Abstract
Although morphology and grain size are important to rice growth and yield, the identity of abundant natural allelic variations that determine agronomically important differences in crops is unknown. Here, we characterized the function of mitogen-activated protein kinase 3 from Oryza officinalis Wall. ex Watt encoded by OrMKK3. Different alternative splicing variants occurred in OrMKK3. Green fluorescent protein (GFP)–OrMKK3 fusion proteins localized to the cell membrane and nuclei of rice protoplasts. Overexpression of OrMKK3 influenced the expression levels of the grain size-related genes SMG1, GW8, GL3, GW2, and DEP3. Phylogenetic analysis showed that OrMKK3 is well conserved in plants while showing large amounts of variation between indica, japonica, and wild rice. In addition, OrMKK3 slightly influenced brassinosteroid (BR) responses and the expression levels of BR-related genes. Our findings thus identify a new gene, OrMKK3, influencing morphology and grain size and that represents a possible link between mitogen-activated protein kinase and BR response pathways in grain growth.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12374-020-09290-2.
Keywords: Oryza officinalis Wall. ex Watt, OrMKK3, Morphology, Grain size
Introduction
Rice is the main cereal crop of over 3.5 billion people (Cheng et al. 2013). Oryza officinalis Wall. ex Watt (CC, 2n = 2x = 24), an important and special wild-rice species, represents a significant source of genes influencing important characteristics such as grain size and strong resistance to abiotic and biotic stresses (Zhang et al. 2019). Recent studies have isolated and characterized several genes for disease resistance from O. officinalis Wall. ex Watt (Jiang et al. 2019), however, the identities of genes for grain size and plant architecture are unknown.
Plant height and tiller number are two major agronomic traits influencing cereal crop architecture and grain yield (Wang et al. 2018a, b) and are always inversely related in rice (Liao et al. 2019), with increased tillers and decreasing plant height of many mutant and transgenic plants (Chen et al. 2012; Choi et al. 2012; Xu et al. 2012; Zhou et al. 2013). While the specific identities of many trait-influencing genes in rice are unknown, many studies have revealed potential genetic influences on trait regulation. For example, Grain number, plant height, and heading date7 (Ghd7), which encodes a CCT-domain-containing transcription factor and has been reported to have major effects on the number of grains per panicle, plant height, and flowering time (Gao et al. 2014). In addition, the QTL DTH8 (QTL for days to heading on chromosome 8) encoding a putative HAP3 subunit of a CCAAT-box-binding transcription factor was shown to regulate heading, plant height, and the number of grains per panicle (Wei et al. 2010). A semi-dwarf and late-flowering mutation altering the function of the WRKY transcription factor Dlf1 caused downregulation of Ehd2/RID1/OsId1 in a signal transduction pathway and influenced height in rice (Cai et al. 2014). OsGA3ox2 encodes a gibberellin (GA) 3β-hydroxylase that catalyzes the conversion of GA20 to GA1FIG. Loss-of-function mutants of OsGA3ox2 show severe dwarfism due to altered GA processing (Itoh et al. 2002). Overall, these studies indicate that multiple pathways influence rice height.
Tiller number is another critical trait of rice that describes the level of grain yield. A comprehensive understanding of the genetic controls of tiller number will enhance knowledge of basic plant development and improve agriculture. Mutations in one gene, This1 that encodes a class-III lipase are associated with high tillering, reduced height, and infertile spikelets (Liu et al. 2013). OsAAP5, an amino acid permease, regulates tiller, and grain yield by affecting the cytokinin levels (Wang et al. 2019). Dwarf27 (D27) is an iron-containing protein involved in the MORE AXILLARY GROWTH 4 (MAX4)/RAMOSUS 1 (RMS1)/DWARF10 (D10) pathway and participates in strigolactone biosynthesis (Lin et al. 2009). OsNPF7.2, a low-affinity nitrate transporter, plays an important role in the control of tiller bud growth and tillering regulation by coordinating the cytokinin and strigolactone pathways (Wang et al. 2018a, b). Tillering is also reported to be inhibited by gibberellin (Duan et al. 2014). Although many studies have been conducted attempting to identify genes that control the tiller development and plant height, only few genes expressed by O. officinalis have been identified.
Rice yield depends on three major factors: grain weight, grain number per panicle, and panicle number per plant. Grain weight is related to grain length, width, and thickness (Duan et al. 2014). The QTL grain length and weight on chromosome 7 (qGLW7) contains a gene encoding a plant-specific transcription factor that positively regulates cell size in the grain hull, resulting in enhanced rice grain length and yield (Si et al. 2016). In addition, GSN1 encodes a mitogen-activated protein kinase phosphatase (OsMKP1) that directly interacts with and inactivates a mitogen-activated protein kinase (OsMPK6) that coordinates the trade-off between grain number and grain size (Tao et al. 2018). Expression levels of GRAIN WIDTH8 (GW8) influence grain, shape, and quality (Wang et al. 2012). Another major QTL regulating grain length in rice, qGL3, suppresses BR signaling (Gao et al. 2019) while DWARF2 reduces grain height and size by regulating gibberellin biosynthesis (Chen et al. 2019). Small grain 11 (SMG11) controls grain size by promoting cell expansion in grain hulls by regulating BR biosynthesis (Fang et al. 2016). Overexpression of constitutively active versions of OsMKKK10 and OsMKKK4 causes production of large and heavy grains, since the OsMKKK10-OsMKK4-OsMitogen-Activated Protein Kinase 6 (MAPK6) cascade exerts control of grain size and weight in rice (Xu et al. 2018). SMG1, encoding MKK4, affects BR responses and thus influences grain size (Duan et al. 2014). Thus, considerable research has revealed that multiple pathways control grain size in rice.
Mitogen-activated protein kinase (Kishi-Kaboshi et al. 2010) cascades transmit exogenous or developmental signals to target molecules through sequential phosphorylation and orchestrate multiple processes related to plant growth, development, and defense response (Andreasson et al. 2010; Pitzschke. 2015; Xu et al. 2015). Mitogen-activated protein kinase 3 (MKK3) mediates resistance to Nilaparvata lugens via phytohormone dynamics (Zhou et al. 2019). MKK3 phosphorylates a basic helix–loop–helix transcription factor, activating the MKK3-mitogen-activated protein kinase 6 (MPK6)-Myelocytomatosis module involved in light signal transduction pathways in Arabidopsis thaliana (Sethi et al. 2014). In addition, mitogen-activated protein kinase (MKKK62)-MKK3-MAPK7/MAPK14 regulates the transcription of OsMFT in rice to mediate seed dormancy (Mao et al. 2019), with MKK3 also regulating seed dormancy in barley (Nakamura et al. 2016).
We previously have identified genes that affect plant height, tillering, and grain growth in O. officinalis Wall. ex Watt. Here, we first describe isolation a gene encoding mitogen-activated protein kinase 3 (OrMKK3) in O. officinalis Wall. ex Watt by homology-based cloning and its subsequent characterization. OrMKK3 is an ortholog of OsMKK3 and appears to influence plant height, tiller, and grain size. Additional results show that OrMKK3 affects BR responses and the expression levels of BR-related genes. Our findings thus indicate that OrMKK3 is a factor influencing plant height, tiller, and grain size in O. officinalis Wall. ex Watt and indicate a possible connection between the MAPK cascades and BRs in grain growth.
Results
OrMKK3 Cloning and Sequence Analysis
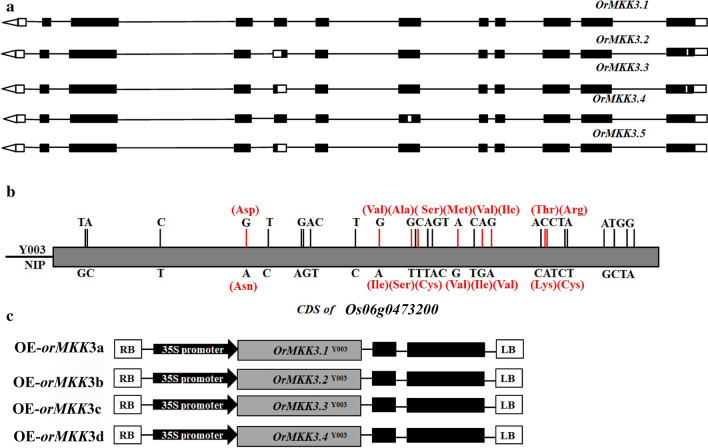
Two pairs of primers (Table S1) were designed based on other available Nipponbare MKK3 nucleotide sequences (Genbank accessions: Os06g0473200) to amplify the OrMKK3 open reading frame from the Y003 line of O. officinalis in Guangxi Province. Five sequences, OrMKK3.1–3.5, were obtained (supplementary materials). Comparisons of nucleotide sequences and gene structures revealed that OrMKK3.2–3.5 were alternative splicing variants of OrMKK3.1. The OrMKK3 alternative splicing events were classified into four categories: alternative exon 5′-ends (5′-AE), exon skipping (Hamel et al. 2006), mutually exclusive exons, and alternative exon 3′-ends (3′-AE). OrMKK3.1 and OrMKK3.5 differed by the presence of a 3′-AE in the 3′-untranslated region (3′-UTR) (Fig. 1a). OrMKK3.2 possessed a 3′-AE and lacked half of exon 8 (Fig. 1a). OrMKK3.3 and OrMKK3.5 both also lacked half of exon 8, with OrMKK3.3 lacked the first half and OrMKK3.5 lacking the latter half (Fig. 1a). Multiple exon skip (ES) and AE events distinguish OrMKK3.4 from OrMKK3.1–3.3 and OrMKK3.5. OrMKK3.4 has a frameshift mutation in the sixth exon away from the 3′ end of the coding sequence (Fig. 1a). In addition, OrMKK3.3 and OrMKK3.5 differed by the presence of a 3′-AE in the 3′-UTR; however, this difference did not cause the production of different proteins. Compared to OrMKK3.1, however, OrMKK3.2, OrMKK3.3, OrMKK3.4, and OrMKK3.5 exhibit a frameshift mutation that is predicted to cause truncated protein production (Fig. 1a).
Fig. 1.
a Five cDNA of alternative splicing in OrMKK3. OrMKK3.1 includes all exons. Alternative 3′ splicing and exon skipping in OrMKK3.2. Alternative 5′ splicing in OrMKK3.3. Exon skipping in OrMKK3.4, Alternative 3′ splicing in OrMKK3.5. b SNP between Nipponbare and Y003 in OrMKK3 CDS. c Complementation vector: OrMKK3a (with CDS of OrMKK3.1), OrMKK3b (with CDS of OrMKK3.2), OrMKK3c (with CDS of OrMKK3.3), and OrMKK3d (with CDS of OrMKK3.4) overexpression vector
From NCBI (https://www.ncbi.nlm.nih.gov/), we found five Nipponbare MKK3 genes named XM_015788456.2, XM_015788457.2, XM_015788458.2, XM_015788459.2, and XM_015788460.2. XM_015788458.2 contained 11 exons, as did OrMKK3.1. XM_015788458.2 and OrMKK3.1 differed by 28 single-nucleotide polymorphisms (SNP) in their open reading frames, although 17 of these SNPs did not affect the encoded proteins while 9 of these SNPs altered the encoded protein sequences. In addition, XM_015788456.2, XM_015788457.2, XM_015788459.2, and XM_015788460.2 each contained the 17 SNPs described above. XM_015788456.2 contained an additional exon beyond exon 8 of OrMKK3.1. XM_015788457.2 contained an additional exon beyond exon 8 and additional 3′-AE events compared to OrMKK3.1. XM_015788459.2 contained two additional exons following exons corresponding to exon 1 and exon 8 of OrMKK3.1. XM_015788459.2 contained an additional exon following exon 1 compared to OrMKK3.1 (Fig. 1b).
OrMKK3 Regulates Height and Tillering
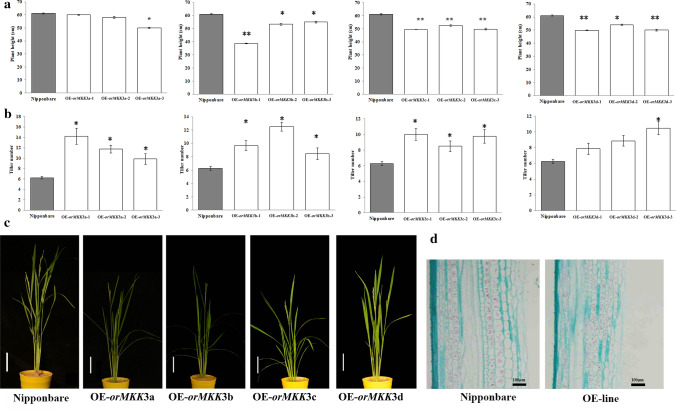
Several experiments were conducted to determine whether the differences in the coding sequences determined OrMKK3 function. First, four overexpression constructs containing OrMKK3.1, OrMKK3.2, OrMKK3.3, and OrMKK3.4 from Y003 driven by the 35S promoter from tobacco cauliflower mosaic virus (CaMV35S) were separately introduced into the Nipponbare cultivar (Fig. 1c). Quantitative real-time polymerase chain reaction (qRT-PCR) results indicated that independent T0 transgenic lines have different expression levels of OrMKK3.1, OrMKK3.2, OrMKK3.3, and OrMKK3.4, with expression levels of these genes also determined in T3 lines using qRT-PCR. T3 generations of three overexpression transgenic lines (OE-lines) of each OrMKK3 splice variant, OE-OrMKK3a, OE-OrMKK3b, OE-OrMKK3c, and OE-OrMKK3d, were used for further analysis (Fig. 2a). At the maturation stage, OE-OrMKK3a, OE-OrMKK3b, OE-OrMKK3c, and OE-OrMKK3d lines were significantly shorter than wild-type plants (Fig. 2a, c). We performed histocytological analysis of the internal structure of internodes using paraffin sections (Fig. 2d) to reveal the cause of OE-line dwarfism (Fig. 2a, c). Results showed that cell sizes were significantly reduced in OE-line internodes, suggesting that cell size was restrained in the OE-lines. These observations also imply that that OrMKK3 may negatively affect cell length, thereby limiting rice plant height.
Fig. 2.
a Plant height of Nipponbare and OE-OrMKK3-lines. b Number of tillers of Nipponbare and OE-OrMKK3-line. c Nipponbare and OE-OrMKK3-line plant at the full heading stage. Bar = 10 cm. d Histocytological analysis on the internal structure of internodes by paraffin section. Cell growth is inhibited in the OE-OrMKK3-line. Bar = 100 µm
OE-OrMKK3a, OE-OrMKK3b, OE-OrMKK3c, and OE-OrMKK3d plants had more tillers than wild-type plants at the full flowering stage (Fig. 2b, c). Tiller number in the OE-lines was significantly more than that in Nipponbare (Fig. 2b, c). In contrast, no significant difference was found between tiller numbers of OE-OrMKK3d and Nipponbare lines (Fig. 2b, c). The overexpression of OrMKK3 thus significantly increased rice tiller number, and the upregulation of OrMKK3 positively influenced tiller shaping.
OrMKK3 Determines Grain Size and Influences 1000 Seed Weight
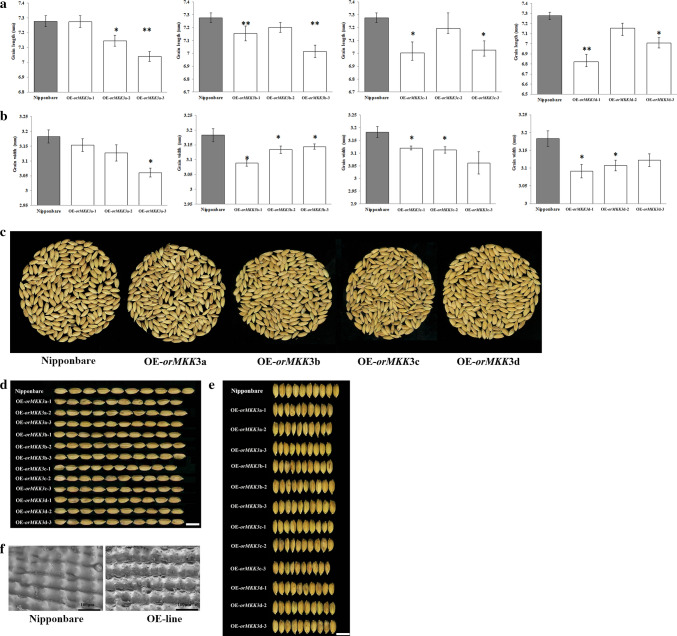
We found that OrMKK3 regulates grain length and width. Grain length and width were reduced in OrMKK3 overexpression transgenic plants (T3 generation; Fig. 3a, d, e), indicating that altered OrMKK3 function results in reduced grain length and width. The spikelet hulls of Y003 plants before fertilization were shorter than those of Nipponbare plants. OE-line plants, in which the OrMKK3 coding sequences were constitutively overexpressed, formed grains that were significantly shorter than those formed by the Nipponbare plants (Fig. 3a, d). We also investigated the 1000-grain weight of Nipponbare and OE-lines. The 1000-grain weight was lower for OE-lines than for Nipponbare lines (Fig. 3c). In addition, histocytological analyses of outer surfaces of husk from OE-lines and Nipponbare revealed that OrMKK3 likely regulates grain size by altering cell numbers. Cell length in either the palea or lemma had little impact on grain size, while total cell numbers of outer and inner epidermal cells in the longitudinal direction in the OE-lines were greater than in Nipponbare (Fig. 3f). These observations suggest that OrMKK3 may inhibit longitudinal growth by increasing cell proliferation. These results indicate that OrMKK3 plays a significant role during the full flowering stage by influencing genes regulating grain size and affecting grain production.
Fig. 3.
a Grain length of Nipponbare and OE-OrMKK3-lines. b Grain width of Nipponbare and OE-OrMKK3-lines. c Two hundred seeds of Nipponbare and OE-OrMKK3-lines. d Grain length of ten seeds of Nipponbare and OE-OrMKK3-lines. e Grain width of ten seeds of Nipponbare and OE-OrMKK3-lines. f Scanning electron microscope images of lemmas of Nipponbare and OE-OrMKK3-line
OrMKK3 Expression Pattern and Subcellular Localization
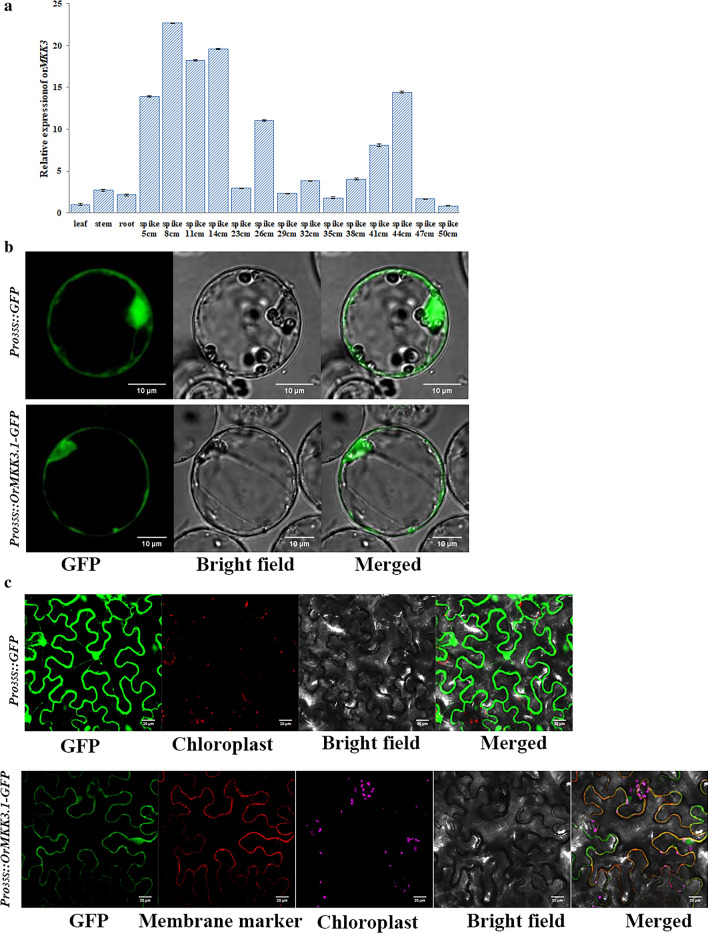
OrMKK3 transcripts were detected in various tissues using qRT-PCR with wild-type Y003 plants to determine the spatial expression profile of OrMKK3. qRT-PCR analysis showed that OrMKK3 was expressed ubiquitously in all examined tissues and organs, including the roots, leaves, stems, internodes, and different developmental stages of panicles (primarily in the rachillae), lemmas, paleae, lodicules, stamens, and pistils (Fig. 4a). Results also revealed that OrMKK3 expression is greater in leaves and young panicles than those in other tissues or organs, with OrMKK3 expression levels gradually increasing during panicle formation.
Fig. 4.
a OrMKK3 expression in different tissues as analysis by qRT-PCR in Y003 line of O. officinalis in Guangxi Province. The OrMKK3 expression in leaf as 1. b Subcellular localization of the OrMKK3 Protein. Pro35S::GFP as control. Scale bar, 20 mm. c Co-localization of 35S: OrMKK3.1-GFP with the plasma lemma marker in tobacco epidermal cells. OrMKK3.1-GFP overlapped with marker at the plasma membrane. Scale bar, 20 mm
Since OrMKK3 contains a kinase domain, we speculated that OrMKK3 is localized to the plasma membrane. We expressed an OrMKK3.1-GFP fusion protein under the control of the 35S promoter in rice leaf protoplasts to determine the subcellular localization of OrMKK3. As shown in Fig. 4b, GFP fluorescence from expression of the 35S:OrMKK3.1-GFP transgene in rice protoplasts was observed exclusively at the cell membrane and nuclei of rice protoplasts (Fig. 4b) and GFP signal was detected at the cell membrane in tobacco epidermal cells (Fig. 4c). These results indicate that OrMKK3 encodes a both membrane-localized and nuclear protein.
OrMKK3 Expression Decreases Cell Proliferation
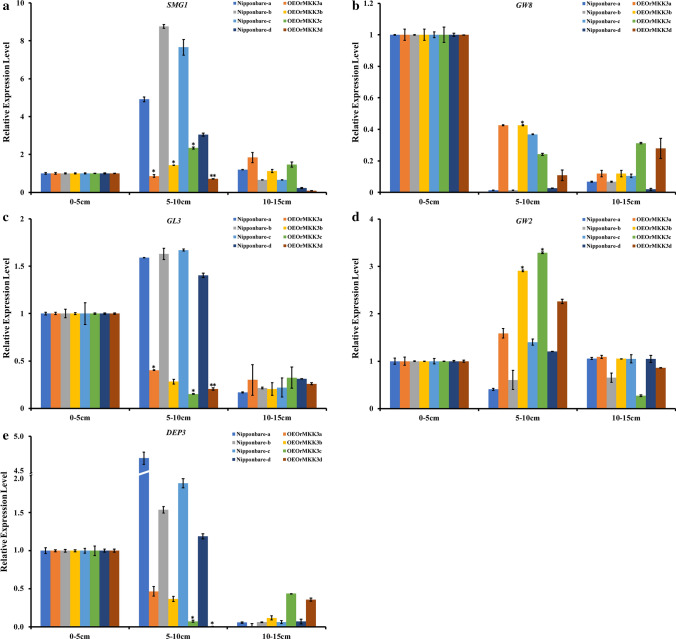
Cell proliferation and expansion significantly influence for organ size (Horiguchi et al. 2006), thus to understand how OrMKK3 affects grains during panicle growth and height development, we examined cell number and size. Histocytological analysis of stem slices and husks were performed with OrMKK3 overexpression and Nipponbare lines. The average length of inner stem cells and outer husk cells of the OE-lines and Nipponbare plants were indistinguishable. This result indicates that OrMKK3 influences cell proliferation rather than cell expansion. Furthermore, we investigated the expression levels of GL3, GRAIN WIDTH2 (GW2), GRAIN WIDTH8 (GW8), SMALL GRAIN 1 (SMG1), and DENSE AND ERECT PANICLE 3 (DEP3), which influences grain size by affecting cell proliferation in young panicles (Table S2) (Yan et al. 2011; Qiao et al. 2011; Wang et al. 2012; Chen et al. 2019; Gao et al. 2019). As shown in Fig. 5, SMG1 expression was not markedly affected by overexpression of OrMKK3 (Fig. 5a). Compared to Nipponbare plants, expression levels of GL3 and DEP3 in 5–10-cm panicles were lower in OE-lines (Fig. 5c, e). Expression levels of GW2 in OE-lines were initially upregulated in 5–10-cm panicles then decreased in 10–15-cm panicles (Fig. 5d). In young panicles, the expression levels of GW8 were significantly reduced in OE-lines and Nipponbare in 10-cm panicles than in 5-cm panicles. These decreases in GL3 and DEP3 expression levels are probably due to the physiological and biochemical activities involved in rice husk shortening. These results indicate that overexpression of OrMKK3 likely inhibits the development of plant cells.
Fig. 5.
Relative expression levels of SMG1, GW8, GL3, GW2 and DEP3 in Nipponbare and OE-OrMKK3-line young panicles. a Relative expression levels of SMG1 in Nipponbare and OE-OrMKK3-line young panicles. b Relative expression levels of GW8 in Nipponbare and OE-OrMKK3-line young panicles. c Relative expression levels of GL3 in Nipponbare and OE-OrMKK3-line young panicles. d Relative expression levels of GW2 in Nipponbare and OE-OrMKK3-line young panicles. e Relative expression levels of DEP3 in Nipponbare and OE-OrMKK3-line young panicles. * and ** indicate a 5% and 1% significance level, respectively, according to the t test. Relative expression levels of genes in 0–5 cm spike as 1, respectively
MKK3 Phylogenetic Analysis
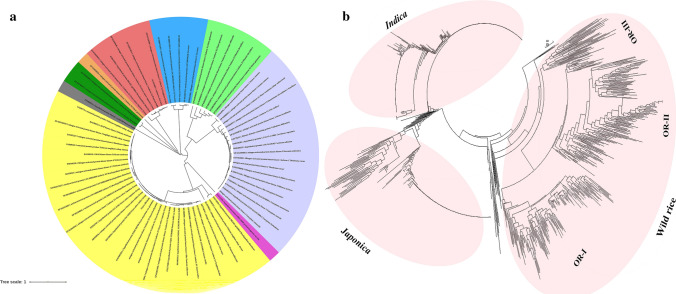
OrMKK3 encodes mitogen-activated protein kinase kinase 3, with OrMKK3a resulting in production of a mutant protein while OrMKK3b, OrMKK3c, and OrMKK3d each contain frameshift mutations. These results thus indicate that OrMKK3.2, OrMKK3.3 and OrMKK3.4 are loss-of-function alleles. We performed phylogenetic analyses using protein sequences of OrMKK3 homologs to examine evolutionary relationships. These homolog sequences were obtained in species including Phoenix dactylifera, Elaeis guineensis, Ananas comosus, Corchorus olitorius, Musa acuminata subsp. malaccensis, Corchorus capsularis, Gossypium, and Hibiscus syriacus. The 73 protein homologs were divided into 9 clades. In protein phylogenetic trees, homologs from O. officinalis, O. indica, O. japonica, Aegilops tauschii, Brachypodium distachyon, O. brachyantha, and O. meyeriana car. granulata were clustered together with Triticum aestivum and Hordeum vulgare (Fig. 6a). This indicates a high level of conserved synteny of MKK3 among multiple plant species. Moreover, these phylogenetic relationships of MKK3 homologs were based on 446 SNPs from wild rice (O. rufipogon) and involved 777 varieties from the world rice core collection. These results indicate that MKK3 sequences are diverse across the range of indica, japonica, and wild-rice varieties. Our phylogenetic trees were divided into five clades. OR-I arose from wild-rice varieties in Guangxi, Yunnan, and Guizhou Provinces while both OR-I and OR-II were widely distributed in east and southeast Asia (Fig. 6b).
Fig. 6.
a Phylogenetic tree of the 73 MKK3 proteins in different species. The phylogenetic tree constructed based on different sequences or indels by MAGE 6.0. Different colors represented different clades. b The phylogenetic tree of in 446 wild rice (O. rufipogon) and 777 varieties from the world rice core collection. The phylogenetic tree constructed based on different functional SNPs or indels by MAGE 6.0. All varieties were categorized by allelic variations in the SNP of OsMKK3
OrMKK3 OE-Lines Show Altered Responses to BR
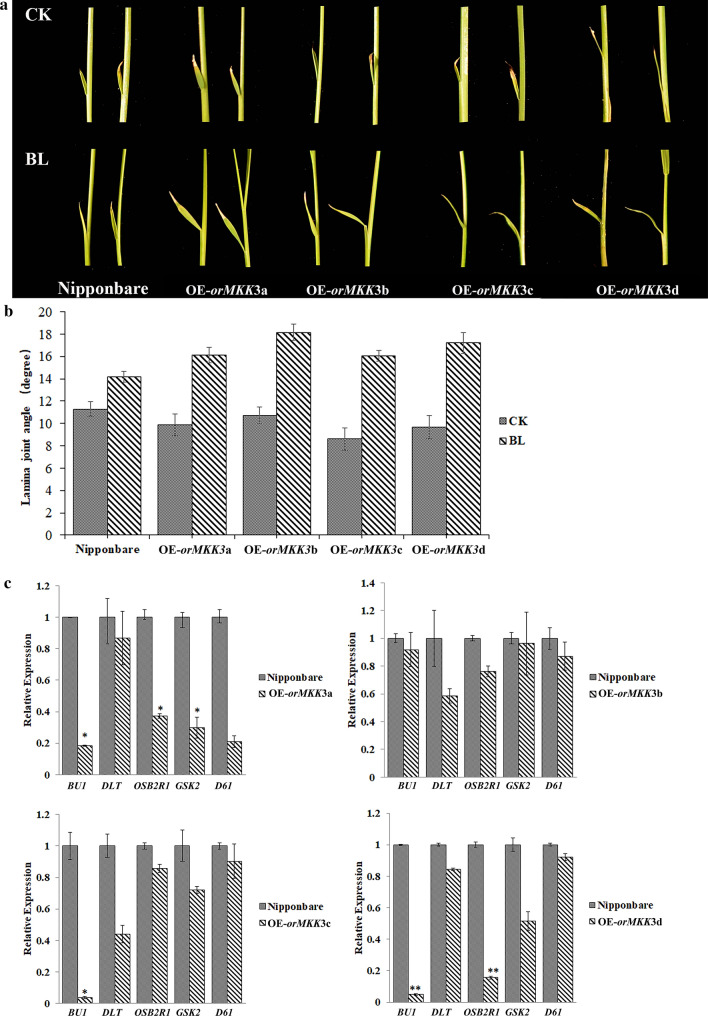
OE-lines and Nipponbare plants were treated with homobrassinolide (homo-BL) to identify potential other BR-related OrMKK3 functions. In germination assays, when treated with 1 µM of homo-BL (a bioactive BR compound), the OE-lines showed larger leaf lamina joint inclination than Nipponbare, indicating enhanced BR sensitivity. In ½ MS medium lacking homo-BL, the OE-line and Nipponbare did not show large leaf lamina joints (Fig. 7a, b). Using qRT-PCR, we compared the expression levels of BR signaling genes including BRASSINOSTEROID INSENSITIVE1 (OsBRI1), glycogen synthase kinase 2 (OsGSK2), BRASSINAZOLE-RESISTANT1 (OsBZR1), BRASSINOSTEROID UPREGULATED1 (BU1), and DWARF AND LOW‐TILLERING (DLT) (Yamamuro et al. 2000; Bai et al. 2007; Tanaka et al. 2009; Zhang et al. 2009; Tong et al. 2009). Expression levels of OsBZR1 and BU1 that positively regulate BR signaling were downregulated in OE-lines compared to in Nipponbare (Fig. 7c). These results supported the hypothesis that OrMKK3 regulates BR biosynthetic genes in rice.
Fig. 7.
OE-OrMKK3-lines have response sensitivity to brassinolide (BL). a Analysis of brassinolide response by the leaf lamina inclination test. b Lamina joint angle show different BR sensitivities. Data were collected from seedlings (n = 30) for each concentration and are shown as means.SE. *P < 0.05; Student’s t test. c Expression levels of BU1, DLT, OSB2R1, GSK2, D61. Five plants from each line were used, and three replicates were performed. Relative expression levels of genes in WT as 1, respectively. SE. *P < 0.05; **P < 0.01; Student’s t test
Discussion
Oryza officinalis Wall. ex Watt is an agriculturally important but seriously endangered species of wild rice. In addition, wild-rice varieties contain abundant genetic diversity and carry many desirable traits, which have proven to be natural genetic reservoirs for the improvement of rice quality and yield. However, comprehensive genetic backgrounds of different Oryza species have been a challenge to work with for rice breeders. Fortunately, this has begun to change, and a series of studies utilizing molecular technology have focused on exploiting important genes.
A transformation-competent artificial chromosome clone harboring a NAM-ATAF1/2-CUC (NAC)-positive genomic fragment was selected based on the presence of a conserved sequences of plant stress-related NAC transcription factors from O. officinalis and integrated into the genome of HJX74 (an indica rice variety). The resulting transgenic lines exhibited high tolerance to drought stress (Liu et al. 2015). In addition, generation-sequencing methods have been used on O. officinalis Wall. ex Watt. A previous study analyzed the leaf transcriptome of O. officinalis [CC] using RNA-Seq and de novo assembly of 68,132 unigenes on the basis of approximately 23 million transcriptome reads and identified a total of 476 unigenes related to disease resistance were identified in O. officinalis (Bao et al. 2015). In this study, we report that OrMKK3 in O. officinalis Wall. ex Watt exhibits alternative splicing. OrMKK3 showed multiple types of alternative splicing and amino acid changes compared to Nipponbare.
Height, tiller, and grain size in crops are important agronomic traits. Several factors that influence height and tiller have been described, including small grain and dwarf 2 (Chen et al. 2019), DTH8 (Wei et al. 2010), and DTH7 (Gao et al. 2014). Several genes that affect grain size have been identified, including GS3, GL3, GS5, GW2, GW8, and SMG1 (Fan et al. 2006; Song et al. 2007; Mao et al. 2010; Li et al. 2011; Wang et al.2012; Qi et al.2012; Duan et al. 2014). Expression levels of SMG1, GW8, GL3, GW2, and DEP3 were significantly between OE-lines and Nipponbare, suggesting that the effect of OrMKK3 on grain size may require SMG1, GW8, GL3, GW2, and DEP3. Xu et al. found that the OsMKKK10-OsMKK4-OsMAPK6 signaling pathway positively regulates grain size and weight in rice (Xu et al. 2018). Sequential phosphorylation activates OsMAPK6 to promote cell proliferation in spikelet hulls, with OsMAPK6 potentially also phosphorylating transcription factors thereby regulating grain size. Knock-out mutants confirmed that MKK3-MAPK6 activates blue light responses in Arabidopsis (Sethi et al. 2014). OsSPL16 (GW8) encodes an SBP-domain transcription factor that binds directly to the GW7 promoter then suppresses GW7 expression to regulate grain width in rice (Hong et al. 2005; Morinaka et al. 2006; Zhang et al. 2012; Fang et al. 2016; Wu et al. 2016; Gao et al. 2019). It will be important to identify and understand relationships between OrMKK3 and transcription factors in rice and evaluate their effects on grain characteristics. However, few such underlying genes from O. officinalis Wall. ex Watt have been identified, and the molecular mechanisms that influence height, tiller, and grain size, including the function of OrMKK3, are still unknown in rice.
Like other MAPKs, MKK3 is involved in various plant physiological–biological activities. Most MKK3 homologs contain a protein kinase domain and NUCLEAR TRANSPORT FACTOR2-like domain in their C-terminal regions (Colcombet et al. 2016). In A. thaliana, an MKK3-based MAPK module acts upstream of reactive oxygen species production (Itoh et al. 2002) and signaling (Takahashi et al. 2011). In addition, MKK3 has been reported to close in response to abscisic acid, ABA activation of MPK7 is blocked in mkk3-1 which expressing a constitutively active version of MKK3 (Huang et al. 2012). Moreover, MKK3 acts in negative regulation in darkness and in light-induced MAPK activation during dark–light transition using knock-out mutants (Lee 2015). MKK3–MPK6 is activated by jasmonic acid and plays a key role in the negative regulation of ATMYC2/JIN1 expression (Takahashi et al. 2007). MKK3 also plays an important role in the ABA-induced MAPK pathway in plant stress signaling (Danquah et al. 2015). MKK3 interacts with MAP kinase kinase kinase 20 (MKKK20) in two non-complementary signaling cascades involved in root cortical microtubule activities (Matton et al. 2018). Studies of wounding, brown planthopper (N. lugens; BPH) infestation (Zhou et al. 2019), and methyl jasmonate or salicylic acid signaling pathways and the expression of MAPK and MAP kinase 3 (MMK3) revealed that OsMKK3-mediated positive regulation of rice resistance to BPH occurs though the phytohormone system (Zhou et al. 2019).
BRs are steroid hormones that regulate fundamental processes of plant growth and development in plants. In rice, BR-deficient mutants (brd2) have small seeds (Hong et al. 2005). This study demonstrated that OE-OrMKK3 lines have larger leaf lamina joint inclination than Nipponbare after BL treatment. In addition, the expression levels of genes related to BR signaling were reduced significantly in OE-OrMKK3 lines compared to Nipponbare. These results thus indicate a possible link between MAPK cascades and BR signaling. In Arabidopsis, BR inhibits stomatal development by suppressing glycogen synthase kinase 3-like kinase BR-insensitive 2 (BIN2) activity and phosphorylating YDA to inhibit phosphorylation of the YDA substrate MKK4 that mediates MAPK kinase kinase (MAPKKK) signaling (Kim et al. 2012). In addition, SMG1 may link MAPK pathways and BRs in grain growth, with OsMKK4/SMG1 influencing BR responses and the expression levels of BR-related genes (Duan et al. 2014). In addition, the receptor-like cytoplasmic kinase BRASSINOSTEROID-SIGNALING KINASE1 interacts with and phosphorylates MAPK kinase kinase5 (MAPKKK5); however, MKK3 and MAPKKK5 were not able to form a protein complex in Arabidopsis based on detection using an anti-Cluc antibody test (Yan et al. 2018). GL3.3 encodes OsPPKL that is similar to Arabidopsis BSU1, which acts in BR signal transduction to promote cell elongation and cell division (Zhang et al. 2012). Gao revealed that qGL3 improves OsGSK3 via dephosphorylation, and that OsGSK3 regulates OsBZR1 phosphorylation and subcellular distribution (Gao et al. 2019). In addition, in rice, 14–3-3 proteins were shown to interact with OsBZR1 using a yeast two-hybrid screen (Bai et al. 2007). These results suggest that BR regulates grain size by promoting seed growth (Morinaka et al. 2006; Wu et al. 2016). In addition, in mice, protein serine/threonine kinase 38-mediated dissociation of 14–3-3 from the ASK1-14–3-3 complex and the increase of interaction between ASK1 and its substrate MKK3 (Ha et al. 2008). EmMKK2 from the fox tapeworm Echinococcus multilocularis closely resembles members of the MKK3/6- and the MEK1/2-MAPKK sub-families that interact with 14–3-3 protein family members within the fox tapeworm (Gelmedin et al. 2010). Collectively, these findings suggest that OrMKK3 regulates plant height, tiller, grain size, 1000-grain weight, and BR responses, representing a possible link between the MAPK cascades and BRs in cell growth. However, further studies are needed to identify the different roles of OrMKK3 in BR signaling and seed size control in rice.
Materials and methods
Plant materials, growth conditions, and phenotyping
Wild-type rice plants (O. officinalis Wall. ex Watt and O. sativa L. spp. japonica) and four OrMKK3 overexpression lines (OE-OrMKK3a, OE-OrMKK3b, OE-OrMKK3c, and OE-OrMKK3d) were used in this study. Different tissues, including mature leaves, stems, and roots of O. officinalis Wall. ex Watt Y003 and O. japonica, were collected and planted in the National Germplasm Nanning Wild Rice Nursery of China. Transgenic lines selected by hygromycin resistance and qRT-PCR after two generations of propagation were cultivated in greenhouses under 16-h light/8-h dark. Three individual overexpression transgenic lines (OE-lines) of OE-OrMKK3a, OE-OrMKK3b, OE-OrMKK3c, and OE-OrMKK3d genotypes at the T3 stage were used for further analysis. The overexpression transgenic lines are different transformed cases with ten plants of OE-OrMKK3a, OE-OrMKK3b, OE-OrMKK3c, and OE-OrMKK3d.
RNA Extraction and qRT-PCR
Total RNA from the above listed tissues were extracted using a Takara minibest plant RNA extraction kit according to the user’s manual (Takara, Catalog no. 9769). Total RNA (2.0 μg) was used for cDNA synthesis with a primeScript™ RT reagent kit with gDNA Eraser (Perfect Real-Time, Takara, Catalog no. RR047B). We performed qRT-PCR using TB Green® Premix Ex Taq™ II and a CFX96 Real-Time system (Bio-RAD, Hercules, CA, USA) following the manufacturer’s instructions. qRT-PCR was performed using 10-µL mixtures: 5 µL of 2 × Green qPCR MasterMix, 1 µL of cDNA, 0.25 µL of each primer (10 µM), and 3.5-µL ddH2O. Amplification steps were 95 °C for 30 s, 40 cycles of 95 °C for 5 s, and 60 °C for 30 s. 65 °C for 5 s, 95 °C for 15 s, 60 °C for 30 s, and 95 °C for 15 s. Each experiment was repeated at least three times. The qRT-PCR analysis was performed using the ΔΔCt method. Details on primers used for detecting the OrMKK3 and OsUbiquitin genes (LOC_Os03g13170) (Chen et al. 2019) and probes are provided in Table S1 (*P < 0.05; ** P < 0.01; Student’s t test).
Plasmid Construction and Plant Transformation
OrMKK3.1, OrMKK3.2, OrMKK3.3, OrMKK3.4, and OrMKK3.5 cDNA sequences from O. officinalis Wall. ex Watt Y003 were amplified using primers OrMKK3-F and OrMKK3-R (Table S1) and products were subcloned into the pEASY®-Blunt Zero cloning vector (Transgen, Catalog no. CB501-01). Next, this cDNA fragment (OrMKK3.1–OrMKK3.4) was cloned into the plant binary vector PMDC32 containing the CaMV 35S promoter. Subsequently, the OrMKK3.1 cDNA fragment was subcloned into the Gateway binary vector PMDC83 containing the CaMV 35S promoter and GFP and digested with PacI-AscI to generate a 35S::OrMKK3.1 construct. All constructs were confirmed by sequencing. 35S::OrMKK3.1, 35S::OrMKK3.2, 35S::OrMKK3.3, and 35S::OrMKK3.4 plasmids were introduced into Agrobacterium tumefaciens strain EHA105 by transformation, and the plasmids transformed into Nipponbare in accordance with a previously published protocol (Hiei et al. 1994).
Phylogenetic Analysis
The DNA or deduced amino acid sequences of OrMKK3 homologs were BLAST searched against the NCBI database (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The phylogenetic tree was constructed with MEGA6 using the neighbor-joining method (Tamura et al. 2013). An alignment of rice MKK3 sequences was constructed using DNAMAN (LynnonBiosoft, V7.0). The SNPs of 777 cultivated rice accessions from 32 countries and 446 wild-rice accessions with publicly available sequencing data from the rice 3 K project (RFGB, http://www.rmbreeding.cn/Index/s) and OryzaGenome (http://viewer.shigen.info/oryzagenome/) were used for phylogenetic analysis (Huang et al. 2012).
BR Treatment
Seeds were surface sterilized by incubation in 20% NaClO for 20 min, washed five times with sterilized water, then germinated in ½ MS medium (hopebio, HB8469-12, China) with 1-μM homo-BL (Sigma-Aldrich Co. LLC., St. Louis, MO, USA) for 10 days in a growth cabinet. Ten-day-old seedlings were used to measure leaf lamina joint inclination.
Subcellular Localization of OrMKK3
The plasmids CaMV35S::GFP and CaMV35S::OrMKK3-GFP were transformed into rice leaf protoplasts as previously described (Bart et al. 2006). After overnight incubation at 25 °C for 24 h in the dark, GFP was detected using a confocal laser-scanning microscope with excitation at 488 nm (Nikon C2-ER). Excitation of the plasma membrane co-localization red fluorescent protein marker mCherry was done at 580 nm and emission light was collected at 610 nm.
Conclusions
OrMKK3 is a negative regulator of plant height and grain shape, a positive regulator of tiller number, and a possible part of the BR signaling pathways. These findings provide information about a new gene and the crucial role of MAPKs in plant growth.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file1 Table S1. Primers for gene amplification. (XLSX 9 KB)
Supplementary file2 Table S2. Primers for qRT-PCR. (DOCX 15 KB)
Acknowledgements
We thank Dr. Zichao Li (China Agricultural University, China) for the constructive suggestions.
Abbreviation
- GFP
Green fluorescent protein
Funding
This study was funded by “Key Project of Guangxi Natural Science Foundation [2018GXNSFDA281053, 2018GXNSFBA281022, 2017GXNSFAA198266]. Open Project of Guangxi Key Laboratory of Rice Genetics and Breeding and Guangxi Major Scientific and Technological Innovation base [2018-05-06-KF08]. National Natural Science Foundation of China [NSFC 31960059, NSFC31960401, NSFC32060476]. National Natural Science Foundation of China (CN) [NSFC32060452]”.
Data Availability Statement
Relevant data are within the paper including CDS of OrMKK3.
Compliance with Ethical Standards
Conflict of Interest
The authors declare no conflict of interest.
Footnotes
Ying Hua Pan, Li Jun Gao and Yun Tao Liang have contributed equally to this work
Contributor Information
Ying Hua Pan, Email: panyinghua2008@163.com.
Gao Xing Dai, Email: dgx@gxaas.net.
Guo Fu Deng, Email: dengguofu163@163.com.
References
- Andreasson E, Ellis B. Convergence and specificity in the Arabidopsis MAPK nexus. Trends Plant Sci. 2010;15(2):106–113. doi: 10.1016/j.tplants.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Bai MY, Zhang LY, Gampala SS, Zhu SW, Song WY, Chong K, Wang ZY. Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc Natl Acad Sci. 2007;104(34):13839–13844. doi: 10.1073/pnas.0706386104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y, Xu S, Jing X, Meng L, Qin ZY. De Novo assembly and characterization of Oryza officinalis leaf transcriptome by using RNA-Seq. Biomed Res Int. 2015;982065:9. doi: 10.1155/2015/982065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bart R, Chern M, Park CJ, Bartley L, Ronald PC. A novel system for gene silencing using siRNAs in rice leaf and stemderived protoplasts. Plant Methods. 2006;2:13. doi: 10.1186/1746-4811-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamman R, Bai FW, Drory SB, Loubert-Hudon A, Ellis B, Matton DP. The Arabidopsis Mitogen-Activated Protein Kinase Kinase Kinase 20 (MKKK20) C-terminal domain interacts with MKK3 and harbors a typical DEF mammalian MAP kinase docking site. Front Plant Sci. 2018;8:1352. doi: 10.3389/fpls.2017.01352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai YH, Chen XJ, Xie K, Xing QK, Wu YW, Li J, Du CH, Sun ZX, Guo ZJ. Dlf1, a WRKY transcription factor, is involved in the control of fowering time and plant height in rice. PLoS ONE. 2014;9:e102529. doi: 10.1371/journal.pone.0102529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WW, Cheng ZJ, Liu LL, Wang M, You XM, Wang J, Zhang F, Zhou CL, Zhang Z, Zhang H, You SM, Wang YP, Luo S, Zhang JH, Wang JL, Wang J, Zhao ZC, Guo XP, Lei CL, Zhang X, Lin QB, Ren YL, Zhu SS, Wan JM. Small Grain and Dwarf 2, encoding an HD-Zip II family transcription factor, regulates plant development by modulating gibberellin biosynthesis in rice. Plant sci. 2019;288:110208. doi: 10.1016/j.plantsci.2019.110208. [DOI] [PubMed] [Google Scholar]
- Chen YN, Fan XR, Song WJ, Zhang YL, Xu GH. Over-expression of OsPIN2 leads to increased tiller numbers, angle and shorter plant height through suppression of OsLAZY1. Plant Biotechnol J. 2012;10:139–149. doi: 10.1111/j.1467-7652.2011.00637.x. [DOI] [PubMed] [Google Scholar]
- Cheng XY, Zhu LL, He GC. Towards understanding of molecular interactions between rice and the brown planthopper. Mol Plant. 2013;6(3):621–634. doi: 10.1093/mp/sst030. [DOI] [PubMed] [Google Scholar]
- Choi MS, Woo MO, Koh EB, Lee JH, Ham TH, Seo HS, Koh HJ. Teosinte Branched 1 modulates tillering in rice plants. Plant Cell Rep. 2012;31:57–65. doi: 10.1007/s00299-011-1139-2. [DOI] [PubMed] [Google Scholar]
- Colcombet J, Sözen C, Hirt H. Convergence of multiple MAP3Ks on MKK3 identifies a set of novel stress MAPK modules. Front Plant Sci. 2016;7:1941. doi: 10.3389/fpls.2016.01941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danquah A, Zelicourt AD, Boudsocq M, Neubauer J, Frey NFD, Leonhardt M, Pateyron S, Gwinner F, Philippe Tamby JP, Ortiz-Masia D, Marcote MJ, Colcombet J. Identification and characterization of an ABA-activated MAP kinase cascade in Arabidopsis thaliana. Plant J. 2015;82(2):232–244. doi: 10.1111/tpj.12808. [DOI] [PubMed] [Google Scholar]
- Duan PG, Rao Y, Zeng DL, Yang YL, Xu R, Zhang BL, Dong GJ, Qian Q, Li YH. SMALL GRAIN 1, which encodes a mitogen-activated protein kinase kinase 4, influences grain size in rice. Plant J. 2014;77:547–557. doi: 10.1111/tpj.12405. [DOI] [PubMed] [Google Scholar]
- Fan CC, Xing YZ, Mao HL, Lu TT, Han B, Xu CG, Li XH, Zhang QF. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor Appl Genet. 2006;112:1164–1171. doi: 10.1007/s00122-006-0218-1. [DOI] [PubMed] [Google Scholar]
- Fang N, Xu R, Huang LJ, Zhang BL, Duan PG, Li N, Luo YH, Li YH. SMALL GRAIN 11 controls grain size, grain number and grain yield in rice. Rice. 2016;9:64. doi: 10.1186/s12284-016-0136-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Jin MN, Zheng XM, Chen J, Yuan DY, Xin YY, Wang MQ, Huang DY, Zhang Z, Zhou KN, Sheng PK, Ma J, Ma WW, Deng HF, Jiang LL, Liu SJ, Wang HY, Wu CY, Yuan LP, Wan JM. Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice. Proc Natl Acad Sci USA. 2014;111(46):16337–16342. doi: 10.1073/pnas.1418204111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao XY, Zhang JQ, Zhang XJ, Zhou J, Jiang ZS, Huang P, Tang ZB, Bao YM, Cheng JP, Tang HJ, Zhang WH, Zhang HS, Huang J. Rice qGL3/OsPPKL1 functions with the GSK3/SHAGGY-Like Kinase OsGSK3 to modulate brassinosteroid signaling. Plant Cell. 2019;31(35):1077–1093. doi: 10.1105/tpc.18.00836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelmedin V, Spiliotis M, Brehm K. Molecular characterisation of MEK1/2- and MKK3/6-like mitogen-activated protein kinase kinases (MAPKK) from the fox tapeworm Echinococcus multilocularis. Int J Parasitol. 2010;40(5):555–567. doi: 10.1016/j.ijpara.2009.10.009. [DOI] [PubMed] [Google Scholar]
- Guo T, Chen K, Dong NQ, Shi CL, Ye WW, Gao JP, Shan JX, Lin HX. GRAIN SIZE AND NUMBER1 negatively regulates the OsMKKK10-OsMKK4-OsMPK6 cascade to cordinate the ntrade-off between grain number per panicle and grain size. Plant Cell. 2018;30:871–888. doi: 10.1105/tpc.17.00959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha H, Jung H, Seong HA. Murine proteine serine/threonine kinase 38 (MPK38) positively regulates apoptosis signal-regulating kinase 1 in a kinase-dependent manner through physical interaction. FASEB J. 2008;22:1. doi: 10.1096/fasebj.22.1_supplement.1050.7. [DOI] [Google Scholar]
- Hamel LP, Nicole MC, Sritubtim S, Morency MJ, Ellis M, Ehlting J, Beaudoin N, Barbazuk B, Klessig D, Lee J, Martin G, Mundy J, Ohashi Y, Scheel D, Sheen J, Xing T, Zhang SQ, Seguin A, Ellis BE. Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci. 2006;11(4):192–198. doi: 10.1016/j.tplants.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994;6:271–282. doi: 10.1046/j.1365-313X.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- Horiguchi G, Ferjani A, Fujikura U, Tsukaya H. Coordination of cell proliferation and cell expansion in the control of leaf size in Arabidopsis thaliana. J. Plant Res. 2006;119:37–42. doi: 10.1007/s10265-005-0232-4. [DOI] [PubMed] [Google Scholar]
- Huang XH, Kurata N, Wei XH, Wang ZX, Wang A, Zhao Q, Zhao Y, Liu KY, Lu HY, Li WJ, Guo YL, Lu YQ, Zhou CC, Fan DL, Weng QJ, Zhu CR, Tao Huang T, Zhang L, Wang YC, Feng L, Furuumi H, Kubo T, Miyabayashi T, Yuan XP, Qun XuQ, Dong GJ, Zhan QL, Li CY, Fujiyama A, Toyoda A, Lu TT, Feng Q, Qian Q, Li JY, Han B. A map of rice genome variation reveals the origin of cultivated rice. Nature. 2012;490:497–501. doi: 10.1038/nature11532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H, Ueguchi-Tanaka M, Sakamoto T, Kayano T, Tanaka H, Ashikari M, Matsuoka M. Modification of rice plant height by suppressing the height-controlling gene, D18, in rice. Breed Sci. 2002;52:215–218. doi: 10.1270/jsbbs.52.215. [DOI] [Google Scholar]
- Jiang CM, Xiao SQ, Li DQ, Ling C, Zhong QF, Yin FY, Yu TQ, Ke X, Zhang DY, Fu J, Chen Y, Wang B, Wang LX, Li EX, Zhang Y, Huang XQ, Cheng ZQ. Identification and expression pattern analysis of bacterial blight resistance genes in Oryza officinalis wall ex watt under Xanthomonas oryzae Pv. oryzae stress. Plant Mol Biol Rep. 2019;37:436–449. doi: 10.1007/s11105-019-01164-3. [DOI] [Google Scholar]
- Kim TW, Michniewicz M, Bergmann DC, Wang ZY. Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature. 2012;482:419–423. doi: 10.1038/nature10794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi-Kaboshi M, Okada K, Kurimoto L, Murakami S, Umezawa T, Shibuya N, Yamane H, Miyao A, Takatsuji H, Takahashi A, Hirochika H. A rice fungal MAMP-responsive MAPK cascade regulates metabolic flow to antimicrobial metabolite synthesis. Plant J. 2010;63:599–612. doi: 10.1111/j.1365-313X.2010.04264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. Mitogen-Activated Protein Kinase Kinase 3 is required for regulation during dark-light transition. Mol Cells. 2015;38:651–656. doi: 10.14348/molcells.2015.0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YB, Fan CC, Xing YZ, Jiang YH, Luo LJ, Sun L, Shao D, Xu CJ, Li XH, Xiao JH, He YQ, Zhang QF. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat Genet. 2011;43:1266–1269. doi: 10.1038/ng.977. [DOI] [PubMed] [Google Scholar]
- Liao ZG, Yu H, Duan JB, Yuan K, Yu CJ, Meng XB, Kou LQ, Chen MJ, Jing YH, Liu GF, Smith SM, Li JY. SLR1 inhibits MOC1 degradation to coordinate tiller number and plant height in rice. Nat Commun. 2019;10:9. doi: 10.1038/s41467-019-10667-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Wang RX, Qian Q, Yan MX, Meng XB, Fu ZM, Yan CY, Jiang B, Su Z, Li JY, Wang YY. DWARF27, an iron-containing protein required for the biosynthesis of srigolactones, regulates rice tiller bud outgrowth. Plant cell. 2009;21:1512–1525. doi: 10.1105/tpc.109.065987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Zhang HH, Chen ZX, Shahid MQ, Fu XL, Liu XD. Drought-tolerant rice germplasm developed from an Oryza officinalis transformation-competent artificial chromosome clone. Genet Mol Res. 2015;14(4):13667–13678. doi: 10.4238/2015.October.28.29. [DOI] [PubMed] [Google Scholar]
- Liu W, Zhang D, Tang MF, Li DY, Zhu YX, Zhu LH, Chen CY. THIS1 is a putative lipase that regulates tillering, plant height, and spikelet fertility in rice. J Exp Bot. 2013;64(14):4389–4402. doi: 10.1093/jxb/ert256. [DOI] [PubMed] [Google Scholar]
- Mao HL, Sun SY, Yao JL, Wang CR, Yu SB, Xu CG, Li XH, Zhang QF. Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. PNAS. 2010;107(45):19579–19584. doi: 10.1073/pnas.1014419107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao XX, Zhang JJ, Liu WG, Yan SJ, Liu Q, Fu H, Zhao JL, Huang WJ, Dong JF, Zhang SH, Yang TF, Yang W, Liu B, Wang F. The MKKK62-MKK3-MAPK7/14 module negatively regulates seed dormancy in rice. Rice. 2019;12:7. doi: 10.1186/s12284-018-0260-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morinaka Y, Sakamoto T, Inukai Y, Agetsuma M, Kitano H, Ashikari M, Matsuoka M. Morphological alteration caused by brassinosteroid insensitivity increases the biomass and grain production of Rice. Plant Physiol. 2006;141:924–931. doi: 10.1104/pp.106.077081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Pourkheirandish M, Morishige H, Kubo Y, Nakamura M, Ichimura K, Seo S, Kanamori H, Wu J, Ando T, Hensel G, Sameri M, Stein N, Sato K, Matsumoto T, Yano M, Komatsuda T. Mitogen-Activated Protein Kinase Kinase 3 regulates seed dormancy in Barley. Curr Biol. 2016;26(6):775–781. doi: 10.1016/j.cub.2016.01.024. [DOI] [PubMed] [Google Scholar]
- Pitzschke A. Modes of MAPK substrate recognition and control. Trends Plant Sci. 2015;20(1):49–55. doi: 10.1016/j.tplants.2014.09.006. [DOI] [PubMed] [Google Scholar]
- Qi P, Lin YS, Song XJ, Shen JB, Huang W, Shan JX, Zhu MZ, Jiang LW, Gao JP, Lin HX. The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3. Cell Res. 2012;22:1666–1680. doi: 10.1038/cr.2012.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao YL, Piao RH, Shi JX, Lee SI, Jiang WZ, Kim BK, Lee J, Han LZ, Ma WB, Koh HJ. Fine mapping and candidate gene analysis of dense and erect panicle 3, DEP3, which confers high grain yield in rice (Oryza sativa L.) Theor Appl Genet. 2011;122:1439–1449. doi: 10.1007/s00122-011-1543-6. [DOI] [PubMed] [Google Scholar]
- Si LZ, Huang XH, Gong H, Luo JH, Hou QQ, Hou TY, Lu TT, Jingjie JJ, Shangguan YY, Chen EW, Gong CX, Zhao Q, Jing YF, Zhao Y, Li Y, Cui LL, Fan DL, Lu YQ, Weng QJ, Wang YC, Zhan QL, Liu KY, Wei XH, An K, An G, Han B. OsSPL13 controls grain size in cultivated rice. Nat Genet. 2016;48:447–456. doi: 10.1038/ng.3518. [DOI] [PubMed] [Google Scholar]
- Song XJ, Huang W, Shi M, Zhu MZ, Lin HX. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet. 2007;39:623–630. doi: 10.1038/ng2014. [DOI] [PubMed] [Google Scholar]
- Takahashi F, Mizoguchi T, Yoshida R, Ichimura K, Shinozaki K. Calmodulin-dependent activation of MAP kinase for ROS homeostasis in Arabidopsis. Mol Cell. 2011;41(6):649–660. doi: 10.1016/j.molcel.2011.02.029. [DOI] [PubMed] [Google Scholar]
- Takahashi F, Yoshida R, Ichimura K, Mizoguchi T, Seo S, Yonezawa M, Maruyama K, Yamaguchi-Shinozaki K, Shinozaki K. The Mitogen-Activated Protein Kinase Cascade MKK3–MPK6 is an important part of the jasmonate signal transduction pathway in Arabidopsis. Plant Cell. 2007;19:805–818. doi: 10.1105/tpc.106.046581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A, NakagawaH TC, Shimatani ZP, Ohtake M, Nomura T, Jiang CJ, Dubouzet JG, Kikuchi S, Sekimoto H, Yokota T, Asami T, Kamakura T, Mori M. BRASSINOSTEROID UPREGULATED1, encoding a helix–loop–helix protein, is a novel gene involved in brassinosteroid signaling and controls bending of the lamina joint in rice. Plant Physiol. 2009;151:669–680. doi: 10.1104/pp.109.140806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong H, Jin Y, Liu W, Li F, Fang J, Yin Y, Qian Q, Zhu L, Chu C. DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J. 2009;58(5):803–816. doi: 10.1111/j.1365-313X.2009.03825.x. [DOI] [PubMed] [Google Scholar]
- Vishmita Sethi V, Raghuram B, Sinha AK, Chattopadhyay S. A Mitogen-Activated Protein Kinase Cascade Module, MKK3-MPK6 and MYC2, is involved in blue light-mediated seedling development in Arabidopsis. Plant Cell. 2014;26:3343–3357. doi: 10.1105/tpc.114.128702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Smith SM, LiJY, Genetic regulation of shoot architecture. Annu Rev Plant Biol. 2018;69:437–468. doi: 10.1146/annurev-arplant-042817-040422. [DOI] [PubMed] [Google Scholar]
- Wang SK, Wu K, Yuan QB, Liu XY, Liu ZB, Lin XY, Zeng RZ, Zhu HT, Dong GJ, Qian Q, Zhang GQ, Fu XD. Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet. 2012;44:950–954. doi: 10.1038/ng.2327. [DOI] [PubMed] [Google Scholar]
- Wang J, Lu K, Nie HP, Zeng QS, Wu BW, Qian JJ, Fang ZM. Rice nitrate transporter OsNPF7.2 positively regulates tiller number and grain yield. Rice. 2018;11:2. doi: 10.1186/s12284-018-0205-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wu BW, Lu K, Wei Q, Qian JJ, Chen YP, Fang ZM. The Amino Acid Permease 5 (OsAAP5) Regulates Tiller Number and Grain Yield in Rice. Plant physiol. 2019;180:1031–1045. doi: 10.1104/pp.19.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Xu J, Guo H, Jiang L, Chen S, Yu C, Zhou Z, Hu P, Zhai H, Wan J. DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol. 2010;153:1747–1758. doi: 10.1104/pp.110.156943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Wang YH, Yu YC, Duan JB, Liao ZG, Xiong GS, Meng XB, Liu GF, Qian Q, Li JY. Degradation of MONOCULM 1 by APC/CTAD1 regulates rice tillering. Nat Commun. 2012;3:750. doi: 10.1038/ncomms1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Zhang SQ. Mitogen-activated protein kinase cascades in signaling plant growth and development. Trends Plant Sci. 2015;20(1):56–64. doi: 10.1016/j.tplants.2014.10.001. [DOI] [PubMed] [Google Scholar]
- Xu R, Duan PG, Yu HY, Zhou ZK, Zhang BL, Wang RC, Li J, Zhang GZ, Zhuang SS, Lyu J, Li N, Chai TY, Tian ZX, Yao SG, Li YH. Control of grain size and weight by the OsMKKK10-OsMKK4-OsMAPK6 signaling pathway in rice. Mol Plant. 2018;11(4):860–873. doi: 10.1016/j.molp.2018.04.004. [DOI] [PubMed] [Google Scholar]
- Yamamuro C, Ihara Y, Wu X, Noguchi T, Fujioka S, Takatsuto S, Ashikari M, Kitano H, Matsuoka M. Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell. 2000;12:1591–1605. doi: 10.1105/tpc.12.9.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan HJ, Zhao YF, Shi H, Li J, Wang YC, Tang DZ. BRASSINOSTEROID-SIGNALING KINASE1 phosphorylates MAPKKK5 to regulate immunity in Arabidopsis. Plant Physiol. 2018;176:2991–3002. doi: 10.1104/pp.17.01757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S, Zou GH, Li SJ, Wang H, Liu HQ, Zhai GW, Guo P, Song HM, Yan CJ, Tao YZ. Seed size is determined by the combinations of the genes controlling different seed characteristics in rice. Theor Appl Genet. 2011;123:1173–1181. doi: 10.1007/s00122-011-1657-x. [DOI] [PubMed] [Google Scholar]
- Yongzhen Wu YZ, Fu Y, Zhao SS, Gu P, Zhu ZF, Sun CQ, Tan LB. CLUSTERED PRIMARY BRANCH 1, a new allele of DWARF11, controls panicle architecture and seed size in rice. Plant Biotechnol J. 2016;14:377–386. doi: 10.1111/pbi.12391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LY, Bai MY, Wu JX, Zhu JY, Wang H, Zhang ZG, Wang WF, Sun Y, Zhao J, Sun XH, Yang HJ, Xu YY, Kim SH, Fujioka S, Lin WH, Chong K, Lu TG, Wang ZY. Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant cell. 2009;21:3767–3780. doi: 10.1105/tpc.109.070441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XJ, Wang JF, Huang J, Lan HX, Wang CL, Yin CF, Wu YY, Tang HJ, Qian Q, Li JY, Zhang HS. Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. PNAS. 2012;109(52):21534–21539. doi: 10.1073/pnas.1219776110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XY, Yin FY, Xiao SQ, Jiang CM, Yu TQ, Chen L, Xue K, Zhong QF, Cheng ZQ, Li WJ. Proteomic analysis of the rice (Oryza officinalis) provides clues on molecular tagging of proteins for brown planthopper resistance. BMC Plant Biol. 2019;19:9. doi: 10.1186/s12870-018-1622-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi Hong Z, Ueguchi-Tanaka M, Fujioka S, Takatsuto S, Yoshida S, Hasegawa Y, Ashikari M, Kitano H, Matsuoka M. The Rice brassinosteroid-deficient dwarf2 Mutant, Defective in the Rice Homolog of Arabidopsis DIMINUTO/DWARF1, is rescued by the endogenously accumulated alternative bioactive brassinosteroid, dolichosterone. Plant Cell. 2005;17:2243–2254. doi: 10.1105/tpc.105.030973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Lin QB, Zhu LH, Ren YL, Zhou KN, Shabek N, Wu FQ, Mao HB, Dong W, Gan L, Ma WW, Gao H, Chen J, Yang C, Wang D, Tan JJ, Zhang X, Guo XP, Wang JL, Jiang L, Liu X, Chen WQ, Chu JF, Yan CY, Ueno K, Ito S, Asami T, Cheng ZJ, Wang J, Lei CL, Zhai HQ, Wu CY, Wang HY, Zheng N, Wan JM. D14–SCFD3-dependent degradation of D53 regulates strigolactone signalling. Nature. 2013;504:406–410. doi: 10.1038/nature12878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou SX, Chen MT, Zhang YB, Gao Q, Noman A, Wang Q, Li H, Chen L, Zhou PY, Lu J, Lou YG. OsMKK3, a stress-responsive protein kinase, positively regulates rice resistance to Nilaparvata lugens via Phytohormone Dynamics. Int J Mol Sci. 2019;20(12):3023. doi: 10.3390/ijms20123023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file1 Table S1. Primers for gene amplification. (XLSX 9 KB)
Supplementary file2 Table S2. Primers for qRT-PCR. (DOCX 15 KB)
Data Availability Statement
Relevant data are within the paper including CDS of OrMKK3.