Abstract
Objective
To identify key genes in ovarian cancer using transcriptome sequencing in two cell lines: MCV152 (benign ovarian epithelial tumour) and SKOV-3 (ovarian serous carcinoma).
Methods
Differentially expressed genes (DEGs) between SKOV-3 and MCV152 were identified. Candidate genes were assessed for enrichment in gene ontology function and Kyoto Encyclopaedia of Genes and Genomes pathway. Candidate gene expression in SKOV-3 and MCV152 cells was validated using Western blots.
Results
A total of 2020 upregulated and 1673 downregulated DEGs between SKOV3 and MCV152 cells were identified that were significantly enriched in the cell adhesion function. Upregulated DEGs, such as angiopoietin 2 (ANGPT2), CD19 molecule (CD19), collagen type IV alpha 3 chain (COL4A3), fibroblast growth factor 18 (FGF18), integrin subunit beta 4 (ITGB4), integrin subunit beta 8 (ITGB8), laminin subunit alpha 3 (LAMA3), laminin subunit gamma 2 (LAMC2), protein phosphatase 2 regulatory subunit Bgamma (PPP2R2C) and spleen associated tyrosine kinase (SYK) were significantly involved in the extracellular matrix-receptor interaction pathway. Downregulated DEGs, such as AKT serine/threonine kinase 3 (AKT3), collagen type VI alpha 1 chain (COL6A1), colony stimulating factor 3 (CSF3), fibroblast growth factor 1 (FGF1), integrin subunit alpha 2 (ITGA2), integrin subunit alpha 11 (ITGA11), MYB proto-oncogene, transcription factor (MYB), phosphoenolpyruvate carboxykinase 2, mitochondrial (PCK2), placental growth factor (PGF), phosphoinositide-3-kinase adaptor protein 1 (PIK3AP1), serum/glucocorticoid regulated kinase 1 (SGK1), toll like receptor 4 (TLR4) and tumour protein p53 (TP53) were involved in PI3K-Akt signalling. Expression of these DEGs was confirmed by Western blot analyses.
Conclusion
Candidate genes enriched in cell adhesion, extracellular matrix–receptor interaction and PI3K-Akt signalling pathways were identified that may be closely associated with ovarian cancer invasion and potential targets for ovarian cancer treatment.
Keywords: Ovarian cancer, transcriptome sequencing, GO function, KEGG pathway, PI3K
This article has been previously published online as a preprint:
Chen J, Peng X and Yu M. Identification of biomarkers associated with ovarian cancer based on transcriptome sequencing. bioRxiv 728618. Preprint published online 7 August 2019. DOI: https://doi.org/10.1101/728618.
Introduction
Ovarian cancer is the seventh most common cancer among women worldwide, and the tenth most common cancer in China,1 with an estimated 239 000 new cases of ovarian cancer and 152 000 deaths worldwide each year.2 Despite improved treatment strategies, the overall survival rate in patients with ovarian cancer remains low, at about 46%, and approximately 5% of female cancer deaths are due to low ovarian cancer survival rates.3 These rates mainly result from the intricate pathogenesis of ovarian cancer, making it difficult to find effective treatment and improve prognosis.4 Therefore, gaining a better understanding of the biological mechanisms associated with ovarian cancer is important for earlier diagnosis and more effective treatment.
Genetic variation has been determined to have functional effects on human cancers,5 and molecular genetic analyses have revealed genetic alterations in several genes related to ovarian cancers, such as tumour protein p53 (p53), erb-b2 receptor tyrosine kinase 2 (ERBB2), MYC proto-oncogene, bHLH transcription factor (c-Myc) and epidermal growth factor receptor (EGFR).6,7 Previous microarray analysis has been applied to profile several genes, such as serpin family B member 2 (SERPINB2), CD24 molecule (CD24) and PDZ domain containing ring finger 3 (SEMACAP3), that may serve as molecular markers for diagnosis and therapy of ovarian cancer.8 In addition to genes, many pathways are also demonstrated to be involved in the progression of ovarian cancer. For instance, the Wnt/β-catenin pathway may mediate the initiation of epithelial ovarian cancer cells by targeting genes that regulate cell proliferation and apoptosis.9 The phosphatidylinositol 3 kinase (PI3K) pathway is found frequently altered in ovarian cancer and may serve as a therapeutic target.10 Despite these findings, improvements in clinical outcomes associated with ovarian cancer remain far from enough. Therefore, there is an urgent need to explore more biomarkers.11
The aim of the present study was to identify differentially expressed genes (DEGs) between ovarian cancer cells and normal controls through transcriptome sequencing of two cell lines, SKOV-3 (derived from an ovarian serous carcinoma) and MCV152 (derived from a benign ovarian epithelial tumour). Function analysis was subsequently performed to investigate potential associated pathways, and expression levels of genes involved in key pathways were validated at protein level.
Materials and methods
Cell lines and cell culture
Two cell lines were investigated in this study: MCV152 cells (controls), derived from SV40-transformed serous papillary cystadenoma cells transfected with telomerase hTERT (Genetics and IVF Institute, Obstetrics and Gynaecology Hospital of Fudan University, Shanghai, China); and SKOV-3 cells (cancer cells), derived from an ovarian serous carcinoma cell line (American Type Tissue Collection; Manassas, VA, USA). Both cell lines were cultured in Eagle’s minimal essential medium supplemented with 10% fetal bovine serum (Tianhang, Zhejiang, China) and maintained at 37 °C in a humidified incubator containing 5% CO2.
RNA extraction, library preparation and sequencing
Total RNA was extracted from cells using TRIzol® reagent (Life Technologies [Thermo Fisher Scientific], Carlsbad, CA, USA) according to the manufacturer’s protocol, and RNA integrity number (RIN) scores were determined using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Poly(A) mRNA was purified from 1 µg total RNA per sample, using Dynal™ oligo(dT)-attached magnetic beads (Invitrogen [Thermo fisher Scientific], Waltham, MA, USA). The mRNA was then cleaved into small fragments (approximately 200 nucleotides) in the presence of fragmentation buffer at 90 °C for 5 min. First strand cDNAs were synthesised using random hexamer-primers (Invitrogen) and cDNA synthesis mix (Invitrogen) at 25 °C for 10 min, followed by 50 °C for 50 min, and terminated at 85 °C for 5 min, and the reaction products were purified using reverse transcriptase (SuperScript® II Reverse Transcriptase, Invitrogen). Second-strand cDNA was then synthesized using Second Strand Reaction Mix, Second Strand Enzyme Mix, and RNase H (all Invitrogen) at 16 °C for 60 min, according to the manufacturer’s instructions. The blunt-end double-stranded cDNA was purified using the PureLink™ PCR Micro Kit (K310010; Invitrogen), according to the manufacturer’s instructions. The 3′ ends of DNA fragments were then adenylated using Klenow fragment of DNA polymerase (TaKaRa Bio, Shiga, Japan) and amplified by polymerase chain reaction. Suitable fragments, as judged by agarose gel electrophoresis, were enriched by PCR amplification to prepare the sequencing library. The cDNA libraries were sequenced using an Illumina HiSeq2000 sequencer (DNA Technologies Core, University of California, Davis, CA, USA) as 50 bp single-end reads (Illumina, Hayward, CA, USA).
Quality control and genome mapping
The raw reads were cleaned by removing adaptor sequences, reads with >2% undefined nucleotides (N), and low-quality reads containing > 20% of bases with qualities of <20, using FastQC software, version 0.11.9. Clean reads were then aligned to the hg38_UCSC_2013 genome using the STAR aligner software, version 2.4.0j, as previously described.5
Data processing and differential expression analysis
Gene expression values were normalized using reads per kilo base million reads (RPKM) and fragments per kilo base million reads (FPKM), then principal component analysis was performed. The DESeq2 algorithm (DESeq2_1.22.2) was applied to filter the DEGs. The significance criteria were: log2 fold change (FC) >2 and false discovery rate (FDR) <0.05. Heatmap plots were drawn in R, based on differential expression analysis, and the colour was determined by the filtering criteria. Volcano plots were also drawn in R, based on analysis of DEGs, and the colour was determined by the filtering criteria.
Gene ontology (GO) and pathway analyses
Gene ontology analysis was performed to elucidate the biological implications of unique genes in the significant or representative DEG profiles.6 GO analysis included biological process (BP), molecular function (MF) and cellular component (CC) analyses. Pathway analysis was used to reveal the significant pathway of the DEGs according to the Kyoto Encyclopaedia of Genes and Genomes (KEGG) database. Fisher’s exact test was applied to identify the significant GO categories and pathways, and FDR was used to correct the P-values.
In addition, significant GO terms (P < 0.05) were selected based on the up- and downregulated DEGs to construct a GO-Tree to summarize the function affected in this study.8 Significant pathways (P < 0.05) enriched by the up- and downregulated DEGs were selected to construct the Path-Act-Network using Cytoscape software, version 3.8.0.12
Western blots
Total protein was extracted from cells using lysis buffer, followed by determination of total protein concentration in the sample. Each protein sample (50 µl) was separated by 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis, and then transferred to polyvinylidene difluoride membranes. Membranes were blocked using 5% non-fat milk for 30 min at room temperature, then incubated with primary antibodies (1:1000 dilution; anti-ANGPT2, CD19, COL4A3, FGF18, ITGB4, ITGB8, LAMA3, LAMC2, PPP2R2C, SGK2, SYK, AKT3, COL6A1, CSF3, FGF1, ITGA2, ITGA11, MYB, PCK2, PGF, PIK3AP1, SGK1, TLR4 and TP53 (all Abcam, Cambridge, UK) at 4 °C overnight. Membranes were washed three times using 1 × Tris-buffered saline-Tween-20 (TBST), and then incubated with the corresponding horseradish peroxidase (HRP)-conjugated secondary antibody (1:5000 dilution; all Abcam) for 1 h at 37 °C, and again washed three times with 1 × TBST. GAPDH was quantified as the loading control. The blots were visualized using enhanced chemiluminescence (ECL) reagent (Cwbiotech, Beijing, China) and Image Lab software, version 5.1 (Bio-Rad Laboratories, Hercules, CA, USA).
Statistical analysis
All data were analysed using GraphPad Prism software, version 5.0 (GraphPad Prism, San Diego, CA, USA). Data are presented as mean ± SD, and differences between three or more groups were analysed using one-way analysis of variance (ANOVA). A P value < 0.05 was considered statistically significant.
Results
Data processing and DEGs selection
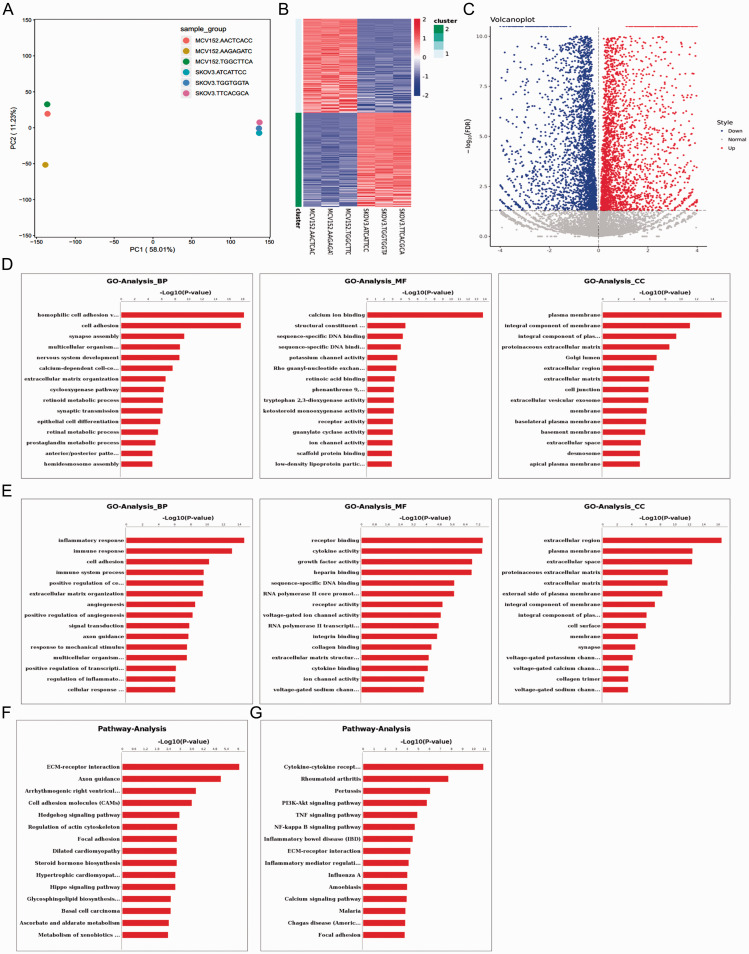
A total of 46 499 161 clean reads were obtained after quality control, and the mapping rate for sequencing data was 87.7%. Principal component analysis showed a good sample correlation between MCV152 and SKOV-3 cell lines (Figure 1A).
Figure 1.
Bioinformatics data analysis of differential gene expression between MCV152 and SKOV-3 cell lines, showing: (A) Principal component analysis by mapping all samples in a two-dimensional diagram composed of the largest component and the second largest component. Gene expression differences were small and correlation was good for RNA-seq samples with short distance; (B) Heatmap of differentially expressed genes (x axis, plotted according to sample name; y axis, plotted according to gene name); (C) Volcano plot of differentially expressed genes (x axis, log2 fold change; y axis, –log10 false discovery rate [FDR]). Red represents upregulated gene, and blue represents downregulated gene; (D) Upregulated genes and (E) downregulated genes in terms of enriched gene ontology (GO) functions, comprising biological process (BP), molecular function (MF) and cellular component (CC); and (F) Upregulated genes and (G) downregulated genes in terms of enriched pathways.
With thresholds of log2 FC >2 and FDR < 0.05, a total of 3 693 DEGs were obtained, including 2 020 that were upregulated (such as tumour associated calcium signal transducer [2TACSTD2], keratin 5 [KRT5], long intergenic non-protein coding RNA 624 [LINC00624], cadherin 16 [CDH16] and protein phosphatase 1 regulatory inhibitor subunit 1B [PPP1R1B]) and 1 673 that were downregulated (such as small nucleolar RNA, H/ACA box 9 [SNORA9], long intergenic non-protein coding RNA 174 [LINC00174], zinc finger protein 215 [ZNF215], PPFIA binding protein 2 [PPFIBP2] and neurotrophic receptor tyrosine kinase 1 [NTRK1]). The summarised DEGs are shown in a heatmap (Figure 1B) and volcano plot (Figure 1C), and the results suggest that DEGs were obviously distinguishable between the two cell lines.
GO and pathway analyses
The upregulated DEGs were significantly enriched in 401 BP terms, 134 MF terms and 66 CC terms, and the top 15 terms for each category are presented in Figure 1D. BP terms were associated with cell adhesion, synapse assembly, and extracellular matrix organization; MF terms were associated with calcium ion binding and structural constituent of the cytoskeleton; and CC terms were related to plasma membrane, and integral component of the membrane. Downregulated DEGs were significantly enriched in 665 BP terms, 139 MF terms and 82 CC terms, and again, the top 15 terms for each category are summarised in Figure 1E. BP terms were associated with inflammatory response, immune response, and cell adhesion; MF terms were related to receptor binding, and cytokine activity; and CC terms were associated with the extracellular region and plasma membrane. Pathway analysis showed that upregulated genes were significantly involved in 33 pathways, such as extracellular matrix–receptor interaction, axon guidance, and arrhythmogenic right ventricular cardiomyopathy, and cell adhesion molecules (Figure 1F). Downregulated DEGs were significantly involved in 75 pathways, such as cytokine–cytokine receptor interaction, rheumatoid arthritis, pertussis, and PI3K-Akt signalling pathways (Figure 1G).
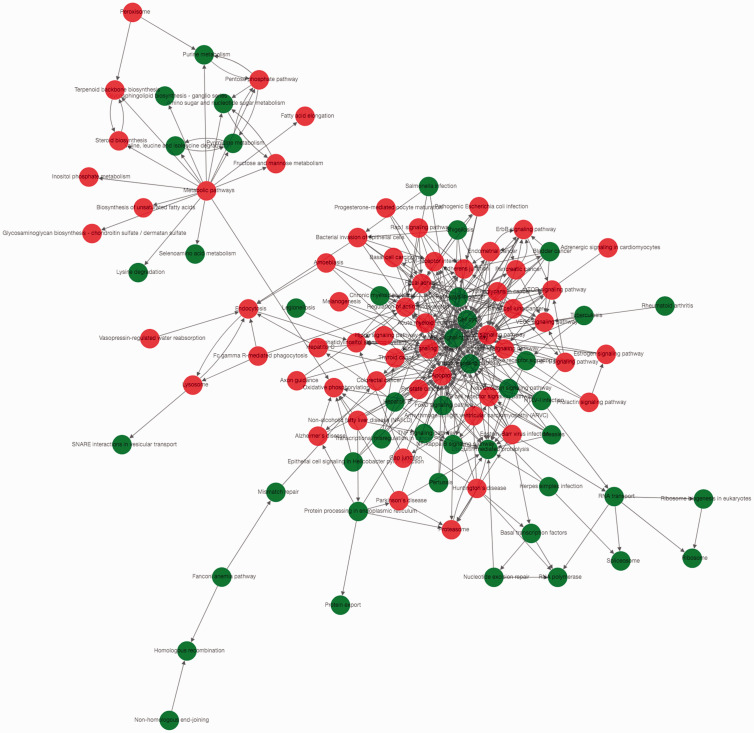
GO and pathway network analyses
The GO-Tree and Path-Act-Network are shown in Figure 2. Cell migration had interactions with six GO terms, such as regulation of cell migration, positive regulation of cell migration, and axon guidance. Axon guidance also had interactions with six GO terms, such as motor neuron axon guidance, axon choice point recognition, and retinal ganglion cell axon guidance. Angiogenesis had interactions with five terms, including patterning of blood vessels, sprouting angiogenesis, and negative regulation of angiogenesis (data not shown). The PI3K-Akt signalling pathway interacted with 25 pathways, such as the Ras signalling pathway, Rap1 signalling pathway, and p53 signalling pathway (Figure 2).
Figure 2.
The Path-Act-Network constructed based on gene ontology (GO) and pathway analysis results. Red represents the significant GO terms or pathways of upregulated genes; Green represents the significant GO terms or pathways of downregulated genes.
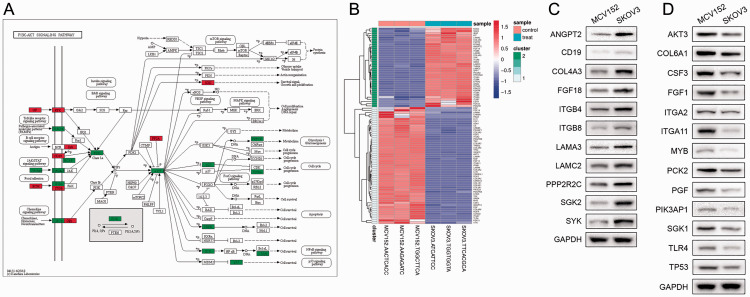
DEGs involved in PI3K-AKT signalling pathway
After analysis of RNA-seq data, the DEGs involved in PI3K-AKT signalling were selected by combining with the KEGG database. PI3K and AKT were inevitable hub nodes with the highest degree of interactions in this signalling pathway. Various signalling pathways, such as toll-like receptor signalling, cell cycle, nuclear factor-κB signalling and p53 signalling pathways also regulated the expression of PI3K and AKT (Figure 3A). Furthermore, DEGs in the PI3K-AKT signalling pathway were clustered by heatmap. For instance, the upregulated genes, nitric oxide synthase 3 (NOS3) and serum/glucocorticoid regulated kinase family member 3 (SGK3), and downregulated genes, fibroblast growth factor 21 (FGF21) and vitronectin (VTN), were clustered among of these DEGs (Figure 3B).
Figure 3.
Differentially expressed genes (DEGs) between MCV152 and SOV-3 cell lines, involved in the phosphatidylinositol 3 kinase (PI3K)-AKT signalling pathway, selected using the Kyoto Encyclopaedia of Genes and Genomes (KEGG) database and analysed by heatmap: (A) Upregulated and down-regulated genes in the PI3K-Akt signalling pathway; (B) Heatmap of genes involved in the PI3K-Akt signalling pathway (x axis, sample name; y axis, gene name). Red represents significant upregulated genes and green represents significant downregulated genes; (C) Representative Western blot showing upregulated DEGs enriched in the PI3K-Akt signalling pathway (angiopoietin 2 [ANGPT2], CD19 molecule [CD19], collagen type IV alpha 3 chain [COL4A3], fibroblast growth factor 18 [FGF18], integrin subunit beta 4 [ITGB4], integrin subunit beta 8 [ITGB8], laminin subunit alpha 3 [LAMA3], laminin subunit gamma 2 [LAMC2], protein phosphatase 2 regulatory subunit Bgamma [PPP2R2C], serum/glucocorticoid regulated kinase 2 [SGK2], and spleen associated tyrosine kinase [SYK]); (D) Representative Western blot showing downregulated DEGs including AKT serine/threonine kinase 3 (AKT3), collagen type VI alpha 1 chain (COL6A1), colony stimulating factor 3 (CSF3), fibroblast growth factor 1 (FGF1), integrin subunit alpha 2 (ITGA2), integrin subunit alpha 11 (ITGA11), MYB proto-oncogene, transcription factor (MYB), phosphoenolpyruvate carboxykinase 2, mitochondrial (PCK2), placental growth factor (PGF), phosphoinositide-3-kinase adaptor protein 1 (PIK3AP1), serum/glucocorticoid regulated kinase 1 (SGK1), toll like receptor 4 (TLR4), and tumour protein p53 (TP53).
In addition, analyses of protein levels by Western blot verified the expression of DEGs enriched in the PI3K-AKT signalling pathway. The results showed that, compared with MCV152 cells, the expression of angiopoietin 2 (ANGPT2), CD19 molecule (CD19), collagen type IV alpha 3 chain (COL4A3), fibroblast growth factor 18 (FGF18), integrin subunit beta 4 (ITGB4), integrin subunit beta 8 (ITGB8), laminin subunit alpha 3 (LAMA3), laminin subunit gamma 2 (LAMC2), protein phosphatase 2 regulatory subunit Bgamma (PPP2R2C) and spleen associated tyrosine kinase (SYK) was upregulated in SKOV3 cells (Figure 3C). The expression of AKT serine/threonine kinase 3 (AKT3), collagen type VI alpha 1 chain (COL6A1), colony stimulating factor 3 (CSF3), fibroblast growth factor 1 (FGF1), integrin subunit alpha 2 (ITGA2), integrin subunit alpha 11 (ITGA11), MYB proto-oncogene, transcription factor (MYB), phosphoenolpyruvate carboxykinase 2, mitochondrial (PCK2), placental growth factor (PGF), phosphoinositide-3-kinase adaptor protein 1 (PIK3AP1), serum/glucocorticoid regulated kinase 1 (SGK1), toll like receptor 4 (TLR4) and tumour protein p53 (TP53) was decreased in SKOV3 cells compared with MCV152 cells (Figure 3D). Western blot results were consistent with the results of bioinformatics analysis.
Discussion
In the present study, 2 020 upregulated and 1 673 downregulated DEGs were revealed between SKOV3 and MCV152 cells, and the upregulated and downregulated DEGs were found to be significantly associated with cell adhesion. Functional analyses indicated that upregulated DEGs were significantly correlated with cell adhesion and extracellular matrix–receptor interaction. Cell–cell adhesion is a fundamental biological process that defines morphogenesis of cells and tissues in multicellular organisms.13 Overexpression of cell adhesion molecules, such as Claudin 3, epithelial discoidin domain-containing receptor 1 (DDR1) and Ep-Cam, are shown to be early events in the development of ovarian cancer.14 In addition, vascular cell adhesion molecule-1 regulates ovarian cancer peritoneal metastasis.15 The signalling conveyed by cell adhesion with the underlying extracellular matrix is closely coordinated with gene regulation during normal tissue homeostasis.16 Disruption of cell adhesion, subsequent changes in adhesion-mediated signal transduction, and an increase in cell motility are characteristics in the development of invasive metastatic cancer.17 Alterations in cell–cell and cell–extracellular matrix interactions have been reported to promote cancer cell invasion and angiogenesis.18,19 Therefore, cell adhesion and extracellular matrix–receptor interaction may play an important role in ovarian cancer invasion.
The PI3K-Akt signalling pathway regulates a variety of cellular processes including cell survival, proliferation, growth, and motility, which are important for tumorigenesis. Inhibition of the receptor tyrosine-protein kinase erbB-3 (HER3)-PI3K-AKT signalling pathway has been shown to enhance apoptosis in ovarian cancer cells.20 Mabuchi et al.21 suggested that the PI3K/AKT/mammalian target of rapamycin (mTOR) pathway could be used as a therapeutic target for ovarian cancer, and mutation or altered expression of components of this pathway is implicated in human cancer.22 Importantly, this pathway was also enriched by DEGs in the present study, consistent with other studies that have demonstrated a pivotal role for the PI3K-Akt signalling pathway in ovarian cancer.21 Based on RNA-seq data and the KEGG database, the expression of DEGs and corresponding proteins associated with PI3K-Akt signalling were validated, including the upregulated ANGPT2, FGF18, ITGB4 and ITGB8 genes, and downregulated AKT3 and PIK3AP1 genes.
Angiopoietin 2 is a member of the human ANGPT-TIE system, which plays an important role in tumour initiation and in increasing the number of tumour vessel sprouts.23 In some human cancers, overexpressed ANGPT2 is correlated with poor prognosis.24 FGF18 is a member of the FGF family, acting as a chemotactic, mitogenic and angiogenic factor,25 and it has been suggested to play a critical role in promoting the progression of ovarian high-grade serous carcinoma.26 ITGB4 and ITGB8 are integrin-family members that control cell attachment to the extracellular matrix and activate intracellular signalling pathways associated with cell survival, growth, differentiation, apoptosis, and migration.27 Increased expression of ITGB4 and ITGB8 has been reported to be associated with tumorigenesis.28 The present results also suggest increased expression of ANGPT2, FGF18, ITGB4 and ITGB8 in SKOV3 cells, which are derived from an ovarian serous carcinoma, compared with that in MCV152 cells (derived from a benign ovarian tumour).
AKT serine/threonine kinase 3, a member of the AKT family, is the key regulator of many cellular processes associated with cancer, including cell proliferation, survival and metastasis.29 Blockade of AKT3 results in a significant reduction in the expression of oestrogen receptor binding site associated antigen 9 (EBAG9), a tumour cell marker, whose expression is closely associated with vascular endothelial growth factor expression in ovarian and uterine tumour samples.30 PIK3AP1 is identified as a tumour suppressor gene and has been reported to be inactivated in head and neck cancers,31 however, its role in ovarian cancer has not been fully investigated. In the present study, AKT3 and PIK3AP1 were demonstrated to be significantly downregulated in SKOV3 cells compared with MCV152 cells, and both genes were involved in various signalling pathways, suggesting that downregulated AKT3 and PIK3AP1 may be associated with ovarian cancer development.
In conclusion, cell adhesion and extracellular matrix–receptor interaction may play an important role in ovarian cancer invasion. The PI3K-Akt signalling pathway may be involved in ovarian cancer progression by upregulating ANGPT2, FGF18, ITGB4 and ITGB8, and downregulating AKT3 and PIK3AP1 gene expression.
Declaration of conflicting interest
The authors declare that there is no conflict of interest.
Funding
This research was supported by a grant from the National Natural Science Foundation of China (81602268).
ORCID iD
Jiazhou Chen https://orcid.org/0000-0003-2969-8652
References
- 1.Reid BM, Permuth JB, Sellers TA. Epidemiology of ovarian cancer: a review. Cancer Biol Med 2017; 14: 9–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Ervik M, et al. GLOBCAN 2012 estimated cancer incidence, mortality and prevalence worldwide in 2012 v1.0: IARC CancerBase No 11. Lyon: International Agency for Research on Cancer, 2013. [Google Scholar]
- 3.Howlader N, Noone AM, Krapcho M, et al. SEER cancer statistics review, 1975–2014. Bethesda: National Cancer Institute, 2017, p.2018. [Google Scholar]
- 4.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin 2017; 67: 7–30. [DOI] [PubMed] [Google Scholar]
- 5.Quitadamo A, Tian L, Hall B, et al. An integrated network of microRNA and gene expression in ovarian cancer. BMC Bioinformatics 2015; 16: S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Welsh JB, Zarrinkar PP, Sapinoso LM, et al. Analysis of gene expression profiles in normal and neoplastic ovarian tissue samples identifies candidate molecular markers of epithelial ovarian cancer. Proc Natl Acad Sci USA 2001; 98: 1176–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nielsen JS, Jakobsen E, HØlund B, et al. Prognostic significance of p53, Her‐2, and EGFR overexpression in borderline and epithelial ovarian cancer. Int J Gynecol Cancer 2004; 14: 1086–1096. [DOI] [PubMed] [Google Scholar]
- 8.Santin AD, Zhan F, Bellone S, et al. Gene expression profiles in primary ovarian serous papillary tumors and normal ovarian epithelium: identification of candidate molecular markers for ovarian cancer diagnosis and therapy. Int J Cancer 2004; 112: 14–25. [DOI] [PubMed] [Google Scholar]
- 9.Arend RC, Londoño-Joshi AI, Straughn JM, Jr, et al. The Wnt/β-catenin pathway in ovarian cancer: a review. Gynecol Oncol 2013; 131: 772–779. [DOI] [PubMed] [Google Scholar]
- 10.Cheaib B, Auguste A, Leary A. The PI3K/Akt/mTOR pathway in ovarian cancer: therapeutic opportunities and challenges. Chin J Cancer 2015; 34: 4–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeng Y. Advances in mechanism and treatment strategy of cancer. Cell Mol Biol (Noisy-le-grand) 2018; 64: 1–3. [PubMed] [Google Scholar]
- 12.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003; 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wickstroem SA, Niessen CM. Cell adhesion and mechanics as drivers of tissue organization and differentiation: local cues for large scale organization. Curr Opin Cell Biol 2018; 54: 89–97. [DOI] [PubMed] [Google Scholar]
- 14.Heinzelmann-Schwarz VA, Margaret GG, Henshall SM, et al. Overexpression of the cell adhesion molecules DDR1, Claudin 3, and Ep-CAM in metaplastic ovarian epithelium and ovarian cancer. Clin Cancer Res 2004; 10: 4427–4436. [DOI] [PubMed] [Google Scholar]
- 15.Slack-Davis JK, Atkins KA, Harrer C, et al. Vascular cell adhesion molecule-1 is a regulator of ovarian cancer peritoneal metastasis. Cancer Res 2009; 69: 1469–1476. [DOI] [PubMed] [Google Scholar]
- 16.Clément R, Dehapiot B, Collinet C, et al. Viscoelastic dissipation stabilizes cell shape changes during tissue morphogenesis. Curr Biol 2017; 27: 3132–3142.e4. [DOI] [PubMed] [Google Scholar]
- 17.Basu S, Cheriyamundath S, Ben-Ze’ev A. Cell–cell adhesion: linking Wnt/β-catenin signaling with partial EMT and stemness traits in tumorigenesis. F1000Res 2018; 7: F1000 Faculty Rev-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng Y, Yao X, Liu X, et al. Anti-angiogenesis triggers exosomes release from endothelial cells to promote tumor vasculogenesis. J Extracell Vesicles 2019; 8: 1629865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sousa B, Pereira J, Paredes J. The crosstalk between cell adhesion and cancer metabolism. Int J Mol Sci 2019; 20: 1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bezler M, Hengstler JG, Ullrich A, et al. Inhibition of doxorubicin-induced HER3-PI3K-AKT signalling enhances apoptosis of ovarian cancer cells. Mol Oncol 2012; 6: 516–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mabuchi S, Kuroda H, Takahashi R, et al. The PI3K/AKT/mTOR pathway as a therapeutic target in ovarian cancer. Gynecol Oncol 2015; 137: 173–179. [DOI] [PubMed] [Google Scholar]
- 22.Vivanco I, Sawyers CL. The phosphatidylinositol 3-kinase–AKT pathway in human cancer. Nat Rev Cancer 2002; 2: 489–501. [DOI] [PubMed] [Google Scholar]
- 23.Hashizume H, Falcón BL, Kuroda T, et al. Complementary actions of inhibitors of angiopoietin-2 and VEGF on tumor angiogenesis and growth. Cancer Res 2010; 70: 2213–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lind AJ, Wikström P, Granfors T, et al. Angiopoietin 2 expression is related to histological grade, vascular density, metastases, and outcome in prostate cancer. Prostate 2005; 62: 394–399. [DOI] [PubMed] [Google Scholar]
- 25.Moore EE, Bendele AM, Thompson DL, et al. Fibroblast growth factor-18 stimulates chondrogenesis and cartilage repair in a rat model of injury-induced osteoarthritis. Osteoarthritis Cartilage 2005; 13: 623–631. [DOI] [PubMed] [Google Scholar]
- 26.El-Gendi S, Abdelzaher E, Mostafa MF, et al. FGF18 as a potential biomarker in serous and mucinous ovarian tumors. Tumour Biol 2016; 37: 3173–3183. [DOI] [PubMed] [Google Scholar]
- 27.Hood JD, Cheresh DA. Role of integrins in cell invasion and migration. Nat Rev Cancer 2002; 2: 91–100. [DOI] [PubMed] [Google Scholar]
- 28.Knyazev EN, Nyushko KM, Alekseev BY, et al. Suppression of ITGB4 gene expression in PC-3 cells with short interfering RNA induces changes in the expression of β-integrins associated with RGD-receptors. Bull Exp Biol Med 2015; 159: 541–545. [DOI] [PubMed] [Google Scholar]
- 29.Bellacosa A, Testa JR, Staal SP, et al. A retroviral oncogene, akt, encoding a serine-threonine kinase containing an SH2-like region. Science 1991; 254: 274–277. [DOI] [PubMed] [Google Scholar]
- 30.Akahira JI, Aoki M, Suzuki T, et al. Expression of EBAG9/RCAS1 is associated with advanced disease in human epithelial ovarian cancer. Br J Cancer 2004; 90: 2197–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guerrero-Preston R, Michailidi C, Marchionni L, et al. Key tumor suppressor genes inactivated by “greater promoter” methylation and somatic mutations in head and neck cancer. Epigenetics 2014; 9: 1031–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]