | 120 | 2.63E-11 | |
| GO:0006261 | DNA-dependent DNA replication | 60 | 5.32E-11 | |
| GO:0002283 | neutrophil activation involved in immune response | 136 | 8.30E-11 | |
| GO:0006260 | DNA replication | 88 | 1.12E-10 | |
| GO:0042119 | neutrophil activation | 137 | 1.30E-10 | |
| GO:0002446 | neutrophil mediated immunity | 137 | 1.30E-10 | |
| GO:0043312 | neutrophil degranulation | 134 | 1.30E-10 | |
| GO:0071900 | regulation of protein serine/threonine kinase activity | 129 | 3.15E-08 | |
| GO:0043405 | regulation of MAP kinase activity | 92 | 9.36E-07 | |
| GO:0044839 | cell cycle G2/M phase transition | 67 | 1.07E-06 | |
| GO:0000086 | G2/M transition of mitotic cell cycle | 62 | 2.16E-06 | |
| GO:0097191 | extrinsic apoptotic signaling pathway | 67 | 3.09E-06 | |
| GO:0140014 | mitotic nuclear division | 77 | 7.42E-06 | |
| GO:0002009 | morphogenesis of an epithelium | 117 | 7.42E-06 | |
| GO-MF terms | GO:0005201 | extracellular matrix structural constituent | 52 | 2.87E-05 |
| GO:0030020 | extracellular matrix structural constituent conferring tensile strength | 20 | 4.43E-05 | |
| GO:0050839 | cell adhesion molecule binding | 115 | 7.55E-04 | |
| GO:0045296 | cadherin binding | 82 | 1.02E-03 | |
| GO:0003697 | single-stranded DNA binding | 35 | 1.24E-03 | |
| GO:0008094 | DNA-dependent ATPase activity | 30 | 2.23E-03 | |
| GO:0003725 | double-stranded RNA binding | 26 | 6.00E-03 | |
| GO:0019838 | growth factor binding | 40 | 6.00E-03 | |
| GO:0004674 | protein serine/threonine kinase activity | 103 | 6.00E-03 | |
| GO:0003688 | DNA replication origin binding | 10 | 6.12E-03 | |
| GO:0002020 | protease binding | 37 | 6.12E-03 | |
| GO:0016887 | ATPase activity | 100 | 7.77E-03 | |
| GO:0004386 | helicase activity | 42 | 8.36E-03 | |
| GO:0061134 | peptidase regulator activity | 56 | 8.91E-03 | |
| GO:0048037 | cofactor binding | 108 | 1.09E-02 | |
| GO-CC terms | GO:0062023 | collagen-containing extracellular matrix | 95 | 1.77E-08 |
| GO:0016323 | basolateral plasma membrane | 67 | 7.50E-08 | |
| GO:0044420 | extracellular matrix component | 26 | 7.50E-08 | |
| GO:0098687 | chromosomal region | 94 | 1.19E-07 | |
| GO:0031012 | extracellular matrix | 118 | 1.60E-06 | |
| GO:0030667 | secretory granule membrane | 80 | 1.60E-06 | |
| GO:0005657 | replication fork | 29 | 2.06E-06 | |
| GO:0070820 | tertiary granule | 50 | 1.25E-05 | |
| GO:0005788 | endoplasmic reticulum lumen | 78 | 1.25E-05 | |
| GO:1905369 | endopeptidase complex | 27 | 1.25E-05 | |
| GO:0000775 | chromosome, centromeric region | 56 | 1.38E-05 | |
| GO:0043596 | nuclear replication fork | 20 | 2.16E-05 | |
| GO:0031983 | vesicle lumen | 84 | 2.42E-05 | |
| GO:0000502 | proteasome complex | 26 | 2.55E-05 | |
| GO:0101002 | ficolin-1-rich granule | 53 | 2.71E-05 | |
| KEGG pathways | hsa03030 | DNA replication | 20 | 1.15E-04 |
| hsa04110 | Cell cycle | 45 | 1.15E-04 | |
| hsa03050 | Proteasome | 21 | 1.34E-03 | |
| hsa03460 | Fanconi anemia pathway | 23 | 1.60E-03 | |
| hsa03440 | Homologous recombination | 18 | 6.30E-03 | |
| hsa04512 | ECM-receptor interaction | 30 | 9.05E-03 | |
| hsa03430 | Mismatch repair | 12 | 9.05E-03 | |
| hsa04657 | IL-17 signaling pathway | 30 | 2.68E-02 | |
| hsa03420 | Nucleotide excision repair | 18 | 2.68E-02 | |
| hsa00071 | Fatty acid degradation | 17 | 2.97E-02 | |
| hsa05222 | Small cell lung cancer | 29 | 2.97E-02 | |
| hsa00380 | Tryptophan metabolism | 16 | 4.16E-02 |
GO-BP: Gene Ontology-Biological Process; GO-MF: Gene Ontology-Molecular Function; GO-CC: Gene Ontology-Cellular Component; KEGG: Kyoto Encyclopedia of Genes and Genomes; FDR: false discovery rate.
DEmRNAs-DEmiRNAs Interactions and Functional Analyses
The miRWalk was used to predict the interactions between DEmRNAs and DEmiRNAs, and obtained 3243 DEmRNAs-DEmiRNAs regulatory pairs (1524 DEmRNAs and 21 DEmiRNAs), including 510 up-regulated DEmiRNAs-down-regulated DEmRNAs pairs and 2733 down-regulated miRNAs-up-regulated mRNAs pairs (Figure 1). Moreover, the GO analysis revealed that these target DEmRNAs were significantly enriched in 308 GO-BP terms, 56 GO-CC terms and 8 GO-MF terms. The top 15 GO-BP terms and GO-CC terms were listed in Table S2 and Figure S5. The top 3 GO-BP terms included DNA replication, regulation of DNA metabolic process and extracellular matrix organization. Membrane ligase complex, membrane microdomain and chromosomal region were top 3 GO-CC terms. The top 3 significantly enriched GO-MF terms were extracellular matrix structural constituent conferring tensile strength, double-stranded RNA binding, and DNA-dependent ATPase activity. These genes were significantly enriched in 9 KEGG pathways, such as hepatitis C, protein digestion and absorption, and TNF signaling pathway (Table S2; Figure S5).
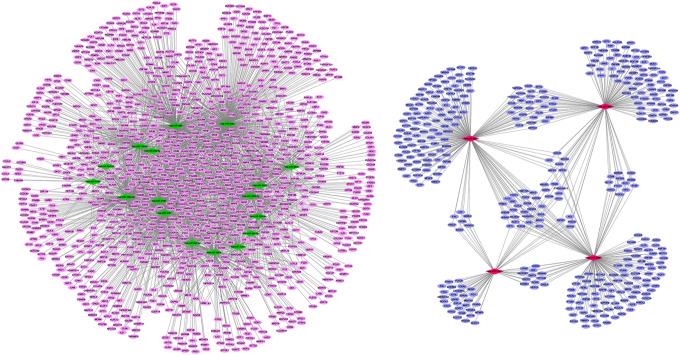
Figure 1.
DEmiRNAs-DEmRNAs regulatory network. The oval and rhombus represents DEmRNAs and DEmiRNAs, respectively. The pink color shows up-regulated mRNA and the blue color shows down-regulated mRNA. The red color shows up-regulated miRNA and the green color shows down-regulated miRNA. DEmRNA: differentially expressed mRNAs; DEmiRNA: differentially expressed miRNAs.
DElncRNAs-DEmRNAs Interactions and Functional Analyses
The target mRNAs of DElncRNAs were predicted by starbase in this study. The results showed that there were 77 DElncRNAs-mRNAs pairs, involving 12 up-regulated lncRNAs, 10 down-regulated lncRNAs, 12 up-regulated mRNAs, 10 down-regulated mRNAs, and 53 predicted mRNAs targets (Figure S6). Figure S7 showed the DElncRNAs-DEmRNAs network, including 6 up-regulated lncRNAs, 6 down-regulated lncRNAs, 12 up-regulated mRNAs, 10 down-regulated mRNAs. Furthermore, the functional analysis results of these target mRNAs showed that they were significantly enriched in 29 GO-BP terms, including chromatin silencing and establishment of protein localization to endoplasmic reticulum, and 19 GO-CC terms, such as cytosolic large ribosomal subunit and myelin sheath (Table S3). However, they were not markedly enriched in any GO-MF terms and KEGG pathways.
DEmiRNAs-DElncRNAs Interactions
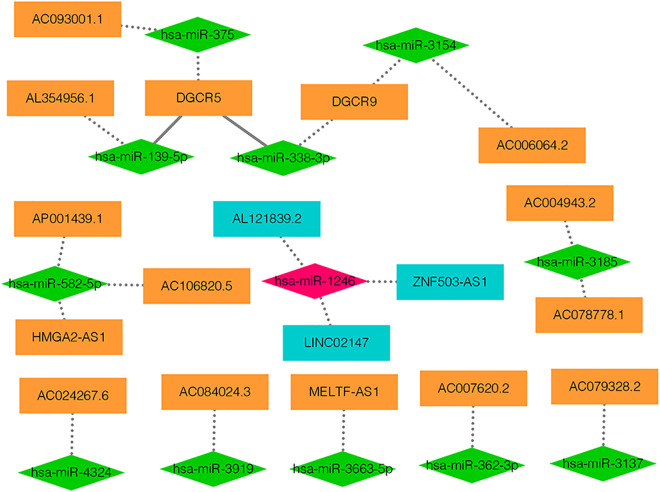
The lncRNAs targeted by DEmiRNAs were predicted by using LncBase database. Figure S8 showed DEmiRNAs-DElncRNAs network, which contained 47 nodes (18 DEmiRNAs and 29 DElncRNAs) and 45 edges. Notably, 43 DEmiRNAs-DElncRNAs pairs were predicted and 2 DEmiRNAs-DElncRNAs pairs (up-regulated DGCR5 and down-regulated miR-338-3p and up-regulated DGCR5-down-regulated has-miR-139-5p) were experimentally validated. Afterward, the opposite expression relationships between DEmiRNAs and DElncRNAs were further extracted and visualized in Figure 2. We found that there were 15 up-regulated lncRNAs, 3 down-regulated lncRNAs, one up-regulated mRNA (has-miR-1246), 11 down-regulated mRNAs in this network (Figure 2).
Figure 2.
DEmiRNAs-DElncRNAs pairs with the opposite expression. The orange rectangle shows the up-regulated lncRNA and dark green rectangle shows the down-regulated lncRNA. The red rhombus shows up-regulated miRNA and green rhombus shows down-regulated miRNA. DElncRNA: differentially expressed lncRNAs; DEmiRNA: differentially expressed miRNAs.
CeRNA Network Construction
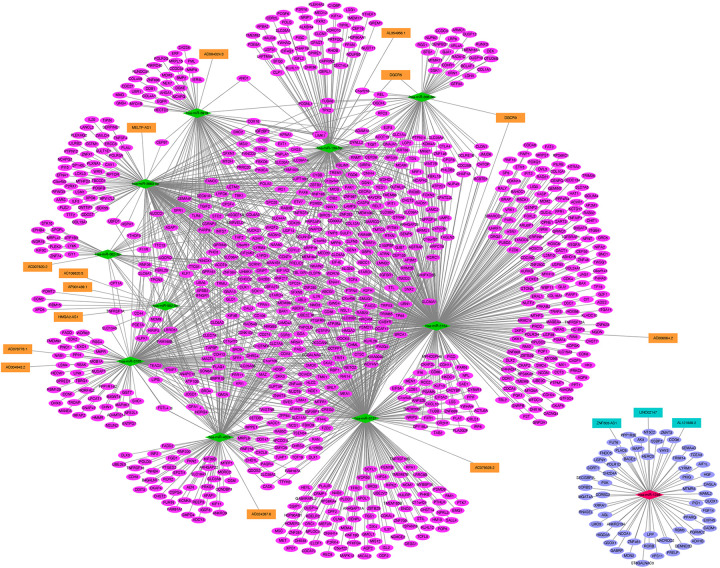
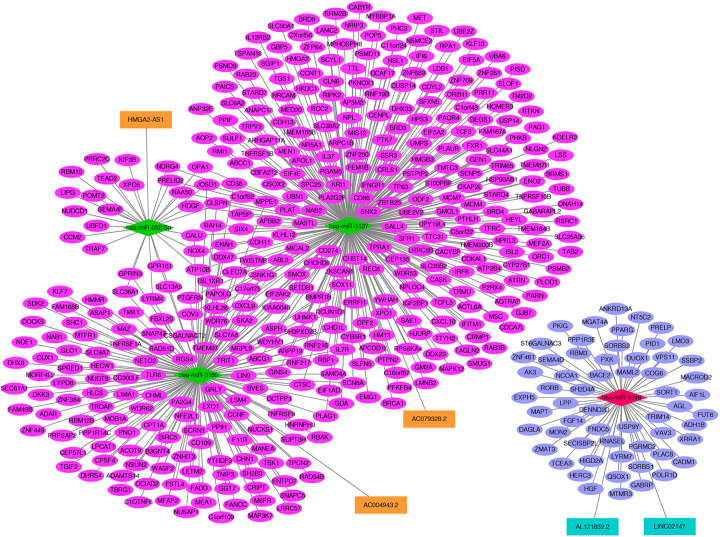
The inverse expression pairs (DEmiRNAs-DElncRNAs pairs and DEmiRNAs-DEmRNAs pairs) were extracted and used for ceRNA network construction. As shown in Figure 3, the network included 995 nodes (11 DEmiRNAs, 17 DElncRNAs and 967 DEmRNAs) and 1733 edges (3 up-regulated DEmiRNAs-down-regulated DElncRNAs pairs, 16 down-regulated DEmiRNAs-up-regulated DElncRNAs pairs, 59 up-regulated DEmiRNAs-down-regulated DEmRNAs pairs, 1655 up-regulated DEmiRNAs-down-regulated DElncRNAs pairs). DGCR5-has-miR-338-3p/has-miR-139-5p was in this ceRNA network. More notably, existing evidence has demonstrated that lncRNAs in the cytoplasm can competitively bind miRNAs by acting as ceRNAs to regulate expression of mRNAs targets. Therefore, the subcellular localization of 17 DElncRNA in ceRNA network was performed by lncLocator. We found that 5 DElncRNAs (down-regulated AL121839.2, down-regulated LINC02147, up-regulated AC079328.2, up-regulated AC004943.2 and up-regulated HMGA2-AS1) may locate in the cytoplasm (Table S4). Subsequently, a DElncRNAs in the cytoplasm-DEmiRNAs-DEmRNAs network was built, including 3 lncRNAs (HMGA2-AS1, AC004943.2 and AC079328.2), 2 down-regulated lncRNAs (AL121839.2 and LINC02147), up-regulated has-miR-1246, 3 down-regulated miRNAs (has-miR-582-5p, has-miR-3137 and has-miR-3185), 404 up-regulated mRNAs and 59 down-regulated mRNAs (Figure 4). Moreover, AL121839.2 and LINC021147 were interacted with has-miR-1246. AC004943.2, AC079328.2 and HMGA2-AS1 targeted has-miR-3185, has-miR-3137 and has-miR-582-5p, respectively (Figure 4).
Figure 3.
DEmiRNAs-DElncRNAs-DEmRNAs regulatory network. The orange rectangle shows the up-regulated lncRNA and dark green rectangle shows the down-regulated lncRNA. The red rhombus shows up-regulated miRNA and green rhombus shows down-regulated miRNA. The rose red oval shows the up-regulated mRNAs and blue oval shows down-regulated mRNAs. DElncRNA: differentially expressed lncRNAs; DEmiRNA: differentially expressed miRNAs; DEmRNA: differentially expressed mRNAs.
Figure 4.
The cytoplasm-DEmiRNAs-DEmRNAs network. The orange rectangle indicates the up-regulated lncRNA and dark green rectangle shows the down-regulated lncRNA. The red rhombus indicates up-regulated miRNA and green rhombus indicates down-regulated miRNA. The rose red oval shows the up-regulated mRNAs and blue oval shows down-regulated mRNAs. DElncRNA: differentially expressed lncRNAs; DEmiRNA: differentially expressed miRNAs; DEmRNA: differentially expressed mRNAs.
Validation the Key DElncRNAs and DEmiRNAs in the TCGA
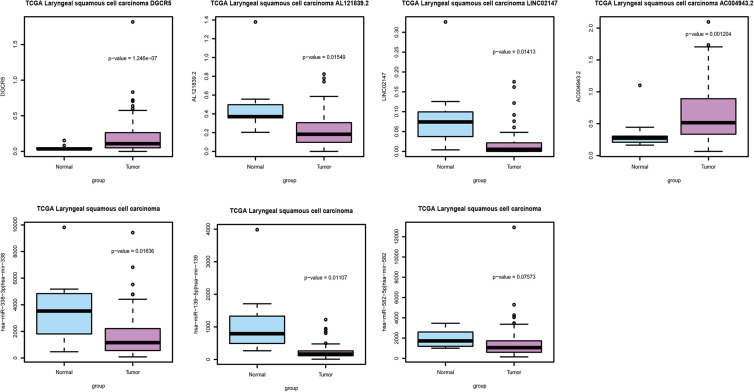
The expression of 4 DElncRNAs (DGCR5, AL121839.2, LINC02147 and AC004943.2) and 3 DEmiRNAs (has-miR-338-3p, has-miR-139-5p and has-miR-582-5p) were verified in TCGA dataset. As presented in Figure 5, the expression patterns of the remaining most of the selected 4 DElncRNAs and 3 DEmiRNAs were consistent with the results of GEO. Two DElncRNAs (AL121839.2 and LINC02147) and 3 DEmiRNAs (has-miR-338-3p, has-miR-139-5p and has-miR-582-5p) were down-regulated while 2 DElncRNAs (DGCR5 and AC004943.2) were up-regulated in LSCC, suggesting that our results were convincing.
Figure 5.
Validation in the TCGA dataset. The x-axis shows normal adjacent (green color) and LSCC (red color) groups and y-axis shows a log2 transformation to the intensities.
ROC Curve and Survival Analysis
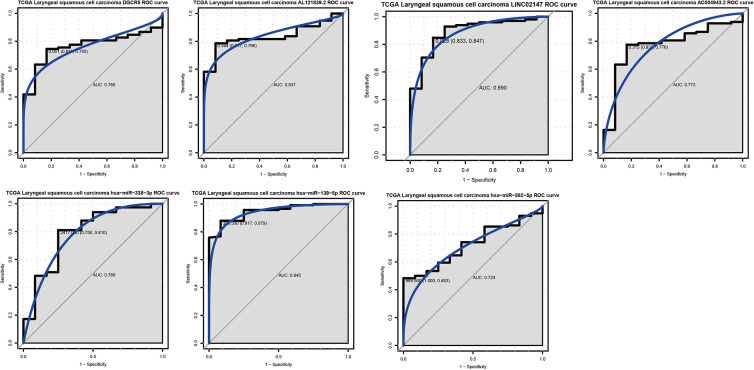
We evaluated the diagnostic value of 4 DElncRNAs (DGCR5, AL121839.2, LINC02147 and AC004943.2) and 3 DEmiRNAs (has-miR-338-3p, has-miR-139-5p and has-miR-582-5p) in LSCC. DGCR5 (AUC = 0.765), AL121839.2 (AUC = 0.837), LINC02147 (AUC = 0.890), AC004943.2 (AUC = 0.773), has-miR-338-3p (AUC = 0.789), has-miR-139-5p (AUC = 0.945) and has-miR-582-5p (AUC = 0.724) were capable of discriminating LSCC and normal controls (Figure 6). In addition, we assessed the prognostic value of these genes in LSCC. Among which, only AL121839.2 was associated with the survival of patients with LSCC. (Figure S9).
Figure 6.
ROC analysis. The AUC was analyzed to evaluate the performance of each DElncRNAs and DEmiRNAs. The x-axis indicated 1-specificity and y-axis indicated sensitivity.
Evaluation the Expression Level and Diagnostic Values of Key DElncRNAs and DEmiRNAs in External Datasets (GSE127165 and GSE133632)
Four key DElncRNAs (DGCR5, AL121839.2, LINC02147 and AC004943.2) were selected to verify in GSE127165. Three key DEmiRNAs (has-miR-338-3p, has-miR-139-5p and has-miR-582-5p) were selected to verify in GSE133632. The gene differential expression analysis found that DGCR5 and AC004943.2 were significantly up-regulated while AL121839.2 and LINC02147, has-miR-338-3p, has-miR-139-5p and has-miR-582-5p were significantly down-regulated, which were consistent with our integration analysis (Figure S10). The ROC results displayed that DGCR5 (AUC = 0.721), AL121839.2 (AUC = 0.936), LINC02147 (AUC = 0.948), AC004943.2 (AUC = 0.729), has-miR-338-3p (AUC = 0.704), has-miR-139-5p (AUC = 0.921) and has-miR-582-5p (AUC = 0.825) were capable of discriminating LSCC and normal controls (Figure S11).
Discussion
Previous evidences have demonstrated that non-coding RNAs, such as miRNAs and lncRNAs, play crucial and complex roles in tumor occurrence and progression.22-24 LncRNAs can regulate mRNA expression by competitively binding to share miRNAs.25 Increasing studies suggest that the lncRNA-miRNA-mRNA axis is involved in the carcinogenesis of LSCC. For example, Wang et al report that lncRNA NEAT1 is highly expressed in LSCC tissues compared with those adjacent non-neoplastic tissues. And experimental data suggests that lncRNA NEAT1 accelerates LSCC progression via regulating miR-107/CDK6 pathway.26 A recent study shows that the expression of lncRNA DLEU2 is significantly increased in LSCC patients and this lncRNA acts as a ceRNA to regulate PIK3CD expression by binding to target miR-30c-5p, thereby activating the Akt signaling pathway.27 Notably, the integrated ceRNA network analysis with large-scale high-throughput sequence data greatly contributes to explore underlying molecular mechanisms of various cancers, including LSCC.12,28
In this current study, we identified 2957 DEmRNAs, 61DElncRNAs and 23 DEmiRNAs, and constructed a lncRNA-miRNA-mRNA ceRNA network by analyzing the expression profiles of LSCC and normal adjacent. Firstly, functional enrichment analysis of DEmRNAs showed that they were mainly enriched in many GO terms associated with extracellular matrix organization, and several significant KEGG pathways such as DNA replication and cell cycle. Overwhelming evidence has indicated that these key biological processes are closely associated with the initiation and development of cancers.29,30 Moreover, functional analyses of the target DEmRNAs of DEmiRNAs revealed that these DEmRNAs also significantly enriched in DNA replication and extracellular matrix organization. Therefore, we inferred that DEmRNAs and DEmiRNAs may be involved in pathogenesis of LSCC by regulating these key biological processes. Notably, 2 pairs (up-regulated lncRNA DGCR5-down-regulated has-miR-338-3p/has-miR-139-5p) in ceRNA network had been experimentally verified. Tang et al suggest that lncRNA DCGR5 is dramatically up-regulated in human LSCC Hep-2 R cells, which is consistent with our results. Besides, they highlight that silencing lncRNA DCGR5 increases the radiosensitivity of human laryngeal carcinoma cells via targeting downstream miR-195.31 Later, this research group investigates the relationship between lncRNA DCGR5 and cancer stem cells (CSC)-like properties of human LSCC lines, and finds that lncRNA DCGR5 can induce CSC-like properties by regulating miR-506 and Wnt signaling pathway in radioresistant laryngeal carcinoma cells.32 Previous studies have demonstrated that has-miR-338-3p is down-regulated in many cancers, such as colorectal cancer cells, breast cancer and gastric cancer,33-35 which is similar to our result. In this study, has-miR-338-3p was significantly down-regulated in LSCC tissues compared to adjacent tissues. We also found that has-miR-139-5p was also reduced in LSCC tissues than adjacent tissues. Luo et al suggest that miR-139 can target CXCR4 and suppress the proliferative, migratory and metastatic ability of LSCC Hep-2 cells.36 Cybula et al previously report that expression of miR-139-3p was down-regulated in laryngeal cancer.37 Therefore, we speculated that lncRNA DCGR5-has-miR-338-3p/has-miR-139-5p axis may be involved the progression of LSCC. However, the detailed functional mechanisms of DCGR5-has-miR-338-3p/has-miR-139-5p axis on LSCC development still need to be elaborated in future.
Existing evidence has shown that numerous lncRNAs exert key biological roles in the nucleus, such as transcriptional regulation and alternative splicing, while many lncRNAs in cytoplasm serve as ceRNAs to control mRNA stability or translation.13,38 In this study, the subcellular localization of lncRNAs in ceRNA network was carried out. Five lncRNAs (down-regulated AL121839.2, down-regulated LINC021147, up-regulated AC004943.2, up-regulated AC079328.2 and up-regulated HMGA2-AS1) were in the cytoplasm. Then, a cytoplasmic lncRNAs-miRNAs-mRNAs regulatory network was extracted. We found that AL121839.2 and LINC02147 were closely interacted with down-regulated has-miR-1246. In addition, AC004943.2, AC079328.2 and HMGA2-AS1 respectively targeted down-regulated has-miR-3185, has-miR-3137 and has-miR-582-5p. Recently, Huang et al show that miR-1246 is up-regulated in LSCC tissues, which is consistent with our result. Besides, they highlight that lack of miR-1246 in Small extracellular vesicle can abrogate the tumorigenesis of LSCC.39 However, few reports have investigated the underlying roles of 5 key cytoplasmic lncRNAs on LSCC. Herein, we preliminarily analyzed the relationships between these key lncRNAs and potential miRNA targets based on the ceRNA network. Whether they act as miRNA sponges to participate in the LSCC progression needs further investigation.
Conclusion
In summary, we identified 2957 DEmRNAs, 61DElncRNAs and 23 DEmiRNAs, and reconstructed an lncRNA-miRNA-mRNA ceRNA network by an integrated analysis. Our findings showed that DEmRNAs were significantly related to many cancer-related pathways, including DNA replication and extracellular matrix organization. LncRNA DCGR5-has-miR-338-3p/has-miR-139-5p axis may be involved in the progression of LSCC. Additionally, 5 cytoplasmic lncRNAs (AL121839.2, LINC021147, AC004943.2, AC079328.2 and HMGA2-AS1) may act as ceRNAs and involve in the pathogenesis of LSCC. DGCR5, AL121839.2, LINC02147, AC004943.2, has-miR-338-3p, has-miR-139-5p and has-miR-582-5p were capable of discriminating LSCC and normal controls. Meanwhile, AL121839.2 had potential prognostic value for LSCC. Although we have identified multiple novel biomarkers associated with diagnostic and prognostic value, there remain limitations in our work. Considering the limitation, we are collecting samples of LSCC, further confirmation experiments will be performed in our following research. Furthermore, the clinical information should be collected to assess the diagnostic value of biomarkers for LSCC patients. Finally, the biological significances of key DElncRNAs and DEmiRNAs will be studied in model systems or cell lines.
Supplemental Material
Supplemental Material, Figure_S1 for LncRNA-Associated ceRNA Network Reveals Novel Potential Biomarkers of Laryngeal Squamous Cell Carcinoma by Zhibin Jing, Sitong Guo, Peng Zhang and Zheng Liang in Technology in Cancer Research & Treatment
Supplemental Material, Figure_S10_-revised for LncRNA-Associated ceRNA Network Reveals Novel Potential Biomarkers of Laryngeal Squamous Cell Carcinoma by Zhibin Jing, Sitong Guo, Peng Zhang and Zheng Liang in Technology in Cancer Research & Treatment
Supplemental Material, Figure_S11-revised for LncRNA-Associated ceRNA Network Reveals Novel Potential Biomarkers of Laryngeal Squamous Cell Carcinoma by Zhibin Jing, Sitong Guo, Peng Zhang and Zheng Liang in Technology in Cancer Research & Treatment
Supplemental Material, Figure_S2 for LncRNA-Associated ceRNA Network Reveals Novel Potential Biomarkers of Laryngeal Squamous Cell Carcinoma by Zhibin Jing, Sitong Guo, Peng Zhang and Zheng Liang in Technology in Cancer Research & Treatment
Supplemental Material, Figure_S3 for LncRNA-Associated ceRNA Network Reveals Novel Potential Biomarkers of Laryngeal Squamous Cell Carcinoma by Zhibin Jing, Sitong Guo, Peng Zhang and Zheng Liang in Technology in Cancer Research & Treatment
Supplemental Material, Figure_S4-revised for LncRNA-Associated ceRNA Network Reveals Novel Potential Biomarkers of Laryngeal Squamous Cell Carcinoma by Zhibin Jing, Sitong Guo, Peng Zhang and Zheng Liang in Technology in Cancer Research & Treatment
Supplemental Material, Figure_S5-revised for LncRNA-Associated ceRNA Network Reveals Novel Potential Biomarkers of Laryngeal Squamous Cell Carcinoma by Zhibin Jing, Sitong Guo, Peng Zhang and Zheng Liang in Technology in Cancer Research & Treatment
Supplemental Material, Figure_S6-revised for LncRNA-Associated ceRNA Network Reveals Novel Potential Biomarkers of Laryngeal Squamous Cell Carcinoma by Zhibin Jing, Sitong Guo, Peng Zhang and Zheng Liang in Technology in Cancer Research & Treatment
Supplemental Material, Figure_S7-revised for LncRNA-Associated ceRNA Network Reveals Novel Potential Biomarkers of Laryngeal Squamous Cell Carcinoma by Zhibin Jing, Sitong Guo, Peng Zhang and Zheng Liang in Technology in Cancer Research & Treatment
Supplemental Material, Figure_S8-revised for LncRNA-Associated ceRNA Network Reveals Novel Potential Biomarkers of Laryngeal Squamous Cell Carcinoma by Zhibin Jing, Sitong Guo, Peng Zhang and Zheng Liang in Technology in Cancer Research & Treatment
Supplemental Material, Figure_S9-revised for LncRNA-Associated ceRNA Network Reveals Novel Potential Biomarkers of Laryngeal Squamous Cell Carcinoma by Zhibin Jing, Sitong Guo, Peng Zhang and Zheng Liang in Technology in Cancer Research & Treatment
Supplemental Material, Supplementary_tables_(1) for LncRNA-Associated ceRNA Network Reveals Novel Potential Biomarkers of Laryngeal Squamous Cell Carcinoma by Zhibin Jing, Sitong Guo, Peng Zhang and Zheng Liang in Technology in Cancer Research & Treatment
Abbreviations
- LSCC
laryngeal squamous cell carcinoma
- lncRNA
long non-coding lncRNA
- miRNAs
microRNA
- ceRNA
competing endogenous RNA
- GEO
Gene Expression Omnibus
- mRNAs
messenger RNAs
- FDR
false discovery rate
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- CSC
cancer stem cells
- TCGA
The Cancer Genome Atlas
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Natural Science Foundation of Tianjin City (Grant No. 17JCYBJC25600).
ORCID iD: Zheng Liang  https://orcid.org/0000-0001-7033-5533
https://orcid.org/0000-0001-7033-5533
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Genden EM, Ferlito A, Silver CE, et al. Evolution of the management of laryngeal cancer. Oral Oncol. 2007;43(5):431–439. [DOI] [PubMed] [Google Scholar]
- 2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- 3. Lu C, Shi X, Wang AY, et al. RNA-Seq profiling of circular RNAs in human laryngeal squamous cell carcinomas. Mol Cancer. 2018;17(1):86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Strojan PH, Haigentz M, Bradford CR, et al. Chemoradiotherapy vs. total laryngectomy for primary treatment of advanced laryngeal squamous cell carcinoma. Oral Oncol. 2013;49(4):283–286. [DOI] [PubMed] [Google Scholar]
- 5. Nicolau-Neto P, Severo Ramundo M, Valverde P, et al. Transcriptome analysis identifies ALCAM overexpression as a prognosis biomarker in laryngeal squamous cell carcinoma. Cancers (Basel). 2020;12(2):470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang Y, Chen Y, Yu JH, Liu GM, Huang ZG. Integrated transcriptome analysis reveals miRNA-mRNA crosstalk in laryngeal squamous cell carcinoma. Genomics. 2014;104(4):249–256. [DOI] [PubMed] [Google Scholar]
- 7. Guan GF, Zheng Y, Wen LJ, et al. Gene expression profiling via bioinformatics analysis reveals biomarkers in laryngeal squamous cell carcinoma. Mol Med Rep. 2015;12(2):2457–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li R, Zhan JD, Li X, et al. Long noncoding RNA FOXD2-AS1 enhances chemotherapeutic resistance of laryngeal squamous cell carcinoma via STAT3 activation. Cell Death Dis. 2020;11(1):41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ren J, Zhu D, Liu M, Sun Y, Tian L. Downregulation of miR-21 modulates RAS expression to promote apoptosis and suppress invasion of laryngeal squamous cell carcinoma. Eur J Cancer. 2010;46(18):3409–3416. [DOI] [PubMed] [Google Scholar]
- 10. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language. Cell. 2011;146(3):353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu Y, Ye F. Construction and integrated analysis of crosstalking ceRNAs networks in laryngeal squamous cell carcinoma. PeerJ. 2019;7:e7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kong X, Qi J, Yan Y, et al. Comprehensive analysis of differentially expressed profiles of lncRNAs, mRNAs, and miRNAs in laryngeal squamous cell carcinoma in order to construct a ceRNA network and identify potential biomarkers. J Cell Biochem. 2019;120(10):17963–17974. [DOI] [PubMed] [Google Scholar]
- 13. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marot G, Foulley JL, Mayer CD, Jaffrézic F. Moderated effect size and P-value combinations for microarray meta-analyses Bioinformatics. 2009;25(20):2692–2699. [DOI] [PubMed] [Google Scholar]
- 15. Yu GC, Wang LG, Han YY, He QY. . clusterProfiler: an R Package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dweep H, Gretz N. . miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12(8):697. [DOI] [PubMed] [Google Scholar]
- 17. Paraskevopoulou MD, Vlachos IS, Karagkouni D, et al. DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucleic Acids Res. 2016;44(Database issue):D231–D238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kong X, Qi J, Yan Y, et al. Comprehensive analysis of differentially expressed profiles of lncRNAs, mRNAs, and miRNAs in laryngeal squamous cell carcinoma in order to construct a ceRNA network and identify potential biomarkers. J Cell Biochem. 2019;120(10):17963–17974. [DOI] [PubMed] [Google Scholar]
- 19. Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52(10):710–718. [DOI] [PubMed] [Google Scholar]
- 20. Cao Z, Pan X, Yang Y, Huang Y, Shen HB. The lncLocator: a subcellular localization predictor for long non-coding RNAs based on a stacked ensemble classifier. Bioinformatics. 2018;34(14):2185–2194. [DOI] [PubMed] [Google Scholar]
- 21. Xia Q, Zhang L, Yan H, Yu L, Shan W, Jiang H. LUCAT1 contributes to MYRF-dependent smooth muscle cell apoptosis and may facilitate aneurysm formation via the sequestration of miR-199a-5p. Cell Biol Int. 2020;44(3):755–763. [DOI] [PubMed] [Google Scholar]
- 22. Li Z, Lu X, Liu YF, et al. Gain of LINC00624 enhances liver cancer progression by disrupting the HDAC6-TRIM28-ZNF354C corepressor complex [published online September 01, 2020]. Hepatology. 2020. [DOI] [PubMed] [Google Scholar]
- 23. Zhou C, Yi C, Yi YX, et al. LncRNA PVT1 promotes gemcitabine resistance of pancreatic cancer via activating Wnt/β-catenin and autophagy pathway through modulating the miR-619-5p/Pygo2 and miR-619-5p/ATG14 axes. Mol Cancer. 2020;19(1):118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wong CM, Tsang FH, Ng IO. Non-coding RNAs in hepatocellular carcinoma: molecular functions and pathological implications. Nat Rev Gastroenterol Hepatol. 2018;15(3):137–151. [DOI] [PubMed] [Google Scholar]
- 25. Militello G, Weirick T, John D, Doring C, Dimmeler S, Uchida S. Screening and validation of lncRNAs and circRNAs as miRNA sponges. Brief Bioinform. 2017;18(5):780–788. [DOI] [PubMed] [Google Scholar]
- 26. Wang P, Wu T, Zhou H, et al. Long noncoding RNA NEAT1 promotes laryngeal squamous cell cancer through regulating miR-107/CDK6 pathway. J Exp Clin Cancer Res. 2016;35:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li XM, Xu F, Meng Q, et al. Long noncoding RNA DLEU2 predicts a poor prognosis and enhances malignant properties in laryngeal squamous cell carcinoma through the miR-30c-5p/PIK3CD/Akt axis. Cell Death Dis. 2020;11(6):472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bai Y, Long J, Liu ZS, et al. Comprehensive analysis of a ceRNA network reveals potential prognostic cytoplasmic lncRNAs involved in HCC progression. J Cell Physiol. 2019;234(10):18837–18848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Walker C, Mojares E, Del Río Hernández A. Role of extracellular matrix in development and cancer progression. Int J Mol Sci. 2018;19(10):3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Boyer AS, Walter D, Sørensen CS. DNA replication and cancer: from dysfunctional replication origin activities to therapeutic opportunities. Semin Cancer Biol. 2016;37-38:16–25. [DOI] [PubMed] [Google Scholar]
- 31. Tang T, Shan G, Zeng F. Knockdown of DGCR5 enhances the radiosensitivity of human laryngeal carcinoma cells via inducing miR-195. J Cell Physiol. 2019;234(8):12918–12925. [DOI] [PubMed] [Google Scholar]
- 32. Tang T, Shan G. DGCR5 promotes cancer stem cell-like properties of radioresistant laryngeal carcinoma cells by sponging miR-506 via Wnt pathway. J Cell Physiol. 2019;234(10):18423–18431. [DOI] [PubMed] [Google Scholar]
- 33. Li W, He Y, Cheng ZL. Long noncoding RNA XIST knockdown suppresses the growth of colorectal cancer cells via regulating microRNA-338-3p/PAX5 axis [published online August 18, 2020]. Eur J Cancer Prev. 2020. [DOI] [PubMed] [Google Scholar]
- 34. He JJ, Wang J, Li SH, Li T, Chen KL, Zhang SJ. Hypoxia-inhibited miR-338-3p suppresses breast cancer progression by directly targeting ZEB2. Cancer Sci. 2020:111(10):3550–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zheng JJ, Que QY, Xu HT, et al. Hypoxia activates SOX5/Wnt/β-catenin signaling by suppressing MiR-338-3p in gastric cancer. Technol Cancer Res Treat. 2020;19:1533033820905825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Luo HN, Wang ZH, Sheng Y, et al. MiR-139 targets CXCR4 and inhibits the proliferation and metastasis of laryngeal squamous carcinoma cells. Med Oncol. 2014;31(1):789. [DOI] [PubMed] [Google Scholar]
- 37. Cybula M, Wieteska Ł, Józefowicz-Korczyńska M, Karbownik MS, Grzelczyk WL, Szemraj J. New miRNA expression abnormalities in laryngeal squamous cell carcinoma. Cancer Biomark. 2016;16(4):559–568. [DOI] [PubMed] [Google Scholar]
- 38. Rashid F, Shah A, Shan G. Long non-coding RNAs in the cytoplasm. Genom Proteom Bioinform. 2016;14(2):73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huang Q, Hsueh CY, Guo Y, Wu XF, Li JY, Zhou L. Lack of miR-1246 in small extracellular vesicle blunts tumorigenesis of laryngeal carcinoma cells by regulating Cyclin G2. IUBMB Life. 2020;72(7):1491–1503. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, Figure_S1 for LncRNA-Associated ceRNA Network Reveals Novel Potential Biomarkers of Laryngeal Squamous Cell Carcinoma by Zhibin Jing, Sitong Guo, Peng Zhang and Zheng Liang in Technology in Cancer Research & Treatment
Supplemental Material, Figure_S10_-revised for LncRNA-Associated ceRNA Network Reveals Novel Potential Biomarkers of Laryngeal Squamous Cell Carcinoma by Zhibin Jing, Sitong Guo, Peng Zhang and Zheng Liang in Technology in Cancer Research & Treatment
Supplemental Material, Figure_S11-revised for LncRNA-Associated ceRNA Network Reveals Novel Potential Biomarkers of Laryngeal Squamous Cell Carcinoma by Zhibin Jing, Sitong Guo, Peng Zhang and Zheng Liang in Technology in Cancer Research & Treatment
Supplemental Material, Figure_S2 for LncRNA-Associated ceRNA Network Reveals Novel Potential Biomarkers of Laryngeal Squamous Cell Carcinoma by Zhibin Jing, Sitong Guo, Peng Zhang and Zheng Liang in Technology in Cancer Research & Treatment
Supplemental Material, Figure_S3 for LncRNA-Associated ceRNA Network Reveals Novel Potential Biomarkers of Laryngeal Squamous Cell Carcinoma by Zhibin Jing, Sitong Guo, Peng Zhang and Zheng Liang in Technology in Cancer Research & Treatment
Supplemental Material, Figure_S4-revised for LncRNA-Associated ceRNA Network Reveals Novel Potential Biomarkers of Laryngeal Squamous Cell Carcinoma by Zhibin Jing, Sitong Guo, Peng Zhang and Zheng Liang in Technology in Cancer Research & Treatment
Supplemental Material, Figure_S5-revised for LncRNA-Associated ceRNA Network Reveals Novel Potential Biomarkers of Laryngeal Squamous Cell Carcinoma by Zhibin Jing, Sitong Guo, Peng Zhang and Zheng Liang in Technology in Cancer Research & Treatment
Supplemental Material, Figure_S6-revised for LncRNA-Associated ceRNA Network Reveals Novel Potential Biomarkers of Laryngeal Squamous Cell Carcinoma by Zhibin Jing, Sitong Guo, Peng Zhang and Zheng Liang in Technology in Cancer Research & Treatment
Supplemental Material, Figure_S7-revised for LncRNA-Associated ceRNA Network Reveals Novel Potential Biomarkers of Laryngeal Squamous Cell Carcinoma by Zhibin Jing, Sitong Guo, Peng Zhang and Zheng Liang in Technology in Cancer Research & Treatment
Supplemental Material, Figure_S8-revised for LncRNA-Associated ceRNA Network Reveals Novel Potential Biomarkers of Laryngeal Squamous Cell Carcinoma by Zhibin Jing, Sitong Guo, Peng Zhang and Zheng Liang in Technology in Cancer Research & Treatment
Supplemental Material, Figure_S9-revised for LncRNA-Associated ceRNA Network Reveals Novel Potential Biomarkers of Laryngeal Squamous Cell Carcinoma by Zhibin Jing, Sitong Guo, Peng Zhang and Zheng Liang in Technology in Cancer Research & Treatment
Supplemental Material, Supplementary_tables_(1) for LncRNA-Associated ceRNA Network Reveals Novel Potential Biomarkers of Laryngeal Squamous Cell Carcinoma by Zhibin Jing, Sitong Guo, Peng Zhang and Zheng Liang in Technology in Cancer Research & Treatment