Abstract
Crop genomics has seen dramatic advances in recent years due to improvements in sequencing technology, assembly methods, and computational resources. These advances have led to the development of new tools to facilitate crop improvement. The study of structural variation within species and the characterization of the pan-genome has revealed extensive genome content variation among individuals within a species that is paradigm shifting to crop genomics and improvement. Here, we review advances in crop genomics and how utilization of these tools is shifting in light of pan-genomes that are becoming available for many crop species.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13059-020-02224-8.
Introduction
Crop improvement is needed now more than ever with challenges associated with feeding an ever-expanding population under increasingly variable growth conditions. The ability to produce crops that meet societal needs is enhanced by a thorough understanding of the genome of a species. Genomic resources expand the toolbox available for plant breeding and crop improvement efforts. Various tools have risen in popularity for plant breeding, in some cases as short-lived bandwagons and others as paradigm shifts in crop improvement [1, 2]. Within crop genomics, advances relevant to crop improvement have primarily been in marker (e.g., Illumina single nucleotide polymorphism (SNP) chips, kompetitive allele-specific PCR (KASP) assays, genotyping-by-sequencing (GBS)) and sequencing (e.g., Illumina, PacBio, Nanopore) technology. Recent innovations are driving a paradigm shift in which the extent and relevance of structural variation within the pan-genome of crop species are now being considered.
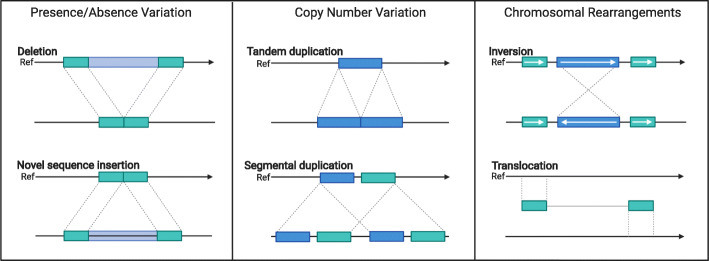
Access to plant genome assemblies in the early 2000s revolutionized thinking about the biology of crops and plant breeding [3–5]. These early assemblies allowed for a deeper understanding of the diversity in plant species, primarily at the level of SNPs [6–9]. However, after a short while, it became obvious that single-reference assemblies represent only a small fraction of species-wide genomic space [10–13]. Extensive structural variants (SVs) (e.g., presence-absence variation (PAV), copy number variation (CNV), and chromosomal rearrangements; Fig. 1) were discovered, with the first two classes contributing to the variation in genome content. Within species, genomes vary in both gene content (e.g., tandem duplicated genes, CNVs dispersed throughout the genome, and PAVs of genes) and repetitive portions of the genome (e.g., transposable elements, knob repeats, centromere repeats). In characterizing this variation, the genomic fraction common to all individuals within a species has been termed the “core” genome and the variable fraction the “dispensable” genome.
Fig. 1.
Diagrams of structural variants that can be found in crop genomes
There are many mechanisms that can generate a structural variation. For example, transposable elements (TEs) can replicate themselves in a genome and can also capture and carry gene sequences to new genomic locations [14–16]. This process can cause significant disruption of the coding portion of the genome [15, 16]. Additionally, structural variation can be introduced through errors during meiotic recombination [17], such as non-allelic homologous recombination (unequal recombination [18]) and double-strand break repair via single-strand annealing [19]. Finally, PAVs can be created, especially in plants, through differential genome fractionation across genotypes following a whole-genome duplication event [20], although in maize, a paleopolyploid, it was shown that this phenomenon played a limited role in creating SVs among elite temperate germplasm [21].
The generation of multiple, reference-quality genome assemblies per crop species is now a reality [22–24]. Our way of thinking about crop genomics is changing as we gain a deeper understanding of the structural variation within the pan-genome. Initial efforts to dissect the genetic architecture of traits (e.g., quantitative trait locus (QTL) mapping and genome-wide association studies (GWAS)) and genomic prediction efforts have relied primarily on SNP markers. The structural variation that has been uncovered in the pan-genome era necessitates a reevaluation of the determinants of phenotype. To date, structural variation has already been associated with environmental adaptation such as tolerance of abiotic and biotic stress [25–28] and flowering time ([29, 30]; for an extensive review, see [31]). Additionally, plant domestication traits such as non-shattering [32] and changes in plant architecture [33, 34] are caused by SVs. For example, a TE insertion ~ 60 kb upstream of the maize tb1 gene played an important role in changing maize architecture during its domestication [35]. In fact, SVs in non-coding regions have been shown in many instances to influence gene expression of nearby genes [36, 37]. Given the breadth of traits affected by SV, their characterization is important for crop domestication and improvement and will facilitate future efforts in these areas.
Crop genomics has transitioned from the era of a single reference genome to a time when we now have access to tens or hundreds of reference-quality genome assemblies within a species (Table 1). This article reviews previous crop genomic efforts relevant to crop improvement and expected advances in light of recent progress in characterizing structural variation at the pan-genome level.
Table 1.
Summary of plant species with pan-genomes currently available
| Species | Estimated mean DNA amount (C-value)a | Method for pan-genome construction | Number of accessions | Sequencing method | Reference |
|---|---|---|---|---|---|
| Brachypodium distachyon |
B. distachyon, 0.32 pg B. stacei, 0.28 pg B. hybridum, 0.63 pg |
De novo assemblyb | 54 | Illumina HiSeq | [38] |
| B. distachyon, Brachypodium hybridum, Brachypodium stacei | De novo assemblyb | 57 |
Illumina HiSeq PacBio |
[39] | |
| Medicago truncatula | 0.47 pg | De novo assemblyb | 15 | Illumina HiSeq | [40] |
| Oryza sativa (Asian rice) |
O. rufipogon, 0.46 pg O. nivara, 0.47 pg O. barthii, 0.60 pg O. glaberrima, 0.53 pg O. sativa, 0.50 pg |
Iterative mapping and assemblyc | 1483 | Illumina HiSeq | [41] |
| O. sativa (Asian rice) | Map to pand | 3010 |
Illumina HiSeq PacBio |
[42] | |
| O. sativa/Oryza rufipogon (Asian and common wild rice) | De novo assemblyb | 66 | Illumina HiSeq | [43] | |
| O. sativa (Asian rice) | De novo assemblyb | 12 | PacBio | [44] | |
| O. rufipogon/O. nivara/O. barthii/O. glaberrima (wild rice and African rice) | De novo assemblyb | 4 | PacBio | [45] | |
| Juglans ssp. (walnuts) | 0.64 pg | De novo assemblyb | 6 | Illumina HiSeq | [46] |
| Malus domestica/M. sieversii/M. sylvestris (apple and wild apple progenitors) |
M. domestica, 0.88 pg M. sieversii, 0.75 pg M. sylvestris, 0.78 pg |
Iterative mapping and assemblyc | 91 |
Illumina HiSeq PacBio |
[47] |
| Brassica oleracea, Brassica macrocarpa (cultivated and wild cabbage) |
B. oleracea, 0.9 pg B. macrocarpa, not available B. napus, 1.10 pg |
Iterative mapping and assemblyc | 10 | Illumina HiSeq | [48] |
| Brassica napus (oilseed) | Map to pand | 9 |
Illumina Hiseq PacBio |
[22] | |
| Solanum lycopersicum (tomato) | 1.06 pg | Map to pand | 725 | Illumina NextSeq | [49] |
| De novo assemblyb | 100 (14 assembled) |
Illumina NextSeq Nanopore |
[23] | ||
| Glycine soja (wild soybean) |
G. soja, 1.10 pg G. max, 1.13 pg |
De novo assemblyb | 7 | Illumina HiSeq | [12] |
| Glycine max (soybean) | De novo assemblyb | 29 |
Illumina HiSeq PacBio |
[24] | |
| Zea mays (maize) | 2.7 pg | Novel transcript assemblye | 503 | Illumina HiSeq | [10] |
| De novo assemblyb | 6 | Illumina HiSeq | [50] | ||
| Capsicum annuum (pepper) | 3.16 pg | Iterative mapping and assemblyc | 383 | Illumina HiSeq | [51] |
| Helianthus annuus (sunflower) | 3.67 pg | Map to pand | 493 | Illumina HiSeq | [52] |
| Triticum aestivum (bread wheat) | 24.65 pg | Iterative mapping and assemblyc | 19 | Illumina HiSeq | [53] |
aThe mean 1C (pg) value was obtained from the plant DNA C-values Database (https://cvalues.science.kew.org), 1 pg = 978 Mb [54]
bAssemble and annotate each genome separately and identify the variable regions
cDe novo assemble individual genome and then compare the assembled genome to the reference genome to capture the gene information that is not present in the reference genome
dMap reads to the reference genome, perform de novo assembly using unmapped reads, and incorporate the information into the reference genome
eDe novo assembly of short reads to capture transcript diversity
Assembly and bioinformatic advances allow characterization of crop pan-genomes
Advances in crop genome assembly technology
Over the last two decades, advances in sequencing technology and assembly algorithms have profoundly affected our understanding of the complexity and structure of genomes. Crops were among the first species with assembled genomes given their economic importance and the relevance of genomic information to breeding. The earliest model crop genomes were assembled with Sanger sequencing, BAC-by-BAC approaches, and overlap-layout-consensus (OLC) assemblers (e.g., rice [3], maize [4], sorghum [55], soybean [56], and grape [57]). Subsequent crop reference genomes increasingly relied on next-generation sequencing (e.g., potato [58]) with some assembled entirely from paired-end and mate-pair Illumina data and de Bruijn graph approaches (e.g., barley, wheat [59, 60]). These crop reference assemblies were, in many cases, rapidly followed by large resequencing studies in which short-read data were generated for additional individuals and mapped to the reference to characterize species-level diversity (e.g., rice [42, 61, 62], maize [6], soybean [63]).
Within the last 5 years, the reduced cost of Illumina sequencing and improved assembly algorithms facilitated de novo assembly of multiple accessions per crop using low-cost short-read data (e.g., maize-PH207 [64], maize-W22 [65], maize-HZS [66], maize-Flint genomes [50], rice genomes [43, 67], soybean genomes [12]). While this approach has generated highly complete and contiguous assemblies of low-copy genic regions, the more repetitive, TE-rich regions of the genome have proven recalcitrant to assembly with short reads, resulting in numerous gaps and partial assembly in these regions.
Recently, the maturation of long-read technology has facilitated much more contiguous and complete assembly of crop genomes [68–72] and, in some cases, multiple long-read-based assemblies within a single species [23, 24]. These assemblies are already facilitating discoveries of the relevance of non-coding and regulatory variation to agronomic traits, among other important discoveries [73, 74]. Sequence data continues to improve rapidly with sequence output increasing steadily and error rates decreasing (e.g., PacBio HiFi libraries), thereby diminishing the cost of assembly and increasing the utility of long-read assemblies for uncovering agronomically relevant variation across lines within crop species.
Characterizing structural variation based on a single reference genome
Methods to detect structural variation began to appear shortly after the publication of the first genome assemblies and have continued to develop as sequencing technologies have advanced (for comprehensive reviews, see [75, 76]). Early efforts to characterize CNV/PAV across species relied on hybridization arrays (e.g., comparative genomic hybridization (CGH)) that were based on probes often designed using only sequence from an initial reference genome assembly [11, 13, 19]. While array-based approaches are relatively inexpensive and high-throughput, they do have limitations. For example, once an array is developed, it is a static instrument, and newly identified loci of interest are not characterized. Additionally, when probes are based on a single reference sequence, ascertainment bias can be observed (i.e., hybridization efficiency diminishes when samples are more divergent from the reference individual).
As short-read resequencing decreased in cost and became commonplace, whole-genome sequencing (WGS) approaches were more frequently used to characterize CNV/PAV in crops [77–79]. These approaches for detecting CNV/PAV fall into three main categories: read depth, read pair, and split read [80]. With read-depth methods, short reads are mapped to a reference, and the relative depth of sequence at a locus serves as a proxy for copy number in a given individual [80]. Read-pair methods identify CNV/PAV based on discrepancies in the distance between paired-end sequences relative to their distance in the reference assembly [80]. Split-read methods detect SVs that interrupt the sequence within short reads [80].
The use of whole-genome sequencing allowed for characterization of a greater breadth of variants than hybridization arrays, but this approach suffers similar limitations: (1) sequence from loci that are missing in the reference genome due to either incomplete assembly or true biological absence does not map and remains uncharacterized, (2) divergent reads map less efficiently, and (3) uneven coverage bias of short-read sequencing can result in inaccuracies [81]. These shortcomings have been addressed to some extent through the assembly of unmapped reads [10, 48] and through the use of pseudo-references in which line-specific SNPs are introduced into the reference to increase mapping efficiency [82]. Characterization of structural variation in the repetitive fraction of the genome is particularly challenging with short-read resequencing data because mapping and assembly of unmapped reads are particularly inefficient and unreliable in these regions.
New approaches have rapidly developed for CNV/PAV characterization that leverage recently developed library preparation techniques and the maturation of single-molecule, long-read sequencing (comprehensively reviewed in [75]). For example, connected-molecule approaches (10x, Hi-C, Strand-Seq) can characterize long-range information using short reads through the development of specialized libraries of linked reads. Single-molecule approaches (optical maps (e.g., Bionano) and long-read sequencing, such as PacBio and Oxford Nanopore Technologies) allow for alignment of sequences from multiple individuals and, because of read length, enable characterization of sequences missing in the reference genome. Both of these approaches allow for the characterization of small- and intermediate-sized SVs. Large SVs (i.e., > 1 Mb) are more effectively characterized using optical maps (e.g., [83]). Collectively, these innovations have led to the most comprehensive characterization of CNV/PAV to date [84, 85]. However, the underlying data are still relatively expensive and must be generated at high depth for confident calls, making them impractical at the scale in which crop improvement programs often operate.
Characterizing structural variation through creation of a pan-genome reference
Access to multiple reference-quality genome assemblies within a species provides opportunities to identify SVs in a non-reference-biased manner. However, a number of challenges arise in such an approach. First, several crop species have large, complex genomes, making numerous assemblies per taxon cost-prohibitive. To overcome this limitation, a small number of breeding program founder individuals, which capture the majority of segregating haplotypes, can be targeted for genome assembly and identification of relevant SVs. Second, while multiple assemblies will reduce reference bias, assembly errors can lead to the detection of false SVs and compromise downstream analysis, particularly when de novo assemblies are generated using different data types or assembly algorithms. A third challenge is the consolidation of pan-genome variation into a single reference or coordinate system, a useful step for the analyses of the biological significance of SVs in crop species including QTL analysis, GWAS, and genomic prediction.
Several methods exist for summarizing SV information in a pan-genome context. One approach is to map resequencing reads to a reference genome, de novo assemble unmapped reads, and add the assembled contigs to the reference assembly (known as the map-to-pan approach) [48, 86]. This strategy can minimize errors by exploiting the information already available from a high-quality reference genome and limit the coordinate consolidation issue, but the genomic locations of newly assembled contigs remain unknown without further analysis. A second alternative is the construction of a graph-based rather than linear reference genome [87]. In this approach, any variant (SNP or SV) is added to the reference as a node at the genomic location where it is discovered [88, 89]. Recently, a hybrid approach between linear and graph-based reference genomes has been developed to build on the strengths of these methods. In this approach, reads are first mapped to a graph-based genome, and haplotypes are associated with one of the reference genomes used to build the graph. Reads are then realigned to this genome leading to more accurate mapping than the graph-based approach alone [90]. For detailed descriptions of each method, and their advantages and disadvantages, see [91, 92].
Relevance of transposable elements to crop improvement
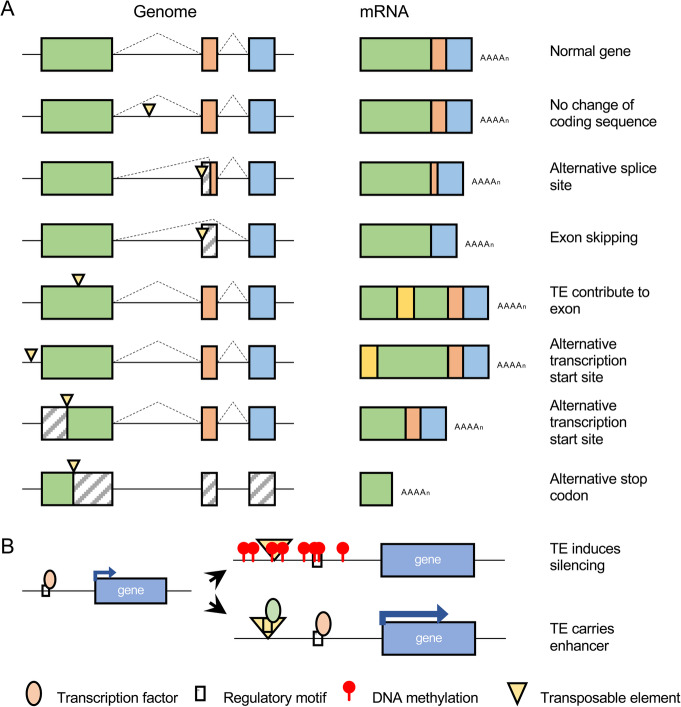
As pan-genomes become widely available for crop species, TEs, a driver of structural variation, will receive increasing attention in crop improvement. Plant genomes (including crop species) are particularly rife with TEs [93], and the relevance of TEs to crop phenotypes has been repeatedly demonstrated. Transposable elements can be functionally relevant in a number of ways including modifying the structure and amount of gene product that is transcribed (Fig. 2 [14, 23, 35, 37, 94–100];). For example, in maize, a Harbinger-like DNA transposon represses the expression of the ZmCCT9 gene to promote flowering under long-day conditions [37]. In rice, a Gypsy retrotransposon has been shown to enhance the expression of the OsFRDL4 gene and promote aluminum tolerance [101]. Two Copia retrotransposons independently inserted into the promoter region of the orange Ruby gene, resulting in its enhanced expression and driving convergent evolution of the blood orange trait [102]. Finally, a Copia retrotransposon Rider has created polymorphism in the SUN locus resulting in the oval shape typical of the Roma tomato variety [103, 104]. Despite their prevalence and relevance to agronomic phenotypes, TEs have, until recently, been largely ignored in crop improvement efforts.
Fig. 2.
Functional consequences of new transposable element insertions. a Possible effects on gene product structure. b Possible effects on gene product abundance
TEs create the majority of insertions and deletions in many crop genomes. For example, > 75% of large InDels (i.e., ≥ 100 bp) in both tomato [23] and soybean [24] pan-genomes consist of at least one TE. Across four maize lines, there is greater than 1.6 Gb of TE sequence that was found to segregate in just this narrow subset of genotypes [105]. Genome-wide variation in TE content at the species level has, until recently, been difficult to characterize because, as described above, the repetitive fraction of genomes has historically been poorly assembled, and there are challenges with accurate read alignment to these regions. Methods to characterize variation in TE content using short-read data [106] and whole-genome comparisons [105] are emerging and will help provide access to a new level of functional variation underlying agronomic phenotypes.
Once TE sequences are captured in de novo genome assemblies, a critical remaining challenge is an accurate annotation to the family level. Three general approaches are used. The first is homology-based using existing TE databases such as Repbase [107] and P-MITE [108]. This approach is quick because it uses annotations from other species, but is limited by the availability of such information and the extent to which TE sequences are conserved across species [109]. The second approach is based on the copy number of sequences [110–112] and is relatively fast and sensitive for the identification of high-copy number repeats. However, the specific annotation of a sequence is unknown (i.e., these could be large gene families, TEs, other types of repeats), and low-copy TEs are often missed. The limited classification information provided by this approach hampers biological inference and utility for crop improvement. The third approach is the de novo identification of TEs based on structural features. Structural annotation does not rely on existing TE libraries and is very sensitive. This method depends critically on knowledge of the diagnostic structural components of TEs and, when this knowledge is incomplete or imprecise, can result in inaccurate annotation [113]. Recently, efforts have been made to combine these approaches into a comprehensive solution for TE annotation. Such pipelines incorporate structural and homology information, repetitiveness, existing TE curations, and extensive filtering to generate high-quality de novo TE annotations. Methods developed based on this approach include EDTA [114] and RepeatModeler2 [112]. Comprehensive TE annotation of high-quality pan-genomes will allow us to further explore their varied roles within crop genomes [115] and to link TE variation, a pervasive form of SV, to phenotypes with agronomic relevance [116].
Advancing QTL mapping and GWAS using crop pan-genomes
Two main approaches are used to identify genomic regions associated with a desired phenotype: QTL mapping with biparental populations and GWAS with panels of diverse individuals. Early crop reference genome assemblies facilitated the development of platforms (e.g., Illumina SNP chips) that allow for rapid, cost-effective genotyping of thousands or millions of SNPs across large sets of individuals. This increase in marker density dramatically increased resolution in mapping studies, which aided in the identification and cloning of QTLs associated with disease resistance, drought tolerance, yield, plant architecture, and other important agronomic traits [117]. With these marker-trait associations identified, breeders can use linked markers to select the best plants in a population without extensive phenotyping, either as functional markers [118] or through marker-assisted selection [119].
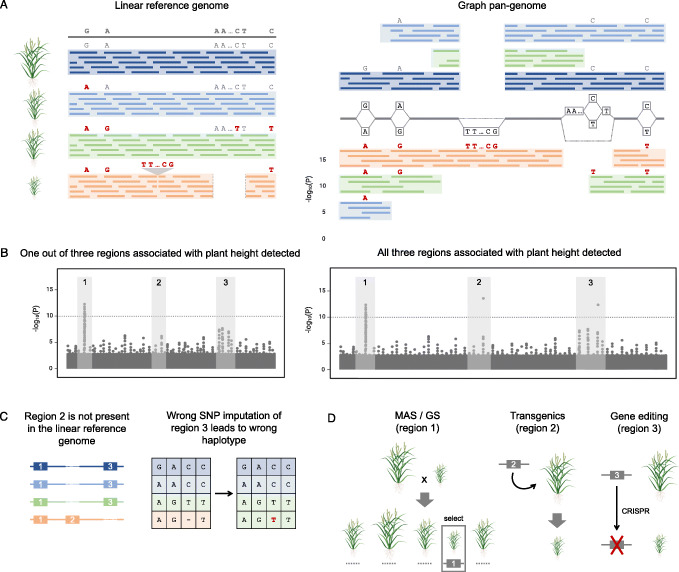
One major concern in QTL mapping or GWAS based on a single reference genome is reference bias [120]. If variants associated with a trait are not present in the reference genome, then QTL mapping or GWAS will not be able to detect them (Fig. 3a, b). For example, a maize gene conferring resistance to sugarcane mosaic virus could be identified by GWAS using markers based on the B73, but not the PH207, genome assembly, because the gene was not present in the PH207 assembly [120]. This situation is further exacerbated with more diverse germplasm (i.e., secondary gene pools), making it difficult to identify causative variation and bring it into the germplasm of breeding programs. A further problem is that true deletions relative to a reference genome are indistinguishable from missing data due to technical problems (e.g., low sequence coverage). Imputation of allelic variants across true deletions can result in decreased power to detect a significant association (Fig. 3c).
Fig. 3.
Impact of pan-genome representation on dissection of quantitative variation and applications to crop improvement. a Mapping reads to a single reference genome assembly (left) or a pan-genome graph (right) that captures structural variation in the species. b Impact of the read mapping method (single reference assembly vs. pan-genome graph) and subsequent variant calling on the ability to dissect the genetic architecture of a trait. c Causes for lack of identification of significant regions of the genome using variants called by mapping reads to a single reference genome assembly. d Methods breeders can utilize to exploit newly identified variants involve marker-assisted selection (MAS) and/or genomic selection (GS), inserting sequence through a transgene, or making other changes to a causative region with genome editing
QTL and GWAS studies have primarily relied on SNP data to date, but other markers have been useful in linking different types of variation to phenotype. For example, GWAS performed with both read depth variants (RDVs, a proxy for SVs), and SNPs in maize demonstrated that RDVs were enriched for significant GWAS results relative to SNPs for traits such as leaf development and disease resistance [77]. Similarly, in a large-scale GWAS using transcript abundance as a marker, gene associations with maize development traits were identified that were not detected by GWAS using SNPs [10]. While read depth and transcript abundance variants were useful in the initial efforts to assess the importance of SVs to phenotypic variation, they do not capture the complete structural variant landscape within a population. For example, read depth variants can only capture SVs that are present in the reference genome (e.g., insertions relative to the reference are not evaluated), leading to a strong reference bias and an incomplete picture of the relationship between SVs and phenotypes. RNA-seq is focused only on transcribed regions, is dependent on what tissues and developmental stages are sampled, and can be driven by both allelic variation in regulatory regions and true structural variation.
As the crop improvement paradigm shifts to a pan-genome perspective, the contribution of SVs to trait variation is becoming clear. Recently in Brassica napus, GWAS was performed with PAVs identified from eight whole-genome assemblies, and causal associations between SVs and silique length, seed weight, and flowering time were discovered that were not captured by SNP-GWAS. Likewise, GWAS based on the graph soybean pan-genome identified a PAV associated with variation in seed luster [24]. In peach, candidate causative SVs for early fruit maturity, flesh color around the stone, fruit shape, and flat shape formation have also been observed [121]. However, our understanding of the importance of SVs to phenotypic trait variation is still in its infancy. As technology and algorithm advancements allow for the complete SV landscape to be characterized at the scale of breeding programs and incorporated into a graph-based framework, it is anticipated that we will see a growing number of SVs underlying phenotypic variation important for crop improvement.
Advancing genomic prediction using crop pan-genomes
A number of important traits for crop improvement are controlled by many QTLs with small effect (e.g., yield). A complex genetic architecture makes it difficult to identify all QTLs underlying a trait, correctly estimate their effects, and introgress them into elite lines using methods such as marker-assisted selection [122–124]. Genomic selection is an alternative approach for complex traits, where marker effects are estimated from a training set, the phenotype of an individual is predicted based on the estimated marker effects (i.e., genomic prediction), and selections are made based on the predicted phenotype [125]. Regression and Bayesian approaches for genomic prediction were first described in the early 2000s and revolutionized animal and plant breeding [126]. Using SNPs as predictors, important agronomic traits such as grain yield, grain moisture, grain quality, biomass traits, and stalk and root lodging have been predicted with fairly high accuracy [127–133].
Traditionally, SNPs identified relative to a single reference genome have been used for genomic selection. However, as described above, there are a number of limitations and biases that are introduced with the use of a single reference for such applications. New approaches for identifying markers within a pan-genome framework are needed to improve prediction accuracy. The Practical Haplotype Graph (PHG) is one such method that successfully deals with the complexity of a species’ pan-genome at the scale necessary for complex traits and plant breeding programs [134]. In the PHG approach, existing genomic resources of breeding program founder lines (e.g., whole-genome resequencing data and/or whole-genome assemblies) are loaded into a graph-pan-genome database. Accurate imputation of low-sequence-coverage individuals (as low as 0.01× coverage) in the breeding population is achieved based on consensus haplotypes derived from the graph-pan-genome database. The PHG is a promising strategy for reducing the costs of genotyping, while also capturing a greater breadth of diversity in large breeding populations.
A major issue in genomic prediction is that genotype by environment (G×E) interactions decrease the prediction accuracy for individuals grown in novel environments. Statistical models that account for G×E have been designed to attempt to overcome this limitation [135–137]. Incorporation of SV data in such prediction models may further help to address issues of G×E in genomic prediction accuracy, because these variants have been shown to play a particularly important role in adaptation across environments. Not all SVs will be tagged by SNPs [70, 77, 138] and phenotypic variation driven by untagged SVs will be missed by prediction models. For example, Lyra et al. found that predictive ability for maize plant height under low nitrogen increased when adding just a few hundred CNVs to an analysis of ~ 20k SNPs [139]. However, while adding these additional markers may result in higher predictive accuracy, their addition may not be practical in breeding programs at the moment, as they require novel data generation and analysis infrastructure. Breeders need to balance the costs of scoring different markers with the increased efficiency of genomic prediction and genetic gain. For the time being, structural variation information from a pan-genome will be most readily used by breeders if existing SNP genotyping technology includes markers in strong linkage disequilibrium (LD) with phenotypically important SVs. For SVs not tagged by SNPs [70, 77, 138], characterization of these variants using novel approaches is only prudent if the genetic gain is large enough to justify the increased cost.
Future challenges and opportunities in applications of pan-genomics for crop improvement
Beyond the promise that recent genomic advances offer for characterizing diversity in model crop systems and for improvement of trait mapping and prediction, they also present opportunities to tackle difficult and understudied crop genomes and could potentially enable novel, gene-editing approaches to breeding.
Complexity of polyploid genomes
Allopolyploidy (the result of interspecific or intergeneric hybridization and chromosome doubling) and autopolyploidy (the result of whole-genome duplication) are particularly common in plant species [140, 141]. In fact, all angiosperms have undergone at least two rounds of polyploidy in their evolutionary history [142]. Many have returned to a diploid state, bearing remnants of this evolutionary history in their genomes [143]. As a natural mechanism, polyploidization can increase allelic diversity, expand the complement of genes, generate novel phenotypic variation, and aid in adaptation to new environments [144, 145]. Taking advantage of this, plant breeders have also generated artificial polyploids resulting in increased grain yield [146], fruit size [147], and seedless fruit [148].
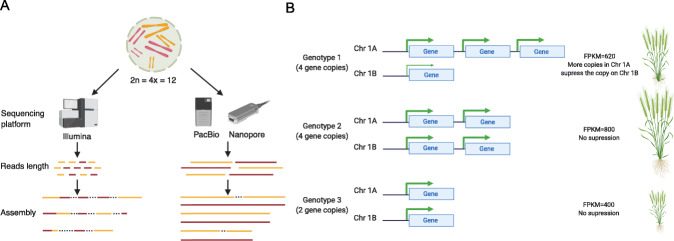
While polyploid crops are vitally important to sustain human life, genomic studies in these species have traditionally been very challenging for a number of reasons. High-quality genome assembly of polyploid species has been difficult to achieve due to their inclusion of multiple, closely related subgenomes and the associated challenges in discriminating homeologous loci and creating non-mosaic subgenome scaffolds (Fig. 4a). Many have resorted to sequencing diploid progenitors [149] or closely related species [58, 150] of polyploid crops in order to reduce genome complexity when generating initial reference assemblies. However, closely related diploids fail to capture lineage-specific SNPs, SVs, and other forms of variation that have accumulated post-polyploidization [39]. Beyond these difficulties in genome assembly, genomic approaches to polyploid crop improvement face further complications: (1) dissection of the genetic architecture of complex traits can be confounded when variants are not mapped to the correct subgenome [151, 152], a technical limitation, and (2) biologically, the more extensive epistatic interactions in polyploids [153, 154] and regulatory feedback between subgenomes can complicate the accurate prediction of phenotype based on genotype [155](Fig. 4b).
Fig. 4.
Impacts of technological advances to facilitate crop improvement in polyploid species. a Impact of sequencing technology on polyploid assembly. b Example of how understanding of a biological process is facilitated by having structural variation within subgenomes resolved beyond simply characterizing the number of copies in the genome
Advances in sequencing technologies and assembly algorithms are already addressing technical challenges in crop genomic research in polyploids [156]. Long-read sequencing with low error rates (e.g., PacBio HiFi reads) has made high-quality polyploid genome assembly possible, with recent assemblies containing fewer gaps and resolved homeologous scaffolds (Fig. 4a). Long-read assemblies now exist for polyploid crop species such as peanut [157], wheat [60], oilseed [22], and strawberry [158]. In some instances (e.g., potato [159]), multiple genome assemblies already exist within species. Nascent polyploid pan-genome studies are uncovering substantial diversity across species. For example, the de novo assembly of a single wheat cultivar captured 107,891 genes, and a map-to-pan assembly of 17 additional cultivars captured ~ 30,000 novel genes [53, 60]. As pan-genomic studies expand in polyploid crop species, we expect that, due to genomic redundancy and complexity, the degree of structural variation within polyploid species will be greater than that observed in diploid species, and SVs may be particularly fruitful markers for genomic approaches to polyploid crop improvement. Technical progress in assembling polyploid genomes (e.g., improvements to haplotype and homeolog phasing) should facilitate basic, biological study of the differences in the genotype-to-phenotype map between diploids and polyploids, knowledge of fundamental importance to the future of polyploid crop improvement.
Genomic resources for understudied crop species
For understudied crops, pan-genome-assisted breeding efforts remain limited due to the small size of the research communities for these species and, in some cases, due to the challenges associated with genome complexity. For the majority of understudied crop species, transcriptome assemblies are currently used as a proxy to the genome for improvement efforts. One such example is Silphium integrifolium, a species with a large genome size (2n = 2x = 14; haploid genome size of ~ 9 Gb [160];) that is currently being domesticated into an oil crop. Through transcriptome assembly and resequencing of 68 wild S. integrifolium accessions, several loci associated with adaptation to different climate conditions were identified [161]. While SNP data helped identify loci under selection, structural variation, an important source of local adaptation, remained uncharacterized. Pennycress (Thlaspi arvense) is another species that is currently being domesticated for use as an oil crop [162]. While it has advanced from an initial transcriptome assembly [163] to a full genome assembly [164], access to pan-genome variation is not yet available, despite the relatively small size (539 Mb) and simple genome structure of the species. Turfgrass and forage crops are further examples of understudied crops with limited genomic resources. Perennial ryegrass (Lolium perenne) has a fragmented draft genome [165], which may not be sufficient to enable pan-genomic research within the species. For other turfgrass species, such as hexaploid hard fescue (Festuca brevipila), long-read sequencing of the transcriptome has been used as a proxy of the reference genome, but it remains difficult to distinguish homeologs using this approach [166].
While pan-genomic studies may be in their infancy in non-model crops, it is anticipated that rapid advances in sequencing, assembly algorithms, and analysis pipelines in model systems and diminishing costs will very quickly enable this research. The time from publication of the first rice genome assembly to release of the first rice pan-genome was over a decade ([3]; Table 1). We anticipate that the development of genomic resources, including pan-genomes, will now be much more rapid. Indeed, pan-genomic studies have already been published in Capsicum (pepper) and Juglans (walnut) species [46, 51], and others will soon follow.
Rapid domestication of new and existing species
The recent availability of high-quality genomes and pan-genomes has enabled a new era of crop domestication. With pan-genome information, breeders can more effectively identify causal genetic variants (e.g., SNPs, CNV, PAV) underlying domestication traits and apply gene-editing tools to rapidly achieve desirable agronomic traits in wild plants. For example, the tomato pan-genome has revealed that variation at the fruit weight QTL fw3.2 is caused by tandem duplication of the cytochrome P450 gene SlKLUH [23] rather than a SNP in the gene’s promoter as proposed earlier [167]. CRISPR/Cas9 gene editing to reduce the copy number of the SKILUH gene successfully altered fruit weight, a crop domestication phenotype [23]. Similarly, by using resequencing data and a map-to-pan approach, Gao et al. conducted a comparative analysis of 725 cultivated tomatoes and close wild relatives, uncovering gene loss during tomato domestication [49]. Further enrichment analysis suggested that defense response genes and nearly 1200 promoter sequences were targeted by selection during domestication and improvement [49]. A non-reference ~ 4 kb substitution in the TomLoxC promoter region was also discovered that modifies fruit flavor [49]. These variants that distinguish crops from their wild relatives are prime targets for gene editing for rapid domestication.
Domestication has greatly reduced the genetic diversity of crops compared to their wild relatives [168]. Identifying and utilizing genetic diversity from crop wild relatives has been a major focus in crop improvement [169, 170]. Together, pan-genome information and CRISPR/Cas9 technologies enable de novo domestication of wild plants and can reduce barriers to the use of genetic variation from secondary and tertiary gene pools (wild relatives) [171, 172]. For example, Zsögön et al. edited six loci in wild tomato (Solanum pimpinellifolium) and significantly increased its yield, productivity, and nutritional value resulting in de novo domestication of tomato [173].
In summary, the complete catalog of variation that has been made possible by recent genomic technology and a pan-genome approach presents a substantial opportunity for crop improvement. We can, not only move beyond single-reference-based resequencing in model crops to a full understanding of structural variation and its link to phenotype, but also tackle complex, polyploid genomes, rapidly move understudied crops into the genomic era, and bring down barriers between crops and their wild relatives so that breeders can more easily expand their tool kit to include exotic germplasm. While further infrastructure and method development is necessary to fully realize this potential, there is a paradigm shift in the making.
Supplementary Information
Acknowledgements
Figures in this manuscript were created in part using BioRender.com.
Peer review information
Kevin Pang was the primary editor of this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Review history
The review history is available as Additional file 1.
Authors’ contributions
RDC, YQ, SO, MBH, and CNH wrote the manuscript. All authors read and approved the final manuscript.
Authors’ information
Twitter handles: @rafael_coletta (Rafael Della Coletta); @YinjieQiu (Yinjie Qiu); @SigmaFacto (Shujun Ou); @mbhufford (Matthew B. Hufford); @HirschCandice (Candice N. Hirsch).
Funding
This work was supported in part by the United States Department of Agriculture (2018-67013-27571) and the National Science Foundation (Grants IOS-1546727, IOS-1934384, IOS-1744001, and IOS-1546719).
Availability of data and materials
Not applicable
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Rafael Della Coletta, Email: della028@umn.edu.
Yinjie Qiu, Email: qiuxx221@umn.edu.
Shujun Ou, Email: oushujun@iastate.edu.
Matthew B. Hufford, Email: mhufford@iastate.edu
Candice N. Hirsch, Email: cnhirsch@umn.edu
References
- 1.Bernardo R. Bandwagons I, too, have known. Theor Appl Genet. 2016;129:2323–2332. doi: 10.1007/s00122-016-2772-5. [DOI] [PubMed] [Google Scholar]
- 2.“Bandwagons I have known”, by N.W. Simmonds - Tropical Agriculture Association. Tropical Agriculture Association. 2019. Available from: https://taa.org.uk/bandwagons-i-have-known-by-n-w-simmonds/. [cited 2020 Jul 20].
- 3.International Rice Genome Sequencing Project The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- 4.Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 5.Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 6.Gore MA, Chia J-M, Elshire RJ, Sun Q, Ersoz ES, Hurwitz BL, et al. A first-generation haplotype map of maize. Science. 2009;326:1115–1117. doi: 10.1126/science.1177837. [DOI] [PubMed] [Google Scholar]
- 7.McNally KL, Childs KL, Bohnert R, Davidson RM, Zhao K, Ulat VJ, et al. Genomewide SNP variation reveals relationships among landraces and modern varieties of rice. Proc Natl Acad Sci U S A. 2009;106:12273–12278. doi: 10.1073/pnas.0900992106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robbins MD, Sim S-C, Yang W, Van Deynze A, van der Knaap E, Joobeur T, et al. Mapping and linkage disequilibrium analysis with a genome-wide collection of SNPs that detect polymorphism in cultivated tomato. J Exp Bot. 2011;62:1831–1845. doi: 10.1093/jxb/erq367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamilton JP, Hansey CN, Whitty BR, Stoffel K, Massa AN, Van Deynze A, et al. Single nucleotide polymorphism discovery in elite North American potato germplasm. BMC Genomics. 2011;12:302. doi: 10.1186/1471-2164-12-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirsch CN, Foerster JM, Johnson JM, Sekhon RS, Muttoni G, Vaillancourt B, et al. Insights into the maize pan-genome and pan-transcriptome. Plant Cell. 2014;26:121–135. doi: 10.1105/tpc.113.119982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Springer NM, Ying K, Fu Y, Ji T, Yeh C-T, Jia Y, et al. Maize inbreds exhibit high levels of copy number variation (CNV) and presence/absence variation (PAV) in genome content. PLoS Genet. 2009;5:e1000734. doi: 10.1371/journal.pgen.1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y-H, Zhou G, Ma J, Jiang W, Jin L-G, Zhang Z, et al. De novo assembly of soybean wild relatives for pan-genome analysis of diversity and agronomic traits. Nat Biotechnol. 2014;32:1045–1052. doi: 10.1038/nbt.2979. [DOI] [PubMed] [Google Scholar]
- 13.Anderson JE, Kantar MB, Kono TY, Fu F, Stec AO, Song Q, et al. A roadmap for functional structural variants in the soybean genome. G3. 2014;4:1307–1318. doi: 10.1534/g3.114.011551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang N, Bao Z, Zhang X, Eddy SR, Wessler SR. Pack-MULE transposable elements mediate gene evolution in plants. Nature. 2004;431:569–573. doi: 10.1038/nature02953. [DOI] [PubMed] [Google Scholar]
- 15.Zhao D, Ferguson AA, Jiang N. What makes up plant genomes: the vanishing line between transposable elements and genes. Biochim Biophys Acta. 1859;2016:366–380. doi: 10.1016/j.bbagrm.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 16.Fedoroff NV. Transposable elements, epigenetics, and genome evolution. Science. 2012;338:758–767. doi: 10.1126/science.338.6108.758. [DOI] [PubMed] [Google Scholar]
- 17.Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009;10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yandeau-Nelson MD, Xia Y, Li J, Neuffer MG, Schnable PS. Unequal sister chromatid and homolog recombination at a tandem duplication of the A1 locus in maize. Genetics. 2006;173:2211–2226. doi: 10.1534/genetics.105.052712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muñoz-Amatriaín M, Eichten SR, Wicker T, Richmond TA, Mascher M, Steuernagel B, et al. Distribution, functional impact, and origin mechanisms of copy number variation in the barley genome. Genome Biol. 2013;14:R58. doi: 10.1186/gb-2013-14-6-r58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freeling M, Woodhouse MR, Subramaniam S, Turco G, Lisch D, Schnable JC. Fractionation mutagenesis and similar consequences of mechanisms removing dispensable or less-expressed DNA in plants. Curr Opin Plant Biol. 2012;15:131–139. doi: 10.1016/j.pbi.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 21.Brohammer AB, Kono TJY, Springer NM, McGaugh SE, Hirsch CN. The limited role of differential fractionation in genome content variation and function in maize (Zea mays L.) inbred lines. Plant J. 2018;93:131–141. doi: 10.1111/tpj.13765. [DOI] [PubMed] [Google Scholar]
- 22.Song J-M, Guan Z, Hu J, Guo C, Yang Z, Wang S, et al. Eight high-quality genomes reveal pan-genome architecture and ecotype differentiation of Brassica napus. Nat Plants. 2020;6:34–45. doi: 10.1038/s41477-019-0577-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alonge M, Wang X, Benoit M, Soyk S, Pereira L, Zhang L, et al. Major impacts of widespread structural variation on gene expression and crop improvement in tomato. Cell. 2020. 10.1016/j.cell.2020.05.021. [DOI] [PMC free article] [PubMed]
- 24.Liu Y, Du H, Li P, Shen Y, Peng H, Liu S, et al. Pan-genome of wild and cultivated soybeans. Cell. 2020. 10.1016/j.cell.2020.05.023. [DOI] [PubMed]
- 25.Knox AK, Dhillon T, Cheng H, Tondelli A, Pecchioni N, Stockinger EJ. CBF gene copy number variation at Frost Resistance-2 is associated with levels of freezing tolerance in temperate-climate cereals. Theor Appl Genet. 2010;121:21–35. doi: 10.1007/s00122-010-1288-7. [DOI] [PubMed] [Google Scholar]
- 26.Maron LG, Guimarães CT, Kirst M, Albert PS, Birchler JA, Bradbury PJ, et al. Aluminum tolerance in maize is associated with higher MATE1 gene copy number. Proc Natl Acad Sci U S A. 2013;110:5241–5246. doi: 10.1073/pnas.1220766110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cook DE, Lee TG, Guo X, Melito S, Wang K, Bayless AM, et al. Copy number variation of multiple genes at Rhg1 mediates nematode resistance in soybean. Science. 2012;338:1206–1209. doi: 10.1126/science.1228746. [DOI] [PubMed] [Google Scholar]
- 28.Sutton T, Baumann U, Hayes J, Collins NC, Shi B-J, Schnurbusch T, et al. Boron-toxicity tolerance in barley arising from efflux transporter amplification. Science. 2007;318:1446–1449. doi: 10.1126/science.1146853. [DOI] [PubMed] [Google Scholar]
- 29.Nitcher R, Distelfeld A, Tan C, Yan L, Dubcovsky J. Increased copy number at the HvFT1 locus is associated with accelerated flowering time in barley. Mol Gen Genomics. 2013;288:261–275. doi: 10.1007/s00438-013-0746-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Würschum T, Boeven PHG, Langer SM, Longin CFH, Leiser WL. Multiply to conquer: copy number variations at Ppd-B1 and Vrn-A1 facilitate global adaptation in wheat. BMC Genet. 2015;16:96. doi: 10.1186/s12863-015-0258-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tao Y, Zhao X, Mace E, Henry R, Jordan D. Exploring and exploiting pan-genomics for crop improvement. Mol Plant. 2019;12:156–169. doi: 10.1016/j.molp.2018.12.016. [DOI] [PubMed] [Google Scholar]
- 32.Lin Z, Li X, Shannon LM, Yeh C-T, Wang ML, Bai G, et al. Parallel domestication of the Shattering1 genes in cereals. Nat Genet. 2012;44:720–724. doi: 10.1038/ng.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan L, Li X, Liu F, Sun X, Li C, Zhu Z, et al. Control of a key transition from prostrate to erect growth in rice domestication. Nat Genet. 2008;40:1360–1364. doi: 10.1038/ng.197. [DOI] [PubMed] [Google Scholar]
- 34.Zhou Y, Zhu J, Li Z, Yi C, Liu J, Zhang H, et al. Deletion in a quantitative trait gene qPE9-1 associated with panicle erectness improves plant architecture during rice domestication. Genetics. 2009;183:315–324. doi: 10.1534/genetics.109.102681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Studer A, Zhao Q, Ross-Ibarra J, Doebley J. Identification of a functional transposon insertion in the maize domestication gene tb1. Nat Genet. 2011;43:1160–1163. doi: 10.1038/ng.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Q, Li Z, Li W, Ku L, Wang C, Ye J, et al. CACTA-like transposable element in ZmCCT attenuated photoperiod sensitivity and accelerated the postdomestication spread of maize. Proc Natl Acad Sci U S A. 2013;110:16969–16974. doi: 10.1073/pnas.1310949110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang C, Sun H, Xu D, Chen Q, Liang Y, Wang X, et al. ZmCCT9 enhances maize adaptation to higher latitudes. Proc Natl Acad Sci U S A. 2018;115:E334–E341. doi: 10.1073/pnas.1718058115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gordon SP, Contreras-Moreira B, Woods DP, Des Marais DL, Burgess D, Shu S, et al. Extensive gene content variation in the Brachypodium distachyon pan-genome correlates with population structure. Nat Commun. 2017;8:2184. doi: 10.1038/s41467-017-02292-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gordon SP, Contreras-Moreira B, Levy JJ, Djamei A, Czedik-Eysenberg A, Tartaglio VS, et al. Gradual polyploid genome evolution revealed by pan-genomic analysis of Brachypodium hybridum and its diploid progenitors. Nat Commun. 2020;11:3670. doi: 10.1038/s41467-020-17302-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou P, Silverstein KAT, Ramaraj T, Guhlin J, Denny R, Liu J, et al. Exploring structural variation and gene family architecture with de novo assemblies of 15 Medicago genomes. BMC Genomics. 2017;18:261. doi: 10.1186/s12864-017-3654-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yao W, Li G, Zhao H, Wang G, Lian X, Xie W. Exploring the rice dispensable genome using a metagenome-like assembly strategy. Genome Biol. 2015;16:187. doi: 10.1186/s13059-015-0757-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang W, Mauleon R, Hu Z, Chebotarov D, Tai S, Wu Z, et al. Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature. 2018;557:43–49. doi: 10.1038/s41586-018-0063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao Q, Feng Q, Lu H, Li Y, Wang A, Tian Q, et al. Pan-genome analysis highlights the extent of genomic variation in cultivated and wild rice. Nat Genet. 2018;50:278–284. doi: 10.1038/s41588-018-0041-z. [DOI] [PubMed] [Google Scholar]
- 44.Zhou Y, Chebotarov D, Kudrna D, Llaca V, Lee S, Rajasekar S, et al. A platinum standard pan-genome resource that represents the population structure of Asian rice. Sci Data. 2020;7:113. doi: 10.1038/s41597-020-0438-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma X, Fan J, Wu Y, Zhao S, Zheng X, Sun C, et al. Whole-genome de novo assemblies reveal extensive structural variations and dynamic organelle-to-nucleus DNA transfers in Asian and African rice. Plant J. 2020. 10.1111/tpj.14946. [DOI] [PMC free article] [PubMed]
- 46.Trouern-Trend AJ, Falk T, Zaman S, Caballero M, Neale DB, Langley CH, et al. Comparative genomics of six Juglans species reveals disease-associated gene family contractions. Plant J. 2020;102:410–423. doi: 10.1111/tpj.14630. [DOI] [PubMed] [Google Scholar]
- 47.Sun X, Jiao C, Schwaninger H, Chao CT, Ma Y, Duan N, et al. Phased diploid genome assemblies and pan-genomes provide insights into the genetic history of apple domestication. Nat Genet. 2020. 10.1038/s41588-020-00723-9. [DOI] [PMC free article] [PubMed]
- 48.Golicz AA, Bayer PE, Barker GC, Edger PP, Kim H, Martinez PA, et al. The pangenome of an agronomically important crop plant Brassica oleracea. Nat Commun 2016. doi: 10.1038/ncomms13390. [DOI] [PMC free article] [PubMed]
- 49.Gao L, Gonda I, Sun H, Ma Q, Bao K, Tieman DM, et al. The tomato pan-genome uncovers new genes and a rare allele regulating fruit flavor. Nat Genet. 2019;51:1044–1051. doi: 10.1038/s41588-019-0410-2. [DOI] [PubMed] [Google Scholar]
- 50.Haberer G, Kamal N, Bauer E, Gundlach H, Fischer I, Seidel MA, et al. European maize genomes highlight intraspecies variation in repeat and gene content. Nat Genet. 2020. 10.1038/s41588-020-0671-9. [DOI] [PMC free article] [PubMed]
- 51.Ou L, Li D, Lv J, Chen W, Zhang Z, Li X, et al. Pan-genome of cultivated pepper (Capsicum) and its use in gene presence--absence variation analyses. New Phytol Wiley Online Library. 2018;220:360–363. doi: 10.1111/nph.15413. [DOI] [PubMed] [Google Scholar]
- 52.Hübner S, Bercovich N, Todesco M, Mandel JR, Odenheimer J, Ziegler E, et al. Sunflower pan-genome analysis shows that hybridization altered gene content and disease resistance. Nat Plants. 2019;5:54–62. doi: 10.1038/s41477-018-0329-0. [DOI] [PubMed] [Google Scholar]
- 53.Montenegro JD, Golicz AA, Bayer PE, Hurgobin B, Lee H, Chan C-KK, et al. The pangenome of hexaploid bread wheat. Plant J. 2017;90:1007–1013. doi: 10.1111/tpj.13515. [DOI] [PubMed] [Google Scholar]
- 54.Pellicer J, Leitch IJ. The plant DNA C-values database (release 7.1): an updated online repository of plant genome size data for comparative studies. New Phytol. 2020;226:301–305. doi: 10.1111/nph.16261. [DOI] [PubMed] [Google Scholar]
- 55.Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- 56.Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- 57.Jaillon O, Aury J-M, Noel B, Policriti A, Clepet C, Casagrande A, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- 58.Potato Genome Sequencing Consortium. Xu X, Pan S, Cheng S, Zhang B, Mu D, et al. Genome sequence and analysis of the tuber crop potato. Nature. 2011;475:189–195. doi: 10.1038/nature10158. [DOI] [PubMed] [Google Scholar]
- 59.Mascher M, Gundlach H, Himmelbach A, Beier S, Twardziok SO, Wicker T, et al. A chromosome conformation capture ordered sequence of the barley genome. Nature. 2017;544:427–433. doi: 10.1038/nature22043. [DOI] [PubMed] [Google Scholar]
- 60.International Wheat Genome Sequencing Consortium (IWGSC), IWGSC RefSeq principal investigators, Appels R, Eversole K, Feuillet C, Keller B, et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science. 2018;361. 10.1126/science.aar7191. [DOI] [PubMed]
- 61.Xu X, Liu X, Ge S, Jensen JD, Hu F, Li X, et al. Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat Biotechnol. 2011;30:105–111. doi: 10.1038/nbt.2050. [DOI] [PubMed] [Google Scholar]
- 62.Huang X, Kurata N, Wei X, Wang Z-X, Wang A, Zhao Q, et al. A map of rice genome variation reveals the origin of cultivated rice. Nature. 2012;490:497–501. doi: 10.1038/nature11532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lam H-M, Xu X, Liu X, Chen W, Yang G, Wong F-L, et al. Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat Genet. 2010;42:1053–1059. doi: 10.1038/ng.715. [DOI] [PubMed] [Google Scholar]
- 64.Hirsch CN, Hirsch CD, Brohammer AB, Bowman MJ, Soifer I, Barad O, et al. Draft assembly of elite inbred line PH207 provides insights into genomic and transcriptome diversity in maize. Plant Cell. 2016;28:2700–2714. doi: 10.1105/tpc.16.00353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Springer NM, Anderson SN, Andorf CM, Ahern KR, Bai F, Barad O, et al. The maize W22 genome provides a foundation for functional genomics and transposon biology. Nat Genet. 2018;50:1282–1288. doi: 10.1038/s41588-018-0158-0. [DOI] [PubMed] [Google Scholar]
- 66.Li C, Song W, Luo Y, Gao S, Zhang R, Shi Z, et al. The HuangZaoSi maize genome provides insights into genomic variation and improvement history of maize. Mol Plant. 2019;12:402–409. doi: 10.1016/j.molp.2019.02.009. [DOI] [PubMed] [Google Scholar]
- 67.Schatz MC, Maron LG, Stein JC, Hernandez Wences A, Gurtowski J, Biggers E, et al. Whole genome de novo assemblies of three divergent strains of rice, Oryza sativa, document novel gene space of aus and indica. Genome Biol. 2014;15:506. doi: 10.1186/s13059-014-0506-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jiao Y, Peluso P, Shi J, Liang T, Stitzer MC, Wang B, et al. Improved maize reference genome with single-molecule technologies. Nature. 2017;546:524–527. doi: 10.1038/nature22971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Belser C, Istace B, Denis E, Dubarry M, Baurens F-C, Falentin C, et al. Chromosome-scale assemblies of plant genomes using nanopore long reads and optical maps. Nat Plants. 2018;4:879–887. doi: 10.1038/s41477-018-0289-4. [DOI] [PubMed] [Google Scholar]
- 70.Yang N, Liu J, Gao Q, Gui S, Chen L, Yang L, et al. Genome assembly of a tropical maize inbred line provides insights into structural variation and crop improvement. Nat Genet. 2019;51:1052–1059. doi: 10.1038/s41588-019-0427-6. [DOI] [PubMed] [Google Scholar]
- 71.VanBuren R, Man Wai C, Wang X, Pardo J, Yocca AE, Wang H, et al. Exceptional subgenome stability and functional divergence in the allotetraploid Ethiopian cereal teff. Nat Commun. 2020;11:884. doi: 10.1038/s41467-020-14724-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu J, Seetharam AS, Chougule K, Ou S, Swentowsky KW, Gent JI, et al. Gapless assembly of maize chromosomes using long read technologies bioRxiv. 2020. p. 2020.01.14.906230. Available from: https://www.biorxiv.org/content/10.1101/2020.01.14.906230v1.full. [cited 2020 Jan 30]. [DOI] [PMC free article] [PubMed]
- 73.Zhou P, Hirsch CN, Briggs SP, Springer NM. Dynamic patterns of gene expression additivity and regulatory variation throughout maize development. Mol Plant. 2019;12:410–425. doi: 10.1016/j.molp.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 74.Song B, Wang H, Wu Y, Rees E, Gates DJ, Burch M, et al. Constrained non-coding sequence provides insights into regulatory elements and loss of gene expression in maize. 2020. p. 2020.07.11.192575. Available from: https://www.biorxiv.org/content/10.1101/2020.07.11.192575v2.full. [cited 2020 Aug 12].
- 75.Ho SS, Urban AE, Mills RE. Structural variation in the sequencing era. Nat Rev Genet. 2020;21:171–189. doi: 10.1038/s41576-019-0180-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kosugi S, Momozawa Y, Liu X, Terao C, Kubo M, Kamatani Y. Comprehensive evaluation of structural variation detection algorithms for whole genome sequencing. Genome Biol. 2019;20:117. doi: 10.1186/s13059-019-1720-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chia J-M, Song C, Bradbury PJ, Costich D, de Leon N, Doebley J, et al. Maize HapMap2 identifies extant variation from a genome in flux. Nat Genet. 2012;44:803–807. doi: 10.1038/ng.2313. [DOI] [PubMed] [Google Scholar]
- 78.Bai Z, Chen J, Liao Y, Wang M, Liu R, Ge S, et al. The impact and origin of copy number variations in the Oryza species. BMC Genomics. 2016;17:261. doi: 10.1186/s12864-016-2589-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mace ES, Tai S, Gilding EK, Li Y, Prentis PJ, Bian L, et al. Whole-genome sequencing reveals untapped genetic potential in Africa’s indigenous cereal crop sorghum. Nat Commun. 2013;4:2320. doi: 10.1038/ncomms3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Alkan C, Coe BP, Eichler EE. Genome structural variation discovery and genotyping. Nat Rev Genet. 2011;12:363–376. doi: 10.1038/nrg2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ross MG, Russ C, Costello M, Hollinger A, Lennon NJ, Hegarty R, et al. Characterizing and measuring bias in sequence data. Genome Biol. 2013;14:R51. doi: 10.1186/gb-2013-14-5-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lemmon ZH, Bukowski R, Sun Q, Doebley JF. The role of cis regulatory evolution in maize domestication. Plos Genet. 2014;10:e1004745. doi: 10.1371/journal.pgen.1004745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Levy-Sakin M, Pastor S, Mostovoy Y, Li L, Leung AKY, McCaffrey J, et al. Genome maps across 26 human populations reveal population-specific patterns of structural variation. Nat Commun. 2019;10:1025. doi: 10.1038/s41467-019-08992-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Elyanow R, Wu H-T, Raphael BJ. Identifying structural variants using linked-read sequencing data. Bioinformatics. 2018;34:353–360. doi: 10.1093/bioinformatics/btx712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sedlazeck FJ, Rescheneder P, Smolka M, Fang H, Nattestad M, von Haeseler A, et al. Accurate detection of complex structural variations using single-molecule sequencing. Nat Methods. 2018;15:461–468. doi: 10.1038/s41592-018-0001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hu Z, Wei C, Li Z. Computational strategies for eukaryotic pangenome analyses. In: Tettelin H, Medini D, editors. The pangenome: diversity, dynamics and evolution of genomes. Cham: Springer; 2020. [PubMed] [Google Scholar]
- 87.Computational Pan-Genomics Consortium Computational pan-genomics: status, promises and challenges. Brief Bioinform. 2018;19:118–135. doi: 10.1093/bib/bbw089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Garrison E, Sirén J, Novak AM, Hickey G, Eizenga JM, Dawson ET, et al. Variation graph toolkit improves read mapping by representing genetic variation in the reference. Nat Biotechnol. 2018;36:875–879. doi: 10.1038/nbt.4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rakocevic G, Semenyuk V, Lee W-P, Spencer J, Browning J, Johnson IJ, et al. Fast and accurate genomic analyses using genome graphs. Nat Genet. 2019;51:354–362. doi: 10.1038/s41588-018-0316-4. [DOI] [PubMed] [Google Scholar]
- 90.Grytten I, Rand KD, Nederbragt AJ, Sandve GK. Assessing graph-based read mappers against a baseline approach highlights strengths and weaknesses of current methods. BMC Genomics. 2020;21:282. doi: 10.1186/s12864-020-6685-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Golicz AA, Batley J, Edwards D. Towards plant pangenomics. Plant Biotechnol J. 2016;14:1099–1105. doi: 10.1111/pbi.12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sherman RM, Salzberg SL. Pan-genomics in the human genome era. Nat Rev Genet. 2020;21:243–254. doi: 10.1038/s41576-020-0210-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Elliott TA, Gregory TR. What’s in a genome? The C-value enigma and the evolution of eukaryotic genome content. Philos Trans R Soc Lond B Biol Sci. 2015;370:20140331. doi: 10.1098/rstb.2014.0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jiang N, Ferguson AA, Slotkin RK, Lisch D. Pack-Mutator-like transposable elements (Pack-MULEs) induce directional modification of genes through biased insertion and DNA acquisition. Proc Natl Acad Sci U S A. 2011;108:1537–1542. doi: 10.1073/pnas.1010814108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wessler SR. The maize transposable Ds1 element is alternatively spliced from exon sequences. Mol Cell Biol. 1991;11:6192–6196. doi: 10.1128/MCB.11.12.6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Piskurek O, Jackson DJ. Transposable elements: from DNA parasites to architects of metazoan evolution. Genes. 2012;3:409–422. doi: 10.3390/genes3030409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chi J-T, Chang HY, Wang NN, Chang DS, Dunphy N, Brown PO. Genomewide view of gene silencing by small interfering RNAs. Proc Natl Acad Sci U S A. 2003;100:6343–6346. doi: 10.1073/pnas.1037853100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lewsey MG, Hardcastle TJ, Melnyk CW, Molnar A, Valli A, Urich MA, et al. Mobile small RNAs regulate genome-wide DNA methylation. Proc Natl Acad Sci U S A. 2016;113:E801–E810. doi: 10.1073/pnas.1515072113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hirsch CD, Springer NM. Transposable element influences on gene expression in plants. Biochim Biophys Acta Gene Regul Mech. 1860;2017:157–165. doi: 10.1016/j.bbagrm.2016.05.010. [DOI] [PubMed] [Google Scholar]
- 100.Makarevitch I, Waters AJ, West PT, Stitzer M, Hirsch CN, Ross-Ibarra J, et al. Transposable elements contribute to activation of maize genes in response to abiotic stress. Plos Genet. 2015;11:e1004915. doi: 10.1371/journal.pgen.1004915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yokosho K, Yamaji N, Fujii-Kashino M, Ma JF. Retrotransposon-mediated aluminum tolerance through enhanced expression of the citrate transporter OsFRDL4. Plant Physiol. 2016;172:2327–2336. doi: 10.1104/pp.16.01214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Butelli E, Licciardello C, Zhang Y, Liu J, Mackay S, Bailey P, et al. Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell. 2012;24:1242–1255. doi: 10.1105/tpc.111.095232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jiang N, Gao D, Xiao H, van der Knaap E. Genome organization of the tomato sun locus and characterization of the unusual retrotransposon Rider. Plant J. 2009;60:181–193. doi: 10.1111/j.1365-313X.2009.03946.x. [DOI] [PubMed] [Google Scholar]
- 104.Xiao H, Jiang N, Schaffner E, Stockinger EJ, van der Knaap E. A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science. 2008;319:1527–1530. doi: 10.1126/science.1153040. [DOI] [PubMed] [Google Scholar]
- 105.Anderson SN, Stitzer MC, Brohammer AB, Zhou P, Noshay JM, O’Connor CH, et al. Transposable elements contribute to dynamic genome content in maize. Plant J. 2019;100:1052–1065. doi: 10.1111/tpj.14489. [DOI] [PubMed] [Google Scholar]
- 106.Nelson MG, Linheiro RS, Bergman CM. McClintock: an integrated pipeline for detecting transposable element insertions in whole-genome shotgun sequencing data. G3. 2017:2763–78. 10.1534/g3.117.043893. [DOI] [PMC free article] [PubMed]
- 107.Bao W, Kojima KK, Kohany O. Repbase update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 2015; 6: 11. Epub 2015/06/06. 10.1186/s13100-015-0041-9 PMID: 26045719. [DOI] [PMC free article] [PubMed]
- 108.Chen J, Hu Q, Zhang Y, Lu C, Kuang H. P-MITE: a database for plant miniature inverted-repeat transposable elements. Nucleic Acids Res. 2014;42:D1176–D1181. doi: 10.1093/nar/gkt1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ou S, Jiang N. LTR_retriever: a highly accurate and sensitive program for identification of long terminal repeat retrotransposons. Plant Physiol. 2018;176:1410–1422. doi: 10.1104/pp.17.01310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Price AL, Jones NC, Pevzner PA. De novo identification of repeat families in large genomes. Bioinformatics. 2005;21(Suppl 1):i351–i358. doi: 10.1093/bioinformatics/bti1018. [DOI] [PubMed] [Google Scholar]
- 111.Girgis HZ. Red: an intelligent, rapid, accurate tool for detecting repeats de-novo on the genomic scale. BMC Bioinformatics. 2015;16:227. doi: 10.1186/s12859-015-0654-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Flynn JM, Hubley R, Goubert C, Rosen J, Clark AG, Feschotte C, et al. RepeatModeler2 for automated genomic discovery of transposable element families. Proc Natl Acad Sci U S A. 2020;117:9451–9457. doi: 10.1073/pnas.1921046117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hoen DR, Hickey G, Bourque G, Casacuberta J, Cordaux R, Feschotte C, et al. A call for benchmarking transposable element annotation methods. Mob DNA. 2015;6:13. doi: 10.1186/s13100-015-0044-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ou S, Su W, Liao Y, Chougule K, Agda JRA, Hellinga AJ, et al. Benchmarking transposable element annotation methods for creation of a streamlined, comprehensive pipeline. Genome Biol. 2019;20:275. doi: 10.1186/s13059-019-1905-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Stitzer MC, Anderson SN, Springer NM, Ross-Ibarra J. The genomic ecosystem of transposable elements in maize. 2019. p. 559922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Domínguez M, Dugas E, Benchouaia M, Leduque B, Jimenez-Gomez J, Colot V, et al. The impact of transposable elements on tomato diversity. 2020. p. 2020.06.04.133835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kumar J, Gupta DS, Gupta S, Dubey S, Gupta P, Kumar S. Quantitative trait loci from identification to exploitation for crop improvement. Plant Cell Rep. 2017;36:1187–1213. doi: 10.1007/s00299-017-2127-y. [DOI] [PubMed] [Google Scholar]
- 118.Liu Y, He Z, Appels R, Xia X. Functional markers in wheat: current status and future prospects. Theor Appl Genet. 2012;125:1–10. doi: 10.1007/s00122-012-1829-3. [DOI] [PubMed] [Google Scholar]
- 119.Collard BCY, Mackill DJ. Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philos Trans R Soc Lond Ser B Biol Sci. 2008;363:557–572. doi: 10.1098/rstb.2007.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gage JL, Vaillancourt B, Hamilton JP, Manrique-Carpintero NC, Gustafson TJ, Barry K, et al. Multiple maize reference genomes impact the identification of variants by genome-wide association study in a diverse inbred panel. Plant Genome. 2019;12. 10.3835/plantgenome2018.09.0069. [DOI] [PubMed]
- 121.Guo J, Cao K, Deng C, Li Y, Zhu G, Fang W, et al. An integrated peach genome structural variation map uncovers genes associated with fruit traits. Genome Biol. 2020;21:258. doi: 10.1186/s13059-020-02169-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dwivedi SL, Crouch JH, Mackill DJ, Xu Y, Blair MW, Ragot M, et al. The molecularization of public sector crop breeding: progress, problems, and prospects. Advances in Agronomy. Academic Press; 2007;95:163–318.
- 123.Xu Y, Crouch JH. Marker-assisted selection in plant breeding: from publications to practice. Crop Sci. 2008;48:391–407. doi: 10.2135/cropsci2007.04.0191. [DOI] [Google Scholar]
- 124.Bernardo R. Molecular markers and selection for complex traits in plants: learning from the last 20 years. Crop Sci. 2008;48:1649–1664. doi: 10.2135/cropsci2008.03.0131. [DOI] [Google Scholar]
- 125.Lorenz AJ, Chao S, Asoro FG, Heffner EL, Hayashi T, Iwata H, et al. Chapter Two - Genomic selection in plant breeding: knowledge and prospects. In: Sparks DL, editor. Advances in Agronomy. Academic Press. 2011;110:77–123.
- 126.Meuwissen TH, Hayes BJ, Goddard ME. Prediction of total genetic value using genome-wide dense marker maps. Genetics. 2001;157:1819–1829. doi: 10.1093/genetics/157.4.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kadam DC, Potts SM, Bohn MO, Lipka AE, Lorenz AJ. Genomic prediction of single crosses in the early stages of a maize hybrid breeding pipeline. G3. 2016;6:3443–3453. doi: 10.1534/g3.116.031286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Albrecht T, Wimmer V, Auinger H-J, Erbe M, Knaak C, Ouzunova M, et al. Genome-based prediction of testcross values in maize. Theor Appl Genet. 2011;123:339–350. doi: 10.1007/s00122-011-1587-7. [DOI] [PubMed] [Google Scholar]
- 129.Massman JM, Gordillo A, Lorenzana RE, Bernardo R. Genomewide predictions from maize single-cross data. Theor Appl Genet. 2013;126:13–22. doi: 10.1007/s00122-012-1955-y. [DOI] [PubMed] [Google Scholar]
- 130.Technow F, Schrag TA, Schipprack W, Bauer E, Simianer H, Melchinger AE. Genome properties and prospects of genomic prediction of hybrid performance in a breeding program of maize. Genetics. 2014;197:1343–1355. doi: 10.1534/genetics.114.165860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.de Oliveira AA, Pastina MM, de Souza VF, da Costa Parrella RA, Noda RW, Simeone MLF, et al. Genomic prediction applied to high-biomass sorghum for bioenergy production. Mol Breed. 2018;38:49. doi: 10.1007/s11032-018-0802-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Heffner EL, Jannink J-L, Iwata H, Souza E, Sorrells ME. Genomic selection accuracy for grain quality traits in biparental wheat populations. Crop Sci. 2011;51:2597–2606. doi: 10.2135/cropsci2011.05.0253. [DOI] [Google Scholar]
- 133.Jarquín D, Kocak K, Posadas L, Hyma K, Jedlicka J, Graef G, et al. Genotyping by sequencing for genomic prediction in a soybean breeding population. BMC Genomics. 2014;15:740. doi: 10.1186/1471-2164-15-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jensen SE, Charles JR, Muleta K, Bradbury PJ, Casstevens T, Deshpande SP, et al. A sorghum practical haplotype graph facilitates genome-wide imputation and cost-effective genomic prediction. Plant Genome. 2020;13:1687. doi: 10.1002/tpg2.20009. [DOI] [PubMed] [Google Scholar]
- 135.Burgueño J, de los Campos G, Weigel K, Crossa J. Genomic prediction of breeding values when modeling genotype × environment interaction using pedigree and dense molecular markers. Crop Sci. 2012;52:707–719. doi: 10.2135/cropsci2011.06.0299. [DOI] [Google Scholar]
- 136.Jarquín D, Crossa J, Lacaze X, Du Cheyron P, Daucourt J, Lorgeou J, et al. A reaction norm model for genomic selection using high-dimensional genomic and environmental data. Theor Appl Genet. 2014;127:595–607. doi: 10.1007/s00122-013-2243-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Heslot N, Akdemir D, Sorrells ME, Jannink J-L. Integrating environmental covariates and crop modeling into the genomic selection framework to predict genotype by environment interactions. Theor Appl Genet. 2014;127:463–480. doi: 10.1007/s00122-013-2231-5. [DOI] [PubMed] [Google Scholar]
- 138.Stuart T, Eichten SR, Cahn J, Karpievitch YV, Borevitz JO, Lister R. Population scale mapping of transposable element diversity reveals links to gene regulation and epigenomic variation. Elife. 2016;5. 10.7554/eLife.20777. [DOI] [PMC free article] [PubMed]
- 139.Lyra DH, Galli G, Alves FC, Granato ÍSC, Vidotti MS, Bandeira E, Sousa M, et al. Modeling copy number variation in the genomic prediction of maize hybrids. Theor Appl Genet. 2019;132:273–288. doi: 10.1007/s00122-018-3215-2. [DOI] [PubMed] [Google Scholar]
- 140.Ramsey J, Schemske DW. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu Rev Ecol Syst. 1998;29:467–501. doi: 10.1146/annurev.ecolsys.29.1.467. [DOI] [Google Scholar]
- 141.Bretagnolle F, Thompson JD. Gametes with the somatic chromosome number: mechanisms of their formation and role in the evolution of autopolyploid plants. New Phytol. 1995;129:1–22. doi: 10.1111/j.1469-8137.1995.tb03005.x. [DOI] [PubMed] [Google Scholar]
- 142.Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473:97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- 143.Wolfe KH. Yesterday’s polyploids and the mystery of diploidization. Nat Rev Genet. 2001;2:333–341. doi: 10.1038/35072009. [DOI] [PubMed] [Google Scholar]
- 144.Crow KD, Wagner GP. What is the role of genome duplication in the evolution of complexity and diversity? Soc Mol Biol Evol. 2005;23:887–892. doi: 10.1093/molbev/msj083. [DOI] [PubMed] [Google Scholar]
- 145.Soltis DE, Visger CJ, Marchant DB, Soltis PS. Polyploidy: pitfalls and paths to a paradigm. Am J Bot. 2016;103:1146–1166. doi: 10.3732/ajb.1500501. [DOI] [PubMed] [Google Scholar]
- 146.Rosyara U, Kishii M, Payne T, Sansaloni CP, Singh RP, Braun H-J, et al. Genetic contribution of synthetic hexaploid wheat to CIMMYT’s spring bread wheat breeding germplasm. Sci Rep. 2019. 10.1038/s41598-019-47936-5. [DOI] [PMC free article] [PubMed]
- 147.Wu J-H, Ferguson AR, Murray BG, Jia Y, Datson PM, Zhang J. Induced polyploidy dramatically increases the size and alters the shape of fruit in Actinidia chinensis. Ann Bot. 2012;109:169–179. doi: 10.1093/aob/mcr256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Varoquaux F, Blanvillain R, Delseny M, Gallois P. Less is better: new approaches for seedless fruit production. Trends Biotechnol. 2000;18:233–242. doi: 10.1016/S0167-7799(00)01448-7. [DOI] [PubMed] [Google Scholar]
- 149.D’Hont A, Denoeud F, Aury J-M, Baurens F-C, Carreel F, Garsmeur O, et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature. 2012;488:213–217. doi: 10.1038/nature11241. [DOI] [PubMed] [Google Scholar]
- 150.Shulaev V, Sargent DJ, Crowhurst RN, Mockler TC, Folkerts O, Delcher AL, et al. The genome of woodland strawberry (Fragaria vesca) Nat Genet. 2011;43:109–116. doi: 10.1038/ng.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Comai L. The advantages and disadvantages of being polyploid. Nat Rev Genet. 2005;6:836–846. doi: 10.1038/nrg1711. [DOI] [PubMed] [Google Scholar]
- 152.Ramírez-González RH, Borrill P, Lang D, Harrington SA, Brinton J, Venturini L, et al. The transcriptional landscape of polyploid wheat. Science. 2018;361. 10.1126/science.aar6089. [DOI] [PubMed]
- 153.Renny-Byfield S, Wendel JF. Doubling down on genomes: polyploidy and crop plants. Am J Bot. 2014;101:1711–1725. doi: 10.3732/ajb.1400119. [DOI] [PubMed] [Google Scholar]
- 154.Bird KA, VanBuren R, Puzey JR, Edger PP. The causes and consequences of subgenome dominance in hybrids and recent polyploids. New Phytol. 2018;220:87–93. doi: 10.1111/nph.15256. [DOI] [PubMed] [Google Scholar]
- 155.Woodhouse MR, Cheng F, Pires JC, Lisch D, Freeling M, Wang X. Origin, inheritance, and gene regulatory consequences of genome dominance in polyploids. Proc Natl Acad Sci U S A. 2014;111:5283–5288. doi: 10.1073/pnas.1402475111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kyriakidou M, Tai HH, Anglin NL, Ellis D, Strömvik MV. Current strategies of polyploid plant genome sequence assembly. Front Plant Sci. 2018;9:1660. doi: 10.3389/fpls.2018.01660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhuang W, Chen H, Yang M, Wang J, Pandey MK, Zhang C, et al. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat Genet. 2019;51:865–876. doi: 10.1038/s41588-019-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Edger PP, Poorten TJ, VanBuren R, Hardigan MA, Colle M, McKain MR, et al. Origin and evolution of the octoploid strawberry genome. Nat Genet. 2019;51:541–547. doi: 10.1038/s41588-019-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kyriakidou M, Anglin NL, Ellis D, Tai HH, Strömvik MV. Genome assembly of six polyploid potato genomes. Sci Data. 2020;7:88. doi: 10.1038/s41597-020-0428-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Van Tassel DL, Albrecht KA, Bever JD, Boe AA, Brandvain Y, Crews TE, et al. Accelerating domestication: an opportunity to develop new crop Ideotypes and breeding strategies informed by multiple disciplines. Crop Sci. 2017;57:1274. doi: 10.2135/cropsci2016.10.0834. [DOI] [Google Scholar]
- 161.Raduski AR, Herman A, Pogoda C, Dorn KM, Van Tassel DL, Kane N, et al. Patterns of genetic variation in a prairie wildflower, Silphium integrifolium, suggest a non-prairie origin and untapped variation available for improved breeding bioRxiv. 2020. p. 2020.06.25.171272. Available from: https://www.biorxiv.org/content/10.1101/2020.06.25.171272v1.abstract. [cited 2020 Jul 14].
- 162.Sedbrook JC, Phippen WB, Marks MD. New approaches to facilitate rapid domestication of a wild plant to an oilseed crop: example pennycress (Thlaspi arvense L.) Plant Sci. 2014;227:122–132. doi: 10.1016/j.plantsci.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 163.Dorn KM, Fankhauser JD, Wyse DL, Marks MD. De novo assembly of the pennycress (Thlaspi arvense) transcriptome provides tools for the development of a winter cover crop and biodiesel feedstock. Plant J. 2013;75:1028–1038. doi: 10.1111/tpj.12267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Dorn KM, Fankhauser JD, Wyse DL, Marks MD. A draft genome of field pennycress (Thlaspi arvense) provides tools for the domestication of a new winter biofuel crop. DNA Res. 2015;22:121–131. doi: 10.1093/dnares/dsu045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Byrne SL, Nagy I, Pfeifer M, Armstead I, Swain S, Studer B, et al. A synteny-based draft genome sequence of the forage grass Lolium perenne. Plant J Wiley Online Library. 2015;84:816–826. doi: 10.1111/tpj.13037. [DOI] [PubMed] [Google Scholar]
- 166.Qiu Y, Yang Y, Hirsch CD, Watkins E. Building a reference transcriptome for the hexaploid hard fescue turfgrass (Festuca brevipila) using a combination of PacBio Isoseq and Illumina sequencing. bioRxiv. 2020. p. 2020.02.26.966952. Available from: https://www.biorxiv.org/content/10.1101/2020.02.26.966952v1.abstract. [cited 2020 Jul 8].
- 167.Chakrabarti M, Zhang N, Sauvage C, Muños S, Blanca J, Cañizares J, et al. A cytochrome P450 regulates a domestication trait in cultivated tomato. Proc Natl Acad Sci U S A. 2013;110:17125–17130. doi: 10.1073/pnas.1307313110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hufford MB, Xu X, van Heerwaarden J, Pyhäjärvi T, Chia J-M, Cartwright RA, et al. Comparative population genomics of maize domestication and improvement. Nat Genet. 2012;44:808–811. doi: 10.1038/ng.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hu B, Wang W, Ou S, Tang J, Li H, Che R, et al. Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat Genet. 2015;47:834–838. doi: 10.1038/ng.3337. [DOI] [PubMed] [Google Scholar]
- 170.Mirzaghaderi G, Mason AS. Broadening the bread wheat D genome. Theor Appl Genet. 2019;132:1295–1307. doi: 10.1007/s00122-019-03299-z. [DOI] [PubMed] [Google Scholar]
- 171.Fernie AR, Yan J. De novo domestication: an alternative route toward new crops for the future. Mol Plant. 2019;12:615–631. doi: 10.1016/j.molp.2019.03.016. [DOI] [PubMed] [Google Scholar]
- 172.Gratacap RL, Wargelius A, Edvardsen RB, Houston RD. Potential of genome editing to improve aquaculture breeding and production. Trends Genet. 2019;35:672–684. doi: 10.1016/j.tig.2019.06.006. [DOI] [PubMed] [Google Scholar]
- 173.Zsögön A, Čermák T, Naves ER, Notini MM, Edel KH, Weinl S, et al. De novo domestication of wild tomato using genome editing. Nat Biotechnol. 2018. 10.1038/nbt.4272. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable