Abstract
Background
Crosstalk between genetic, epigenetic, and immune alterations in upper tract urothelial carcinomas and their role in shaping muscle invasiveness and patient outcome are poorly understood.
Results
We perform an integrative genome- and methylome-wide profiling of diverse non-muscle-invasive and muscle-invasive upper tract urothelial carcinomas. In addition to mutations of FGFR3 and KDM6A, we identify ZFP36L1 as a novel, significantly mutated tumor suppressor gene. Overall, mutations of ZFP36 family genes (ZFP36, ZFP36L1, and ZFP36L2) are identified in 26.7% of cases, which display a high mutational load. Unsupervised DNA methylation subtype classification identifies two epi-clusters associated with distinct muscle-invasive status and patient outcome, namely, EpiC-low and EpiC-high. While the former is hypomethylated, immune-depleted, and enriched for FGFR3-mutated, the latter is hypermethylated, immune-infiltrated, and tightly associated with somatic mutations of SWI/SNF genes.
Conclusions
Our study delineates for the first time the key role for convergence between genetic and epigenetic alterations in shaping clinicopathological and immune upper tract urothelial carcinoma features.
Keywords: Upper tract urothelial carcinomas, ZFP36L1, DNA methylation, SWI/SNF gene mutations, Sequencing, Immunity, Epigenetics
Introduction
Urothelial carcinoma is considered the fifth most common cancer in Western countries, and it is divided at the pathological level into two groups: non-muscle-invasive (NMI) and muscle-invasive (MI) tumors, according to the level of invasion of the detrusor muscle. NMI represents 75% of tumors, while MI represents the latter 25% [1]. NMI tumors are costly to treat as they often recur, and 10–15% of those patients progress to an MI state [1]. Upper tract urothelial carcinomas represent 5–10% among all urothelial carcinomas, and they can arise within the ureter or the renal pelvis, which are derived from different embryonic tissues as compared to the bladder urothelium [2, 3]. Such differences might dictate whether patients with UTUC display a higher incidence of invasive disease at diagnosis as compared to patients with bladder carcinomas [3].
While genome-wide genetic alterations of muscle-invasive bladder carcinomas have been extensively studied by The Cancer Genome Atlas (TCGA) project and others, genetic alterations occurring in UTUC are limited [4, 5]. Recent studies using whole-exome sequencing and/or targeted sequencing of UTUC samples identified recurrent mutations of genes known to be altered in bladder carcinomas, although with different frequencies (e.g., HRAS) [6, 7]. At the transcriptomic level, UTUCs were mostly found to be luminal papillary and displayed T cell-depleted immune contexture, possibly related to the FGFR3 overexpression [5]. However, to our knowledge, epigenetic alterations in UTUC, in particular, DNA methylation, and their crosstalk with genetic and transcriptomic clinicopathological tumor features are unknown.
Although the epigenetic alterations of MI bladder carcinomas have been extensively studied by TCGA and other groups, comparative analysis of epigenetic alterations in MI relative to NMI bladder carcinomas has been limited, using often selected cancer-related genes or supervised analysis [8]. In fact, the recent remarkable study exploring genome-wide alterations of NMI bladder carcinomas did not include DNA methylation profiling [9].
To fill this knowledge gap, we decided to investigate the putative contribution of both genetic and epigenetic alterations in dictating muscle invasiveness, a key predictor of poor outcome in UTUC patients. We identified different key findings, including the discovery of novel mutations affecting the zinc-finger RNA-binding protein ZFP36L1 in 20% of cases, a rate by far the highest among cancer subtypes profiled to date. In addition, we identified two methylome clusters, EpiC-low and EpiC-high, associated with distinct clinicopathological and genetic tumor features, as well as patient survival. While the EpiC-low cluster was hypermethylated, immune-inflamed, enriched in MI cases, and harbored a high rate of somatic mutations of SWI/SNF genes, the EpiC-high cluster was hypomethylated, immune-desert, enriched for NMI cases, and harbored a high rate of FGFR3 mutations. These data pave the way for therapeutic interventions in the most threatening subgroup of UTUC, providing a rational for personalizing therapies in this setting.
Results
Samples, clinical data, and analytic approach
Forty fresh-frozen surgically resected primary UTUCs, including 20 NMI and 20 MI cases, were collected retrospectively from the pathology biobank at Pitié-Salpêtrière Hospital and re-evaluated histopathologically by one expert pathologist (E.C.). According to the European Association of Urology guidelines, all samples had been tested for microsatellite instability status; all were microsatellite stable. Overall, DNA and RNA of good quality and quantity were obtained for 40 and 20 cases, respectively. Matched germline DNA from adjacent bladder tissues were also collected in 30 cases. Whole-exome sequencing (WES) was performed for 30-paired UTUCs and adjacent normal tissues. Targeted FGFR3 sequencing was performed for 35 cases. RNA sequencing was performed on 20 UTUC cases; in addition, DNA methylation was analyzed using Infinium EPIC arrays on 35 UTUC cases and 8 normal adjacent bladder tissues. Detailed clinical and pathologic characteristics of the cohort are reported in Additional file 1: Table S1. The median follow-up time for the 40 patients was 53 months (range 2–98); 18 patients had recurred, and 12 patients have died from disease progression at the last follow-up. Among the clinical variables, muscle invasion status, age, gender, pathological stage, localization, and grade were not associated with progression-free survival or with overall survival (Additional file 1: Table S2).
Landscape of somatic mutations and focal copy number alterations
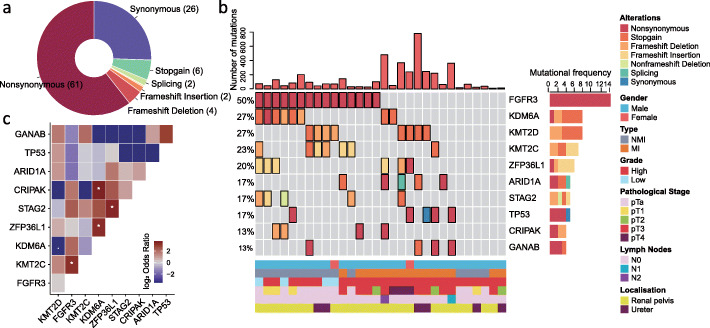
The 30 UTUC samples studied by WES included 15 NMI and 15 MI cases. We identified 4239 putative somatic mutations, including 2569 missense mutations, 1081 silent mutations, and 272 indels, with an average of 2.9 ± 3.5 mutations per megabase (Additional file 1: Table S3). Non-synonymous single-nucleotide variants were the most frequent mutations identified (61%) (Fig. 1a). The median mutational load per megabase was 1.5 (range 0.3–15.7). Using MutSigCV, significantly mutated genes (SMGs) (false discovery rate [FDR] < 0.05) were FGFR3 (50%, FDR = 3.15 × 10−3), KDM6A (27%, FDR = 1.03 × 10−3), and ZFP36L1 (20%, FDR = 2.62 × 10−3) (Fig. 1b, Additional file 1: Table S4). ZFP36L1 was not previously reported to be mutated in UTUC; as a member of ZFP36 family genes, ZFP36L1 is a zinc-finger RNA-binding protein that regulates several cytoplasmic AU-rich element (ARE)-containing mRNA transcripts by favoring their poly (A) tail removal or deadenylation, leading to the attenuation of protein synthesis. Mutations of FGFR3, KDM6A, and ZFP36L1 alterations were all verified by Sanger sequencing. Other frequent mutations (> 10%) with a trend of significance (P < 0.05, FDR > 0.05) involved MLL2 (P = 0.037), KMT2C (P = 0.02), STAG2 (P = 8.3 × 10−7), ARID1A (P = 0.01), TP53 (P = 0.018), CRIPAK (P = 0.0001), and GANAB (P = 0.04) (Fig. 1c). Mutations of FGFR3 and KMT2C co-occur mutually (log2 odd ratio = 3.1, P = 0.04; Fig. 1c, Additional file 1: Table S5); this was also validated in the MSKCC UTUC cohort (log2 odd ratio = 2.9, P < 0.001; FDR < 0.001) [7]. In addition, we observed a mutual co-occurrence of mutations affecting ZFP36L1 with both KDM6A (log2 odd ratio = 3.1; P = 0.03) and STAG2 (log2 odd ratio = 3.29, P = 0.04; Additional file 1: Table S5). Mutations of MLL2 (P = 0.04), ARID1A (P = 0.01), and GANAB (P = 0.02) showed significant association with a higher mutation load, as well as a tendency for ZFP36L1 (P = 0.06).
Fig. 1.
Genetic landscape of somatic mutations identified by whole-exome sequencing of 30 upper tract urothelial carcinomas (UTUC), including 15 muscle-invasive tumors and 15 non-muscle-invasive tumors. a Distribution of somatic mutations showing that non-synonymous mutations are the most frequent type. b OncoPrint depicting mutational load and most frequently mutated genes identified (≥ 10% of cases) in the whole UTUC cohort. c Heatmap for genes with mutual exclusivity or co-occurrence in the whole UTUC cohort. Stars refer to correlations that are statistically significant
Among genes with mutations arising in > 10% of samples, only FGFR3 mutations were significantly associated with improved overall survival (OS) (P = 0.005) and progression-free survival (PFS) at distant sites (P = 0.007). We then looked for an association between gene mutations and muscle-invasive status; as expected, we found that FGFR3 mutations were enriched in NMI as compared to MI tumors (73% vs 27%; P = 0.027, FDR = 0.2); conversely, ARID1A mutations were enriched in MI tumors (33% vs 0%; P = 0.042, FDR = 0.2; Additional file 1: Table S6).
We then analyzed statistically significant focal copy number variation (CNV) changes using the GISTIC algorithm. Considering focal peaks, 1p36.33, 9p21.3, and 11p15.1 were lost (Additional file 2: Fig. S1a). Notably, 9p21.3 encompass CDKN2A and CDKN2B tumor suppressor genes. The analysis revealed six additional regions with gains at a significant frequency at 1q23.3, 6p21.33, 8p11.23, 8q22.3, 12q15, and 19q12 regions. Several of these events involve known cancer-related oncogenes, such as YWHAZ (8q22.3), MDM2 (12q15), and CCNE1 (19q12) (Additional file 2: Fig. S1b). We then analyzed the association between CNV, somatic mutations, and clinicopathological tumor features and somatic mutations. We identified significant enrichment in MI cases for 8q22.3 gain, 12q15 gain, and 11p15.11 loss (Additional file 2: Fig. S1c). We also observed mutual exclusivity of 8q22.3 gain with FGFR3 mutations (P = 0.03), as well as mutual occurrence with TP53 mutations (P = 0.02) (Additional file 2: Fig. S1d).
Mutational frequency comparison between UTUC and BLCA according to muscle invasiveness status
We hypothesized that UTUC and BLCA may show different mutational frequencies according to muscle invasiveness status. To compare mutational frequency regarding NMI, we retrieved mutation data of 24 NMI-BLCA from the Hurst cohort and compared it with our NMI-UTUCs (n = 15), and we found KMT2C (P = 0.08) was more likely to mutate in NMI-UTUCs (n = 15) whereas PIK3CA mutations were significantly enriched in NMI-BLCAs (P = 0.0001) (Additional file 2: Fig. S2a). We then compared the most frequently mutated genes between MI-UTUC (n = 15) and TCGA MI-BLCA (n = 412). We found that MI-UTUC showed significantly more mutations of GANAB (P = 0.021), CRIPAK (P = 0.022), and ZFP36L1 (P = 0.1) (Additional file 2: Fig. S2b).
Prevalence of mutations affecting ZFP36 genes family
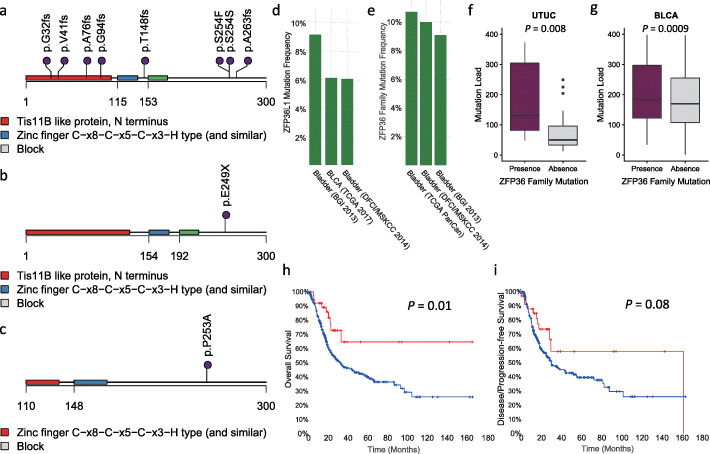
As ZFP36L1 mutations have not been previously reported to be altered in UTUC, we thus investigated their significance. Overall, eight mutations were identified in a total of six cases. Strikingly, six out of eight ZFP36L1 mutations were frameshift insertions or deletions, and two were a non-synonymous mutation predicted to be deleterious by Poly2phen and SIFT (Fig. 2a). When we explored mutations of other members of the ZFP36 family, we identified one additional UTUC case with a ZFP36L2 stop-gain mutation (E249X) and another with a deleterious ZFP36 (p.P253A) mutation, both already existing in COSMIC (Fig. 2b, c). Notably, ZFP36L2 E249X was identified as a hotspot for mutations in the bladder TCGA, consolidating our findings about the relevance of this mutation in urothelium carcinogenesis (Additional file 2: Fig. S3a). Thus, overall mutations of ZFP36 family genes represent 26.7% (n = 8/30) among all UTUC cases. Mutations of ZFP36 family genes were not associated with clinicopathological tumor features and patient overall survival (not shown).
Fig. 2.
Distribution of mutations affecting ZFP36 family genes in UTUC and several public bladder carcinoma datasets. Lollipops represent mutations of a ZFP36L1, b ZFP36L2, and c ZFP36 genes. d Distribution of mutation frequency of ZFP36L1 in different public bladder carcinoma cohorts using the cBioPortal database. e Distribution of mutation frequency of ZFP36 family in different bladder carcinoma cohorts using the cBioPortal database. f Box plot representing the mutational load in UTUC samples according to ZFP36 family gene mutations. g Box plot representing mutational load in bladder carcinoma TCGA cohort according to ZFP36 family mutations. h Kaplan-Meier curves for overall survival of patients with bladder carcinomas in TCGA cohort according to ZFP36 family gene mutations status. i Kaplan-Meier curves for progression-free survival of patients with bladder carcinomas in TCGA cohort according to ZFP36 family mutations
Frequency of ZFP36 family gene mutations across cancer subtypes
We then decided to analyze the frequency of ZFP36L1 mutations in 10,967 cancer samples related to 32 different histopathological cancer subtypes analyzed by TGCA. Overall, 120 (1.09%) samples were identified, and the highest frequency was observed in bladder cancer (n = 30/410; 7.3%) indicating the importance of this gene both in UTUC and in bladder carcinogenesis (Additional file 2: Fig. S3b). When we combined mutations of ZFP36L2 and ZFP36 to ZFP36L1, the frequency of ZFP36 gene family mutations reached 3% (n = 297) in all TCGA cohorts, with the highest frequencies observed in bladder cancers (Additional file 2: Fig. S3c). Strikingly, ZFP36L1 mutations were consistently altered in three independent studies exploring the genetic landscape of bladder carcinomas (TCGA, DFCI, and BGI), with frequencies ranging from 6 to 8.5% (Fig. 2d). The frequency of ZFP36 gene family mutations ranged between 9.1 and 10%, among the three bladder carcinoma cohorts (Fig. 2e); notably, mutations of ZFP36L1 and ZFP36L2 were mutually co-occurring (P = 0.02; FDR = 0.06). This was also significant using all TCGA samples (P < 0.001; FDR = 0.002). UTUC samples with ZFP36 gene family mutations harbored a higher mutational load as compared to others (P = 0.008; Fig. 2f). This observation was validated in the TCGA bladder cancer dataset (P = 0.0009; Fig. 2g).
Next, we then looked at the expression of ZFP36L1 in the FANTOM5 dataset and Human Protein Atlas and discovered that the bladder urothelium is one of the tissues with the highest mRNA and protein expression levels, highly suggesting the functional significance of this gene (Additional file 2: Fig. S4a-b).
Finally, a pooled analysis of 562 patients in three bladder cohorts (TCGA, DFCI, and BGI) showed that bladder carcinomas with ZFP36 gene family mutations were associated with better overall survival as compared to others (P = 0.01; Fig. 2h); we also observed a trend toward improved progression-free survival in these patients (P = 0.08; Fig. 2i).
Functional analysis of ZFP36L1 knockdown
To investigate the biological role of ZFP36L1, we performed loss-of-function experiments of ZFP36L1 using siRNA in the TCCSUP bladder cancer cell line (Additional file 2: Fig. S5a). Light microscopy images revealed disruption of the cell to cell junctions and clear change to spindle-shaped morphology in the cells with ZFP36L1 knockdown (Additional file 2: Fig. S5b). This was associated with loss of E-cadherin expression, consistent with the epithelial-mesenchymal transition (EMT) (Additional file 2: Fig. S5a). While no effect of the ZFP36L1 knockdown was observed regarding cell proliferation and apoptosis (Additional file 2: Fig. S5c-d), transwell assay showed a significant increase in cell migration in ZFP36L1-depleted cells as compared to control cells (Additional file 2: Fig. S5e).
Mutational signature profiles of UTUC
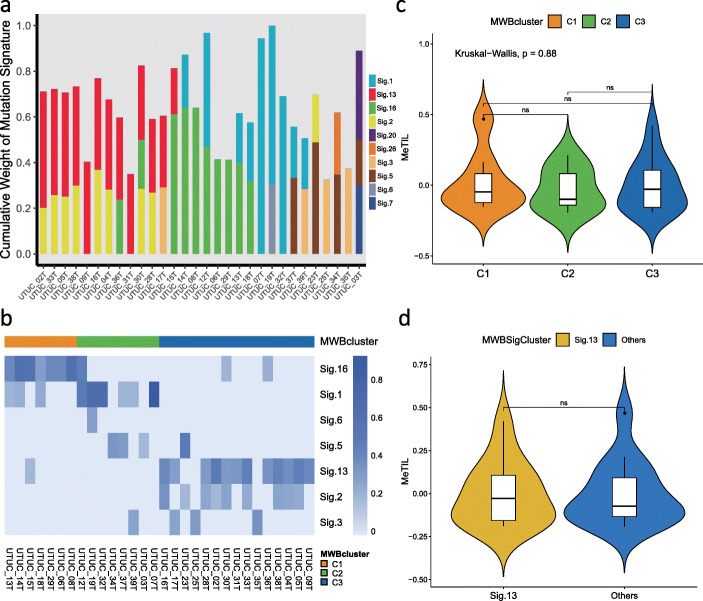
We then sought to decipher the heterogeneity of UTUC by characterizing the mutational signatures [10]. We found high variability between samples with signatures 1, 13, and 16 being the predominant ones (Fig. 3a). We applied a non-negative matrix factorization (NMF) that identified three robust mutation weight (MW)-based clusters considering different mutational signatures and determined signature contributors to each cluster (Fig. 3b). MWBcluster C1 (n = 7) was characterized by signature 16, of which etiology remains unknown. C2 (n = 8) was characterized by enrichment of signature 1, linked to the endogenous mutational process initiated by spontaneous deamination of 5-methylcytosine. Finally, C3 was enriched (n = 15) by the presence of mutational signatures 13 and 2, related to the activity of apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like (APOBEC). As mutation signatures of APOBEC cytidine deaminase were associated with high rates of somatic mutations, and likely increased tumor-infiltrating lymphocytes (TILs), we asked whether either these three clusters or signature 13 was associated with TILs, inferred from DNA methylation (MeTIL) analysis for samples for which DNA methylation and WES were available. We did not find any association (Fig. 3c, d). Notably, UTUC tumors with FGFR3 mutation showed a significantly lower MeTIL score as compared to others (P = 7.8 × 10−5; Additional file 2: Fig. S6a). This was also validated in the TCGA bladder carcinoma cohort (P = 9.0 × 10−6; Additional file 2: Fig. S6b). The immune infiltration derived by FGFR3 mutation was independent from muscle invasiveness status (P = 0.008 for MI; P = 0.024 for NMI). We also found that UTUC MI samples tended to present with a higher MeTIL score relative to NMI ones (P = 0.089; Additional file 2: Fig. S6c).
Fig. 3.
Analysis of mutational signatures and correlation with immunity. a Mutation signature analysis of upper tract urothelial carcinomas showing that signatures 1, 13, and 16 have the highest cumulative weight among the 30 signatures identified by Alexandrov. b Non-negative matrix factorization (NMF) showing three robust clusters based on the mutation weight matrix of different mutational signatures. MWBcluster C1 (n = 7), C2 (n = 8), and C3 (n = 15) were characterized by signature 16, signature 1, and signatures 13, respectively. c Violin plot showing no difference of DNA methylation tumor-infiltrating lymphocyte (MeTIL) score between the identified three clusters. d Violin plot showing no difference of DNA methylation tumor-infiltrating lymphocyte (MeTIL) score between signature 13 and other signatures. MWBcluter: mutation weight-based cluster
Detection of fusion transcripts and unsupervised clustering of gene expression
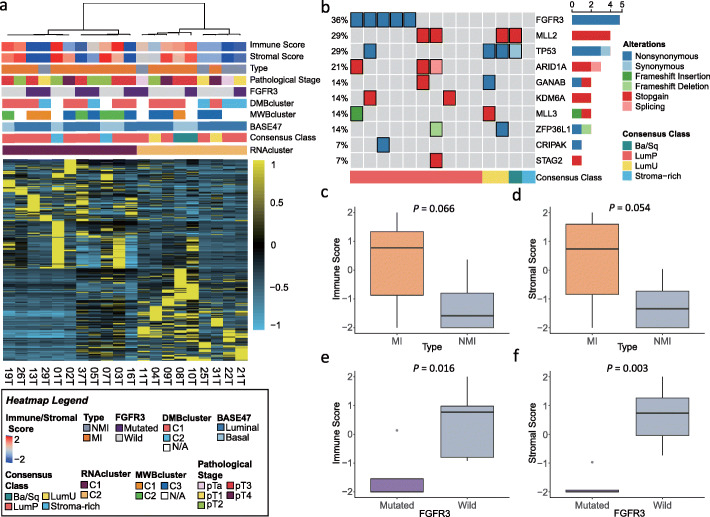
We first analyzed 20 UTUC (15 MI and 5 NMI) for fusion transcripts and detected only one case (UTUC-11) with the oncogenic FGFR3-TACC3 fusion. No other fusions were identified. Unsupervised consensus hierarchical clustering identified two clusters (Fig. 4a). There was no difference observed between C1 and C2 regarding clinicopathological tumor features, probably due to the small cohort size (Additional file 1: Table S7). To better understand the biology of UTUC relative to bladder carcinomas, we applied the recently identified consensus molecular classification of muscle-invasive bladder carcinomas to our cohort. Fourteen cases (70%) were classified as luminal papillary, including all NMI cases (Additional file 1: Table S8). For the remaining cases, two were classified as luminal unstable, two as stromal-rich, and two as basal/squamous. None of the stromal enrich or basal squamous harbored FGFR3 mutations (Fig. 4b). We also performed clustering of the gene expression using the BASE47 signature. We observed that the majority of our cases were “luminal-like” (n = 14), while 6 cases were classified as “basal-like” (Fig. 4a, Additional file 2: Fig. S7). Although the number was small, MI tumors tended to show higher enrichment for immune and stromal scores as compared to NMI tumors (P = 0.066 for immune scores; P = 0.054 for stromal scores; Fig. 4c, d). FGFR3 mutations and/or fusions were associated with significantly lower immune and stromal scores (P = 0.016 for immune scores; P = 0.003 for stromal scores; Fig. 4e, f).
Fig. 4.
Analysis of upper tract urothelial carcinomas (UTUC) transcriptome. a Unsupervised mRNA clustering of 20 UTUC cases showing two clusters, with no clear association identified between them and clinical tumor features. b OncoPrint depicting UTUC subtypes according to the molecular consensus classification of muscle-invasive bladder cancers. Recurrent somatic mutations in those UTUC cases with available genetic data are also shown. c, d Immune and stromal gene expression scores in muscle-invasive (MI) and non-muscle-invasive (NMI) UTUC samples. e, f Immune and stromal gene expression scores in FGFR3-mutated and FGFR3 wild-type UTUC samples. DMBcluster: DNA methylation-based epi-cluster; MWBcluster: mutation weight-based cluster; RNAcluster: RNA expression-based cluster
DNA methylation subtype classification
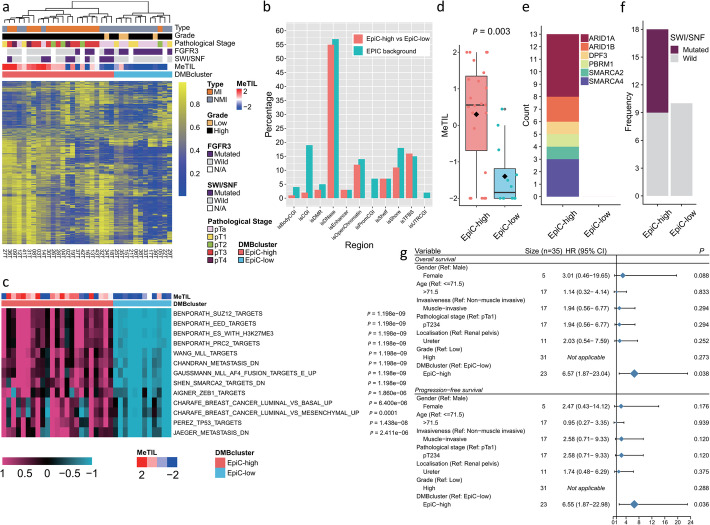
To obtain subtype classifications of UTUC samples (n = 35), we performed unsupervised hierarchical clustering using the 1% most variable probes, after excluding probes with no available values and those located on sex chromosomes (n = 836,691). We identified two robust DNA methylation-based (DMB) epi-clusters: EpiC-C1 (n = 23; 65.7%) and EpiC-C2 (n = 12; 34.3%) (Fig. 5a). Heterogeneity within the two clusters is also supported by principal component analysis (PCA) (Additional file 2: Fig. S8). We did not observe enrichment for specific regions in the genome, which were more susceptible to DNA methylation changes (Fig. 5b). Notably, those occurred mainly outside the promoter CpG islands (Fig. 5b). To determine the differentially methylated probes (DMPs) and differentially methylated regions (DMRs) between the two identified epi-clusters, we harnessed the chip analysis methylation pipeline (ChAMP) using default parameters of EPIC arrays. Using the most stringent criteria, we picked up 14,243 significantly hypermethylated probes from a total of 242,687 DMPs. Among these, EpiC-C1 presented frequent hypermethylation, since a total of 14,209 probes were significantly hypermethylated as compared to EpiC-C2, whereas only 34 probes gained methylation in EpiC-C2. In this manner, we re-designated EpiC-C1 as EpiC-high and EpiC-C2 as EpiC-low accordingly. GSEA demonstrated that DMRs were enriched for pathways related to the polycomb repressive complex 2 (PRC2) and MLL targets. Interestingly, we also observed enrichment for SMARCA2 target genes and ZEB1 targets (Fig. 5c). Notably, the EpiC-low UTUC subtype harbored a significantly lower MeTIL score as compared to the MeTIL high subtype (P = 0.003) (Fig. 5d).
Fig. 5.
Analysis of upper tract urothelial carcinomas (UTUC) DNA methylation. a Unsupervised clustering of most variable DNA methylation probes in UTUC showing two epi-clusters: EpiC-C1(high) and EpiC-C2(low). b Percentage and distribution of differentially DNA methylation probes (blue) between EpiC-high and EpiC-low subgroups across different genomic regions annotated by EPIC arrays (red). Note that there is no enrichment identified in specific genomic regions. c Single-sample Gene Set Enrichment Analysis (ssGSEA) showing pathways enriched in EpiC-high relative to EpiC-low cluster. d Boxplot showing significantly higher MeTIL score in EpiC-high subgroup than that in EpiC-low subgroup (P = 0.003). e Count of mutations in SWI/SNF family genes in EpiC-high and EpiC-low clusters. f Frequency of mutations affecting SWI/SNF family genes in EpiC-high versus EpiC-low UTUC subgroups. Note that 9 out of 18 (50%) UTUC samples in the EpiC-high subgroup harbored mutations as compared to none in the EpiC-low subgroup. g Forest plot showing that the DMBcluster of EpiC-high and EpiC-low is a significant prognostic factor as compared to other major clinicopathological features for either overall survival or progression-free survival
Association between DNA methylation classification, genetic and clinicopathological tumor, and patient features
Integrative analysis of EpiC-low and EpiC-high UTUC samples with matched genetic landscape showed that EpiC-high UTUC samples were highly enriched for mutations in SWI/SNF genes (50% vs 0%, P = 0.01, FDR = 0.054; Fig. 5e, f), while samples classified as EpiC-low were enriched for FGFR3 mutations (90% vs 28%, P = 0.004, FDR = 0.044; Fig. 5a, Additional file 1: Table S9).
In addition, EpiC-high UTUC samples were enriched for MI tumors (n = 16/23, 69.6%) in contrast to EpiC-low UTUC samples, which were enriched for NMI tumors (11/12, 91.7%, P = 0.0009; Additional file 1: Table S10). No other differences according to age and gender were found (Additional file 1: Table S10). We then analyzed the associations between clinicopathological tumor features, UTUC DMB epi-clusters, and patient survival (Fig. 5g). We found that patients with tumors belonging to the EpiC-high cluster had shorter overall survival as compared to those belonging to the EpiC-low subtype (P = 0.038; HR = 6.57; 95% CI 1.87–23.04; Additional file 2: Fig. S9a). Likewise, they had shorter distant metastasis-free survival as compared to those belonging to the EpiC-low subtype (P = 0.036; HR = 6.55; 95% CI 1.87–22.98; Additional file 2: Fig. S9b). No association with survival was identified for other clinicopathological tumor variables, which highlights the prognostic value of epi-clusters in UTUC.
Hypomethylation of FGFR3-mutated UTUC
We then decided to assess how the epigenetic landscape of EpiC-high and EpiC-low tumors diverge from the normal urothelium. To do so, we performed unsupervised hierarchical clustering of UTUC (n = 35) and adjacent normal samples (n = 8); as expected, normal samples gathered together as compared to those from UTUC, which clustered into two groups (Additional file 2: Fig. S11). One UTUC cluster was FGFR3-enriched (n = 15/24) and the other was FGFR3 wild-type (n = 0/11) (P = 0.0008), consistent with the notion that FGFR3 mutation might be associated with distinct epigenome alterations. In this context, we looked for differentially methylated probes between FGFR3-mutated and wild-type UTUC samples. Overall, 84,717 (10.1%) out of 836,691 EPIC probes were differentially methylated (Δβ value ≥ 0.2 or ≤ − 0.2, FDR < 0.05), the majority being hypomethylated in FGFR3-mutated tumors (n = 82,991; 97.8%). The hypomethylated probes were mildly enriched in enhancers (3.8% vs 3.2%) (P < 0.001) and DNAse hypersensitive sites (63.7% vs 57.4%) (P < 0.001) (Additional file 2: Fig. S11a); in addition, those probes were overall related to 8136 differentially methylated regions (DMRs) (Additional file 1: Table S11). Among the top DMRs, we note many overlapping with genes known to be involved in bladder cancer, such as FGFR3, GATA3, KRT15, and KRT5 (Additional file 2: Fig. S11b). GSEA found that those DMR were enriched for polycomb targets, as well as for several FGFR signaling pathways (Additional file 2: Fig. S11b).
Subtype classification of bladder cancer cell lines according to DNA methylation
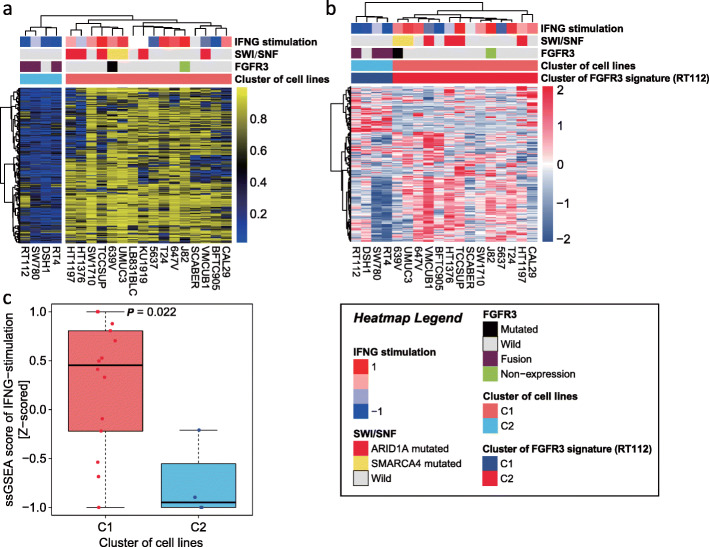
To answer the question whether FGFR3 somatic mutations can modulate DNA methylation in urothelial carcinomas, we thus decided to analyze the DNA methylation landscape of 20 bladder cancer cell lines. Notably, supervised clustering using the 14,209 hypermethylated probes derived from UTUC EpiC-high vs EpiC-low identified two subgroups tightly associated with FGFR3 and SWI/SNF gene mutations (Fig. 6a). Notably, all but one cell line (DSH1) in the C2 (75%) cluster harbored FGFR3 fusions with no SWI/SNF gene mutations. Conversely, only 2 out of 16 cell lines (12.5%) in the C1 cluster had FGFR3 mutations; interestingly, FGFR3 mutation is non-functional in J82 cell lines (no expression of FGFR3) and co-occurred with SMARCA4 mutation in the 639V cell line. Supervised clustering of FGFR3 gene expression signature showed similar findings (expression data was available for 18 cases) (Fig. 6b). Notably, expression of interferon-γ stimulation signature was upregulated in the C1 as compared to the C2 clusters (P = 0.022).
Fig. 6.
a Supervised clustering using hypermethylated probes derived from UTUC EpiC-high vs EpiC-low clusters and showing two subgroups of bladder cancer cell lines linked with SWI/SNF mutations (C1) and FGFR3 translocation status (C2). Note that the probes depicted in the heatmap were those that were statistically significant. b Supervised clustering extracted from Mahe of bladder cancer cell lines according to FGFR3 gene expression signature (using RT112 bladder cancer cell line). Note that although the DSH1 cell line does not harbor any FGFR3 mutation, it clusters with the three other cell lines harboring FGFR3 fusions and shares a similar transcriptomic program. c Box plot for interferon-γ stimulation signature score using single-sample gene set enrichment analysis (ssGSEA) in C1 and C2 clusters
Methylation of promoter genes in UTUC
We then looked at promoter genes located in the CpG islands, which were methylated in UTUC tumor samples with a frequency higher than 10%. We identified 905 genes methylated in our cohort (Additional file 1: Table S12). Notably, ACTL6B, a member of SWI/SNF, was frequently methylated (20% in tumors vs 0% in normal samples). Functional annotation analysis using DAVID identified that those are enriched for homeobox genes (P = 3.2 × 10−40), embryonic morphogenesis (P = 3.9 × 10−13), mesenchymal cell development (P = 1.8 × 10−6), and urogenital system development (P = 1.1 × 10−4). We then looked if there were any genes differentially methylated between MI and NMI tumors from one side, as well as FGFR3- versus non-FGFR3-mutated tumors from the other side. No statistically significant enriched promoter was found, suggesting that the majority of changes between FGFR3-mutated and FGFR3 wild-type FGFR3 tumors, as well as between MI and NMI tumors, arose outside promoters. No promoter methylation of ZFP36L1 was identified in our cohort.
Association of epigenetic UTUC two subtypes’ signatures with TCGA bladder muscle-invasive cancer cohort
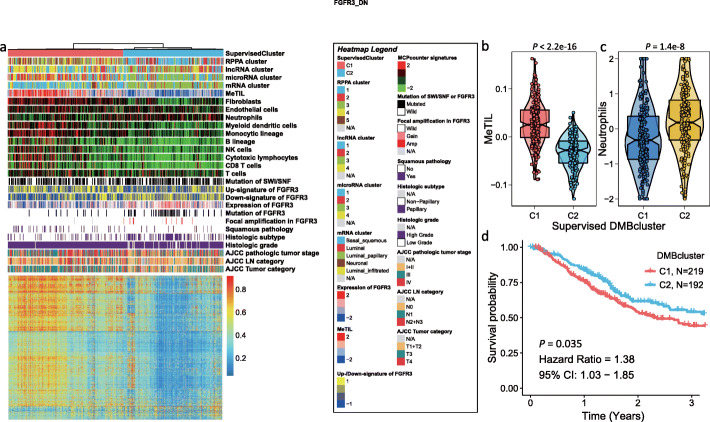
We further decided to apply the 14,209 hypermethylated probes derived from UTUC EpiC-high vs EpiC-low to TCGA muscle-invasive bladder cancer cohort, where we identified through supervised consensus clustering two subgroups (Fig. 7a). Similar to the UTUC cohort, the BLCA-C1 presented a dramatically high methylation level and a higher MeTIL score as compared to BLCA-C2 (P < 2.2 × 10−16; Fig. 7b). While BLCA-C2 was enriched for tumors with low grade, low stage, and papillary differentiation, BLCA-C1 was enriched for tumors with high grade, high stage, and non-papillary differentiation (Additional file 1: Table S12). Notably, our classification correlates with mRNA, miRNA, RPPA, and DNA methylation subtype TCGA classification (Additional file 1: Table S13). Additionally, BLCA-C2 was enriched for FGFR3 mutations (Additional file 1: Table S14), along with focal FGFR3 amplification and higher expression of FGFR3 (Fig. 7a). Notably, BLCA-C1 presented a significantly higher MeTIL score (P < 2.2 × 10−16) as compared to the BLCA-C2 cluster (Fig. 7b). We thus decided to use MCP-counter to deconvolute populations of immune and stromal cells in both groups; we found enrichment of fibroblasts, myeloid dendritic cells, monocytic cells, B cells, T cells, and cytotoxic CD8 T cells in the BLCA-C1 cluster, while BLCA-C2 was significantly enriched for neutrophils (P = 1.4 × 10−8; Fig. 7b, c). Finally, patients with tumors belonging to the BLCA-C1 subgroup had a poor median OS as compared to those belonging to the BLCA-C2 subgroup (P = 0.035; HR = 1.38; 95% CI 1.03–1.85). The median OS for patients in BLCA-C1 was 2.4 years (95% CI 1.9–3.7) and 5.4 years (95% CI 2.5–8.7) for those in the BLCA-C2 subgroup (Fig. 7d).
Fig. 7.
a Supervised consensus clustering of TCGA bladder cancers using hypermethylated probes derived from UTUC EpiC-high vs EpiC-low. Note the presence of two subgroups: BLCA-C1 and BLCA-C2. While the earlier is hypermethylated and immune enriched, the latter is enriched for FGFR mutations and FGFR amplification and displayed FGFR3 overexpression. In addition, BLCA-C2 is an immune desert with no infiltration by T cells and fibroblasts. b Box plot showing the MeTIL score distribution according to EpiC signature. Note that BLCA-C1 harbored a higher score than the BLCA-C2 subgroup. c Box plot showing the neutrophils score distribution according to EpiC signature. Note that BLCA-C2 harbored a higher score than the BLCA-C1 subgroup. d Kaplan-Meier curves for the overall survival in the TCGA-BLCA cohort showing that the BLCA-C1 subgroup harbored a poor outcome as compared to the BLCA-C2 subgroup
Integrative clustering based on multi-omics data
To understand the crosstalk between genetic and epigenetic profiling on the 28 UTUC samples for which WES and DNA methylation profiling were available, we performed integrative clustering of somatic copy number variation (SCNV), mutation, and methylation data using iClusterBayes. All mutations of SWI/SNF genes, including ARID1A, were combined for integrative clustering. Two iClusters (i.e., iCluster-high [n = 17] and iCluster-low [n = 9]) were identified and distinguished EpiC-high and EpiC-low clusters (P = 4.9 × 10−5). Four mutation contributors were identified with a posterior probability greater than 0.5 (i.e., FGFR3 [0.99], SWI/SNF [0.63], KMT2D [0.63], and TP53 [0.56]) (Additional file 2: Fig. S12a). Among which, FGFR3 (P = 0.011, FDR = 0.047) and KMT2D (P = 0.028, FDR = 0.057) were significantly enriched in iCluster-low, whereas SWI/SNF (P = 0.098, FDR = 0.13) show a tendency for being enriched in iCluster-high (Additional file 1: Table S15). Additionally, 6971 methylation probes and four copy number alterations made contributions to the clustering process (Additional file 2: Fig. S12b-c). Copy number alteration contributors comprise deletion of both 1p36.33 (0.66) and 9p21.3 (0.54) and amplification of both 8p11.23 (0.63) and 8q22.3 (0.85), where amplification of 8q22.3 was significantly associated with iCluster-high (P = 0.023, FDR = 0.093) (Additional file 2: Fig. S12d, Additional file 1: Table S15). Notably, 8q22.3 contains genes for which amplification was recently shown to define aggressive bladder cancer [11].
Discussion
We performed the first comprehensive genomic and epigenomic profiling of UTUC with the goal to identify molecular underpinnings of muscle invasiveness and determine novel key drivers linked with clinicopathological tumor features and patient outcome. We identified two UTUC methylome subtypes (i.e., EpiC-low and EpiC-high), which were enriched for NMI and MI samples, respectively. While the former one was hypomethylated, immune-depleted, and enriched for FGFR3 mutations, the latter one was hypermethylated, immune-infiltrated, and tightly associated with somatic mutations of SWI/SNF genes. Notably, we observed similar findings in bladder carcinoma cell lines, suggesting that FGFR3 alterations (i.e., fusions, mutations) might alter the methylome via direct or indirect mechanisms. This is also consistent with the notion that loss of function of SWI/SNF genes leads to the inability of the complex to counteract polycomb [12], which controls DNA methylation [13].
In addition to other mutations known to be altered in UTUC, we discovered for the first time a high rate of mutations of the ZFP36L1 gene. The majority of those were truncating mutations and likely oncogenic. If we consider mutations affecting all ZFP36 family genes, those were present in more than one quarter among UTUC cases. These results have not been identified previously in two different cohorts [4, 5]. This might be related to either the low number size of the cohorts analyzed or to the limited accurate detection in WES of InDels, often more challenging than SNVs [14]. Future studies are needed to clarify the true incidence of those mutations and their clinical relevance. As a zinc-finger RNA-binding protein regulating several cytoplasmic AU-rich elements (ARE), ZFP36L1 ultimately attenuates protein synthesis through the degradation of several mRNA transcripts. Using loss-of-function experiments, we observed increased cell motility of a bladder cancer cell line, likely through EMT, although we did not detect any effect on cell proliferation and apoptosis. These data are consistent with recently reported functional analysis using in vitro and in vivo experiments, demonstrating that ZFP36L1 suppress hypoxia and cell cycle signaling in bladder carcinomas [15]. Thus, ZFP36L1 mutations might lead to an increase in cell migration, bolstering tumor progression. In addition, UTUC cases harboring ZFP36 family gene mutations displayed higher tumor mutational loads, an observation which we validated in the TCGA bladder carcinoma dataset. These data are consistent with the notion that a higher tumor mutational load is positively associated with a better survival prognosis in numerous cancer types [16].
Previous studies investigated mutations of UTUC and identified a small proportion of cases with germline mutations in MMR genes, with the majority of tumors occurring sporadically [6]. Consistent with these recent findings, none of our sporadic UTUC has a mutator phenotype, and the majority of cases have luminal papillary-enriched transcriptome signatures, although caution might be considered in interpreting the results, as almost half of the cases assessed had FGFR3 mutations and were NMI tumors.
Besides genetic alterations, we investigated for the first time, to our knowledge, DNA methylation landscapes of both muscle-invasive and non-muscle-invasive UTUCs using an epigenome-wide approach. We discovered that DNA methylation is capable of distinguishing two distinct epigenetic UTUC subtypes, linked with muscle invasiveness status. This classification might be used in the future, if validated, to predict outcomes of resected UTUC patients and help stratify those that may benefit from preoperative chemotherapy or adjuvant FGFR3-targeted agents [17]. The discovery of an FGFR3-mutated UTUC subtype which is hypomethylated and immune-depleted, as compared to FGFR3 wild-type tumors, is novel. Future mechanistic data are needed to better understand if the epigenetic remodeling occurs directly or indirectly. Another observation that also deserves to be discussed is the link between immunity and SWI/SNF genetic tumor alterations which we showed to be associated with higher TILs in UTUC. In addition, bladder cell lines without FGFR3 alterations and which harbored SWI/SNF gene mutations showed higher interferon-γ stimulation signature. However, whether this association could be explained mechanistically by SWI/SNF mutations is unclear. Further studies are needed to clarify these findings in UTUC, given that the link between mutations in SWI/SNF genes and immunity is contradictory in different cancer subtypes [18, 19].
In colorectal cancer, a positive correlation was found between oncogenic BRAF mutations and hypermethylation of multiple promoter CpG islands, known as CpG island methylator phenotype [20]. A high level of mutations of SWI/SNF genes in FGFR3 wild-type UTUC tumors might explain their CpG island hypermethylation, as SWI/SNF genes have been shown to antagonize the polycomb repressive complex 2 (PRC2) [12].
From a therapeutic standpoint, our data suggest that EpiC-Low UTUC might benefit from the combination of FGFR3 inhibitors with PD-1/PD-L1 inhibitors as a targeted therapeutic strategy to modulate the T cell-depleted phenotype, consistent with the results of a recent study [5]. Indeed, erdafitinib was granted accelerated approval by the FDA in relapsed/refractory metastatic bladder cancer on the basis of phase 2 trial results, showing a response rate of 40% in 90 patients with tumors that harbored actionable FGFR alterations [21]. For the remaining tumors with SWI/SNF mutations, EZH2 inhibition with and without cisplatin might represent an effective option, as it has been recently demonstrated in bladder cancer cells and xenografts, through a mechanism that activates a natural killer (NK) cell-based immune response [22].
Our study is the first to propose two distinct routes of UTUC carcinogenesis through a crosstalk between genetic and epigenetic alterations. We used multi-omics data of a very well-annotated monocentric cohort with comprehensive integration of a wealth of data. However, our study has some limitations, including a small number of cases analyzed due to the relative rarity of tumor type, although we have been able to validate some of our observations in cohorts of bladder carcinomas. Further confirmation of our findings in larger UTUC cohorts is warranted. Future studies also need to examine the stability of the molecular profiles we identified in UTUC across matched primary and metastatic tumors.
In summary, our findings define the foundation of the molecular basis of UTUC heterogeneity. We also provide a roadmap for the rational clinical development of targeted and immunotherapeutic strategies that are specific to UTUC, but also potentially applicable to other tumor types harboring FGFR3-activating molecular alterations.
Experimental models and subjects details
Sample collection and histopathological analysis
Fresh-frozen UTUC of 40 cases was collected retrospectively from the Pitié-Salpetrière Hospital Biobank and re-evaluated histopathologically by one expert pathologist (E.C.). Overall, DNA and RNA of good quality and quantity were obtained for 40 and 20 cases, respectively. Detailed clinical and pathologic characteristics of the cohort are reported in Additional file 1: Table S1. Matched germline DNA from adjacent bladder tissues was also collected in 30 cases. Whole-exome sequencing (WES) was performed for 30-paired UTUCs and adjacent normal tissue. RNA sequencing was performed on 20 UTUC cases; in addition, DNA methylation was analyzed using Infinium EPIC arrays on 35 UTUC cases and 8 normal adjacent bladder tissues.
Ethical approval
All patients had previously provided written informed consent for tumor collection and subsequent analysis. The collection and use of tissues followed the procedures that are in accordance with the ethical standards formulated in the Declaration of Helsinki. The study has been approved by the ethical committee of the Pitié-Salpêtrière Hospital (IDF-6, Ile de France).
Nucleic acid extraction
DNA extraction was performed using the DNeasy Blood & Tissue Kit (Qiagen) according to the manufacturer’s instructions. RNA extraction was performed using the RNeasy Kit (QiagenQIAGEN) according to the manufacturer’s instructions. Quality control of extracted nucleic acids was done using an Agilent 2100 Bioanalyzer.
Whole-exome sequencing and somatic mutation detection
Exome capture was performed using Agilent SureSelect Human All Exon 50 Mb according to the manufacturer’s instructions. Briefly, 3 mg of DNA from each sample was used to prepare the sequencing library through shearing of the DNA, followed by ligation of sequencing adaptors. Whole-exome sequencing was performed, and paired-end sequencing (2 × 76 bp) was carried out using the Illumina HiSeq 2000; the resulting data were analyzed with the Illumina pipeline to generate raw FASTQ files. The coverage of our germline samples and tumor samples varied between 43–80× and 79–158×, respectively. The technical details and mutation detection were done according to the pipeline we previously reported [23]. We filtered out all known single-nucleotide variants (SNVs)/indels in the UCSC dbSNP 135 and 1000 Human Genome Project SNP databases, and kept any mutations, which are in the Catalogue of Somatic Mutations in Cancer (COSMIC) database, curated by the Wellcome Trust Sanger Institute. The variation classification for each mutation was annotated by ANNOVAR. Each somatic mutation or indel was annotated with its functional effect by SIFT to determine whether a mutation candidate was synonymous or nonsynonymous (benign or deleterious). Mutations that failed to be annotated by ANNOVAR, labeled with “unknown,” were removed first for downstream analysis. We used MutSigCV_v1.41 (www.broadinstitute.org) to infer the most significantly mutated genes across these samples, and 10 frequently mutated (> 10%) genes that past test of significance were identified (P < 0.05) [24]. The mutation landscape for those 10 significantly mutated genes across 30 UTUC samples was shown by OncoPrint using R package ComplexHeatmap [25]. The tumor mutational load per megabase was computed by summing all types of mutations, divided by 50 MB. We harnessed mutation signatures to decipher samples that shared similar mutational spectra. All non-single-base substitutions (e.g., insertions, deletions, and complex multi-base substitutions) were filtered out of the table, leaving single-base substitution mutations annotated as nonsense, missense, or coding silent substitutions [26]. Mutational signatures described by Alexandrov et al. [10] and curated at http://cancer.sanger.ac.uk/cosmic/signatures were evaluated using R package deconstructSigs [27] with the following parameters: “exome2genome” trinu-cleotide-count normalization and signature cutoff at 0.2. Unknown signatures were subsequently discarded. We used non-negative matrix factorization (NMF) by R package NMF with the method of “lee” and rank number of three to deconvolute the mutational signature landscape and determine the signature contributor for each class where contributors were identified by extractFeatures() with “max” method [28, 29].
Validation of somatic mutations by Sanger sequencing
Validation of selected somatic variants in three genes, including FGFR3, KDM6A, and ZFP36L1, was performed on DNA extracted from UTUC and normal adjacent samples analyzed by WES. In addition, FGFR3 hotspot mutations (S249C and Y373C) were investigated in the remaining samples not analyzed by WES (n = 5). The genomic region surrounding the putative mutations was amplified with polymerase chain reaction (PCR) using specific primer pairs designed with the Primer Express 3.0 Software (Applied Biosystem). PCR products were then purified with the Qiaquick PCR purification kit (Qiagen, Milan, Italy) and sequenced on both strands using the Big Dye Terminator v1.1 Cycle Sequencing kit (Applied Biosystems). Sanger sequencing was performed on ABI 3730 Genetic Analyzer (Applied Biosystems).
RNA sequencing
Total RNA for 20 UTUC samples was converted into a library of template molecules for sequencing on the Illumina HiSeq 2000 according to the NuGen Ovation RNA-Seq System V2 protocol. In brief, first, single-stranded cDNA was synthesized from 100 ng of DNase1-treated total RNA using a mix of DNA/RNA chimeric primers that hybridize to both the 50 portions of the poly (A) sequence and randomly across the transcript. Second, strand synthesis produced double-stranded cDNA, which was amplified using single-primer isothermal strand displacement amplification. The resultant cDNA was fragmented to 200 bp (mean fragment size) with the S220 Focused-ultrasonicator (Covaris) and used to make barcoded sequencing libraries on the SPRI-TE Nucleic Acid Extractor (Beckman-Coulter). Libraries were quantitated by qPCR (KAPA Systems), multiplexed, and sequenced, four samples per lane, on the HiSeq2000 using 75-bp paired-end sequencing. The resulting data were analyzed with the current Illumina pipeline to generate raw FASTQ files. The raw, paired-end reads were aligned to the human reference genome, GRCh37/hg19, using the MOSAIK alignment software. MOSAIK works with paired-end reads from Illumina HiSeq 2000 and uses both a hashing scheme and the Smith-Waterman algorithm to produce gapped optimal alignments and to map exon junction-spanning reads with a local alignment option for RNA-seq. The resulting alignments were then saved as a standard bam file. We then counted the mapped reads in mRNA annotated in GENCODE15 to generate the raw counts for each gene using the HTSeq-count script distributed with the HTSeq package. We calculated the number of fragments per kilobase of non-overlapped exon per million fragments mapped (FPKM) [30]. To reduce noise, we filtered out low expressed mRNA, if its FPKM value is lower than 1 in at least 90% of the samples.
Unsupervised clustering for UTUC mRNA profile
Implementation of FPKM transformation and the filter procedure resulted in 12,492 unique genes with reliably measured expression. The gene expression data were then median centered in both directions and log2 transformed prior to clustering. Then, we performed unsupervised hierarchical clustering with k = 2 as the number of clusters by basically using the hclust() R function with Ward’s clustering method and Manhattan distance measures for each run. The consensus process was set to 80% of features and samples re-sampling with 500 perturbations. The final hierarchical clustering based on the consensus matrix used a distance measurement of Manhattan with Ward’s clustering method. Differential expression analysis was executed by the use of the edgeR R package, fitting a negative binomial generalized log-linear model [31].
DNA methylation and bioinformatics analysis
Global DNA methylation was assessed using the Infinium HumanMethylation850 (EPIC) BeadChip Array. Briefly, genomic DNA (500–1000 ng) was bisulfite-converted using the Zymo EZ DNA methylation kit (Zymo Research, Irvine, CA) according to the manufacturer’s recommendations. The amount of bisulfite-converted DNA and the completeness of the bisulfite conversion for each sample were assessed using a panel of MethyLight-based real-time PCR quality control assays. Bisulfite-converted DNA was then used as a substrate for the Illumina EPIC BeadArrays, as recommended by the manufacturer. Specifically, each sample was whole-genome amplified (WGA) and then enzymatically fragmented. Samples were then hybridized overnight to an 8-sample BeadArray, in which the WGA-DNA molecules annealed to locus-specific DNA oligomers linked to individual bead types. After the chemical processes, BeadArrays were scanned and the raw signal intensities were extracted from the *.IDAT files using the “noob” function in the minfi R package. The “noob” function corrects for background fluorescence intensities and red-green dye bias. The beta (β) value for each probe was calculated as M/(M + U), in which M and U respectively refer to the (pre-processed) mean methylated and unmethylated probe signal intensities. Probes with measurements in which the fluorescent intensity was not statistically significantly above the background signal (detection P value > 0.05) were removed from the dataset.
Pattern discovery of the methylation EPIC profile was performed in 35 UTUC samples. Any methylation probe that was located in a sex chromosome or that had at least one NA value was removed out of the total 866,091 probes. In those filtered (836,691), we picked the top 1% highly variable DNA methylation probes and performed unsupervised hierarchical clustering using the Euclidean distance and Ward’s clustering methods. Clusters (k = 2) were generated to methylation C1 and C2 based on the cutree() function. Differentially methylated probes (DMPs) and differentially methylated regions (DMRs) were achieved through the standard process of ChAMP with arraytype of EPIC [32]. To be specific, we adjusted the P value threshold for DMP detection, set to 0.05, and used the Benjamin-Hochberg and DMRcate methods to define DMRs. Let us denote as the mean β value of probe k in C1 and as that in C2. We determined probe k as the significantly hypermethylated probe in C1 if , , and FDR < 0.05. Using the above threshold, we identified 14,209 significantly hypermethylated probes in C1.
Data acquisition of external BLCA and UTUC cohorts
We extracted three omics data from the TCGA-BLCA cohort where gene expression and DNA methylation data were downloaded from UCSC Xena (https://xena.ucsc.edu/), and somatic mutation data and detailed clinicopathological information were obtained from cBioPortal (http://www.cbioportal.org/) under the archive of Bladder Cancer [33]. Overall, 412 muscle-invasive bladder carcinoma samples were used for DNA methylation and somatic mutation analysis, and 407 samples for expression data. Second, we included the Hurst cohort [9] which contains somatic SNVs and small insertions/deletions identified by whole-exome sequencing for 24 TaG2 bladder tumors. Third, we collected omics data from urinary tract cell lines. Corresponding DNA methylation profiling for 20 cell lines and expression array data for 18 cell lines (2 out of 20 have no data for expression) were retrieved from Genomics of Drug Sensitivity in Cancer (GDSC, https://www.cancerrxgene.org/). Specific mutation of FGFR3 and SWI/SNF pathway was assessed from cBioPortal (https://www.cbioportal.org/) under the archive of Cancer Cell Line Encyclopedia (Broad 2019). Only SWI/SNF gene mutations considered as oncogenic drivers were included. FGFR3 signature was collected from differential expression analysis of RT112 cell line treated with FGFR3 siRNA according to the literature [34]. Interferon-γ stimulation signature was retrieved from a previous study [35].
Subtype inference for UTUC and BLCA tumors
We used R package consensusMIBC [36], which implements a nearest centroid transcriptomic classifier that assigned class labels according to the consensus molecular classification of MIBC to infer consensus subtype of UTUC and TCGA-BLCA. Use function of getConsensusClass() with normalized expression data and parameters by default, one of the following six molecular classes will be assigned due to the highest correlation coefficient, including luminal papillary (LumP), luminal non-specified (LumNS), luminal unstable (LumU), stroma-rich, basal/squamous (Ba/Sq), and neuroendocrine-like (NE-like). We also inferred the BASE47 subtypes for our UTUCs based on a 47-gene signature according to the literature [37] by using unsupervised hierarchical clustering with distance measurement of 1-Pearson’s coefficient and linkage function of Ward.D2.
Supervised clustering of BLCA methylation 450k profile
BLCA methylation data contains DNA methylation β value of 433 BLCA samples with 412 tumor samples and 21 normal samples assessed by TCGA using the Illumina Infinium HumanMethylation450 platform. Out of 14,209 hypermethylated probes derived from UTUC EpiC-high vs EpiC-low, a total of intersected 3790 hypermethylated probes was used for supervised clustering of the TCGA-BLCA methylation 450k profile. To assess the stability of the discovered clusters, we performed a consensus hierarchical clustering. We conducted 500 runs of hierarchical clustering on the resampled data. For each run, 80% samples and 80% features were randomly chosen. The distance measurement was set as Manhattan, and the linkage function was set as Ward.D2. Based on the 500 runs, a consensus was obtained by taking the average over the connectivity matrices of every perturbed dataset. Then, we carried out hierarchical clustering with the consensus matrix as a similarity matrix, with a distance measurement of 1-Pearson’s coefficient and linkage function of Ward.D2.
Detection of frequently methylated genes
For genes having more than one probe mapping to its promoter, the median β value was considered to get 12,066 methylation genes. To minimize the influence of normal tissue contamination in DNA methylation data, we excluded methylation genes found in more than 50% normal samples with a β value ≥ 0.2 or genes with a median β value ≥ 0.2 in normal samples, and 11,346 methylation genes remained. We determined a gene’s methylation status by a β value cutoff of 0.3 and calculated its methylation percentage across all UTUC samples. A frequently methylated gene was defined if the corresponding percentage was greater than 10%.
Gene set enrichment analysis
For gene set enrichment analysis based on gene expression data, R package clusterProfiler was used for the pre-ranked gene list (descending ordered log2FoldChange value) derived from differential expression analysis [38, 39]. Molecular Signature Database gene sets were tested by using the gene set of msigdb.v6.0.symbols.gmt downloaded from the GSEA website (http://software.broadinstitute.org/gsea/). GSEA for DNA methylation were executed by the embedded champ.GSEA() function in ChAMP with the typical Fisher’s method. The enrichment scores of molecular pathways were evaluated using the gene set variation analysis method via R package GSVA [40, 41].
Integrative analysis of genetic and epigenetic profiling
Bayesian integrative clustering was performed by R package iClusterPlus by using the iCluster Bayesian method [42]. We applied this method on three available data types: genetic mutation, DNA methylation, and copy number alteration data with 26 shared samples. Basically, the mutation data contains 10 significantly mutated genes that were revealed by the MutSigCV algorithm with a P value of less than 0.05 and a mutational frequency greater than three cases. We further curated the mutation data by removing ARID1A but attaching SWI/SNF pathway mutation. We used the top 1% most variable probes as the methylation dataset. An under a confidence level of 0.75, amplification or deletion of arm with a q value less than 0.25 were selected, including 7 amplification peaks and 3 deletion peaks. For the sake of integrative analysis, the copy number was transformed into a binary term (e.g., 1 for deletion or amplification and 0 for no change). The parameter list was set as K = 1 (two clusters), n.burnin = 18,000, n.draw = 12,000, prior.gamma = 0.5 for the indicator variable gamma of each data set, sdev = 0.05, and thin = 3. We fitted binomial distribution for mutation data and copy number data and Gaussian for methylation data. The clustering contributor was considered if its corresponding posterior probability was greater than 0.5.
Copy number variation analysis
Recurrent focal somatic copy number alterations were detected and localized using GISTIC2.0 [43, 44] with the thresholds of copy number amplifications/deletions being equal to ± 0.15 and q value threshold being equal to 0.2.
Quantify the immune and stromal level of UTUC samples
The population abundance of tissue-infiltrating immune and stromal cell populations was estimated by R package MCPcounter per sample using normalized count data [45]. The presence of infiltrating immune/stromal cells in tumor tissue was estimated by R package ESTIMATE [46]. DNA methylation-based immune infiltration scores (MBII scores) were extracted by TCGA previous work [47]. Additionally, the individual DNA methylation of tumor-infiltrating lymphocyte (MeTIL) score was calculated according to the literature [48].
Cell line culture
TCCSUP cell line was purchased from ATCC and authenticated. Cells were tested and were verified for free of Mycoplasma contamination. These cells were grown in (EAGLE, 10% FCS, 0.1 mM AANE, 1 mM sodium pyruvate, 40 μg/ml gentamicin) media.
ZFP36L1 knockdown
3 × 105 cells were plated in each well of a 6-well plate. After 24 h, cells were transfected with selected siRNA. For transfection, 4 μl of 5 nM of Human ZFP36L1 (#L-011816-00-0005) siRNA smart pool or non-silencing pool siRNA (Cont siRNA) (#L-011816-00-0005) (Dharmacon) was diluted in 100 μl of OptiMEM media. In a separate tube, 8 μl of Lipofectamine RNAi Max reagent (# 2185383, Invitrogen™) was diluted in 100 μl of OptiMEM media. After 5 min of incubation, both the solutions were mixed and kept for 15 min incubation at room temperature. Later, the mixture was added drop wide to each well. The pictures of cells after 48 h from transfection were taken with macro-microscope with × 10 magnification. Later, the transfected cells were used for the Western blots and functional assays.
Western blots
After 48 h of transfection, the cells were scrapped from the plate in a tube and washed with PBS (1×). After washing, the cells were centrifuged, and the supernatant was discarded. The pellet was suspended in twice the volume of the LSDB buffer and 1× protease inhibitor (# 40091500, Sigma-Aldrich) and kept in liquid nitrogen for 2 min. Later, the sample was transferred in a water bath at 37 °C for 2 min. The incubation steps were repeated twice, and then the sample was centrifuged at 4 °C. After centrifugation, the supernatant was transferred to another tube. The total protein concentration was determined by dissolving 1 μl of total protein extract in 1× protein assay dye (# 5000006, Bio-Rad) and quantifying the amount by the Bradford dye-binding method. The protein samples were loaded in NuPAGE (4–12% bis-tris Gel, # 20070610, Invitrogen™) for electrophoresis and later transferred on the PVDF. The membrane first was blocked with 5% milk and then probed with the primary antibody overnight. The next day, the membrane was washed with PBST (1× PBS, 0.01% Tween 20) and then blocked with the secondary antibody. After incubation, the membrane was washed with the PBST, and full blots were treated with ECL (# UC180107, Protein biology) for the acquisition of the signal using the imager system. The Primary antibody used were ZFP36L1 (1:1000; #2119, Cell Signaling Technology), E-cadherin (1:1000; #9782, Cell Signaling Technology), and actin (1:5000).
Cell proliferation
The cell proliferation was determined using Invitrogen™ CellTrace™ (#2161821) according to the manufacturer’s instructions. 3 × 105 cells were plated in a 6-well plate. The next day, the cells were transfected with either si ZFP36L1 or si control, and after 7 h, they were treated with cell trace violet dye for 20 min. The dye was replaced with (EAGLE, 10% FCS, 0.1 mM AANE, 1 mM sodium pyruvate, 40 μg/ml gentamicin) media. After 72 h transfection, the cell was trypsinized and centrifuged. The pellet was dissolved in PBS, and cells were used to analyze the cell cycle distribution using flow cytometry (BD. FORTRESSA X20). The data obtained was analyzed using the FlowJo software. The numbers are expressed as mean ± SEM of the percentage of low-proliferating cells, and three independent experiments were performed.
Cell apoptosis
After 24 h from plating, 3 × 105 cells were transfected with either the si ZFP36L1 or si control. After 72 h transfection, the cells were trypsinized and centrifuged at 4 °C. The pellets were treated with 100 μl of BD cytofix/cytoplasm fixation permeabilization solution (Kit #0071517, BD. Biosciences) and kept in ice for 20 min. Later, the cells were washed with the washing buffer (1× BD wash buffer in 1% BSA solution) and centrifuged at 4 °C. The supernatant was discarded, and the pellet was treated with PE rabbit anti-active caspase-3 (# 55082, BD. Biosciences) for 30 min kept in dark at room temperature. The final washing was done with the washing buffer (1× BD wash buffer in 1% BSA solution) and centrifuged at 4 °C. The supernatant was discarded, and the pellet was dissolved in PBS and analyzed for cascade 3 activity using flow cytometry (BD. FORTRESSA X20). The data obtained was analyzed using the FlowJo software. The numbers are expressed as mean ± SEM of % of apoptotic cells, and three independent experiments were performed.
Transwell assay
The transfected TCCSUP cells with either si ZFP36L1 or si control after 72 h were used for the transwell assay. The transfected cells (5 × 104) were taken in 200 μl (EAGLE, 0.01% FCS, 0.1 mM AANE, 1 mM sodium pyruvate, 40 μg/ml gentamicin) media and plated on top of the 8-μm Transwell filter membrane in a 24-well plate. Using a pipette, 600 μl (EAGLE, 10% FCS, 0.1 mM AANE, 1 mM sodium pyruvate, 40 μg/ml gentamicin) media was added in the lower chamber of the 24-well plate. After 72 h of incubation, the membranes were washed with PBS (1×) and were fixed in 3.4% formaldehyde by adding 600 μl in the lower chamber and 200 μl in the upper chamber. The membranes were washed with PBS (1×) and then incubated in DAPI (1 μg, 20 μl/ml) for 30 min. After incubation, the cells from the above chamber were removed carefully with the cotton bud and washed with PBS (1×). The membranes were later viewed under the inverted microscope (ZEISS observer), and pictures were taken to count the number of cells. The counts of migrated cells are expressed as mean ± SEM of at least three independent experiments, each performed in triplicate.
Statistical analyses
All statistical tests were executed by R/3.6.2, including Fisher’s exact test for categorical data, a two-sample Mann-Whitney test for continuous data, a log-rank test Kaplan-Meier curve, and Cox proportional hazards regression for hazard ratio (HR) with 95% confidence interval (95% CI) [49, 50]. Survival analysis was performed by R package survival. Mutual exclusivity analysis was done using one-side Fisher’s exact test, where there was an alternative parameter of “less” for mutual exclusivity and “greater” for co-occurrence analysis. For unadjusted comparisons, a two-sided P < 0.05 was considered statistically significant.
Supplementary Information
Additional file 1: Table S1. Clinicopathological tumor features of upper-tract urothelial carcinomas samples used for the study. Table S2. Association between clinical variables and patients progression-free and overall survival. Table S3. List of somatic mutations identified in the whole-exome sequencing of upper-tract urothelial carcinomas (n = 30). Table S4. List of significantly mutated genes identified by the MutSigCV algorithm. Table S5. Mutual exclusivity and co-occurrence analysis for the 10 most significantly mutated genes using the one-side Fisher’s exact test. Table S6. Association between top 10 frequently significantly mutated UTUC genes and muscle-invasive status. Table S7. Association between UTUC RNA unsupervised clustering and clinical variables. Table S8. Subtypes of UTUC samples using consensus molecular classification of muscle-invasive bladder cancer. Table S9. Association between top 10 frequently significantly mutated genes and DNA methylation-based clusters in UTUC samples. Table S10. Association between clinical variables and DNA methylation UTUC based clustering in samples with EpiC-low and EpiC-high signatures. Table S11. List of differentially methylated regions (DMR) using hypomethylated probes in FGFR3-mutated UTUC cases as compared to FGFR-wild type samples. Table S12. List of genes with promoter CpG islands methylation in UTUC tumor samples. Only those with a frequency higher than 10% are reported. Table S13. Association between UTUC EpiC signature and clinical variables in TCGA bladder carcinomas cohort. Table S14. Association between significant mutations of UTUC and supervised TCGA DNA methylation based clustering. Table S15. Association between mutation and copy number contributors and iClusters.
Additional file 2: Fig. S1. Copy number variants analysis. Fig. S2. Comparative analysis of the frequency of most frequent mutations in upper-tract urothelial carcinomas (UTUC) as compared to urothelial bladder carcinomas (UBC). Fig. S3. ZFP36 family mutation in diverse histopathological cancer subtypes. Fig. S4. a) Human protein atlas and FANTOM5 datasets showing that protein expression and RNA expression of ZFP36L1 is the highest and the most expressed in the urinary bladder as compared to other tissues. Fig. S5. ZFP36L1 inhibition increases cell motility in vitro. Fig. S6. Distribution of MeTIL score between FGFR3-mutant and wild type of UTUC and UBC, and between muscle-invasive and non-muscle invasive UTUC samples. Fig. S7. Heatmap of BASE47 bladder signature in UTUC samples showing two subgroups of “luminal-like” and “basal-like”. Fig. S8. Principal component analysis showing heterogeneity of the two epi-clusters EpiC-C1 and EpiC-C2, obtained through unsupervised clustering of most variable DNA methylation probes. Fig. S9. Kaplan-Meier curves for progression-free survival regarding UTUC epi-clusters. Fig. S10. Unsupervised clustering of DNA methylation using most variable probes in the whole dataset encompassing UTUC (n = 35) samples and adjacent normal urothelium (n = 8). Fig. S11. Distribution of DNA methylation probes across different genomic regions significantly hypomethylated in FGFR3-mutated tumors versus FGFR3-wild type as compared to EPIC arrays. Heatmap showing differentially methylated probes located in top ranked DMR and corresponding ssGSEA results. Fig. S12. Integrative clustering analysis of multi-omics data. Full Western blots containing the entire ladder for loss-of-function experiments of ZFP36L1 using siRNA in TCCSUP bladder cancer cell line.
Additional file 3. Review history (LOG 180 bytes).
Acknowledgments
Review history
The review history is available as Additional file 3.
Peer review information
Anahita Bishop was the primary editor of this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Authors’ contributions
Conceptualization: X.S., F.Y., E.C., M.R., J-P. S., D.K., and G.G.M. Methodology: X.S., X.L., H.Y., R.M., and I.D. Formal computational analysis: X.S., X.L., H.Y., and F.Y. Investigation: X.S., X.L., S.K.B., E.C., R.M., H.Y. M.R., J-P.S., D.K., I.D., N.M.T., F.Y., and G.G.M. Mechanistic studies on ZFP36L1: S.K.B. and I.D. Original draft: X.S., X.L., F.Y., and G.G.M. Writing—review and editing: X.S., X.L., S.K.B., E.C., H.Y., R.M., M.R., J-P.S., D.K., I.D., N.M.T., F.Y., and G.G.M. Resources and data collection: E.C., R.M., M.R., J-P.S., D.K., and G.G.M. Supervision: X.S., F.Y., and G.G.M. The authors read and approved the final manuscript.
Funding
This work was supported in part by grants from the Foundation AVEC (G.M), Pfizer research grant (G.M.), the National Key R&D Program of China (2019YFC1711000) (F.Y.), the National Natural Science Foundation of China (81973145) (F.Y.), the “Double First-Class” University project (CPU2018GY09) (F.Y.), MD Anderson Leukemia SPORE grant P50 CA100632 (X.S.), UPR/MDACC Partnership for Excellence in Cancer Research grant U54 CA096300 (X.S.), and the University of Texas MD Anderson Cancer Center support grant P30 CA016672 (X.S.).
Availability of data and materials
RNA sequencing data generated in this paper have been deposited at the NCBI Sequence Read Archive (SRA) hosted by the NIH (SRA accession: PRJNA678814) [51]. DNA methylation data generated have been submitted to GEO (GEO accession: GSE161651) [52]. WES sequencing data have been deposited at gated repository [53]; WES dataset requires access control, and there is a corresponding Data Access Committee (DAC) who determines access permissions. Data access requests are reviewed by DAC [53]. Gene expression and DNA methylation data from the TCGA-BLCA cohort were downloaded from UCSC Xena (https://xena.ucsc.edu/), and somatic mutation data and detailed clinicopathological information were obtained from cBioPortal (http://www.cbioportal.org/) under the archive of Bladder Cancer [33]. DNA methylation and RNA-seq expression of bladder cancer cell lines were retrieved from Genomics of Drug Sensitivity in Cancer (GDSC, https://www.cancerrxgene.org/).
Competing interests
The authors declare no conflicts of interest regarding the current work.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xiaoping Su, Xiaofan Lu, and Sehrish Khan Bazai contributed equally to the work.
Fangrong Yan and Gabriel G. Malouf contributed equally to the work and should be considered as co-corresponding authors.
Contributor Information
Fangrong Yan, Email: f.r.yan@163.com.
Gabriel G. Malouf, Email: maloufg@igbmc.fr
Supplementary Information
The online version contains supplementary material available at 10.1186/s13059-020-02230-w.
References
- 1.Babjuk M, Burger M, Comperat EM, Gontero P, Mostafid AH, Palou J, van Rhijn BWG, Roupret M, Shariat SF, Sylvester R, et al. European Association of Urology Guidelines on Non-muscle-invasive Bladder Cancer (TaT1 and Carcinoma In Situ) - 2019 update. Eur Urol. 2019;76:639–657. doi: 10.1016/j.eururo.2019.08.016. [DOI] [PubMed] [Google Scholar]
- 2.Roupret M, Babjuk M, Comperat E, Zigeuner R, Sylvester RJ, Burger M, Cowan NC, Gontero P, BWG VR, Mostafid AH, et al. European Association of Urology Guidelines on Upper Urinary Tract Urothelial Carcinoma: 2017 update. Eur Urol. 2018;73:111–122. doi: 10.1016/j.eururo.2017.07.036. [DOI] [PubMed] [Google Scholar]
- 3.Leow JJ, Chong KT, Chang SL, Bellmunt J. Upper tract urothelial carcinoma: a different disease entity in terms of management. ESMO Open. 2016;1:e000126. doi: 10.1136/esmoopen-2016-000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moss TJ, Qi Y, Xi L, Peng B, Kim TB, Ezzedine NE, Mosqueda ME, Guo CC, Czerniak BA, Ittmann M, et al. Comprehensive genomic characterization of upper tract urothelial carcinoma. Eur Urol. 2017;72:641–649. doi: 10.1016/j.eururo.2017.05.048. [DOI] [PubMed] [Google Scholar]
- 5.Robinson BD, Vlachostergios PJ, Bhinder B, Liu W, Li K, Moss TJ, Bareja R, Park K, Tavassoli P, Cyrta J, et al. Upper tract urothelial carcinoma has a luminal-papillary T-cell depleted contexture and activated FGFR3 signaling. Nat Commun. 2019;10:2977. doi: 10.1038/s41467-019-10873-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Audenet F, Isharwal S, Cha EK, Donoghue MTA, Drill EN, Ostrovnaya I, Pietzak EJ, Sfakianos JP, Bagrodia A, Murugan P, et al. Clonal relatedness and mutational differences between upper tract and bladder urothelial carcinoma. Clin Cancer Res. 2019;25:967–976. doi: 10.1158/1078-0432.CCR-18-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sfakianos JP, Cha EK, Iyer G, Scott SN, Zabor EC, Shah RH, Ren Q, Bagrodia A, Kim PH, Hakimi AA, et al. Genomic characterization of upper tract urothelial carcinoma. Eur Urol. 2015;68:970–977. doi: 10.1016/j.eururo.2015.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolff EM, Chihara Y, Pan F, Weisenberger DJ, Siegmund KD, Sugano K, Kawashima K, Laird PW, Jones PA, Liang G. Unique DNA methylation patterns distinguish noninvasive and invasive urothelial cancers and establish an epigenetic field defect in premalignant tissue. Cancer Res. 2010;70:8169–8178. doi: 10.1158/0008-5472.CAN-10-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hurst CD, Alder O, Platt FM, Droop A, Stead LF, Burns JE, Burghel GJ, Jain S, Klimczak LJ, Lindsay H, et al. Genomic subtypes of non-invasive bladder cancer with distinct metabolic profile and female gender Bias in KDM6A mutation frequency. Cancer Cell. 2017;32:701–715. doi: 10.1016/j.ccell.2017.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu CC, Li CF, Chen IH, Lai MT, Lin ZJ, Korla PK, Chai CY, Ko G, Chen CM, Hwang T, et al. YWHAZ amplification/overexpression defines aggressive bladder cancer and contributes to chemo-/radio-resistance by suppressing caspase-mediated apoptosis. J Pathol. 2019;248(4):476-87. [DOI] [PMC free article] [PubMed]
- 12.Bracken AP, Brien GL, Verrijzer CP. Dangerous liaisons: interplay between SWI/SNF, NuRD, and Polycomb in chromatin regulation and cancer. Genes Dev. 2019;33:936–959. doi: 10.1101/gad.326066.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 14.Hasan MS, Wu X, Zhang L. Performance evaluation of indel calling tools using real short-read data. Hum Genomics. 2015;9:20. doi: 10.1186/s40246-015-0042-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loh XY, Sun QY, Ding LW, Mayakonda A, Venkatachalam N, Yeo MS, Silva TC, Xiao JF, Doan NB, Said JW, et al. RNA-binding protein ZFP36L1 suppresses hypoxia and cell-cycle signaling. Cancer Res. 2020;80:219–233. doi: 10.1158/0008-5472.CAN-18-2796. [DOI] [PubMed] [Google Scholar]
- 16.Wang X, Li M. Correlate tumor mutation burden with immune signatures in human cancers. BMC Immunol. 2019;20:4. doi: 10.1186/s12865-018-0285-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Birtle A, Johnson M, Chester J, Jones R, Dolling D, Bryan RT, Harris C, Winterbottom A, Blacker A, Catto JWF, et al. Adjuvant chemotherapy in upper tract urothelial carcinoma (the POUT trial): a phase 3, open-label, randomised controlled trial. Lancet. 2020;395(10232):1268-77. [DOI] [PMC free article] [PubMed]
- 18.Li J, Wang W, Zhang Y, Cieslik M, Guo J, Tan M, Green MD, Wang W, Lin H, Li W, et al. Epigenetic driver mutations in ARID1A shape cancer immune phenotype and immunotherapy. J Clin Invest. 2020;130:2712–2726. doi: 10.1172/JCI134402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen J, Ju Z, Zhao W, Wang L, Peng Y, Ge Z, Nagel ZD, Zou J, Wang C, Kapoor P, et al. ARID1A deficiency promotes mutability and potentiates therapeutic antitumor immunity unleashed by immune checkpoint blockade. Nat Med. 2018;24:556–562. doi: 10.1038/s41591-018-0012-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toyota M, Ohe-Toyota M, Ahuja N, Issa JP. Distinct genetic profiles in colorectal tumors with or without the CpG island methylator phenotype. Proc Natl Acad Sci U S A. 2000;97:710–715. doi: 10.1073/pnas.97.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loriot Y, Necchi A, Park SH, Garcia-Donas J, Huddart R, Burgess E, Fleming M, Rezazadeh A, Mellado B, Varlamov S, et al. Erdafitinib in locally advanced or metastatic urothelial carcinoma. N Engl J Med. 2019;381:338–348. doi: 10.1056/NEJMoa1817323. [DOI] [PubMed] [Google Scholar]
- 22.Ramakrishnan S, Granger V, Rak M, Hu Q, Attwood K, Aquila L, Krishnan N, Osiecki R, Azabdaftari G, Guru K, et al. Inhibition of EZH2 induces NK cell-mediated differentiation and death in muscle-invasive bladder cancer. Cell Death Differ. 2019;26:2100–2114. doi: 10.1038/s41418-019-0278-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malouf GG, Su X, Yao H, Gao J, Xiong L, He Q, Comperat E, Couturier J, Molinie V, Escudier B, et al. Next-generation sequencing of translocation renal cell carcinoma reveals novel RNA splicing partners and frequent mutations of chromatin-remodeling genes. Clin Cancer Res. 2014;20:4129–4140. doi: 10.1158/1078-0432.CCR-13-3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 26.Jarvis MC, Ebrahimi D, Temiz NA, Harris RS. Mutation signatures including APOBEC in cancer cell lines. JNCI cancer spectrum. 2018;2:pky002. doi: 10.1093/jncics/pky002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenthal R, McGranahan N, Herrero J, Taylor BS, Swanton C. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. 2016;17:31. doi: 10.1186/s13059-016-0893-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee DD, Seung HS. Algorithms for non-negative matrix factorization. In Adv Neural Inf Process Syst. Cambridge: MIT Press; 2001. p. 556–6.
- 29.Gaujoux R, Seoighe C. A flexible R package for nonnegative matrix factorization. BMC bioinformatics. 2010;11:367. doi: 10.1186/1471-2105-11-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, Van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson MD, Mccarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tian Y, Morris TJ, Webster AP, Yang Z, Beck S, Feber A, Teschendorff AE. ChAMP: updated methylation analysis pipeline for Illumina BeadChips. Bioinformatics. 2017;33:3982–3984. doi: 10.1093/bioinformatics/btx513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robertson AG, Kim J, Alahmadie H, Bellmunt J, Guo G, Cherniack AD, Hinoue T, Laird PW, Hoadley KA, Akbani R. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell. 2017;171(3):540-556.e25. [DOI] [PMC free article] [PubMed]
- 34.Mahe M, Dufour F, Neyret-Kahn H, Moreno-Vega A, Beraud C, Shi M, Hamaidi I, Sanchez-Quiles V, Krucker C, Dorland-Galliot M. An FGFR 3/MYC positive feedback loop provides new opportunities for targeted therapies in bladder cancers. EMBO molecular medicine. 2018;10:e8163. doi: 10.15252/emmm.201708163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia-Diaz A, Shin DS, Moreno BH, Saco J, Escuin-Ordinas H, Rodriguez GA, Zaretsky JM, Sun L, Hugo W, Wang X. Interferon receptor signaling pathways regulating PD-L1 and PD-L2 expression. Cell Rep. 2017;19:1189–1201. doi: 10.1016/j.celrep.2017.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamoun A, de Reyniès A, Allory Y, Sjödahl G, Robertson AG, Seiler R, Hoadley KA, Groeneveld CS, Al-Ahmadie H, Choi W. A consensus molecular classification of muscle-invasive bladder cancer. Eur Urol. 2020;77:420–433. doi: 10.1016/j.eururo.2019.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Damrauer JS, Hoadley KA, Chism DD, Fan C, Tiganelli CJ, Wobker SE, Yeh JJ, Milowsky MI, Iyer G, Parker JS. Intrinsic subtypes of high-grade bladder cancer reflect the hallmarks of breast cancer biology. Proc Natl Acad Sci. 2014;111:3110–3115. doi: 10.1073/pnas.1318376111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics: a journal of integrative biology. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eide PW, Bruun J, Lothe RA, Sveen A. CMScaller: an R package for consensus molecular subtyping of colorectal cancer pre-clinical models. Sci Rep. 2017;7:16618. doi: 10.1038/s41598-017-16747-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mo Q, Shen R, Guo C, Vannucci M, Chan KS, Hilsenbeck SG. A fully Bayesian latent variable model for integrative clustering analysis of multi-type omics data. Biostatistics. 2017;19:71–86. doi: 10.1093/biostatistics/kxx017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Becht E, Giraldo NA, Lacroix L, Buttard B, Elarouci N, Petitprez F, Selves J, Laurent-Puig P, Sautès-Fridman C, Fridman WH. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17:218. doi: 10.1186/s13059-016-1070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW, Levine DA. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Yang T-HO, Porta-Pardo E, Gao GF, Plaisier CL, Eddy JA. The immune landscape of cancer. Immunity. 2018;48:812–830. e814. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jeschke J, Bizet M, Desmedt C, Calonne E, Dedeurwaerder S, Garaud S, Koch A, Larsimont D, Salgado R, Van den Eynden G. DNA methylation–based immune response signature improves patient diagnosis in multiple cancers. J Clin Invest. 2017;127:3090–3102. doi: 10.1172/JCI91095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bland JM, Altman DG. Survival probabilities (the Kaplan-Meier method) Bmj. 1998;317:1572–1580. doi: 10.1136/bmj.317.7172.1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fox J, Weisber S. Cox Proportional-Hazards Regression for Survival Data in R. In: An Appendix to An R Companion to Applied Regression. Second. 2011. p. 1–20.
- 51.Su X, Lu X, Bazai SK, Compérat E, Mouawad R, Yao H, Rouprêt M, Spano JP, Khayat D, Davidson I, Tannir NM, Yan F et al. RNA sequencing study of upper-tract urothelial carcinomas https://www.ncbi.nlm.nih.gov/bioproject/PRJNA678814. [DOI] [PMC free article] [PubMed]
- 52.Su X, Lu X, Bazai SK, Compérat E, Mouawad R, Yao H, Rouprêt M, Spano JP, Khayat D, Davidson I, Tannir NM, Yan F et al. Epigenome analysis of upper-tract urothelial carcinomas samples and normal adjacent tissue (NAT) https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE161651. [DOI] [PMC free article] [PubMed]
- 53.Su X, Lu X, Bazai SK, Compérat E, Mouawad R, Yao H, Rouprêt M, Spano JP, Khayat D, Davidson I, Tannir NM, Yan F et al. Exome-sequencing of 30 upper-tract urothelial carcinomas and their matched normal. https://odin.mdacc.tmc.edu/~xsu1/UTUC_Data.html. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Clinicopathological tumor features of upper-tract urothelial carcinomas samples used for the study. Table S2. Association between clinical variables and patients progression-free and overall survival. Table S3. List of somatic mutations identified in the whole-exome sequencing of upper-tract urothelial carcinomas (n = 30). Table S4. List of significantly mutated genes identified by the MutSigCV algorithm. Table S5. Mutual exclusivity and co-occurrence analysis for the 10 most significantly mutated genes using the one-side Fisher’s exact test. Table S6. Association between top 10 frequently significantly mutated UTUC genes and muscle-invasive status. Table S7. Association between UTUC RNA unsupervised clustering and clinical variables. Table S8. Subtypes of UTUC samples using consensus molecular classification of muscle-invasive bladder cancer. Table S9. Association between top 10 frequently significantly mutated genes and DNA methylation-based clusters in UTUC samples. Table S10. Association between clinical variables and DNA methylation UTUC based clustering in samples with EpiC-low and EpiC-high signatures. Table S11. List of differentially methylated regions (DMR) using hypomethylated probes in FGFR3-mutated UTUC cases as compared to FGFR-wild type samples. Table S12. List of genes with promoter CpG islands methylation in UTUC tumor samples. Only those with a frequency higher than 10% are reported. Table S13. Association between UTUC EpiC signature and clinical variables in TCGA bladder carcinomas cohort. Table S14. Association between significant mutations of UTUC and supervised TCGA DNA methylation based clustering. Table S15. Association between mutation and copy number contributors and iClusters.
Additional file 2: Fig. S1. Copy number variants analysis. Fig. S2. Comparative analysis of the frequency of most frequent mutations in upper-tract urothelial carcinomas (UTUC) as compared to urothelial bladder carcinomas (UBC). Fig. S3. ZFP36 family mutation in diverse histopathological cancer subtypes. Fig. S4. a) Human protein atlas and FANTOM5 datasets showing that protein expression and RNA expression of ZFP36L1 is the highest and the most expressed in the urinary bladder as compared to other tissues. Fig. S5. ZFP36L1 inhibition increases cell motility in vitro. Fig. S6. Distribution of MeTIL score between FGFR3-mutant and wild type of UTUC and UBC, and between muscle-invasive and non-muscle invasive UTUC samples. Fig. S7. Heatmap of BASE47 bladder signature in UTUC samples showing two subgroups of “luminal-like” and “basal-like”. Fig. S8. Principal component analysis showing heterogeneity of the two epi-clusters EpiC-C1 and EpiC-C2, obtained through unsupervised clustering of most variable DNA methylation probes. Fig. S9. Kaplan-Meier curves for progression-free survival regarding UTUC epi-clusters. Fig. S10. Unsupervised clustering of DNA methylation using most variable probes in the whole dataset encompassing UTUC (n = 35) samples and adjacent normal urothelium (n = 8). Fig. S11. Distribution of DNA methylation probes across different genomic regions significantly hypomethylated in FGFR3-mutated tumors versus FGFR3-wild type as compared to EPIC arrays. Heatmap showing differentially methylated probes located in top ranked DMR and corresponding ssGSEA results. Fig. S12. Integrative clustering analysis of multi-omics data. Full Western blots containing the entire ladder for loss-of-function experiments of ZFP36L1 using siRNA in TCCSUP bladder cancer cell line.
Additional file 3. Review history (LOG 180 bytes).
Data Availability Statement
RNA sequencing data generated in this paper have been deposited at the NCBI Sequence Read Archive (SRA) hosted by the NIH (SRA accession: PRJNA678814) [51]. DNA methylation data generated have been submitted to GEO (GEO accession: GSE161651) [52]. WES sequencing data have been deposited at gated repository [53]; WES dataset requires access control, and there is a corresponding Data Access Committee (DAC) who determines access permissions. Data access requests are reviewed by DAC [53]. Gene expression and DNA methylation data from the TCGA-BLCA cohort were downloaded from UCSC Xena (https://xena.ucsc.edu/), and somatic mutation data and detailed clinicopathological information were obtained from cBioPortal (http://www.cbioportal.org/) under the archive of Bladder Cancer [33]. DNA methylation and RNA-seq expression of bladder cancer cell lines were retrieved from Genomics of Drug Sensitivity in Cancer (GDSC, https://www.cancerrxgene.org/).