Abstract
Circular RNAs (circRNAs), which are single-stranded closed-loop RNA molecules lacking terminal 5′ caps and 3′ poly(A) tails, are attracting increasing scientific attention for their crucial regulatory roles in the occurrence and development of various diseases. With the rapid development of high-throughput sequencing technologies, increasing numbers of differentially expressed circRNAs have been identified in bladder cancer (BCa) via exploration of the expression profiles of BCa and normal tissues and cell lines. CircRNAs are critically involved in BCa biological behaviours, including cell proliferation, tumour growth suppression, cell cycle arrest, apoptosis, invasion, migration, metastasis, angiogenesis, and cisplatin chemoresistance. Most of the studied circRNAs in BCa regulate cancer biological behaviours via miRNA sponging regulatory mechanisms. CircRNAs have been reported to be significantly associated with many clinicopathologic characteristics of BCa, including tumour size, grade, differentiation, and stage; lymph node metastasis; tumour numbers; distant metastasis; invasion; and recurrence. Moreover, circRNA expression levels can be used to predict BCa patients’ survival parameters, such as overall survival (OS), disease-free survival (DFS), and progression-free survival (PFS). The abundance, conservation, stability, specificity and detectability of circRNAs render them potential diagnostic and prognostic biomarkers for BCa. Additionally, circRNAs play crucial regulatory roles upstream of various signalling pathways related to BCa carcinogenesis and progression, reflecting their potential as therapeutic targets for BCa. Herein, we briefly summarize the expression profiles, biological functions and mechanisms of circRNAs and the potential clinical applications of these molecules for BCa diagnosis, prognosis, and targeted therapy.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12943-020-01300-8.
Keywords: Circular RNA, Bladder cancer, ceRNA, Biomarker, Targeted therapy
Background
Circular RNAs (circRNAs) are single-stranded closed-loop RNA molecules without terminal 5′ caps and 3′ poly(A) tails [1]. Although circRNAs were first discovered in viruses in 1976, they were initially regarded as functionless by-products of aberrant RNA splicing and consequently did not receive considerable scientific attention for decades [2, 3]. CircRNAs are produced from precursor mRNAs mainly by lariat-driven circularization and intron pairing-driven circularization, resulting in three types of circRNAs: exonic circRNAs (ecRNAs), exon-intron circRNAs (elciRNAs), and intronic circRNAs (ciRNAs) [4–7]. With the rapid development of high-throughput sequencing technologies, increasing numbers of differentially expressed circRNAs have been identified in normal and malignant human cells [8]. Numerous circRNAs exist widely in tissues, serum, and urine, and their expression profiles are cell type-specific, tissue-specific, or developmental stage-specific [9–12]. Increasing evidence suggests that circRNAs are involved in the occurrence and development of various diseases, such as cardiovascular diseases [13], diabetes [14], neurological dysfunction [15], and cancer [16–19]. In particular, circRNAs have been reported to play pivotal roles in the development and progression of cancer and might function as cancer biomarkers and novel therapeutic targets [20, 21]. CircRNAs perform regulatory roles at the transcriptional and posttranscriptional levels; for example, they modulate gene transcription [6], act as microRNA (miRNA) sponges [22], interact with RNA-binding proteins (RBPs) [23], and can be translated into peptides [24].
Bladder cancer (BCa) is the most common malignant tumour of the urinary system, and its incidence is increasing worldwide [25]. BCa is divided into three main pathological types: bladder urothelial carcinoma (BUC), squamous cell carcinoma and adenocarcinoma, with BUC accounting for > 90% of all cases of BCa [26]. To assign risk, BUC can be further categorized into muscle-invasive BCa (MIBC) and non-muscle-invasive BCa (NMIBC), with NMIBC accounting for approximately 75% of all cases [27]. Recently, the treatment of BCa has achieved great advances worldwide. Apart from traditional surgical resection, chemotherapy, and radiotherapy, immunotherapy is a promising method for BCa treatment [28, 29]. However, postoperative recurrence and distant metastasis make five-year survival rates for advanced BCa still low [30, 31]. Therefore, identifying potential therapeutic targets and biomarkers for BCa is of great importance.
An increasing number of studies have shown that differential expression of circRNAs is associated with the carcinogenesis and progression of BCa. In this review, we summarize the expression profiles, biological functions and mechanisms, and clinical significance of BCa-related circRNAs.
Biogenesis, function, and study approaches of circRNAs
Biogenesis of circRNAs
CircRNAs are produced from pre-mRNAs and are thought to be the result of exon-skipping events. Although the specific mechanism of circRNA biogenesis is still unidentified, two widely accepted models of circRNA circularization can explain the back-splicing processes known as lariat-driven circularization and intron pairing-driven circularization [4]. In the lariat-driven circularization model, circularization requires covalent binding between the splicing donor and splicing acceptor to form an exon-containing lariat, resulting in the formation of ecRNAs [32]. In the intron pairing-driven circularization model, circularization is generated by base pairing between reverse complementary sequences. Alu repeats, originally characterized by the action of the Arthrobacterluteus restriction endonuclease, are repetitive complementary sequences located in flanking introns and are highly abundant and exist in more than 10% of the human genome [4, 33]. Introns consisting of Alu repeats are more likely to pair with each other, leading to circularization of exons and production of diverse circRNAs [34]. Unlike ecRNAs, elciRNAs retain introns that are not spliced out completely [6]. CiRNAs are generated from intron lariats that escape the process of intron debranching and degradation [5]. In addition, circRNA biogenesis has been reported to be regulated by a number of proteins, such as RBPs [23], enzymes [35], and transcription factors [36]. RBPs are crucial regulatory factors that interact with specific binding sites in flanking intronic sequences of precursor mRNAs to promote or suppress circRNA formation. For example, quaking (QKI) is an RBP that induces exon circularization and then facilitates the biogenesis of circRNAs when it binds to intronic QKI binding motifs [23]. In addition, another RBP, muscleblind (MBL), has been reported to interact with its own pre-mRNA, leading to the formation of circMBL [37]. Adenosine deaminase acting on RNA (ADAR1), a kind of RNA-editing enzyme, was reported to negatively regulate the formation of circRNAs by reducing the RNA pairing structure of flanking introns and backsplicing [35]. Moreover, the nuclear RNA helicase DHX9 can reduce the formation of circRNAs by downregulating Alu element-induced intron pairing [38]. Finally, the transcription factor Twist1 was found to bind the Cul2 promoter to selectively promote the expression of Cullin2 (Cul2) circular RNA during the epithelial–mesenchymal transition in hepatocellular carcinoma [36]. In brief, the biogenesis of circRNAs and the regulatory mechanisms involved in circularization remain vague. More research is needed to help us understand the circRNA circularization processes in depth.
Functions of circRNAs
CircRNAs were initially regarded as functionless by-products of aberrant RNA splicing [2, 3]. With the rapid development of high-throughput sequencing technologies, an increasing number of circRNAs have been found to be involved in physiological and pathological processes by acting as miRNA sponges [22], interacting with RBPs [23], regulating transcription or splicing [37, 39], and translating proteins [24]. Among these biological processes, circRNAs most commonly exert their function by sponging miRNAs in tumour cells. For example, ciRS-7 (circ_Cdr1as) serves as an miRNA sponge of miR-7, resulting in decreased miR-7 function and upregulation of miR-7 target genes [40]. In addition to acting as miRNA sponges, some circRNAs may also serve as protein sponges or decoys to interact with RBPs. For instance, circ_Foxo3 was found to block cell cycle progression by binding to the cell cycle proteins cyclin-dependent kinase 2 (CDK2) and cyclin-dependent kinase inhibitor 1 (p21) [41]. In addition, circ-PABPN1 was found to bind to human antigen R/ELAV-like protein 1 (HuR) and prevent HuR from binding to PABPN1 mRNA, resulting in the inhibition of PABPN1 translation [42]. Some circRNAs have also been identified to regulate gene transcription or selective splicing. Circ_EIF3J and circ_PAIP2 have been reported to promote the transcription of PAIP2 and EIF3J by interacting with U1 snRNPs [6]. Additionally, circ_Mbl was reported to compete with linear MBL mRNA for selective splicing [37]. Finally, increasing evidence has demonstrated that some circRNAs can exert their functions by translating proteins. Due to the absence of 5′-cap and 3′-poly(A) structures, circRNAs were initially considered to be untranslatable [43]. Recently, translatable circRNAs containing internal ribosome entry sites (IRESs) were found to be translated into proteins in a cap-independent manner [44–46]. For example, circ-ZNF609 was reported to be translated into a protein that controls myoblast proliferation [24]. In addition, circFXBW7 can be translated into a novel 21-kDa protein to suppress the tumorigenesis of glioma [47].
Approaches for circRNA studies
To date, genome-wide annotation of circRNAs, experimental validation of circRNAs, and overexpression/suppression of circRNAs are the main approaches to explore the functional implications of circRNAs. First, ribo-RNA-seq profiles rRNA-depleted total RNAs, including both poly(A) (linear) and nonpoly(A) (circular) RNAs. In addition, p(A)- RNA-seq profiles only non-poly(A) RNA. Ribo-RNA-seq or p(A)-RNA-seq combined with RNase R, which digests linear RNAs and preserves circRNAs, is more suitable for biochemical enrichment detection of circRNAs [48]. In addition, bioinformatic mapping was used to identify RNA-seq reads uniquely mapped to back-splice junctions (BSJs) via a number of algorithms [49]. In addition to RNA-seq profiling, microarray technology is also used for circRNA annotation [50]. Second, a series of experimental approaches, including PCR [51], northern blotting [5], and RNA fluorescence in situ hybridization (FISH) [6], are widely used to validate the existence of circRNAs [49]. Finally, overexpression/suppression of circRNAs are gain/loss of function used to annotate circRNAs’ function. Overexpression of circRNAs can be achieved in trans by overexpression plasmids, which contain circRNA-producing exons and their flanking intronic sequences with intronic complementary sequences [52]. In theory, manipulation of the endogenous promoter with the CRISPR/Cas9 genome engineering system or replacement of the weak intronic RNA pair with a strong one can lead to overexpression of both circular and linear RNAs from a gene locus in cis [49]. RNAi-mediated degradation [53] and the RNA-guided, RNA-targeting Cas13 system [54] represent strategies for circRNA knockdown. The CRISPR/Cas9 genome engineering system targeting circRNA-forming exons or disrupting intronic RNA pairs are strategies for circRNA knockout [55, 56]. In conclusion, improvements in methods to study circRNAs without affecting their residing genes and the wide employment of improved experimental approaches will be able to provide new insights into the biogenesis and functional implications of circRNAs in the future.
Research on and discovery of circRNAs in BCa
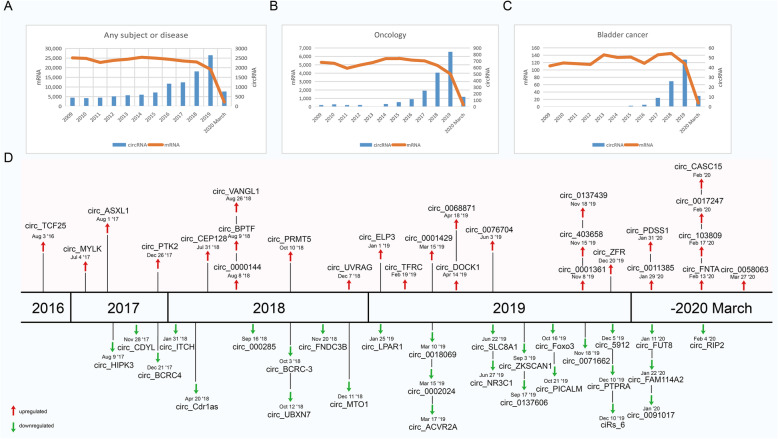
A full review was performed using Web of Science to search for reports with the key words (“circular RNA” or “circRNA”) and (“bladder cancer” or “bladder urothelial carcinoma” or “bladder neoplasm” or “bladder tumor” or “bladder tumour”) published over the past 10 years (January 2009–March 2020). Research regarding the discovery and characterization of circRNAs has increased annually, while protein-coding gene (mRNA) discovery research has remained stable (Fig. 1a). Similar trends are observed in the contexts of oncology in general (Fig. 1b) and BCa specifically (Fig. 1c). These findings suggest a growing focus on circRNAs and their roles in tumorigenesis. Collectively, related research has resulted in the validation of 55 BCa-related circRNAs (27 upregulated and 28 downregulated) in the past 10 years (Fig. 1d).
Fig. 1.
Research on and discovery of circRNAs in BCa. The amount of research, as quantified by the annual number of peer-reviewed publications, has been relatively stable for mRNAs (orange line) but not for circRNAs (blue bars) in the following categories: a overall, for any subject or disease; b oncology; and c BCa. d Increasing numbers of novel circRNAs were identified from 2016 to March 2020
A variety of methods have been developed to study the structures and functions of circRNAs. RNA sequencing (RNA-seq) [32] and microarray technology [50] are widely used for the identification of new circRNAs and the quantification of circRNA expression. Reverse transcription-polymerase chain reaction (RT-PCR) [51] and Northern blot [57] are two approaches used to further verify circRNAs. In addition, FISH can be used to analyse the subcellular localization of circRNAs [58].
To better study the biological functions and applications of circRNAs, numerous circRNA-associated public databases have recently been developed to facilitate circRNA analyses. These online databases are currently used for circRNA identification, prediction, localization, characterization and functional analysis and provide tools for investigating the interactions of circRNAs with targets. For example, CircBase contains circRNA information from different species [59]. CircRNADisease and Circ2Traits provide disease-associated circRNA annotations [60, 61]. Other databases and their common uses are listed in Table S1.
CircRNA expression profiles in BCa
With the rapid development of high-throughput sequencing technologies, a large number of novel dysregulated circRNAs have been identified in BCa cell lines and tissues, most of which are differentially expressed between BCa tissues and adjacent normal tissues, indicating the important roles of these circRNAs in BCa development and progression. Primary expression profiles obtained via ribosomal RNA-depleted RNA-seq and circRNA microarrays have been widely employed for the discovery of novel circRNAs [32, 59, 62]. For example, according to circRNA microarray data from 4 paired BCa tissues and adjacent normal bladder tissues, Zhong et al. identified 3243 circRNAs in total and 469 circRNAs that were differentially expressed in BCa compared with normal tissues, 285 of which were significantly upregulated, while 184 were downregulated [63]. Li et al. identified 16,353 circRNAs that were expressed in 3 paired BCa and adjacent normal tissues, 571 of which were differentially expressed; 47 circRNAs were significantly upregulated, and 524 circRNAs were downregulated [64]. Zheng et al. identified 67,358 circRNAs that were expressed in 1 pair of BCa and adjacent normal bladder tissues, and circ_HIPK3 was verified to directly bind to miR-124 and inhibit miR-124 activity [65]. In another study, RNA-seq data from BCa tissues of 9 different grades and adjacent normal bladder tissues revealed a total of 316 (205 upregulated and 111 downregulated) and 244 (109 upregulated and 135 downregulated) dysregulated circRNAs in high-grade BCa vs. normal tissue and in high-grade BCa vs. low-grade BCa, respectively; 42 of these circRNAs overlapped [66]. In addition, secondary bioinformatic analyses based on Gene Expression Omnibus (GEO) databases are frequently performed to identify differentially expressed circRNAs [67]. By performing a comprehensive bioinformatics analysis of RNA-seq data from 457 NMIBC samples, Okholm et al. identified 15,223 unique circRNAs that were supported by at least two reads in at least two different samples, and 113 abundant circRNAs were differentially expressed between high- and low-risk tumour subtypes; furthermore, the expression of 13 circRNAs correlated with progression [68]. Among 11 studies, the microarray dataset GSE92675 from the platform GPL19978 was the most commonly used database for secondary bioinformatic analyses intended to identify novel circRNAs for further research [69–79]. BCa-related circRNAs identified by RNA-seq and microarray analyses are listed in Table 1.
Table 1.
Overview of circRNAs identifed by RNA sequencing and microarrays in BCa
| Sample | Special treatment | Detection Method | GEO database | Data source | total circRNA | Number of circRNA differently expressed (fold change ≥2) | circRNAs validated by qRT-PCR | Ref./PMID |
|---|---|---|---|---|---|---|---|---|
| 4 paird BCa and BCN tissues | RNAse R | CircRNA microarray | GSE92675 | CircRNA microarray | 3243 | 469 (285 upregulated, 184 downregulated) | 6 | 27484176 |
| 3 paird BCa and BCN tissues | rRNA-depleted and RNase R | RNA-seq | GSE97239 | RNA-seq | 16,353 (6154) | 571 (47 upregulated, 524 downregulated) | circ_HIPK3 | 28794202 |
| 1 paird BCa and BCN tissues | rRNA-depleted | RNA-seq | GSE77661 | RNA-seq | 67,358(27296) | / | circ_HIPK3 | 27050392 |
| 4 paird BC and BCN tissues | / | RNA-seq | / | RNA-seq | / | 59(7 upregulated, 52 downregulated) | hsa circ 0018069 | 30984788 |
| 2 paird BCa and BCN tissues | RNAse R | RNA-seq | / | RNA-seq | 6834 | 567 | 40 | 30745833 |
| 5 paird BCa and BCN tissues | rRNA and linear RNA-depleted | RNA-seq | / | RNA-seq | 88,732(62,788) | 56 (14 upregulated, 42 downregulated) | 2 | 29151929 |
| 4 paird BCa and BCN tissues | rRNA-depleted | RNA-seq | / | RNA-seq | / | 118 (34 upregulated, 84 downregulated) | 3 | 30025927 |
| 9 different grades of BCa and BCN tissues | / | RNA-seq | / | RNA-seq | / | 244 (H vs L), 316 (H vs N), 42 circRNAs overlapped | 7 | 31545480 |
| BCa 5637, T24 and SV-HUC-1 cell lines | / | CircRNA microarray | / | CircRNA microarray | / | / | circ_CASC15 | 31072448 |
| 10 paird BCa and HC urine samples | Rnase R | CircRNA microarray | / | CircRNA microarray | / | 86 (53 upregulated, 33 downregulated) | circ_0137439 | 31777254 |
| 3 pairs of BCSCs and BCNSCs samples | / | CircRNA microarray | / | CircRNA microarray | 4451 | 127 (113 upregulated, 14 downregulated) | circ_103809 | 32065779 |
| 3 paird BCa and BCN tissues | / | CircRNA microarray | / | CircRNA microarray | / | 734 (478 upregulated, 256 downregulated) | 8 | 30983072 |
| 3 paird BCa and BCN tissues | Rnase R | CircRNA microarray | GSE112719 | CircRNA microarray | / | 80 (37 upregulated, 43 downregulated) | circ_101320 | 30305293 |
| 3 paird BCa and BCN tissues | / | CircRNA microarray | / | CircRNA microarray | 1038 | / | 7 | 29558461 |
| 457 NMIBC samples | / | RNA-Seq | / | Bioinformatics analysis | 15,223 | / | 13 | 29263845 |
| 4 paird BCa and BCN tissues | / | CircRNA microarray | GSE92675 | Bioinformatics analysis | / | 469 (285 upregulated, 184 downregulated) | circ_MYLK | 28687357 |
| 3 paird BCa and BCN tissues | / | / | GSE97239, GSE92675 | Bioinformatics analysis | / | 18 (5 upregulated, 13 downregulated) | 3 | 31169020 |
| 4 paird BCa and BCN tissues | / | CircRNA microarray | GSE92675 | Bioinformatics analysis | / | 200 | hsa_circ_0000144 | 30098434 |
| 4 paird BCa and BCN tissues | / | CircRNA microarray | GSE92675 | Bioinformatics analysis | / | 408 | circ_0058063 | 30362519 |
| 4 paird BCa and BCN tissues | / | CircRNA microarray | GSE92675 | Bioinformatics analysis | / | 89 (66 upregulated, 23 downregulated) | circ_0001429 | 30909190 |
| 4 paird BCa and BCN tissues | / | CircRNA microarray | GSE92675 | Bioinformatics analysis | / | 433 (264 upregulated, 169 downregulated) | circ_CEP128 | 30134837 |
| 4 paird BCa and BCN tissues | / | CircRNA microarray | GSE92675 | Bioinformatics analysis | 3423 | 433 (264 upregulated, 169 downregulated) | circ_CEP128 | 30939216 |
| 4 paird BCa and BCN tissues | / | CircRNA microarray | GSE92675 | Bioinformatics analysis | / | 312 (195 upregulated, 117 downregulated) | circ_0058063 | 32181485 |
| 4 paird BCa and BCN tissues | / | CircRNA microarray | GSE92675 | Bioinformatics analysis | / | / | circ_VANGL1 | 30146736 |
| 4 paird BCa and BCN tissues | / | CircRNA microarray | GSE92675 | Bioinformatics analysis | 3243 | 469 (285 upregulated, 184 downregulated) | / | 27363013 |
| 4 paird BCa and BCN tissues | / | CircRNA microarray | GSE92675 | Bioinformatics analysis | / | 428 (261 upregulated, 167 downregulated) | hsa_circ_0011385 | 32015691 |
circRNAs circular RNAs, BCa bladder cancer, BCN bladder cancer tissues paired adjacent normal bladder tissues, H high-grade bladder cancer, L low-grade bladder cancer, N normal tissue, HC healthy controls, BCSCs bladder cancer stem cells, BCNSCs bladder cancer non-stem cells, NMIBC non muscle-invasive bladder cancer
For microarray or RNA-seq data analyses, paired t tests were performed to analyse significant differences. The false discovery rate (FDR) was applied to determine the P-value threshold, and an FDR < 0.05 was recommended. CircRNAs (fold changes ≥ 2.0 and P-values < 0.05) have been reported to be significantly differentially expressed [63, 64]. For RT-PCR or Northern blotting, β-actin or GAPDH was used as a reference gene. Mean values, median expression levels, or concrete data are used as cut-off values [80–83]. No unified standards are available to determine thresholds for circRNA detection.
Biological functions and molecular mechanisms of circRNAs in BCa
CircRNAs regulate the hallmarks of cancer
In 2011, Hanahan and Weinberg proposed ten hallmarks of cancer that result in the progressive conversion of normal cells into cancerous cells [84]. Here, we briefly summarize the well-known circRNAs involved in the essential stages of tumourigenesis and progression in BCa to examine the correlations between circRNAs and the hallmark features of cancer (Fig. 2a).
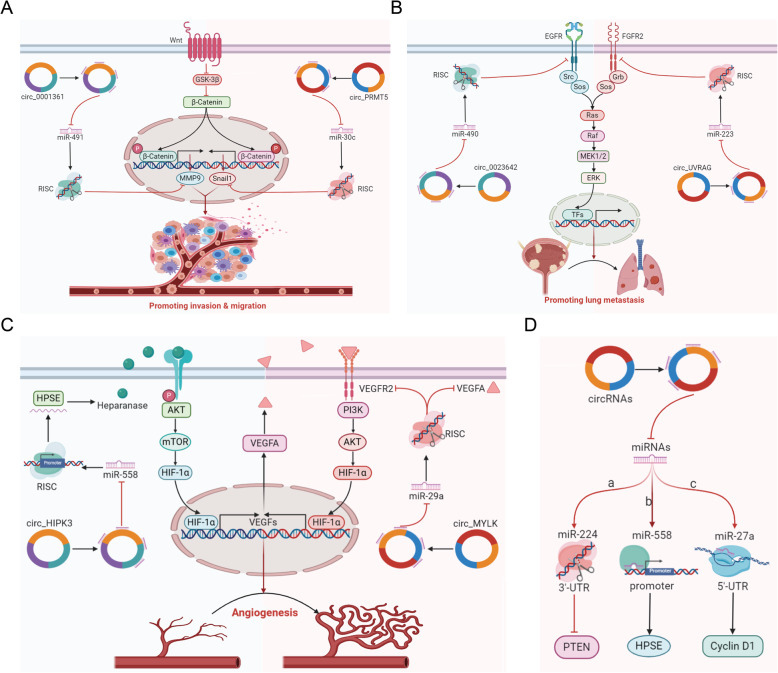
Fig. 2.
The relationship between circRNAs and BCa. a CircRNAs are associated with the hallmarks of BCa. b CircRNAs regulate cell proliferation, apoptosis, invasion, migration and metastasis, angiogenesis, and cisplatin chemoresistance in BCa cells
Recently, several oncogenic and antioncogenic circRNAs have been discovered to regulate cell proliferation, tumour growth suppression, cell cycle arrest, apoptosis, invasion, migration, metastasis, angiogenesis, and cisplatin chemoresistance in BCa cells (Fig. 2b and Table 2).
Table 2.
Dysregulated circRNAs in BCa
| Name | CircBase ID | Sponge target | Gene | Function | Types of BCa tissues and BCa cell lines | Ref./PMID |
|---|---|---|---|---|---|---|
| upregulated | ||||||
| circ_0058063 | hsa_circ_0058063 | miR-486-3p | FOXP4 | promoted cell proliferation, invasion, and inhibited apoptosis | 94 cases of BCa and the non-cancerous normal tissues; 5637, BIU-87 and RT-112 cell lines | 32181485 |
| circ_0058063 | hsa_circ_0058063 | miR-145-5p | CDK6 | promoted cell proliferation and migration, inhibited cell apoptosis, and decreased cell cycle arrest | 25 pairs of BCa tissues and adjacent normal tissues; T24 and J82 cell lines | 30362519 |
| circ_0017247 | hsa_circ_0017247 | / | Wnt/β-catenin | promoted cell proliferation and tumor formation, inhibited apoptosis, and decreased cell cycle arrest | 50 BCa tissues and adjacent tissues; UM-UC3, SW780, BIU and J82 cell lines | 32096177 |
| circ_103809 | hsa_circ_0072088 | miR-511 | / | promoted the self-renewal, migration and invasion | 3 pairs of BCa tissues and adjacent normal bladder tissues; T24 and EJ cell lines | 32065779 |
| circ_FNTA | hsa_circ_0084171 | miR-370-3p | FNTA | promoted invasion and decreased chemosensitivity to cisplatin | 41 BCa tissues and adjacent normal bladder tissues; T24, J82, 5637, and UMUC3 cell lines | 32052578 |
| circ_PDSS1 | hsa_circ_0093398 | miR-16 | / | promoted cell proliferation, invasion and migration | 72 BCa and adjacent healthy tissues; HT-1197 and UMUC3 cells | 31868205 |
| circ_403658 | hsa_circ_0004383 | / | LDHA | promoted cell growth and invasion | 123 BCa tissues and the matched adjacent tissues; SW780, 5637, T24, J82 and RT4 cell lines | 31814891 |
| circ_0137439 | hsa_circ_0137439 | miR-142-5p | MTDH | promoted cell proliferation, migration, and metastasis | 116 bladder cancer urine samples and 30 normal samples; T24 and 5637 cell lines | 31777254 |
| circ_VANGL1 | hsa_circ_0002623 | miR-605-3p | VANGL1 | promoted cell proliferation, migration, and invasion | 87 BCa tissues and 37 normal adjacent tissues; T24 and EJ cells | 30146736 |
| circ_VANGL1 | hsa_circ_0002623 | miR-1184 | IGFBP2 | promoted cell proliferation, migration, and invasion | 60 BCa and corresponding paracancerous tissue; J82, T24, EJ, RT-4, UM-UC-3, and TCC cell lines | 31758655 |
| circ_ZFR | hsa_circ_0072088 | miR-377 | ZEB2 | promoted cell growth, migration and invasion, and decreased cell cycle arrest and apoptosis | 104 pairs of BCa tissues and adjacent normal tissues; UMUC3, T24, J82, 5637, SW780, EJ and BIU87 cell lines | 31746333 |
| circ_0001361 | hsa_circ_0001361 | miR-491-5p | MMP9 | promoted cell invasion and metastasis | 69 pairs of BCa tissues and matched adjacent normal bladder epithelial tissues; SV-HUC-1, EJ, UMUC3, RT4, and 5637 cell lines | 31705065 |
| circ_UVRAG | hsa_circ_0023642 | miR-223 | FGFR2 | promoted proliferation, migration, tumor formation, and metastasis | T24, EJ, J82, UM-UC-3, TCC, and RT-4 cell lines | 30387298 |
| circ_0023642 | hsa_circ_0023642 | miR-490-5p | EGFR | promoted cell invasion and metastasis | J82 and UMUC3 cell lines | 31455760 |
| circ_CASC15 | hsa_circ_0075828 | miR-1224-5p | CREB1 | promoteed cell proliferation | 67 pair BCa tissues and matched para-carcinoma tissues; 5637, and T24 cell lines | 31072448 |
| circ_0068871 | hsa_circ_0068871 | miR-181a-5p | FGFR3 | promoted cell proliferation and migration | 32 BCa and adjacent normal tissue; T24, UMUC3, EJ, and J82 cell lines | 30999937 |
| circ_DOCK1 | hsa_circ_0020394 | miR-132-3p | Sox5 | promoted cell proliferation, migration, and tumour growth | 23 BC tissue specimens and 32 normal bladder tissues; BIU-87, EJ-m3, T24 and 5673 cell lines | 30983072 |
| circ_CEP128 | hsa_circ_0102722 | miR-145-5p | SOX11 | promoted cell proliferation, and decreased cell apoptosis and cell cycle arrest | 10 pairs of BCa tissues and adjacent bladder tissues; RT-112, 5637, BIU-87, TCCSUP and HEK293T cell lines | 30134837 |
| circ_CEP128 | hsa_circ_0102722 | miR-145-5p | Myd88 | promoted cell proliferation and migration, and decreased cell apoptosis and cell cycle arrest | 40 BCa specimens and blood samples; 293T, J82 and T24 cell lines | 30939216 |
| circ_0001429 | hsa_circ_0001429 | miR-205-5p | VEGFA | enhanced cell propagation and metastasis, reduced cell apoptosis, and promoted tumor growth and lung metastasis | 20 pairs of BCa tissues and paired adjacent normal bladder tissues; T24 cells and 5637 cells | 30909190 |
| circ_TFRC | has_circ_0001445 | miR-107 | TFRC | promoted the invasion, proliferation and tumor growth, contributed to an EMT phenotype | 57 BCa patients tissues compared with adjacent normal patients tissues, EJ, T24, 5637, UMUC3, BIU87, J82, and SW780 cell lines | 30782157 |
| circ_ELP3 | hsa_circ_0001785 | / | / | promoted cell proliferation, and reduced apoptosis and chemosensitivity to cisplatin | 18 pairs of tissue samples and 30 bladder cancer samples; T24 and 5647 cells | 30745833 |
| circ_PRMT5 | hsa_circ_0031250 | miR-30c | SNAIL1/E-cadherin | promoted cell EMT | 119 UCB tissues with matched adjacent normal bladder tissues; T24, TCC-SUP, 5637, and UM-UC-3 cell lines | 30305293 |
| circ_BPTF | hsa_circ_0000799 | miR-31-5p | RAB27A | promoted cell migration and invasive, and tumor growth | 72 pairs of specimens of BCa tissues and adjacent noncancerous tissues; UM-UC-3 and T24 cell lines | 30103209 |
| circ_0000144 | hsa_circ_0000144 | miR-217 | RUNX2 | promoted cell proliferation and invasion | 21 pairs of BCa tissues and adjacent normal tissues; T24, EJ, UMUC3, RT4 and 253J cell lines | 30098434 |
| circ_PTK2 | hsa_circ_0003221 | / | / | promoted the proliferation and migration | 40 pairs of BCa tissue and blood samples. T24 and 5637 cell lines | 29125888 |
| circ_MYLK | hsa_circ_0002768 | miR-29a | VEGFA/VEGFR2 | accelerated cell proliferation, migration, tube formation, and promoted EMT | 32 pairs bladder carcinomas and matched para-carcinomas; EJ, T24, 5673 and BIU-87 cell lines | 28687357 |
| circ_TCF25 | hsa_circ_0041103 | miR-103a-3p/miR-107 | CDK6 | promoted proliferation and migration | 40 pairs bladder carcinoma tissue and matched para-carcinoma tissues; T24 and EJ cell lines | 27484176 |
| downregulated | ||||||
| circ_FUT8 | hsa_circ_0003028 | miR-570-3p | KLF10 | inhibited migration, invasion, and metastasis | 145 BCa tissues and 50 matched adjacent normal bladder tissues; T24, SV-HUC-1, and UM-UC-3 cell lines | 32072011 |
| circ_RIP2 | hsa_circ_0005777 | miR-1305 | Tgf-β2/smad3 | promoted migration, invasion, clone formation and EMT | 45 paired BCa and the adjacent normal tissues, 58 bladder cancer tissues, 5637 and UM-UC-3 cell lines | 32019579 |
| circ_FAM114A2 | hsa_circ_0001546 | miR-762 | ΔNP63/TP63 | inhibited migration, invasion and proliferation | 31 BCa tissues and paired adjacent noncancer tissues; T24, J82, 5637, and 293T cell lines | 31969560 |
| circ_0091017 | hsa_circ_0091017 | miR-589-5p | / | inhibited cell proliferation, migration and invasiveness | 40 pairs of BCa tissues and normal adjacent tissues; 5637, EJ, T24, UMUC-3, and RT4 cell lines | 31957821 |
| ciRs_6 | hsa_circ_0006260 | miR-653 | March1 | suppressed cell growth and increased cell cycle arrest | 45 paired bladder cancer and the adjacent normal tissues, 58 bladder cancer tissues; 5637 and UM-UC-3 cell lines | 31819015 |
| circ_5912 | hsa_circ_0005912 | / | TGF-β signaling | suppressesed cell proliferation, invasion and migration | 58 BCa tissues and the matched adjacent tissues; T24 and SW780 cell lines | 31808751 |
| circ_PTPRA | hsa_circ_0006117 | miR-636 | KLF9 | inhibited cell proliferation and tumor growth | 104 BCa specimens, 64 matched BC and adjacent normal specimens; T24 and UM-UC-3 cell lines | 31821171 |
| circ_Foxo3 | hsa_circ_0006404 | miR-191-5p | / | promoted cell apoptosis | 30 BCa tissues and adjacent normal bladder tissues; T24, UM-UC-3 and J82 cell lines | 31802888 |
| circ_0071662 | hsa_circ_0071662 | miR-146b-3p | HPGD/NF2 | suppressed cell proliferation and invasion | 97 BCa tissues and matched adjacent normal tissues; BIU-87, T-24, EJ-28 and J82 cell lines | 31757227 |
| circ_PICALM | hsa_circ_0023919 | miR-1265 | STEAP4 | inhibited cell invasion and metastasis | 168 BCa samples and 40 corresponding adjacent normal tissue samples; T24, UM-UC-3, J82, RT-4, and HEK-293T cell lines | 31648990 |
| circ_0137606 | hsa_circ_0137606 | miR-1231 | PHLPP2 | suppressed cell proliferation and metastasis | 13 high-grade BCa, low-grade BCa and a normal controlpatients tissues; T24 and SV-HUC-1 cell lines | 31545480 |
| circ-ZKSCAN1 | hsa_circ_0001727 | miR-1178-3p | p21 | inhibited cell proliferation, migration, invasion and metastasis | 68 BCa tissues and the matched normal tissues; T24, UM-UC-3, 5637, and EJ cell lines | 31481066 |
| circ_NR3C1 | hsa_circ_0001543 | miR-27a-3p | cyclin D1 | inhibited cell proliferation, cell cycle progression, and tumor growth | 42 pairs of BCa tissues and adjacent normal bladder tissues; T24, EJ, UMUC3, J82, and 5637 cell lines | 31255724 |
| circ_SLC8A1 | hsa_circ_0000994 | miR-130b/miR-494 | PTEN | inhibited cell migration, invasion and proliferation | 70 pairs of human bladder cancer tissues compared with their adjacent normal tissues; 5637, T24, J82, EJ, UMUC, and RT4 cell lines | 31228937 |
| circ_Cdr1as | hsa_circ_0001946 | miR-1270 | APAF1 | induced cell apoptosis and enhanced chemosensitivity to cisplatin | 160 BCa tissues; TCCSUP, 5367, T24 and EJ cell lines | 31131537 |
| circ_Cdr1as | hsa_circ_0001946 | miR-135a | / | inhibited cell proliferation, invasion and migration, and tumour growth | 94 pairs of BCa tissues and adjacent normal tissues; EJ and T24 cell lines | 29694981 |
| circ_ACVR2A | hsa_circ_0001073 | miR-626 | EYA4 | suppressed cell proliferation and metastasis | 50BC tissues and matched adjacent normal epithelial tissues; T24, UM-UC-3, RT4, J82, 5637, HT-1376, and TCCSUP cell lines | 31101108 |
| circ_0002024 | hsa_circ_0002024 | miR-197-3p | / | suppressed cell proliferation, migration, and invasion | 20 BCa and normal samples; EJ, 5637, T24, and UMUC-2 cell lines | 30972190 |
| circ_CDYL | hsa_circ_0008285 | / | C-MYC | inhibited cell growth and migration | 30 pairs of BCa tissues and paired surrounding normal bladder tissues; EJ and T24T cells | 30968727 |
| circ_LPAR1 | hsa_circ_0087960 | miR-762 | / | inhibited cell invasion and metastasis | 125 BCa tissues and 68 paired cancer tissues and adjacent non-tumorous tissues; 5637 and T24 cell lines | 30867795 |
| circ_MTO1 | hsa_circ_0007874 | miR-221 | / | inhibited cell EMT and metastasis | 117 bladder cancer tissues and the matched adjacent tissues; UMUC3, SVHUC1, T24, J82 and 5637 cell lines | 30551873 |
| circ_FNDC3B | hsa_circ_0006156 | miR-1178-3p | G3BP2 | inhibited cell proliferation, migration, invasion, tumorigenesis and metastasis | 82 BCa tissues and 56 pairs of BCa tissues and adjacent noncancerous tissues; T24 and UM-UC-3 cell lines | 30458784 |
| circ_UBXN7 | hsa_circ_0001380 | miR-1247-3p | B4GALT3 | inhibited cell proliferation, migration, invasion, and tumor growth | 84 cases of BCa tissues including 30 paired BC tissues and adjacent nontumor tissues; SV-HUC-1, T24 and UM-UC-3 cell lines | 30312173 |
| circ_HIPK3 | hsa_circ_0000284 | miR-558 | HPSE | inhibited cell migration, invasion, and angiogenesis, tumor growth, and metastasis | 44 pairs of bladder cancer tissues and paired adjacent normal bladder tissues; UMUC3, and T24 cell lines | 28794202 |
| circ_BCRC3 | hsa_circ_0001110 | miR-182-5p | p27 | inhibited cell proliferation, and impairs tumor growth, and increased cell cycle arrest | 47 BCa tissues and their adjacent normal bladder tissues; EJ,T24, and SV-HUC-1 cell lines | 30285878 |
| circ_BCRC4 | hsa_circ_0001577 | miR-101 | EZH2 | promoted cell apoptosis and inhibited cell viability | 24 pairs of fresh bladder cancer tissues and surrounding normal adjacent bladder tissues; UMUC3 and T24 cell lines | 29270748 |
| circ_ITCH | hsa_circ_0001141 | miR-17, miR-224 | p21, PTEN | inhibited cell proliferation, migration, invasion and metastasis, induced cell cycle arrest and cell apoptosis | 72 pairs of BCa tumor and normal tissues; UMUC3, T24, J82, 353J, 5637, TCC, EJ and RT4 cell lines | 29386015 |
circRNAs circular RNAs, BCa bladder cancer, EMT epithelial to mesenchymal transition
Cell proliferation
Tumour cells can sustain active proliferative states via activation of cell proliferation signalling pathways [84]. The PI3K/Akt/CREB signalling pathway is an important regulatory pathway of cell proliferation [85]. Circ_CASC15, derived from the CASC15 gene, promotes cell proliferation by acting as a miR-1224 sponge to activate oncogenic CREB1 expression in BCa [86]. Phosphatase and tensin homologue (PTEN), a negative regulator of the PI3K/Akt pathway, is highly involved in BCa progression [87]. Lu et al. found that circ_SLC8A1 inhibits BCa cell proliferation, migration, and invasion by upregulating PTEN expression [88]. Circ_ITCH, generated from several exons of itchy E3 ubiquitin protein ligase, suppresses cell proliferation by sponging miR-224 to increase the expression of PTEN in BCa [89] (Fig. 3a). Consistent with the results regarding BCa, circ_ITCH has also been reported to be downregulated and to suppress cell proliferation by inhibiting the Wnt/beta-catenin pathway in lung cancer [90], colorectal cancer [91], and oesophageal squamous cell carcinoma [92]. Hsa_circ_0000144, which is produced through back-splicing of the SLAMF6 first intron, facilitates BCa cell proliferation by upregulating the expression of RUNX2, which promotes cellular malignancy in BCa [71, 93]. Liang et al. also suggested that circ_0058063 facilitates BCa cell proliferation and invasion via the circ_0058063/miR-486-3p/FOXP4 axis [76]. Circ_0071662 has been identified to suppress BCa cell proliferation and invasion by upregulating the tumour suppressor genes HPGD and NF2 [94]. Song et al. revealed that the hsa_circ_0137439/miR-142-5p/MTDH axis contributes to the promotion of BCa cell proliferation and migration [81]. In addition, hsa_circ_0091017 has been found to inhibit BCa cell proliferation and migration [95]. Yu et al. proposed that circ_PDSS1 may promote proliferation, invasion and migration by inhibiting the tumour suppressor miR-16 [96]. Circ_PTK2 has also been reported to promote BCa cell proliferation and migration [97].
Fig. 3.
CircRNAs regulate cell proliferation, tumour growth suppression, cell cycle arrest, and apoptosis in BCa cells. a Roles of circRNAs in cell proliferation. b Roles of circRNAs in tumour growth suppression. c Roles of circRNAs in cell cycle arrest. d Roles of circRNAs in apoptosis
Apart from sustaining proliferative signals, cancer cells continuously evade the growth-suppressive effects of the tumour suppressor pathway [84]. TP63 is a member of the p53 family of transcription factors, and its ΔNp63 isoform is the major tumour-suppressing isoform in BUC cell lines and primary tumours [98]. Liu et al. reported that circ_FAM114A2 inhibits tumour growth by regulating ΔNP63 [99]. In addition, the tumour suppressor Krüppel-like factor 9 (KLF9) has been reported to inhibit tumour growth by modulating p53 [100]. In one investigation, He et al. found that circ_PTPRA inhibits BCa cell proliferation in vitro and tumour growth in vivo by upregulating KLF9 [101] (Fig. 3b). Circ_BCRC3 has also been identified to function as a tumour suppressor, inhibiting BCa tumour growth through the miR-182-5p/p27 axis [102]. Su et al. showed that ciRs-6 suppresses BCa growth by elevating the expression of March1, a tumour suppressor gene that encodes an E3 ubiquitin-ligating enzyme [103]. Furthermore, ZEB2 has been reported to play oncogenic roles in BCa [104, 105]. According to Zhang et al., circ_ZFR facilitates BCa cell growth, migration and invasion by upregulating the expression of this gene [82].
Additionally, dysregulation of cell cycle regulators contributes to limitless tumour cell growth and proliferation [84]. C-MYC and C-MYC-induced genes play crucial roles in cell cycle control and cell growth [106]. For example, circ_CDYL induces cell cycle arrest by downregulating C-MYC and C-MYC-induced gene expression in BCa cells [107]. Cyclin D1, a key cell cycle-related protein, is believed to regulate the G1-to-S phase transition [108]. As reported by Zheng et al., circ_NR3C1 can induce G0/G1 arrest by suppressing cyclin D1 expression and subsequently inhibits cell cycle progression in BCa [109]. P21, a direct regulator of the cell cycle, plays a vital role in inducing growth arrest in the G1 phase by suppressing the activity of cyclin D-CDK2/4 complexes [110]. Bi et al. proposed that circ_ZKSCAN1 acts as a tumour suppressor to promote cell cycle arrest via the circ_ZKSCAN1/miR-1178-3p/p21 axis [111]. Circ_Cdr1as has also been found to mediate cell cycle arrest to exert anti-oncogenic functions in BCa cells by restoring p21 activity [112] (Fig. 3c). Among cell cycle-related CDKs, CDK6 has been identified as a major oncogenic driver of progression from G1 phase to S phase [113]. Circ_TCF25 promotes proliferation and migration by increasing CDK6 expression [63]. Sun et al. also discovered that circ_0058063 enhances BCa cell proliferation and migration abilities via the circ_0058063/miR-145-5p/CDK6 pathway in BCa [72].
Apoptosis
Apoptosis, autophagy, and necrosis are major mechanisms leading to controlled cell death that are strictly controlled by tumour cells [114]. Tumour cells can evade apoptosis, enabling them to achieve immortality. Apoptosis protease-activating factor-1 (APAF-1) is a key regulatory factor that interacts with cytochrome c released from the mitochondria, thus activating the caspase cascade to execute apoptosis [115]. Circ_Cdr1as, also known as ciRS-7 or CDR1NAT, has been identified to induce apoptosis of BCa cells by elevating APAF1 expression [116]. Forkhead box transcription factor class O3 (FOXO3) is another key factor that participates in apoptotic processes [117]. Wang et al. showed that circ_Foxo3 facilitates FOXO3-mediated apoptosis through miR-191-5p signalling [118]. Proapoptotic effects of circ_Foxo3 have also been observed in breast carcinoma biopsies and in cancer cell lines [119]. Consistent with these findings, Li et al. found by KEGG analysis that hsa_circ_0018069 may mediate the Foxo signalling pathway to exert anticancer effects [80] (Fig. 3d). In contrast, Wu et al. showed that circ_CEP128 promotes cell proliferation and suppresses apoptosis in the context of BCa by targeting SOX11 [74]. In another study, circ_CEP128 was illustrated to increase cell proliferation and inhibit apoptosis via the miR-145-5p/MYD88/MAPK signalling pathway [75]. According to Li et al., circ_BCRC4 enhances apoptosis through miR-101/EZH2 signalling [120].
Invasion, migration and metastasis
Invasion, migration and metastasis of tumour cells into lymphatic and blood vessels for dissemination into the circulation eventually results in tumour colonization of distant organs [121]. MMP9, a member of the zinc-dependent endopeptidase family, plays crucial roles in invasion and migration by degrading the extracellular matrix in BCa [122, 123]. Liu et al. reported that circ_0001361, which is derived from two exons of the FNDC3B gene, increases MMP9 expression to promote BCa cell invasion and metastasis [124]. In addition, epithelial-mesenchymal transition (EMT) is an important mechanism for tumour invasion and metastasis [125]. Chen et al. revealed that circPRMT5 regulates the SNAIL1/E-cadherin-induced EMT pathway to promote BUC cell invasion and migration [62] (Fig. 4a). Su et al. indicated that circ_5912 suppresses the invasion and migration of BCa cells via the TGF-β2-induced EMT signalling pathway [126]. He et al. further revealed that circ_FUT8 suppresses the invasion and migration of BCa cells by regulating Slug and EMT [127]. Moreover, circ_RIP2 promotes BCa proliferation, invasion and migration by inducing EMT via activation of the miR-1305/TGF-β2/smad3 pathway [128]. Circ_TFRC has been reported to upregulate the proliferative and invasive abilities of BCa cells by activating the EMT signalling pathway [129]. In addition, FOXP4 promotes the migration and invasion of breast cancer cells via EMT [130]. Consistent with these findings, Liang et al. illustrated that circ_0058063 promotes BCa cell proliferation and invasion by upregulating FOXP4 expression [76]. G3BP2, a member of the Ras-GTPase-activating protein (RasGAP) SH3 domain-binding protein (G3BP) family, is significantly overexpressed in multiple types of human tumours and contributes to tumour invasion [131, 132]. Circ_FNDC3B has been found to inhibit BCa cell proliferation, migration and invasion by suppressing the G3BP2 and SRC/FAK signalling pathways [133]. In contrast, androgen receptor (AR) has been found to mediate BCa development and progression [134, 135]. Chen et al. confirmed that AR-mediated circ_FNTA activity can promote BCa cell invasion via miR-370-3p/FNTA/KRAS signals [136]. Notably, the critical roles of cancer stem cells (CSCs) or cancer-initiating cells in tumorigenesis have attracted increasing scientific attention [137, 138]. Circ_103809 has been identified to be highly expressed in bladder CSCs and to promote the self-renewal, migration and invasion of BCa by sponging miR-511 [139]. Insulin-like growth factor-binding protein 2 (IGFBP2) has been found to be related to cell migration and invasion [140]. Yang et al. discovered that circ_VANGL1 accelerates BCa cell invasion, migration and proliferation by increasing IGFBP2 expression [141]. In another study, circ_VANGL1 was found to accelerate BCa cell invasion, migration and proliferation by increasing VANGL1 expression [77]. Liu et al. proposed that circ_DOCK1 increases the proliferation and migration potential of BCa cells via the circDOCK1/hsa-miR-132-3p/Sox5 signalling pathway [142]. Lin et al. demonstrated that circ_LPAR1 reduces invasion and metastasis via miR-762 [143]. Finally, Liu et al. verified that circ_UBXN7 suppresses cell growth and invasion by upregulating B4GALT3 [144].
Fig. 4.
CircRNAs regulate invasion, migration and metastasis and angiogenesis in BCa cells and the molecular mechanisms of circRNAs in BCa. a Roles of circRNAs in invasion and migration. b Roles of circRNAs in lung metastasis. c Roles of circRNAs in angiogenesis. d Molecular mechanisms of circRNAs in BCa
Increasing evidence has revealed that circRNAs can act as metastasis activators or suppressors in BCa. The Wnt/β-catenin signalling pathway is highly involved in tumour metastasis [145, 146]. Han et al. illustrated that hsa_circ_0017247 enhances BCa cell metastasis by activating the Wnt/β-catenin signalling pathway [147]. In addition, Li et al. proposed that hsa_circ_0137606 can suppress BCa cell proliferation and metastasis via the hsa_circ_0137606/miR-1231/PHLPP2 axis [66]. Circ_ACVR2A has been found to significantly suppress the proliferation and metastasis of BCa by targeting the miR-626/EYA4 axis [148]. According to Wu et al. [149], circ_0023642 suppresses BCa cell invasion and metastasis by modulating the circ_0023642/miR-490-5p/EGFR signalling pathway. In addition, circ_UVRAG suppresses BCa cell proliferation and metastasis by targeting the miR-223/FGFR2 axis [150] (Fig. 4b). Li et al. found that circ_MTO1 inhibits BCa cell EMT and metastasis by sponging miR-221 [151]. FAK, a well-known tyrosine kinase, is closely related to metastasis in cancer [152]. Finally, circ_PICALM, which is generated from exons 9–12 of PICALM, has been identified to inhibit metastasis of BCa by modulating FAK activation and EMT [153].
Angiogenesis
Angiogenesis, the process by which rapidly growing malignant tissues are continuously supplied with nutrients and oxygen and cleared of metabolic wastes, is essential for tumour growth and progression. Without angiogenesis, tumours are unlikely to grow beyond a size of 100–200 μm [154]. Vascular endothelial growth factor (VEGF), a marker gene of angiogenesis, plays a key role in inducing angiogenesis during tumour growth and metastasis [155]. VEGFA, the expression of which is strongly induced by hypoxia, is one of the most potent inducers of angiogenesis [156]. VEGFR2, the primary VEGFA receptor, is the key molecule for VEGF signalling in tumour angiogenesis [157]. Circ_0001429 has been reported to induce angiogenesis to promote BCa cell growth and metastasis by increasing VEGFA expression [73]. As illustrated by Zhong et al. [69], circRNA_MYLK promotes angiogenesis by increasing the levels of VEGFA and the activity of VEGFR2. Circ_HIPK3 has also been demonstrated to inhibit angiogenesis of BCa cells by sponging miR-558 to reduce heparanase (HPSE) expression [64] (Fig. 4c). In addition, circ_403658, which is induced by HIF-1α, increases the expression of VEGFR and EGFR [158]. In addition to the VEGF family, the fibroblast growth factor (FGF) family is the other best-studied family of angiogenic growth factors. These factors could promote the proliferation, differentiation and migration of endothelial cells during angiogenesis by interacting with their corresponding receptors [159, 160]. According to Mao et al., the hsa_circ_0068871/miR-181a-5p/FGFR3 axis may play a vital role in the progression of BCa [161].
Cisplatin chemoresistance
Although BCa is relatively sensitive to chemotherapy, decreasing cisplatin chemoresistance is a crucial therapeutic strategy for MIBC [162, 163]. RAB27A, a member of the Rab family, plays pivotal roles in multiple processes of tumourigenesis via protein transport and small GTPase-mediated signal transduction [164]. Moreover, upregulation of RAB27A expression promotes proliferation and chemoresistance in BCa [165]. Consistent with these findings, Bi et al. found that circ-BPTF promotes BCa progression by increasing RAB27A expression [166]. APAF-1, a major apoptosis-regulating factor, has also been found to modulate cisplatin sensitivity [167–169]. Notably, Yuan et al. revealed that circ_Cdr1as may increase the cisplatin-induced chemosensitivity of BCa cells through the circ_Cdr1as/miR-1270/APAF1 axis [116]. Hypoxia also enhances resistance to therapy, thus playing critical roles in cancer biology [170]. Furthermore, cancer stem-like cells have been reported to contribute to cisplatin resistance in BCa [171]. Su et al. identified a specific hypoxia-elevated circRNA, circ_ELP3, that promotes cisplatin resistance in BCa by targeting cancer stem-like cells [172]. AR has also been found to mediate cisplatin sensitivity and thereby suppress BCa cell growth [173]. Indeed, AR-mediated circ_FNTA activity decreases cisplatin sensitivity via miR-370-3p/FNTA/KRAS signals [136].
Molecular mechanisms of circRNAs in BCa
CircRNAs perform regulatory roles mainly by acting as miRNA sponges [22], interacting with RBPs [23], and being translated into peptides [24]. Most circRNAs can regulate BCa-related signalling pathways via ceRNA-related regulatory mechanisms. The ceRNA hypothesis specifies that circRNAs can act as molecular sponges that compete with mRNAs for binding to miRNAs, thus inhibiting the activities of the corresponding miRNAs. miRNAs exert their functions through three mechanisms: (1) suppression of translation by binding to the 3’UTRs of target genes, (2) activation of translation by binding to the promoters of target genes, and (3) activation of translation by binding to the 5’UTRs of target genes. For example, circ_ITCH acts as a molecular sponge for miR-224 in BCa; as miR-224 normally inhibits PTEN expression by targeting its 3’UTR, circ_ITCH-mediated sponging ultimately leads to upregulation of PTEN expression in BCa [89]. In addition, circ_HIPK3 sponges miR-558, which normally directly binds to the promoter of the HPSE gene and increases its mRNA expression; thus, circ_HIPK3-mediated sponging in BCa ultimately negatively regulates HPSE expression [64]. Finally, Zheng et al. revealed that circ_NR3C1 directly sponges miR-27a-3p; as miR-27a-3p typically interacts with the cyclin D1 mRNA 5’UTR to facilitate nearby initiating ribosome binding, circ_NR3C1-mediated sponging downregulates cyclin D1 expression [109] (Fig. 4d). Similarly, circ_FNDC3B has been reported to suppress G3BP2 expression by sponging miR-1178-3p, which binds to the 5’UTR of G3BP2 [133]. It should be noted that the miRNA sponge function of circRNAs also depends on the abundance of miRNAs/circRNAs [174] and the number of binding sites for miRNAs contained in each cell [175]. CircRNAs containing many competing binding sites are more likely to have miRNA sponge functions [176, 177]. The most well-characterized circRNA is ciRS-7, which contains more than 70 miR-7 binding sites. It serves as a miR-7 sponge, leading to decreased miR-7 activity and accelerated expression of miR-7-targeted transcripts [178]. Many other circRNAs containing fewer miRNA binding sites can also serve as miRNA sponges. However, their miRNA sponging ability may be limited. Thus, it seems unlikely that all circRNAs can function as miRNA sponges.
Clinical significance of circRNAs in BCa
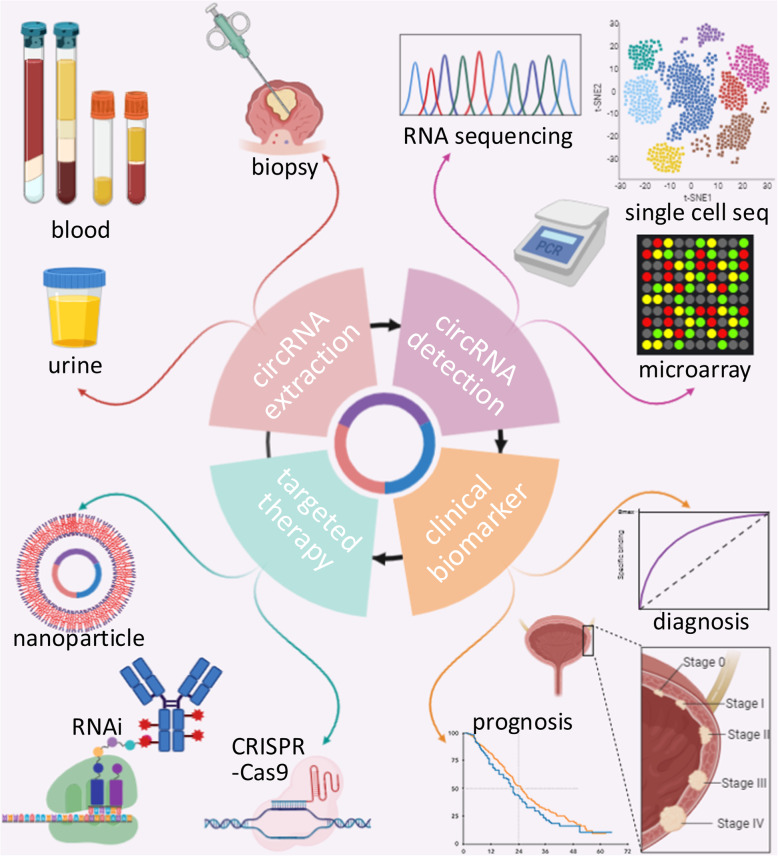
The high incidence and mortality of BCa reflect the need for strategies to improve its early diagnosis, prognosis, and effective treatment. CircRNAs show considerable potential for use as diagnostic and prognostic biomarkers in BCa. First, circRNAs, as unique endogenous noncoding RNAs, are highly conserved and broadly expressed in various tissues, including human BCa and normal bladder tissues [37, 64, 179]. Second, circRNAs are characterized by high stability due to their covalently closed loop structures and by resistance to RNA exonucleases or RNase R [180]. Third, the expression profiles of circRNAs are cell type-specific, tissue-specific, or developmental stage-specific [9, 50]. Finally, apart from solid tissues, BCa-related circRNAs can be detected in blood and urine [81, 97]. RNA-seq [32], circRNA microarrays [50], PCR [51], and Northern blot analysis [57] are widely used methods for circRNA detection and identification. CircRNAs with potential diagnostic, prognostic and predictive value in BCa are summarized in Table 3. As mentioned above, circRNAs play crucial regulatory roles in BCa and are involved in various signalling pathways in BCa, including pathways related to cell proliferation, tumour growth suppression, cell cycle arrest, apoptosis, invasion, migration, metastasis, angiogenesis, and cisplatin chemoresistance. Thus, overexpression or knockdown of related circRNAs might be an effective intervention strategy for BCa progression. RNA interference (RNAi) [181–183], CRISPR/Cas9 editing [55], plasmid transfection [184], and lentiviral vector infection [185] are methods that can be used to decrease or increase circRNA levels. Additionally, nanoparticles can be loaded with exogenous circRNAs and used to carry them for targeted therapy [186] (Fig. 5).
Table 3.
Utility of circRNAs for the clinical management of BCa
| circRNA name | circBase ID | Cilinical Sample | Utility | Ref./PMID | ||
|---|---|---|---|---|---|---|
| Diagnostic | Prognostic | Predictive | ||||
| circRNA-MYLK | hsa_circ_0002768 | tissue | √ | 28687357 | ||
| Circ_0058063 | hsa_circ_0058063 | tissue | √ | 32181485 | ||
| hsa_circ_0076704 | hsa_circ_0076704 | tissue | √ | 31169020 | ||
| hsa_circ_0000144 | hsa_circ_0000144 | tissue | √ | 30098434 | ||
| circUVRAG | hsa_circ_0023642 | tissue | √ | 30387298 | ||
| circ_0071662 | hsa_circ_0071662 | tissue | √ | 31757227 | ||
| circ-ITCH | hsa_circ_0001141 | tissue | √ | 29386015 | ||
| circLPAR1 | hsa_circ_0087960 | tissue | √ | 30867795 | ||
| circPTPRA | hsa_circ_0006117 | tissue | √ | 31821171 | ||
| circUBXN7 | hsa_circ_0001380 | tissue | √ | 30312173 | ||
| ciRs-6 | hsa_circ_0006260 | tissue | √ | 31819015 | ||
| circ_FAM114A2 | hsa_circ_0001546 | tissue | √ | 31969560 | ||
| circ_SLC8A1 | hsa_circ_0000994 | tissue | √ | 31228937 | ||
| circ_0068871 | hsa_circ_0068871 | tissue | √ | 30999937 | ||
| CEP128 | hsa_circ_0102722 | tissue | √ | 30134837 | ||
| circPTK2 | hsa_circ_0003221 | tissue and blood | √ | 29125888 | ||
| circCDYL | hsa_circ_0008285 | tissue | √ | √ |
29263845, 30968727 |
|
| circHIPK3 | hsa_circ_0000284 | tissue | √ | √ | 29263845, 28794202 | |
| circFUT8 | hsa_circ_0003028 | tissue | √ | √ | 32072011 | |
| circRNA_403658 | hsa_circ_0004383 | tissue | √ | √ | 31814891 | |
| circRNA_000285 | hsa_circ_0000285 | tissue and serum | √ | √ | 30509102 | |
| circPICALM | hsa_circ_0023919 | tissue | √ | √ | 31648990 | |
| circ0001361 | hsa_circ_0001361 | tissue | √ | √ | 31705065 | |
| circRIP2 | hsa_circ_0005777 | tissue | √ | √ | 32019579 | |
| cTFRC | has-circ-0001445 | tissue | √ | √ | 30782157 | |
| circ-VANGL1 | hsa_circ_0002623 | tissue | √ | √ | 30146736 | |
| circ5912 | hsa_circ_0005912 | tissue | √ | √ | 31808751 | |
| circFNDC3B | hsa_circ_0006156 | tissue | √ | √ | 30458784 | |
| circ-ZKSCAN1 | hsa_circ_0001727 | tissue | √ | √ | 31481066 | |
| circMTO1 | hsa_circ_0007874 | tissue | √ | √ | 30551873 | |
| circCASC15 | hsa_circ_0075828 | tissue | √ | √ | 31072448 | |
| Circ-BPTF | hsa_circ_0000799 | tissue | √ | √ | 30103209 | |
| circPRMT5 | hsa_circ_0031250 | tissue, serum and urine | √ | √ | 30305293 | |
| hsa circ 0018069 | hsa circ_0018069 | tissue | √ | √ | 30984788 | |
| circZFR | hsa_circ_0072088 | tissue | √ | √ | √ | 31746333 |
| circASXL1 | hsa_circ_0001136 | tissue | √ | √ | √ | 31966702 |
| circ_0137439 | hsa_circ_0137439 | urine | √ | √ | √ | 31777254 |
Fig. 5.
Clinical applications of circRNAs in BCa
Relationships between circRNA levels and clinicopathologic characteristics in BCa
CircRNAs have been reported to be significantly associated with many clinicopathologic characteristics in BCa, including tumour size, grade, differentiation, and stage; lymph node metastasis (LNM); tumour numbers; distant metastasis (DM); invasion; and recurrence. Li et al. observed that circ_0018069 is significantly downregulated in BCa tissues and in T24 and Biu-87 cells and that circ_0018069 levels are correlated with tumour grade, tumour stage, and muscular invasion depth in the context of BCa [80]. Circ_0137439 has also been reported to be significantly upregulated in urine samples from individuals with BCa. Moreover, hsa_circ_0137439 levels are correlated with tumour stage and grade, LNM, and history of MIBC [81]. Furthermore, circASXL1 is evidently upregulated in tissues obtained from BCa patients, and its levels are significantly associated with tumour grade, tumour stage, lymph node invasion, and DM [187]. Similarly, circ_ZFR is significantly upregulated in BCa tissues, and its levels are positively correlated with tumour stage, tumour grade, and LNM [82]. Chi et al. demonstrated that hsa_circ_0000285 levels are significantly reduced in BCa tissues and serum compared to adjacent tissues and serum from healthy controls and that this downregulation is associated with cisplatin resistance, tumour size, differentiation, LNM, DM, and TNM stage [83]. Circ_0001361 has been shown to be overexpressed in BCa tissues and cell lines, and its levels in BCa tissues are correlated with pathologic grade and muscle invasion [124]. Similarly, hsa_circ_0068871 is overexpressed in BCa tissues and cell lines, and its levels in BCa tissues are correlated with T stage and N stage [161]. In contrast, circ_0071662 is downregulated in BCa tissues and cell lines, and its expression levels are significantly associated with LNM and DM [94]. Zhuang et al. revealed that high hsa_circ_0075828 expression in BCa tissues and cells is associated with tumour stage [86]. According to Su and colleagues, circ_5912 is significantly downregulated in BCa tissues compared with normal control tissues, and its levels are correlated with BCa grade, stage, and metastasis [126]. The expression levels of circ_BPTF have been found to be increased in BCa tissues and cell lines compared with noncancerous tissues and cell lines, and high levels of circ_BPTF are positively associated with tumour grade [166]. Sun et al. suggested that circ_CDYL is expressed at low levels in BCa tissues and cell lines and that its expression levels are negatively correlated with BCa pathological stage [107]. In contrast, circ_CEP128 is significantly upregulated in BCa tissues, and its levels correlate positively with tumour size, TNM stage and LNM [74]. Circ_FAM114A2 has been identified to be downregulated in both BUC tissue specimens and cell lines, and high circ_FAM114A2 expression levels are negatively associated with pathological TNM stage and grade [99]. Similarly, circ_FNDC3B is downregulated in BC tissues, and its levels correlate with pathological T stage, grade, and LNM [133]. In addition, other circRNAs, such as circ_FUT8 [127], circ_HIPK3 [64, 68], circ_ITCH [89], circ_MTO1 [151], circ_PICALM [153], circ_PRMT5 [62], circ_PTK2 [97], circ_PTPRA [101], circ_RIP2 [128], hsa_circ_0058063 [76], circ_403658 [158], circ_SLC8A1 [88], circ_TFRC [129], circ_UBXN7 [144], circ_VANGL1 [77], circ_ZKSCAN1 [111], and ciRs_6 [103], are also detectable in BCa tissue or blood and are associated with various clinicopathologic characteristics in BCa (Table 4). Most studies have reported that there is no relationship between circRNA levels and gender in BCa. It should be noted that epidemiological studies show obvious gender differences in the incidence and prognosis of BCa [134]. The aetiology of this gender difference has been linked to sex hormones and their receptors, including estrogen receptor (ER) and AR [188, 189]. Circ_0023642 and circ_FNTA are estrogen receptor- and androgen receptor-mediated circRNAs, respectively. Wu et al. found that estrogen receptor alpha (ERα) decreased circ_0023642 levels and subsequently increased miR-490-5p expression, resulting in decreased EGFR expression to suppress BCa cell invasion [149]. Similarly, Chen et al. reported that the AR-regulated circular RNA circFNTA competes with the microRNA miR-370-3p to increase the expression of its host gene FNTA, which then activates KRAS signalling to promote BCa cell invasion and resistance to cisplatin [136].
Table 4.
Relationship between circRNAs level and clinicopathologic characteristics in BCa
| Study | circRNA Name | CircBase ID | Host gene | Position | Cilinical Sample | Detection method | Cut-off value | Reference gene | Dysregulation | Number of patients | Gender | Age | Tumor size | Grade | DF | Stage | LMN | Number of tumors | DM | Invasion | Recurrence | Potential function | AUC | Sensitivity and specifcity | OS | DFS/RFS | PFS | Ref./PMID |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Li et al. | circ_0018069 | hsa circ_0018069 | KIAA1462 | chr10:30315031–30318795 | tissues | qPCR | 0.0007 | β-actin | downregulated | 41 | No | No | No | Yes | – | Yes | No | – | – | Yes | – | Diagnosis | 0.709 | 97.6, 46.3% | – | – | – | 30984788 |
| Song et al. | circ_0137439 | hsa_circ_0137439 | MTDH | chr8:98673299–98708521 | urine | qPCR | 1.36 | GAPDH | upregulated | 116 | No | No | – | Yes | – | Yes | Yes | – | – | Yes | – | Diagnosis | 0.89 | 87.93, 80.06% | Yes | Yes | – | 31777254 |
| Tang et al. | circ_ASXL1 | hsa_circ_0001136 | ASXL1 | chr20:30954186–30956926 | tissues | qPCR | – | U6 | upregulated | 61 | No | No | No | Yes | – | Yes | Yes | No | Yes | No | – | Diagnosis | 0.77 | 68.6, 76.9% | Yes | – | – | 31966702 |
| Zhang et al. | circ_ZFR | hsa_circ_0072088 | ZFR | chr5:32379220–32388780 | tissues | qPCR | MEL | GAPDH | upregulated | 104 | No | No | No | Yes | – | Yes | Yes | – | – | No | Yes | Diagnosis | 0.8216 | – | Yes | – | Yes | 31746333 |
| Huang et al. | circ_0000144 | hsa_circ_0000144 | SLAMF6 | chr1:160472466–160472794 | tissues | qPCR | – | U6 | upregulated | 69 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | Yes | – | – | 30098434 |
| Chi et al. | circ_000285 | hsa_circ_0000285 | HIPK3 | chr11:33362513–33363232 | tissues, serum | qPCR | MV | β-actin | downregulated | 197 | No | No | Yes | – | Yes | Yes | Yes | – | Yes | – | – | – | – | – | Yes | – | – | 30509102 |
| Liu et al. | circ_0001361 | hsa_circ_0001361 | FNDC3B | chr3:171830241–171851336 | tissues | qPCR | – | GAPDH | upregulated | 69 | No | No | No | Yes | – | Yes | No | – | – | Yes | – | – | – | – | Yes | – | – | 31705065 |
| Mao et al. | circ_0068871 | hsa_circ_0068871 | FGFR3 | chr4:1801473–1804791 | tissues | qPCR | – | GAPDH | upregulated | 32 | No | No | No | – | – | Yes | – | – | No | – | – | – | – | – | – | – | – | 30999937 |
| Abulizi et al. | circ_0071662 | hsa_circ_0071662 | TPPP | chr5:659976–693510 | tissues | qPCR | – | – | downregulated | 158 | No | No | Yes | – | – | Yes | – | – | – | – | – | – | – | – | Yes | – | – | 31757227 |
| Zhuang et al. | circ_CASC15 | hsa_circ_0075828 | CASC15 | chr6:22020567–22020771 | tissues | qPCR | – | GAPDH | upregulated | 67 | No | No | No | – | – | Yes | No | – | – | – | – | – | – | – | Yes | Yes | – | 31072448 |
| Su et al. | circ_5912 | hsa_circ_0005912 | FIP1L1 | chr4:54265896–54294350 | tissues | qPCR | – | GAPDH | downregulated | 58 | No | No | – | Yes | – | Yes | Yes | No | – | – | – | – | – | Yes | – | – | 31808751 | |
| Bi et al. | circ_BPTF | hsa_circ_0000799 | BPTF | chr17:65941524–65972074 | tissues | qPCR | MEL | – | upregulated | 72 | No | No | No | Yes | – | No | No | – | Yes | Yes | – | – | – | Yes | – | – | 30103209 | |
| Sun et al. | circ_CDYL | hsa_circ_0008285 | CDYL | chr6:4891946–4892613 | tissues | qPCR | – | GAPDH | downregulated | 30 | No | No | – | No | – | Yes | No | – | No | – | – | – | – | – | – | 30968727 | ||
| Okholm et al. | circ_CDYL | hsa_circ_0008285 | CDYL | chr6:4891946–4892613 | tissues | qPCR | MEL | – | downregulated | 457 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | Yes | 29263845 |
| Wu et al. | circ_CEP128 | hsa_circ_0102722 | CEP128 | chr14:81209418–81304622 | tissues | qPCR | – | GAPDH | upregulated | 10 | No | No | Yes | – | – | Yes | Yes | – | – | – | – | – | – | – | – | – | 30134837 | |
| Liu et al. | circ_FAM114A2 | hsa_circ_0001546 | FAM114A2 | chr5:153413351–153414527 | tissues | qPCR | NAT | GAPDH | downregulated | 31 | No | No | No | Yes | – | Yes | No | – | – | – | – | – | – | – | – | – | 31969560 | |
| Liu et al. | circ_FNDC3B | hsa_circ_0006156 | FNDC3B | chr3:171965322–171969331 | tissues | qPCR | – | GAPDH | downregulated | 82 | No | No | No | Yes | – | Yes | Yes | – | – | No | No | – | – | – | Yes | – | – | 30458784 |
| He et al. | circ_FUT8 | hsa_circ_0003028 | FUT8 | chr14:66028054–66028484 | tissues | qPCR | – | GAPDH | downregulated | 145 | No | No | No | Yes | – | No | Yes | No | – | – | – | – | – | – | Yes | – | – | 32072011 |
| Li et al. | circ_HIPK3 | hsa_circ_0000284 | HIPK3 | chr11:33307958–33309057 | tissues | qPCR | – | GAPDH | downregulated | 44 | No | No | – | Yes | – | Yes | Yes | – | – | Yes | – | – | – | – | – | – | – | 28794202 |
| Okholm et al. | circ_HIPK3 | hsa_circ_0000284 | HIPK3 | chr11:33307958–33309057 | tissues | qPCR | MEL | – | downregulated | 457 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | Yes | 29263845 |
| Yang et al. | circ_ITCH | hsa_circ_0001141 | ITCH | chr20:33001547–33037285 | tissues | qPCR | – | β-actin | downregulated | 70 | No | No | No | Yes | – | No | – | – | – | – | – | – | – | – | Yes | – | – | 29386015 |
| Lin et al. | circ_LPAR1 | hsa_circ_0087960 | LPAR1 | chr9:113734352–113735838 | tissues | qPCR | – | β-actin | upregulated | 125 | No | No | No | No | – | No | – | No | – | – | No | – | – | – | – | – | – | 30867795 |
| Li et al. | circ_MTO1 | hsa_circ_0007874 | MTO1 | chr6:74175931–74176329 | tissues | qPCR | – | β-actin | downregulated | 117 | – | – | – | – | – | – | Yes | – | – | – | – | – | – | – | Yes | Yes | – | 30551873 |
| Yan et al. | circ_PICALM | hsa_circ_0023919 | PICALM | chr11:85707868–85714494 | tissues | qPCR | – | GAPDH | downregulated | 168 | No | No | No | Yes | – | Yes | Yes | – | – | – | – | – | – | – | Yes | – | – | 31648990 |
| Chen et al. | circ_PRMT5 | hsa_circ_0031250 | PRMT5 | chr14:23395341–23396023 | tissues | qPCR | – | GAPDH | upregulated | 119 | No | No | – | No | – | Yes | – | – | – | – | – | – | – | – | – | Yes | – | 30305293 |
| Xu et al. | circ_PTK2 | hsa_circ_0003221 | PTK2 | chr8:141856358–141900868 | tissue, blood | qPCR | – | GAPDH | upregulated | 40 | – | – | – | – | Yes | Yes | Yes | – | – | – | – | – | – | – | – | – | – | 29125888 |
| He et al. | circ_PTPRA | hsa_circ_0006117 | PTPRA | chr20:2944917–2945848 | tissues | qPCR | – | GAPDH | downregulated | 104 | No | No | Yes | No | – | Yes | No | No | – | – | – | – | – | – | Yes | – | – | 31821171 |
| Su et al. | circ_RIP2 | hsa_circ_0005777 | RGNEF | chr5:73136304–73136585 | tissues | qPCR | – | GAPDH | downregulated | 58 | No | No | – | Yes | – | Yes | Yes | No | – | – | – | – | – | – | Yes | – | – | 32019579 |
| Liang et al. | circ_0058063 | hsa_circ_0058063 | ATIC | chr2:216177220–216213972 | tissues | qPCR | – | GAPDH | upregulated | 94 | No | No | – | Yes | – | Yes | – | No | – | – | – | – | – | – | Yes | – | – | 32181485 |
| Wei et al. | circ_403658 | hsa_circ_0004383 | ZNF292 | chr6:87920168–87928449 | tissues | qPCR | – | GAPDH | upregulated | 123 | No | No | Yes | – | – | Yes | No | – | Yes | – | – | – | – | – | Yes | – | – | 31814891 |
| Lu et al. | circ_SLC8A1 | hsa_circ_0000994 | SLC8A1 | chr2:40655612–40657444 | tissues | qPCR | – | GAPDH | downregulated | 70 | No | No | – | Yes | – | Yes | No | – | – | – | – | – | – | – | – | – | – | 31228937 |
| Su et al. | circ_TFRC | has_circ_0001445 | TFRC | chr3:195785154–195,787,118 | tissues | qPCR | – | GAPDH | upregulated | 220 | No | No | – | Yes | – | Yes | Yes | No | – | – | – | – | – | – | Yes | – | – | 30782157 |
| Liu et al. | circ_UBXN7 | hsa_circ_0001380 | UBXN7 | chr3:196118683–196129890 | tissues | qPCR | – | GAPDH | downregulated | 84 | No | No | No | Yes | – | Yes | No | – | – | No | – | – | – | – | Yes | – | – | 30312173 |
| Zeng et al. | circ_VANGL1 | hsa_circ_0002623 | VANGL1 | chr1:116202261–116206889 | tissues | qPCR | – | GAPDH | upregulated | 43 | No | No | – | Yes | – | Yes | Yes | – | – | – | – | – | – | – | Yes | – | – | 30146736 |
| Bi et al. | circ_ZKSCAN1 | hsa_circ_0001727 | ZKSCAN1 | chr7:99621041–99621930 | tissues | qPCR | – | GAPDH | downregulated | 137 | No | No | No | Yes | – | Yes | Yes | – | – | – | Yes | – | – | – | Yes | Yes | – | 31481066 |
| Su et al. | ciRs_6 | hsa_circ_0006260 | SLC41A2 | chr12:105321750–105322472 | tissues | qPCR | – | GAPDH | downregulated | 58 | No | No | – | Yes | – | Yes | No | No | – | – | – | – | – | – | Yes | – | – | 31819015 |
| Liu et al. | circ_0076704 | hsa_circ_0076704 | CD2AP | chr6:47471015–47522502 | tissues | qPCR | – | β-actin | upregulated | 70 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | Yes | – | – | 31169020 |
| Yang et al. | circ_UVRAG | hsa_circ_0023642 | UVRAG | chr11:75727858–75728024 | tissues | qPCR | – | – | upregulated | 402 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | NO | – | – | 30387298 |
| Zhong et al. | circ_MYLK | hsa_circ_0002768 | MYLK | chr3:123471177–123512691 | tissues | qPCR | – | – | upregulated | 32 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | Yes | – | 28687357 |
circRNAs circular RNAs, BCa bladder cancer, MV mean value, MEL median expression level, LNM lymph nodes metastasis, NAT adjacent noncancerous tissues, DF differentiation, LNM lymph node metastasis, DM distant metastasis, AUC area under the curve, OS over survival, DFS disease-free survival, RFS recurrence-free survival, PFS progression-free survival
CircRNAs as diagnostic biomarkers for BCa
The clinical value of circRNAs as diagnostic biomarkers has been explored in many studies. The area under the receiver operating characteristic (ROC) curve (AUC) of circ_0018069 for BCa diagnosis is 0.709, and the sensitivity and specificity are 97.6 and 46.3%, respectively [80]. The AUC of a ROC curve generated for urinary cell-free hsa_circ_0137439 levels is 0.890, with a sensitivity and specificity of 87.93 and 80.06%, respectively [81]. The AUC for circASXL1 in tumour invasion (T2-T4 tumour) diagnosis is 0.770, with a sensitivity and specificity of 68.6 and 76.9%, respectively [187]. The AUC for circ_ZFR in BCa diagnosis is 0.8216 [82] (Table 4).
CircRNAs as prognostic biomarkers for BCa
CircRNA levels can also be used to predict patient survival parameters, such as overall survival (OS), disease-free survival (DFS), and progression-free survival (PFS). To further analyse the prognostic value of circRNAs in BCa, we collected information from studies reporting survival information and evaluated the associations between circRNA expression levels and OS, DFS, and PFS. Fourteen upregulated circRNAs were reported to predict poor OS [69–71, 76, 77, 81, 82, 86, 124, 129, 150, 158, 166, 187], while thirteen downregulated circRNAs were reported to predict poor OS [83, 89, 94, 101, 103, 111, 126–128, 133, 144, 151, 153]. Kaplan-Meier survival analysis indicated that higher expression of circ_0137439, circ_CASC15, and circPRMT5 was associated with poorer DFS [62, 81, 86]. Two studies revealed that higher expression of circ_ZKSCAN1 and circ_MTO1 was associated with longer DFS [111, 151]. A study by Zhang et al. revealed a significantly elevated risk of progression for patients with high circ_ZFR expression levels [82]. In addition, patients with high circ_CDYL and circ_HIPK3 expression were reported to have a reduced risk of progression [68].
Conclusion
Over the past 10 years, the importance of elucidating circRNA biology to our understanding of tumorigenesis has become evident. As outlined in this review, considerable evidence indicates that circRNAs play key roles in BCa. To date, fifty-five circRNAs among hundreds of aberrantly expressed circRNAs have been identified to be specifically associated with BCa. Notably, BCa-related circRNAs have been discovered to regulate cancer-related biological behaviours via ceRNA regulatory mechanisms. Existing reports feature methodologies and study designs that others can use for further investigation of circRNAs of interest. CircRNAs have been reported to be significantly associated with many clinicopathologic characteristics of BCa and with BCa patient survival parameters, and the abundance, conservation, stability, specificity and detectability of circRNAs render them potential diagnostic and prognostic biomarkers for BCa. Additionally, circRNAs play crucial regulatory roles upstream of various signalling pathways related to BCa carcinogenesis and progression, reflecting their potential as therapeutic targets for BCa.
Some limitations of previous research on circRNAs in BCa should be noted. First, the biogenesis of circRNAs and the regulatory mechanisms involved in circularization remain vague. More research is needed to help us understand the circRNA circularization processes in depth. Second, no unified standards are available to determine thresholds for circRNA detection. Third, previous studies on circRNAs in BCa lacked circRNAs with BCa specificity. More circRNAs with relative bladder cancer specificity may be further characterized in future studies. Fourth, almost all reported circRNAs in BCa exert functions via miRNA sponge mechanisms. The other three classical mechanisms, including sponging of RBPs, regulation of transcription and translation into peptides or proteins, have rarely been studied in BCa. Fifth, all circRNAs reported in BCa are currently in the basic research stage. Further investigation of circRNAs as diagnostic biomarkers, prognostic biomarkers, or targeted therapy for BCa in well-designed multicentre cohort studies is necessary.
Supplementary Information
Additional file 1: Table S1. Database for circRNA research
Acknowledgements
We thank all other researchers in our laboratory.
Abbreviations
- circRNAs
Circular RNAs
- BCa
Bladder cancer
- OS
Overall survival
- DFS
Disease-free survival
- PFS
Progression-free survival
- ecRNAs
Exonic circRNAs
- elciRNAs
Exon-intron circRNAs
- ciRNAs
Intronic circRNAs
- miRNA
microRNA
- RBPs
RNA-binding proteins
- BUC
Bladder urothelial carcinoma
- MIBC
Muscle-invasive bladder cancer
- NMIBC
Non-muscle-invasive bladder cancer
- QKI
Quaking
- MBL
Muscleblind
- ADAR1
Adenosine deaminase acting on RNA
- Cul2
Cullin2
- HuR
Human antigen R/ELAV-like protein 1
- IRESs
Internal ribosome entry sites
- BSJs
Back-splice junctions
- FISH
Fluorescence in situ hybridization
- RNA-seq
RNA sequencing
- RT-PCR
Reverse transcription-polymerase chain reaction
- GEO
Gene Expression Omnibus
- FDR
False discovery rate
- PTEN
Phosphatase and tensin homologue
- KLF9
Krüppel-like factor 9
- APAF-1
Apoptosis protease-activating factor-1
- FOXO3
Forkhead box transcription factor class O3
- EMT
Epithelial-mesenchymal transition
- G3BP
Ras-GTPase-activating protein SH3 domain-binding protein
- AR
Androgen receptor
- CSCs
Cancer stem cells
- IGFBP2
Insulin-like growth factor-binding protein 2
- VEGF
Vascular endothelial growth factor
- HPSE
Heparanase
- FGF
Fibroblast growth factor
- RNAi
RNA interference
- LNM
Lymph node metastasis
- DM
Distant metastasis
- ER
Estrogen receptor
- ERα
Estrogen receptor alpha
- ROC
Receiver operating characteristic
- AUC
Area under the receiver operating characteristic curve
Authors’ contributions
XY, TY, HL, PL, CD, XW, KJ and HL collected the related papers. KT, XY, and ZY contributed to the conception and design of the review. KT and XY contributed to all tables and figures and drafted the manuscript. DX, EP, ZC, and ZY revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81900645).
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors agreed to publish this manuscript.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73:3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cocquerelle C, Mascrez B, Hetuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7:155–160. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- 4.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 6.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 7.Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18:5–18. doi: 10.1038/nrc.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patop IL, Wust S, Kadener S. Past, present, and future of circRNAs. EMBO J. 2019;38:e100836. doi: 10.15252/embj.2018100836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen X, Fan S, Song E. Noncoding RNAs: new players in cancers. Adv Exp Med Biol. 2016;927:1–47. doi: 10.1007/978-981-10-1498-7_1. [DOI] [PubMed] [Google Scholar]
- 12.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 13.Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, Zhou LY, Sun T, Wang M, Yu T, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. 2016;37:2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 14.Stoll L, Sobel J, Rodriguez-Trejo A, Guay C, Lee K, Veno MT, Kjems J, Laybutt DR, Regazzi R. Circular RNAs as novel regulators of beta-cell functions in normal and disease conditions. Mol Metab. 2018;9:69–83. doi: 10.1016/j.molmet.2018.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akhter R. Circular RNA and Alzheimer's Disease. Adv Exp Med Biol. 2018;1087:239–243. doi: 10.1007/978-981-13-1426-1_19. [DOI] [PubMed] [Google Scholar]
- 16.Geng Y, Jiang J, Wu C. Function and clinical significance of circRNAs in solid tumors. J Hematol Oncol. 2018;11:98. doi: 10.1186/s13045-018-0643-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu W, Bi ZY, Chen ZL, Liu C, Li LL, Zhang F, Zhou Q, Zhu W, Song YY, Zhan BT, et al. Emerging landscape of circular RNAs in lung cancer. Cancer Lett. 2018;427:18–27. doi: 10.1016/j.canlet.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 18.Sun J, Li B, Shu C, Ma Q, Wang J. Functions and clinical significance of circular RNAs in glioma. Mol Cancer. 2020;19:34. doi: 10.1186/s12943-019-1121-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shan C, Zhang Y, Hao X, Gao J, Chen X, Wang K. Biogenesis, functions and clinical significance of circRNAs in gastric cancer. Mol Cancer. 2019;18:136. doi: 10.1186/s12943-019-1069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou R, Wu Y, Wang W, Su W, Liu Y, Wang Y, Fan C, Li X, Li G, Li Y, et al. Circular RNAs (circRNAs) in cancer. Cancer Lett. 2018;425:134–142. doi: 10.1016/j.canlet.2018.03.035. [DOI] [PubMed] [Google Scholar]
- 21.Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, Lo-Coco F, Tay Y, Beck AH, Pandolfi PP. Oncogenic role of fusion-circRNAs derived from Cancer-associated chromosomal translocations. Cell. 2016;166:1055–1056. doi: 10.1016/j.cell.2016.07.035. [DOI] [PubMed] [Google Scholar]
- 22.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 23.Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 24.Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in Myogenesis. Mol Cell. 2017;66:22–37. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamat AM, Hahn NM, Efstathiou JA, Lerner SP, Malmstrom PU, Choi W, Guo CC, Lotan Y, Kassouf W. Bladder cancer. Lancet. 2016;388:2796–2810. doi: 10.1016/S0140-6736(16)30512-8. [DOI] [PubMed] [Google Scholar]
- 26.Fleshner NE, Herr HW, Stewart AK, Murphy GP, Mettlin C, Menck HR. The National Cancer Data Base report on bladder carcinoma. The American College of Surgeons Commission on Cancer and the American Cancer Society. Cancer. 1996;78:1505–1513. doi: 10.1002/(SICI)1097-0142(19961001)78:7<1505::AID-CNCR19>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 27.Kirkali Z, Chan T, Manoharan M, Algaba F, Busch C, Cheng L, Kiemeney L, Kriegmair M, Montironi R, Murphy WM, et al. Bladder cancer: epidemiology, staging and grading, and diagnosis. Urology. 2005;66:4–34. doi: 10.1016/j.urology.2005.07.062. [DOI] [PubMed] [Google Scholar]
- 28.Lobo N, Mount C, Omar K, Nair R, Thurairaja R, Khan MS. Landmarks in the treatment of muscle-invasive bladder cancer. Nat Rev Urol. 2017;14:565–574. doi: 10.1038/nrurol.2017.82. [DOI] [PubMed] [Google Scholar]
- 29.Felsenstein KM, Theodorescu D. Precision medicine for urothelial bladder cancer: update on tumour genomics and immunotherapy. Nat Rev Urol. 2018;15:92–111. doi: 10.1038/nrurol.2017.179. [DOI] [PubMed] [Google Scholar]
- 30.Nadal R, Bellmunt J. Management of metastatic bladder cancer. Cancer Treat Rev. 2019;76:10–21. doi: 10.1016/j.ctrv.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 31.Alifrangis C, McGovern U, Freeman A, Powles T, Linch M. Molecular and histopathology directed therapy for advanced bladder cancer. Nat Rev Urol. 2019;16:465–483. doi: 10.1038/s41585-019-0208-0. [DOI] [PubMed] [Google Scholar]
- 32.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 34.Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 35.Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M, Dieterich C, Rajewsky N. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10:170–177. doi: 10.1016/j.celrep.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 36.Meng J, Chen S, Han JX, Qian B, Wang XR, Zhong WL, Qin Y, Zhang H, Gao WF, Lei YY, et al. Twist1 regulates Vimentin through Cul2 circular RNA to promote EMT in hepatocellular carcinoma. Cancer Res. 2018;78:4150–4162. doi: 10.1158/0008-5472.CAN-17-3009. [DOI] [PubMed] [Google Scholar]
- 37.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 38.Aktas T, Avsar Ilik I, Maticzka D, Bhardwaj V, Pessoa Rodrigues C, Mittler G, Manke T, Backofen R, Akhtar A. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature. 2017;544:115–119. doi: 10.1038/nature21715. [DOI] [PubMed] [Google Scholar]
- 39.Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 40.Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609–5612. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 41.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S, Kim J, Noh JH, Kim KM, Martindale JL, Gorospe M. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14:361–369. doi: 10.1080/15476286.2017.1279788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilusz JE. A 360 degrees view of circular RNAs: from biogenesis to functions. Wiley Interdiscip Rev RNA. 2018;9:e1478. doi: 10.1002/wrna.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wesselhoeft RA, Kowalski PS, Anderson DG. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat Commun. 2018;9:2629. doi: 10.1038/s41467-018-05096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 1995;268:415–417. doi: 10.1126/science.7536344. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015;21:172–179. doi: 10.1261/rna.048272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, Huang N, Yang X, Zhao K, Zhou H, et al. Novel role of FBXW7 circular RNA in repressing Glioma tumorigenesis. J Natl Cancer Inst. 2018;110:304–15. [DOI] [PMC free article] [PubMed]
- 48.Zhang XO, Dong R, Zhang Y, Zhang JL, Luo Z, Zhang J, Chen LL, Yang L. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016;26:1277–1287. doi: 10.1101/gr.202895.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71:428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 50.Li S, Teng S, Xu J, Su G, Zhang Y, Zhao J, Zhang S, Wang H, Qin W, Lu ZJ, et al. Microarray is an efficient tool for circRNA profiling. Brief Bioinform. 2019;20:1420–1433. doi: 10.1093/bib/bby006. [DOI] [PubMed] [Google Scholar]
- 51.Panda AC, Gorospe M. Detection and analysis of circular RNAs by RT-PCR. Bio Protoc. 2018;8:e2775. [DOI] [PMC free article] [PubMed]
- 52.Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233–2247. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, Kjems J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DBT, Kellner MJ, Regev A, et al. RNA targeting with CRISPR-Cas13. Nature. 2017;550:280–284. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Piwecka M, Glazar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Cerda Jara CA, Fenske P, et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017;357:eaam8526. [DOI] [PubMed]
- 56.Zhang Y, Xue W, Li X, Zhang J, Chen S, Zhang JL, Yang L, Chen LL. The biogenesis of nascent circular RNAs. Cell Rep. 2016;15:611–624. doi: 10.1016/j.celrep.2016.03.058. [DOI] [PubMed] [Google Scholar]
- 57.Schneider T, Schreiner S, Preusser C, Bindereif A, Rossbach O. Northern blot analysis of circular RNAs. Methods Mol Biol. 1724;2018:119–133. doi: 10.1007/978-1-4939-7562-4_10. [DOI] [PubMed] [Google Scholar]
- 58.Zirkel A, Papantonis A. Detecting circular RNAs by RNA fluorescence in situ hybridization. Methods Mol Biol. 1724;2018:69–75. doi: 10.1007/978-1-4939-7562-4_6. [DOI] [PubMed] [Google Scholar]
- 59.Glazar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20:1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao Z, Wang K, Wu F, Wang W, Zhang K, Hu H, Liu Y, Jiang T. circRNA disease: a manually curated database of experimentally supported circRNA-disease associations. Cell Death Dis. 2018;9:475. doi: 10.1038/s41419-018-0503-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ghosal S, Das S, Sen R, Basak P, Chakrabarti J. Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front Genet. 2013;4:283. doi: 10.3389/fgene.2013.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen X, Chen RX, Wei WS, Li YH, Feng ZH, Tan L, Chen JW, Yuan GJ, Chen SL, Guo SJ, et al. PRMT5 circular RNA promotes metastasis of Urothelial carcinoma of the bladder through sponging miR-30c to induce epithelial-Mesenchymal transition. Clin Cancer Res. 2018;24:6319–6330. doi: 10.1158/1078-0432.CCR-18-1270. [DOI] [PubMed] [Google Scholar]
- 63.Zhong Z, Lv M, Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci Rep. 2016;6:30919. doi: 10.1038/srep30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang C, Liu D, Wang M, Wang L, Zeng F, Jiang G. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017;18:1646–1659. doi: 10.15252/embr.201643581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li W, Li Y, Sun Z, Zhou J, Cao Y, Ma W, Xie K, Yan X. Comprehensive circular RNA profiling reveals the regulatory role of the hsa_circ_0137606/miR1231 pathway in bladder cancer progression. Int J Mol Med. 2019;44:1719–1728. doi: 10.3892/ijmm.2019.4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Szabo L, Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet. 2016;17:679–692. doi: 10.1038/nrg.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Okholm TLH, Nielsen MM, Hamilton MP, Christensen LL, Vang S, Hedegaard J, Hansen TB, Kjems J, Dyrskjot L, Pedersen JS. Circular RNA expression is abundant and correlated to aggressiveness in early-stage bladder cancer. NPJ Genom Med. 2017;2:36. doi: 10.1038/s41525-017-0038-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhong Z, Huang M, Lv M, He Y, Duan C, Zhang L, Chen J. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–317. doi: 10.1016/j.canlet.2017.06.027. [DOI] [PubMed] [Google Scholar]
- 70.Liu L, Wu SQ, Zhu X, Xu R, Ai K, Zhang L, Zhao XK. Analysis of ceRNA network identifies prognostic circRNA biomarkers in bladder cancer. Neoplasma. 2019;66:736–745. doi: 10.4149/neo_2019_190107N25. [DOI] [PubMed] [Google Scholar]
- 71.Huang W, Lu Y, Wang F, Huang X, Yu Z. Downregulation of circular RNA hsa_circ_0000144 inhibits bladder cancer progression via stimulating miR-217 and suppressing RUNX2 expression. Gene. 2018;678:337–342. doi: 10.1016/j.gene.2018.08.036. [DOI] [PubMed] [Google Scholar]
- 72.Sun M, Zhao W, Chen Z, Li M, Li S, Wu B, Bu R. Circ_0058063 regulates CDK6 to promote bladder cancer progression by sponging miR-145-5p. J Cell Physiol. 2019;234:4812–4824. doi: 10.1002/jcp.27280. [DOI] [PubMed] [Google Scholar]
- 73.Cao W, Zhao Y, Wang L, Huang X. Circ0001429 regulates progression of bladder cancer through binding miR-205-5p and promoting VEGFA expression. Cancer Biomark. 2019;25:101–113. doi: 10.3233/CBM-182380. [DOI] [PubMed] [Google Scholar]
- 74.Wu Z, Huang W, Wang X, Wang T, Chen Y, Chen B, Liu R, Bai P, Xing J. Circular RNA CEP128 acts as a sponge of miR-145-5p in promoting the bladder cancer progression via regulating SOX11. Mol Med. 2018;24:40. doi: 10.1186/s10020-018-0039-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sun M, Zhao W, Chen Z, Li M, Li S, Wu B, Bu R. Circular RNA CEP128 promotes bladder cancer progression by regulating Mir-145-5p/Myd88 via MAPK signaling pathway. Int J Cancer. 2019;145:2170–2181. doi: 10.1002/ijc.32311. [DOI] [PubMed] [Google Scholar]
- 76.Liang H, Huang H, Li Y, Lu Y, Ye T. CircRNA_0058063 functions as a ceRNA in bladder cancer progression via targeting miR-486-3p/FOXP4 axis. Biosci Rep. 2020;40:BSR20193484. [DOI] [PMC free article] [PubMed]
- 77.Zeng Z, Zhou W, Duan L, Zhang J, Lu X, Jin L, Yu Y. Circular RNA circ-VANGL1 as a competing endogenous RNA contributes to bladder cancer progression by regulating miR-605-3p/VANGL1 pathway. J Cell Physiol. 2019;234:3887–3896. doi: 10.1002/jcp.27162. [DOI] [PubMed] [Google Scholar]
- 78.Huang M, Zhong Z, Lv M, Shu J, Tian Q, Chen J. Comprehensive analysis of differentially expressed profiles of lncRNAs and circRNAs with associated co-expression and ceRNA networks in bladder carcinoma. Oncotarget. 2016;7:47186–47200. doi: 10.18632/oncotarget.9706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lu HC, Yao JQ, Yang X, Han J, Wang JZ, Xu K, Zhou R, Yu H, Lv Q, Gu M. Identification of a potentially functional circRNA-miRNA-mRNA regulatory network for investigating pathogenesis and providing possible biomarkers of bladder cancer. Cancer Cell Int. 2020;20:31. doi: 10.1186/s12935-020-1108-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li M, Wang Y, Liu Y, Zhang X, Liu J, Wang P. Low expression of hsa_circ_0018069 in human bladder Cancer and its clinical significance. Biomed Res Int. 2019;2019:9681863. doi: 10.1155/2019/9681863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Song Z, Zhang Q, Zhu J, Yin G, Lin L, Liang C. Identification of urinary hsa_circ _0137439 as potential biomarker and tumor regulator of bladder cancer. Neoplasma. 2020;67:137–146. doi: 10.4149/neo_2018_181214N970. [DOI] [PubMed] [Google Scholar]
- 82.Zhang WY, Liu QH, Wang TJ, Zhao J, Cheng XH, Wang JS. CircZFR serves as a prognostic marker to promote bladder cancer progression by regulating miR-377/ZEB2 signaling. Biosci Rep. 2019;39:BSR20192779. [DOI] [PMC free article] [PubMed]
- 83.Chi BJ, Zhao DM, Liu L, Yin XZ, Wang FF, Bi S, Gui SL, Zhou SB, Qin WB, Wu DM, Wang SQ. Downregulation of hsa_circ_0000285 serves as a prognostic biomarker for bladder cancer and is involved in cisplatin resistance. Neoplasma. 2019;66:197–202. doi: 10.4149/neo_2018_180318N185. [DOI] [PubMed] [Google Scholar]
- 84.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 85.Yu M, Li W, Luo S, Zhang Y, Liu H, Gao Y, Wang X, Wilson JX, Huang G. Folic acid stimulation of neural stem cell proliferation is associated with altered methylation profile of PI3K/Akt/CREB. J Nutr Biochem. 2014;25:496–502. doi: 10.1016/j.jnutbio.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 86.Zhuang C, Huang X, Yu J, Gui Y. Circular RNA hsa_circ_0075828 promotes bladder cancer cell proliferation through activation of CREB1. BMB Rep. 2020;53:82–87. doi: 10.5483/BMBRep.2020.53.2.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Morgensztern D, McLeod HL. PI3K/Akt/mTOR pathway as a target for cancer therapy. Anti-Cancer Drugs. 2005;16:797–803. doi: 10.1097/01.cad.0000173476.67239.3b. [DOI] [PubMed] [Google Scholar]
- 88.Lu Q, Liu T, Feng H, Yang R, Zhao X, Chen W, Jiang B, Qin H, Guo X, Liu M, et al. Circular RNA circSLC8A1 acts as a sponge of miR-130b/miR-494 in suppressing bladder cancer progression via regulating PTEN. Mol Cancer. 2019;18:111. doi: 10.1186/s12943-019-1040-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang C, Yuan W, Yang X, Li P, Wang J, Han J, Tao J, Li P, Yang H, Lv Q, Zhang W. Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol Cancer. 2018;17:19. doi: 10.1186/s12943-018-0771-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wan L, Zhang L, Fan K, Cheng ZX, Sun QC, Wang JJ. Circular RNA-ITCH suppresses lung Cancer proliferation via inhibiting the Wnt/beta-catenin pathway. Biomed Res Int. 2016;2016:1579490. doi: 10.1155/2016/1579490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huang G, Zhu H, Shi Y, Wu W, Cai H, Chen X. cir-ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/beta-catenin pathway. PLoS One. 2015;10:e0131225. doi: 10.1371/journal.pone.0131225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li F, Zhang L, Li W, Deng J, Zheng J, An M, Lu J, Zhou Y. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget. 2015;6:6001–6013. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhao X, Ji Z, Xie Y, Liu G, Li H. MicroRNA-154 as a prognostic factor in bladder cancer inhibits cellular malignancy by targeting RSF1 and RUNX2. Oncol Rep. 2017;38:2727–2734. doi: 10.3892/or.2017.5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Abulizi R, Li B, Zhang CG. Circ_0071662, a novel tumor biomarker, suppresses bladder cancer cell proliferation and invasion by sponging miR-146b-3p. Oncol Res. 2019. 10.3727/096504019X15740729375088. [DOI] [PubMed]
- 95.Zhang L, Xia HB, Zhao CY, Shi L, Ren XL. Cyclic RNA hsa_circ_0091017 inhibits proliferation, migration and invasiveness of bladder cancer cells by binding to microRNA-589-5p. Eur Rev Med Pharmacol Sci. 2020;24:86–96. doi: 10.26355/eurrev_202001_19898. [DOI] [PubMed] [Google Scholar]
- 96.Yu Q, Liu P, Han G, Xue X, Ma D. CircRNA circPDSS1 promotes bladder cancer by down-regulating miR-16. Biosci Rep. 2020;40:BSR20191961. [DOI] [PMC free article] [PubMed] [Retracted]
- 97.Xu ZQ, Yang MG, Liu HJ, Su CQ. Circular RNA hsa_circ_0003221 (circPTK2) promotes the proliferation and migration of bladder cancer cells. J Cell Biochem. 2018;119:3317–3325. doi: 10.1002/jcb.26492. [DOI] [PubMed] [Google Scholar]
- 98.Koga F, Kawakami S, Fujii Y, Saito K, Ohtsuka Y, Iwai A, Ando N, Takizawa T, Kageyama Y, Kihara K. Impaired p63 expression associates with poor prognosis and uroplakin III expression in invasive urothelial carcinoma of the bladder. Clin Cancer Res. 2003;9:5501–5507. [PubMed] [Google Scholar]
- 99.Liu T, Lu Q, Liu J, Xie S, Feng B, Zhu W, Liu M, Liu Y, Zhou X, Sun W, et al. Circular RNA FAM114A2 suppresses progression of bladder cancer via regulating NP63 by sponging miR-762. Cell Death Dis. 2020;11:47. doi: 10.1038/s41419-020-2226-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sun J, Wang B, Liu Y, Zhang L, Ma A, Yang Z, Ji Y, Liu Y. Transcription factor KLF9 suppresses the growth of hepatocellular carcinoma cells in vivo and positively regulates p53 expression. Cancer Lett. 2014;355:25–33. doi: 10.1016/j.canlet.2014.09.022. [DOI] [PubMed] [Google Scholar]
- 101.He Q, Huang L, Yan D, Bi J, Yang M, Huang J, Lin T. CircPTPRA acts as a tumor suppressor in bladder cancer by sponging miR-636 and upregulating KLF9. Aging (Albany NY) 2019;11:11314–11328. doi: 10.18632/aging.102530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xie F, Li Y, Wang M, Huang C, Tao D, Zheng F, Zhang H, Zeng F, Xiao X, Jiang G. Circular RNA BCRC-3 suppresses bladder cancer proliferation through miR-182-5p/p27 axis. Mol Cancer. 2018;17:144. doi: 10.1186/s12943-018-0892-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Su Y, Feng W, Zhong G, Ya Y, Du Z, Shi J, Chen L, Dong W, Lin T. ciRs-6 upregulates March1 to suppress bladder cancer growth by sponging miR-653. Aging (Albany NY). 2019;11:11202–23:BSR20181436. [DOI] [PMC free article] [PubMed]
- 104.Wang S, Zhang G, Zheng W, Xue Q, Wei D, Zheng Y, Yuan J. MiR-454-3p and miR-374b-5p suppress migration and invasion of bladder cancer cells through targetting ZEB2. Biosci Rep. 2018;38:BSR20181436. [DOI] [PMC free article] [PubMed]
- 105.Liu Q, Liu H, Cheng H, Li Y, Li X, Zhu C. Downregulation of long noncoding RNA TUG1 inhibits proliferation and induces apoptosis through the TUG1/miR-142/ZEB2 axis in bladder cancer cells. Onco Targets Ther. 2017;10:2461–2471. doi: 10.2147/OTT.S124595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kress TR, Sabo A, Amati B. MYC: connecting selective transcriptional control to global RNA production. Nat Rev Cancer. 2015;15:593–607. doi: 10.1038/nrc3984. [DOI] [PubMed] [Google Scholar]
- 107.Sun J, Zhang H, Tao D, Xie F, Liu F, Gu C, Wang M, Wang L, Jiang G, Wang Z, Xiao X. CircCDYL inhibits the expression of C-MYC to suppress cell growth and migration in bladder cancer. Artif Cells Nanomed Biotechnol. 2019;47:1349–1356. doi: 10.1080/21691401.2019.1596941. [DOI] [PubMed] [Google Scholar]
- 108.MacLachlan TK, Sang N, Giordano A. Cyclins, cyclin-dependent kinases and cdk inhibitors: implications in cell cycle control and cancer. Crit Rev Eukaryot Gene Expr. 1995;5:127–156. doi: 10.1615/CritRevEukarGeneExpr.v5.i2.20. [DOI] [PubMed] [Google Scholar]
- 109.Zheng F, Wang M, Li Y, Huang C, Tao D, Xie F, Zhang H, Sun J, Zhang C, Gu C, et al. CircNR3C1 inhibits proliferation of bladder cancer cells by sponging miR-27a-3p and downregulating cyclin D1 expression. Cancer Lett. 2019;460:139–151. doi: 10.1016/j.canlet.2019.06.018. [DOI] [PubMed] [Google Scholar]
- 110.Cheng WL, Lin TY, Tseng YH, Chu FH, Chueh PJ, Kuo YH, Wang SY. Inhibitory effect of human breast cancer cell proliferation via p21-mediated G1 cell cycle arrest by araliadiol isolated from Aralia cordata Thunb. Planta Med. 2011;77:164–168. doi: 10.1055/s-0030-1250177. [DOI] [PubMed] [Google Scholar]
- 111.Bi J, Liu H, Dong W, Xie W, He Q, Cai Z, Huang J, Lin T. Circular RNA circ-ZKSCAN1 inhibits bladder cancer progression through miR-1178-3p/p21 axis and acts as a prognostic factor of recurrence. Mol Cancer. 2019;18:133. doi: 10.1186/s12943-019-1060-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li P, Yang X, Yuan W, Yang C, Zhang X, Han J, Wang J, Deng X, Yang H, Li P, et al. CircRNA-Cdr1as exerts anti-oncogenic functions in bladder Cancer by sponging MicroRNA-135a. Cell Physiol Biochem. 2018;46:1606–1616. doi: 10.1159/000489208. [DOI] [PubMed] [Google Scholar]
- 113.Choi YJ, Anders L. Signaling through cyclin D-dependent kinases. Oncogene. 2014;33:1890–1903. doi: 10.1038/onc.2013.137. [DOI] [PubMed] [Google Scholar]
- 114.Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kluck RM, Bossy-Wetzel E, Green DR, Newmeyer DD. The release of cytochrome c from mitochondria: a primary site for Bcl-2 regulation of apoptosis. Science. 1997;275:1132–1136. doi: 10.1126/science.275.5303.1132. [DOI] [PubMed] [Google Scholar]
- 116.Yuan W, Zhou R, Wang J, Han J, Yang X, Yu H, Lu H, Zhang X, Li P, Tao J, et al. Circular RNA Cdr1as sensitizes bladder cancer to cisplatin by upregulating APAF1 expression through miR-1270 inhibition. Mol Oncol. 2019;13:1559–1576. doi: 10.1002/1878-0261.12523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhang Y, Gan B, Liu D, Paik JH. FoxO family members in cancer. Cancer Biol Ther. 2011;12:253–259. doi: 10.4161/cbt.12.4.15954. [DOI] [PubMed] [Google Scholar]
- 118.Wang C, Tao W, Ni S, Chen Q. Circular RNA circ-Foxo3 induced cell apoptosis in urothelial carcinoma via interaction with miR-191-5p. Onco Targets Ther. 2019;12:8085–8094. doi: 10.2147/OTT.S215823. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 119.Du WW, Fang L, Yang W, Wu N, Awan FM, Yang Z, Yang BB. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24:357–370. doi: 10.1038/cdd.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Li B, Xie F, Zheng FX, Jiang GS, Zeng FQ, Xiao XY. Overexpression of CircRNA BCRC4 regulates cell apoptosis and MicroRNA-101/EZH2 signaling in bladder cancer. J Huazhong Univ Sci Technolog Med Sci. 2017;37:886–890. doi: 10.1007/s11596-017-1822-9. [DOI] [PubMed] [Google Scholar]
- 121.Polacheck WJ, Zervantonakis IK, Kamm RD. Tumor cell migration in complex microenvironments. Cell Mol Life Sci. 2013;70:1335–1356. doi: 10.1007/s00018-012-1115-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kader AK, Liu J, Shao L, Dinney CP, Lin J, Wang Y, Gu J, Grossman HB, Wu X. Matrix metalloproteinase polymorphisms are associated with bladder cancer invasiveness. Clin Cancer Res. 2007;13:2614–2620. doi: 10.1158/1078-0432.CCR-06-1187. [DOI] [PubMed] [Google Scholar]
- 124.Liu F, Zhang H, Xie F, Tao D, Xiao X, Huang C, Wang M, Gu C, Zhang X, Jiang G. Hsa_circ_0001361 promotes bladder cancer invasion and metastasis through miR-491-5p/MMP9 axis. Oncogene. 2020;39:1696–1709. doi: 10.1038/s41388-019-1092-z. [DOI] [PubMed] [Google Scholar]
- 125.Wang L, Wu H, Wang L, Zhang H, Lu J, Liang Z, Liu T. Asporin promotes pancreatic cancer cell invasion and migration by regulating the epithelial-to-mesenchymal transition (EMT) through both autocrine and paracrine mechanisms. Cancer Lett. 2017;398:24–36. doi: 10.1016/j.canlet.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 126.Su Y, Du Z, Zhong G, Ya Y, Bi J, Shi J, Chen L, Dong W, Lin T. circ5912 suppresses cancer progression via inducing MET in bladder cancer. Aging (Albany NY) 2019;11:10826–10838. doi: 10.18632/aging.102464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.He Q, Yan D, Dong W, Bi J, Huang L, Yang M, Huang J, Qin H, Lin T. circRNA circFUT8 Upregulates Krupple-like factor 10 to inhibit the metastasis of bladder Cancer via sponging miR-570-3p. Mol Ther Oncolytics. 2020;16:172–187. doi: 10.1016/j.omto.2019.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Su Y, Feng W, Shi J, Chen L, Huang J, Lin T. circRIP2 accelerates bladder cancer progression via miR-1305/Tgf-beta2/smad3 pathway. Mol Cancer. 2020;19:23. doi: 10.1186/s12943-019-1129-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Su H, Tao T, Yang Z, Kang X, Zhang X, Kang D, Wu S, Li C. Circular RNA cTFRC acts as the sponge of MicroRNA-107 to promote bladder carcinoma progression. Mol Cancer. 2019;18:27. doi: 10.1186/s12943-019-0951-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ma T, Zhang J. Upregulation of FOXP4 in breast cancer promotes migration and invasion through facilitating EMT. Cancer Manag Res. 2019;11:2783–2793. doi: 10.2147/CMAR.S191641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Guitard E, Parker F, Millon R, Abecassis J, Tocque B. G3BP is overexpressed in human tumors and promotes S phase entry. Cancer Lett. 2001;162:213–221. doi: 10.1016/S0304-3835(00)00638-8. [DOI] [PubMed] [Google Scholar]
- 132.Zhang H, Zhang SH, He HW, Zhang CX, Yu DK, Shao RG. Downregulation of G3BPs inhibits the growth, migration and invasion of human lung carcinoma H1299 cells by suppressing the Src/FAK-associated signaling pathway. Cancer Gene Ther. 2013;20:622–629. doi: 10.1038/cgt.2013.62. [DOI] [PubMed] [Google Scholar]
- 133.Liu H, Bi J, Dong W, Yang M, Shi J, Jiang N, Lin T, Huang J. Invasion-related circular RNA circFNDC3B inhibits bladder cancer progression through the miR-1178-3p/G3BP2/SRC/FAK axis. Mol Cancer. 2018;17:161. doi: 10.1186/s12943-018-0908-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Dobruch J, Daneshmand S, Fisch M, Lotan Y, Noon AP, Resnick MJ, Shariat SF, Zlotta AR, Boorjian SA. Gender and bladder Cancer: a collaborative review of etiology, biology, and outcomes. Eur Urol. 2016;69:300–310. doi: 10.1016/j.eururo.2015.08.037. [DOI] [PubMed] [Google Scholar]
- 135.Miyamoto H, Yang Z, Chen YT, Ishiguro H, Uemura H, Kubota Y, Nagashima Y, Chang YJ, Hu YC, Tsai MY, et al. Promotion of bladder cancer development and progression by androgen receptor signals. J Natl Cancer Inst. 2007;99:558–568. doi: 10.1093/jnci/djk113. [DOI] [PubMed] [Google Scholar]
- 136.Chen J, Sun Y, Ou Z, Yeh S, Huang CP, You B, Tsai YC, Sheu TJ, Zu X, Chang C. Androgen receptor-regulated circFNTA activates KRAS signaling to promote bladder cancer invasion. EMBO Rep. 2020;21:e48467. doi: 10.15252/embr.201948467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Vermeulen L. de Sousa e Melo F, Richel DJ, Medema JP. The developing cancer stem-cell model: clinical challenges and opportunities. Lancet Oncol. 2012;13:e83–e89. doi: 10.1016/S1470-2045(11)70257-1. [DOI] [PubMed] [Google Scholar]
- 138.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 139.Tao T, Yuan S, Liu J, Shi D, Peng M, Li C, Wu S. Cancer stem cell-specific expression profiles reveal emerging bladder cancer biomarkers and identify circRNA_103809 as an important regulator in bladder cancer. Aging (Albany NY) 2020;12:3354–3370. doi: 10.18632/aging.102816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Patil SS, Railkar R, Swain M, Atreya HS, Dighe RR, Kondaiah P. Novel anti IGFBP2 single chain variable fragment inhibits glioma cell migration and invasion. J Neuro-Oncol. 2015;123:225–235. doi: 10.1007/s11060-015-1800-7. [DOI] [PubMed] [Google Scholar]
- 141.Yang D, Qian H, Fang Z, Xu A, Zhao S, Liu B, Li D. Silencing circular RNA VANGL1 inhibits progression of bladder cancer by regulating miR-1184/IGFBP2 axis. Cancer Med. 2020;9:700–710. doi: 10.1002/cam4.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Liu P, Li X, Guo X, Chen J, Li C, Chen M, Liu L, Zhang X, Zu X. Circular RNA DOCK1 promotes bladder carcinoma progression via modulating circDOCK1/hsa-miR-132-3p/Sox5 signalling pathway. Cell Prolif. 2019;52:e12614. doi: 10.1111/cpr.12614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lin G, Sheng H, Xie H, Zheng Q, Shen Y, Shi G, Ye D. circLPAR1 is a novel biomarker of prognosis for muscle-invasive bladder cancer with invasion and metastasis by miR-762. Oncol Lett. 2019;17:3537–3547. doi: 10.3892/ol.2019.9970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Liu H, Chen D, Bi J, Han J, Yang M, Dong W, Lin T, Huang J. Circular RNA circUBXN7 represses cell growth and invasion by sponging miR-1247-3p to enhance B4GALT3 expression in bladder cancer. Aging (Albany NY) 2018;10:2606–2623. doi: 10.18632/aging.101573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Song J, Shu H, Zhang L, Xiong J. Long noncoding RNA GAS5 inhibits angiogenesis and metastasis of colorectal cancer through the Wnt/beta-catenin signaling pathway. J Cell Biochem. 2019;120:6937–51. [DOI] [PubMed]
- 146.He YM, Xiao YS, Wei L, Zhang JQ, Peng CH. CUL4B promotes metastasis and proliferation in pancreatic cancer cells by inducing epithelial-mesenchymal transition via the Wnt/beta-catenin signaling pathway. J Cell Biochem. 2018;119:5308–5323. doi: 10.1002/jcb.26643. [DOI] [PubMed] [Google Scholar]
- 147.Han CT, Bao QY, Cheng SJ, Liu M, Qian HN, Li D. Circular RNA hsa_circ_0017247 acts as an oncogene in bladder cancer by inducing Wnt/beta-catenin signaling pathway. Eur Rev Med Pharmacol Sci. 2020;24:1081–1087. doi: 10.26355/eurrev_202002_20158. [DOI] [PubMed] [Google Scholar]
- 148.Dong W, Bi J, Liu H, Yan D, He Q, Zhou Q, Wang Q, Xie R, Su Y, Yang M, et al. Circular RNA ACVR2A suppresses bladder cancer cells proliferation and metastasis through miR-626/EYA4 axis. Mol Cancer. 2019;18:95. doi: 10.1186/s12943-019-1025-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wu L, Zhang M, Qi L, Zu X, Li Y, Liu L, Chen M, Li Y, He W, Hu X, et al. ERalpha-mediated alterations in circ_0023642 and miR-490-5p signaling suppress bladder cancer invasion. Cell Death Dis. 2019;10:635. doi: 10.1038/s41419-019-1827-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yang C, Wu S, Wu X, Zhou X, Jin S, Jiang H. Silencing circular RNA UVRAG inhibits bladder cancer growth and metastasis by targeting the microRNA-223/fibroblast growth factor receptor 2 axis. Cancer Sci. 2019;110:99–106. doi: 10.1111/cas.13857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Li Y, Wan B, Liu L, Zhou L, Zeng Q. Circular RNA circMTO1 suppresses bladder cancer metastasis by sponging miR-221 and inhibiting epithelial-to-mesenchymal transition. Biochem Biophys Res Commun. 2019;508:991–996. doi: 10.1016/j.bbrc.2018.12.046. [DOI] [PubMed] [Google Scholar]
- 152.Luo M, Guan JL. Focal adhesion kinase: a prominent determinant in breast cancer initiation, progression and metastasis. Cancer Lett. 2010;289:127–139. doi: 10.1016/j.canlet.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yan D, Dong W, He Q, Yang M, Huang L, Kong J, Qin H, Lin T, Huang J. Circular RNA circPICALM sponges miR-1265 to inhibit bladder cancer metastasis and influence FAK phosphorylation. EBioMedicine. 2019;48:316–331. doi: 10.1016/j.ebiom.2019.08.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Folkman J. Tumor angiogenesis: therapeutic implications. N Engl J Med. 1971;285:1182–1186. doi: 10.1056/NEJM197108122850711. [DOI] [PubMed] [Google Scholar]
- 155.Tischer E, Gospodarowicz D, Mitchell R, Silva M, Schilling J, Lau K, Crisp T, Fiddes JC, Abraham JA. Vascular endothelial growth factor: a new member of the platelet-derived growth factor gene family. Biochem Biophys Res Commun. 1989;165:1198–1206. doi: 10.1016/0006-291X(89)92729-0. [DOI] [PubMed] [Google Scholar]
- 156.Kurihara T, Westenskow PD, Friedlander M. Hypoxia-inducible factor (HIF)/vascular endothelial growth factor (VEGF) signaling in the retina. Adv Exp Med Biol. 2014;801:275–281. doi: 10.1007/978-1-4614-3209-8_35. [DOI] [PubMed] [Google Scholar]
- 157.Dvorak HF. Vascular permeability factor/vascular endothelial growth factor: a critical cytokine in tumor angiogenesis and a potential target for diagnosis and therapy. J Clin Oncol. 2002;20:4368–4380. doi: 10.1200/JCO.2002.10.088. [DOI] [PubMed] [Google Scholar]
- 158.Wei Y, Zhang Y, Meng Q, Cui L, Xu C. Hypoxia-induced circular RNA has_circRNA_403658 promotes bladder cancer cell growth through activation of LDHA. Am J Transl Res. 2019;11:6838–6849. [PMC free article] [PubMed] [Google Scholar]
- 159.Presta M, Dell'Era P, Mitola S, Moroni E, Ronca R, Rusnati M. Fibroblast growth factor/fibroblast growth factor receptor system in angiogenesis. Cytokine Growth Factor Rev. 2005;16:159–178. doi: 10.1016/j.cytogfr.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 160.Husseman J, Palacios SD, Rivkin AZ, Oehl H, Ryan AF. The role of vascular endothelial growth factors and fibroblast growth factors in angiogenesis during otitis media. Audiol Neurootol. 2012;17:148–154. doi: 10.1159/000333805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Mao W, Huang X, Wang L, Zhang Z, Liu M, Li Y, Luo M, Yao X, Fan J, Geng J. Circular RNA hsa_circ_0068871 regulates FGFR3 expression and activates STAT3 by targeting miR-181a-5p to promote bladder cancer progression. J Exp Clin Cancer Res. 2019;38:169. doi: 10.1186/s13046-019-1136-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Massari F, Santoni M, Ciccarese C, Brunelli M, Conti A, Santini D, Montironi R, Cascinu S, Tortora G. Emerging concepts on drug resistance in bladder cancer: implications for future strategies. Crit Rev Oncol Hematol. 2015;96:81–90. doi: 10.1016/j.critrevonc.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 163.Chen J, Wang L, Tang Y, Gong G, Liu L, Chen M, Chen Z, Cui Y, Li C, Cheng X, et al. Maspin enhances cisplatin chemosensitivity in bladder cancer T24 and 5637 cells and correlates with prognosis of muscle-invasive bladder cancer patients receiving cisplatin based neoadjuvant chemotherapy. J Exp Clin Cancer Res. 2016;35:2. doi: 10.1186/s13046-015-0282-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Aizawa T, Komatsu M. Rab27a: a new face in beta cell metabolism-secretion coupling. J Clin Invest. 2005;115:227–230. doi: 10.1172/JCI24269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Liu J, Gong X, Zhu X, Xue D, Liu Y, Wang P. Rab27A overexpression promotes bladder cancer proliferation and chemoresistance through regulation of NF-kappaB signaling. Oncotarget. 2017;8:75272–75283. doi: 10.18632/oncotarget.20775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bi J, Liu H, Cai Z, Dong W, Jiang N, Yang M, Huang J, Lin T. Circ-BPTF promotes bladder cancer progression and recurrence through the miR-31-5p/RAB27A axis. Aging (Albany NY) 2018;10:1964–1976. doi: 10.18632/aging.101520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kamarajan P, Sun NK, Sun CL, Chao CC. Apaf-1 overexpression partially overcomes apoptotic resistance in a cisplatin-selected HeLa cell line. FEBS Lett. 2001;505:206–212. doi: 10.1016/S0014-5793(01)02817-4. [DOI] [PubMed] [Google Scholar]
- 168.Del Bello B, Valentini MA, Comporti M, Maellaro E. Cisplatin-induced apoptosis in melanoma cells: role of caspase-3 and caspase-7 in Apaf-1 proteolytic cleavage and in execution of the degradative phases. Ann N Y Acad Sci. 2003;1010:200–204. doi: 10.1196/annals.1299.034. [DOI] [PubMed] [Google Scholar]
- 169.Zang YS, Zhong YF, Fang Z, Li B, An J. MiR-155 inhibits the sensitivity of lung cancer cells to cisplatin via negative regulation of Apaf-1 expression. Cancer Gene Ther. 2012;19:773–778. doi: 10.1038/cgt.2012.60. [DOI] [PubMed] [Google Scholar]
- 170.Vaupel P, Mayer A. Hypoxia in cancer: significance and impact on clinical outcome. Cancer Metastasis Rev. 2007;26:225–239. doi: 10.1007/s10555-007-9055-1. [DOI] [PubMed] [Google Scholar]
- 171.Zhang Y, Wang Z, Yu J, Shi J, Wang C, Fu W, Chen Z, Yang J. Cancer stem-like cells contribute to cisplatin resistance and progression in bladder cancer. Cancer Lett. 2012;322:70–77. doi: 10.1016/j.canlet.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 172.Su Y, Yang W, Jiang N, Shi J, Chen L, Zhong G, Bi J, Dong W, Wang Q, Wang C, Lin T. Hypoxia-elevated circELP3 contributes to bladder cancer progression and cisplatin resistance. Int J Biol Sci. 2019;15:441–452. doi: 10.7150/ijbs.26826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kashiwagi E, Ide H, Inoue S, Kawahara T, Zheng Y, Reis LO, Baras AS, Miyamoto H. Androgen receptor activity modulates responses to cisplatin treatment in bladder cancer. Oncotarget. 2016;7:49169–49179. doi: 10.18632/oncotarget.9994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Cheng DL, Xiang YY, Ji LJ, Lu XJ. Competing endogenous RNA interplay in cancer: mechanism, methodology, and perspectives. Tumour Biol. 2015;36:479–488. doi: 10.1007/s13277-015-3093-z. [DOI] [PubMed] [Google Scholar]
- 175.Ala U, Karreth FA, Bosia C, Pagnani A, Taulli R, Leopold V, Tay Y, Provero P, Zecchina R, Pandolfi PP. Integrated transcriptional and competitive endogenous RNA networks are cross-regulated in permissive molecular environments. Proc Natl Acad Sci U S A. 2013;110:7154–7159. doi: 10.1073/pnas.1222509110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Denzler R, Agarwal V, Stefano J, Bartel DP, Stoffel M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol Cell. 2014;54:766–776. doi: 10.1016/j.molcel.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17:272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 178.Zheng XB, Zhang M, Xu MQ. Detection and characterization of ciRS-7: a potential promoter of the development of cancer. Neoplasma. 2017;64:321–328. doi: 10.4149/neo_2017_301. [DOI] [PubMed] [Google Scholar]
- 179.Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 180.Suzuki H, Tsukahara T. A view of pre-mRNA splicing from RNase R resistant RNAs. Int J Mol Sci. 2014;15:9331–9342. doi: 10.3390/ijms15069331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Rao DD, Vorhies JS, Senzer N, Nemunaitis J. siRNA vs. shRNA: similarities and differences. Adv Drug Deliv Rev. 2009;61:746–759. doi: 10.1016/j.addr.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 182.Cooper DA, Cortes-Lopez M, Miura P. Genome-wide circRNA profiling from RNA-seq data. Methods Mol Biol. 1724;2018:27–41. doi: 10.1007/978-1-4939-7562-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Wang L, Luo T, Bao Z, Li Y, Bu W. Intrathecal circHIPK3 shRNA alleviates neuropathic pain in diabetic rats. Biochem Biophys Res Commun. 2018;505:644–650. doi: 10.1016/j.bbrc.2018.09.158. [DOI] [PubMed] [Google Scholar]
- 184.Tatomer DC, Liang D, Wilusz JE. Inducible expression of eukaryotic circular RNAs from plasmids. Methods Mol Biol. 1648;2017:143–154. doi: 10.1007/978-1-4939-7204-3_11. [DOI] [PubMed] [Google Scholar]
- 185.Wang ST, Liu LB, Li XM, Wang YF, Xie PJ, Li Q, Wang R, Wei Q, Kang YH, Meng R, Feng XH. Circ-ITCH regulates triple-negative breast cancer progression through the Wnt/beta-catenin pathway. Neoplasma. 2019;66:232–239. doi: 10.4149/neo_2018_180710N460. [DOI] [PubMed] [Google Scholar]
- 186.Freimann K, Arukuusk P, Kurrikoff K, Parnaste L, Raid R, Piirsoo A, Pooga M, Langel U. Formulation of stable and homogeneous cell-penetrating peptide NF55 nanoparticles for efficient gene delivery in vivo. Mol Ther Nucleic Acids. 2018;10:28–35. doi: 10.1016/j.omtn.2017.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Tang G, Xie W, Qin C, Zhen Y, Wang Y, Chen F, Du Z, Wu Z, Zhang B, Shen Z, et al. Expression of circular RNA circASXL1 correlates with TNM classification and predicts overall survival in bladder cancer. Int J Clin Exp Pathol. 2017;10:8495–8502. [PMC free article] [PubMed] [Google Scholar]
- 188.Hsu I, Vitkus S, Da J, Yeh S. Role of oestrogen receptors in bladder cancer development. Nat Rev Urol. 2013;10:317–326. doi: 10.1038/nrurol.2013.53. [DOI] [PubMed] [Google Scholar]
- 189.Chang C, Lee SO, Yeh S, Chang TM. Androgen receptor (AR) differential roles in hormone-related tumors including prostate, bladder, kidney, lung, breast and liver. Oncogene. 2014;33:3225–3234. doi: 10.1038/onc.2013.274. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Database for circRNA research
Data Availability Statement
Not applicable.