To the Editor:
Recent advances in single-cell omics have provided increasing insights into the pathogenesis of human diseases, including those affecting the lung (1–7). The density of omics data relevant to lung biology and diseases is increasing exponentially through the work of research consortia and individual investigators (1, 3, 8–12). Discerning the best way to optimize the use of these rich datasets, integrate multiomics data, extract biologically meaningful knowledge, and make that knowledge available to the research community in a user-friendly manner is a challenging opportunity. With support from the National Heart, Lung, and Blood Institute (NHLBI) “LungMAP” (Lung Map) consortium, we developed the Lung Gene Expression Analysis (LGEA) database and web portal to facilitate access and visualization of extensive bulk, sorted, single-cell transcriptomic and image data from human and mouse lungs at different stages of development and disease (13, 14). Data hosted on LGEA are primarily produced by LungMAP research centers. We process and interpret the data and make it available to all investigators before its publication (8). LGEA has been widely used by researchers from more than 130 institutions from 52 different countries and has been cited in more than 130 scientific publications. The newly updated LGEA version 3 introduces a new featured web toolset, “lung-at-a-glance,” for exploring and understanding complex multiomics and imaging data, providing an interactive web interface to bridge lung anatomic ontology classifications to lung structure, histology, and immunofluorescence confocal images and cell type–specific gene expression.
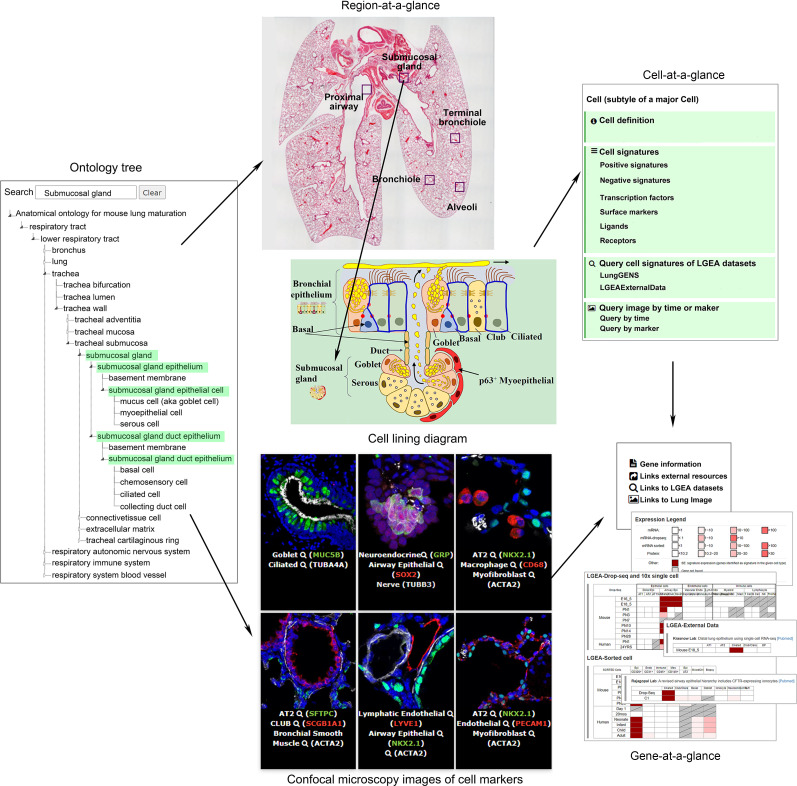
Lung-at-a-glance consists of “region,” “cell,” and “gene,” three interactive components all designed to provide data access with a single click on the icons (https://research.cchmc.org/pbge/lunggens/tools/lung_at_glance.html). We name the toolset as “lung-at-a-glance” because it provides the first comprehensive lung anatomic ontology tree along the proximal–distal axis of the organ, including epithelial, stromal, vascular, neural, and immunologic components provide a “head-to-toe” view of the lung. Major anatomical regions, cells within each region, and gene markers associated with each cell, provide an “inside-out” view of the lung. The at-a-glance toolset provides a collection of comprehensive interrelated data and knowledge resources with an intuitive and interactive web interface for data analysis, integration, and visualization. The anatomic ontology for human and mouse lungs was developed by the National Heart, Lung, and Blood Institute LungMAP Consortium Ontology Subcommittee using web ontology language. This is the first comprehensive anatomic ontology of the lung organized along the proximal–distal axis of the lung into epithelial, stromal, vascular, neural, and immunologic components, containing ∼300 terms for fetal and postnatal structures, tissues, and cells, which were identified for each species (15). We converted the abstract version of anatomic ontology terms into searchable, clickable, and expandable web-tree structures on the lung-at-a-glance home page, serving as an interactive bridge to connect lung images and lung gene expression (Figure 1). Investigators can navigate the hierarchical structure of the anatomical tree or use the search box to directly locate regions or cells of interest.
Figure 1.
“Lung-at-a-glance” consists of the following three interactive and interconnected components: “region-at-a-glance,” “cell-at-a-glance,” and “gene-at-a-glance.” Region-at-a-glance enables users to search a specific lung region using the interactive ontology tree navigation or by clicking the boxes in the lung image to explore cells residing in the selected region. Cell-at-a-glance offers a collection of information on queried cell types, including cell definition, cell type–specific positive and negative markers, transcription factors, ligand receptors, and hyperlinks that are mapped to the chosen cell type in all datasets in the Lung Gene Expression Analysis (LGEA) web portal and immunofluorescence confocal images of the chosen cell type. Gene-at-a-glance allows users to query a gene of interest to obtain associated gene information, including hyperlinks to external knowledge bases, expression patterns in LGEA datasets, and immunofluorescence confocal images associated with the gene marker. A two-dimensional heatmap organized by developmental times and cell types was used to summarize expression patterns and statistics of queried genes in all datasets available in LGEA databases. Screen images are taken from the LGEA web portal (https://research.cchmc.org/pbge/lunggens/mainportal.html). Adapted from Reference 16. LYVE1 = lymphatic vessel endothelial hyaluronan receptor 1; NKX2.1 = NK2 homeobox 1; PECAM1 = platelet and endothelial cell adhesion molecule 1; SOX2 = SRY-box transcription factor; TUBA4A = tubulin alpha 4a; TUBB3 = tubulin beta 3 class III.
“Region-at-a-glance” enables users to search a specific lung region using the interactive navigation tool or by clicking one of the annotated lung regions (e.g., “proximal airway,” “submucosal gland,” “bronchiole,” “terminal bronchiole,” and “alveoli”) on the hematoxylin and eosin stained lung image. Users can explore cells within selected regions using interactive mouse hover features (embedded in the anatomical ontology tree), images, and diagrams.
“Cell-at-a-glance” can be activated by clicking the cell name or image on the “cell-at-a-glance” page or by clicking a cell of interest from selected regional diagrams on the region-at-a-glance page. Cell-at-a-glance offers a collection of information related to the queried cell type, including cell definition, cell type–specific positive and negative markers, transcription factors, ligand receptors predicted by our group (https://github.com/xu-lab/LGEA_Cell_Signature), and hyperlinks to all datasets in LGEA and immunofluorescence confocal images of the chosen cell type. Approximately 40 cell types are available for study in the current cell-at-a-glance.
“Gene-at-a-glance” enables users to query a gene of interest and obtain RNA/protein expression patterns in all LGEA datasets in a two-dimensional heatmap. Hyperlinks to external knowledgebase and immunofluorescence confocal images of relevant cell markers are provided in the gene-at-a-glance page. The three components of lung-at-a-glance are interconnected, offering users a one-stop bioinformatics tool for lung research (Figure 1). For example, investigators can start their search at specific anatomic regions, explore a particular cell type within the region, and identify cell-specific markers, ligand receptors, and transcription factors expressed in the cell type of interest across lung developmental stages.
In addition to lung-at-a-glance, the LGEA version 3 new release represents a significant update of the previous version, expanding to 10 functional query panels from three panels in the previous version (13, 14). In addition to the transcriptomic data from normal lung developmental studies, the current LGEA web portal extends the scope to include proteomics, epigenetic, lung ontology, and lung disease data (https://research.cchmc.org/pbge/lunggens/mainportal.html). To facilitate the use and integration of these data resources, we developed several bioinformatics tools in the “LGEA ToolBox” for investigators to compare and integrate their own gene list of interest with LGEA datasets. The complete functional panels and their functionality are described in Appendix I in the online data supplement. To facilitate the training and usage of lung-at-a-glance and other tools of the LGEA web portal, we have provided online tutorials and user case examples on the LGEA home page and in Appendix II in the online data supplement.
In summary, the LGEA web portal is designed for intuitive and practical interrogation of comprehensive omics data obtained during normal lung morphogenesis and diseases by research investigators with various levels of experience and training in computational approaches. The new LGEA release provides improved interactive, graphical web interfaces for search, visualization, and secondary analyses, from which outputs can be readily visualized, interpreted, and downloaded. The featured toolset of lung-at-a-glance offers end-to-end web functions to access and search lung anatomic ontology terms and to explore the corresponding structure and morphology of tissue regions, cells, and marker gene expression patterns in all LGEA datasets. To our knowledge, this is the first web application connecting anatomic ontology terms to lung structure and histology with single-cell expression data. These tools and enriched data resources can be used to enhance hypothesis generation and scientific discovery. The LGEA database will be continually updated with more omics data generated in LungMAP phase 2 and other laboratories interested in having their data hosted on the website. New query functions will be developed to enhance data and knowledge interrogation. Data are made available and synchronized on the LungMAP website (https://www.lungmap.net/). The LGEA web portal version 3 (http://research.cchmc.org/pbge/lunggens/mainportal.html) and lung-at-a-glance toolset (https://research.cchmc.org/pbge/lunggens/tools/lung_at_glance.html) are freely available for noncommercial use, and data are readily integrated with omics data and lung image data from other research centers at the BREATH (Bioinformatics REsource ATlas for the Healthy lung) database and are displayed on the LungMAP website (https://www.lungmap.net/).
This study has been previously reported in the form of a preprint (16).
Supplementary Material
Acknowledgments
Acknowledgment
The authors thank Dr. Sara Lin (Program Director) and all members of the LungMAP research consortium.
Footnotes
Supported by U.S. National Institutes of Health grants U01HL122642, U01HL148856, U01HL134745, and P30 DK117467 and the Chan Zuckerberg Foundation (Human Cell Atlas Lung Seed Network).
Author Contributions: Y.D., M.G., and Y.X. conceived and designed the web application. Y.D. developed the database and web application of Lung Gene Expression Analysis web portal. W.O. developed the web application of Lung Gene Expression Analysis lung ontology. Y.D. and W.O. developed the lung-at-a-glance toolsets. J.A.K. and J.A.W. designed and developed the web application of lung image. Y.D., M.G., S.Z., and Y.X. contributed to data analysis and interpretation. Y.D., J.A.W., and Y.X. wrote the manuscript. All authors contributed to the manuscript editing and approved the final manuscript.
This letter has a data supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Author disclosures are available with the text of this letter at www.atsjournals.org.
References
- 1.Goldfarbmuren KC, Jackson ND, Sajuthi SP, Dyjack N, Li KS, Rios CL, et al. Dissecting the cellular specificity of smoking effects and reconstructing lineages in the human airway epithelium. Nat Commun. 2020;11:2485. doi: 10.1038/s41467-020-16239-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reyfman PA, Walter JM, Joshi N, Anekalla KR, McQuattie-Pimentel AC, Chiu S, et al. Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am J Respir Crit Care Med. 2019;199:1517–1536. doi: 10.1164/rccm.201712-2410OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schiller HB, Montoro DT, Simon LM, Rawlins EL, Meyer KB, Strunz M, et al. The human lung cell atlas: a high-resolution reference map of the human lung in health and disease. Am J Respir Cell Mol Biol. 2019;61:31–41. doi: 10.1165/rcmb.2018-0416TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vieira Braga FA, Kar G, Berg M, Carpaij OA, Polanski K, Simon LM, et al. A cellular census of human lungs identifies novel cell states in health and in asthma. Nat Med. 2019;25:1153–1163. doi: 10.1038/s41591-019-0468-5. [DOI] [PubMed] [Google Scholar]
- 5.Raredon MSB, Adams TS, Suhail Y, Schupp JC, Poli S, Neumark N, et al. Single-cell connectomic analysis of adult mammalian lungs. Sci Adv. 2019;5:eaaw3851. doi: 10.1126/sciadv.aaw3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo M, Yu JJ, Perl AK, Wikenheiser-Brokamp KA, Riccetti M, Zhang EY, et al. Identification of the lymphangioleiomyomatosis cell and its uterine origin [preprint] bioRxiv. 2019 [accessed 2019 Oct 8]. Available from: https://www.biorxiv.org/content/10.1101/784199v1. [Google Scholar]
- 7.Xu Y, Mizuno T, Sridharan A, Du Y, Guo M, Tang J, et al. Single-cell RNA sequencing identifies diverse roles of epithelial cells in idiopathic pulmonary fibrosis. JCI Insight. 2016;1:e90558. doi: 10.1172/jci.insight.90558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ardini-Poleske ME, Clark RF, Ansong C, Carson JP, Corley RA, Deutsch GH, et al. LungMAP Consortium. LungMAP: the molecular atlas of lung development program. Am J Physiol Lung Cell Mol Physiol. 2017;313:L733–L740. doi: 10.1152/ajplung.00139.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han X, Wang R, Zhou Y, Fei L, Sun H, Lai S, et al. Mapping the mouse cell atlas by microwell-seq. Cell. 2018;172:1091–1107, e17. doi: 10.1016/j.cell.2018.02.001. [Published erratum appears in Cell 173:1307.] [DOI] [PubMed] [Google Scholar]
- 10.Regev A, Teichmann SA, Lander ES, Amit I, Benoist C, Birney E, et al. Human Cell Atlas Meeting Participants. The human cell atlas. Elife. 2017;6:e27041. doi: 10.7554/eLife.27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peyser R, MacDonnell S, Gao Y, Cheng L, Kim Y, Kaplan T, et al. Defining the activated fibroblast population in lung fibrosis using single-cell sequencing. Am J Respir Cell Mol Biol. 2019;61:74–85. doi: 10.1165/rcmb.2018-0313OC. [DOI] [PubMed] [Google Scholar]
- 12.Regan EA, Hersh CP, Castaldi PJ, DeMeo DL, Silverman EK, Crapo JD, et al. Omics and the search for blood biomarkers in chronic obstructive pulmonary disease: insights from COPDGene. Am J Respir Cell Mol Biol. 2019;61:143–149. doi: 10.1165/rcmb.2018-0245PS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du Y, Guo M, Whitsett JA, Xu Y. ‘LungGENS’: a web-based tool for mapping single-cell gene expression in the developing lung. Thorax. 2015;70:1092–1094. doi: 10.1136/thoraxjnl-2015-207035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du Y, Kitzmiller JA, Sridharan A, Perl AK, Bridges JP, Misra RS, et al. Lung Gene Expression Analysis (LGEA): an integrative web portal for comprehensive gene expression data analysis in lung development. Thorax. 2017;72:481–484. doi: 10.1136/thoraxjnl-2016-209598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan H, Deutsch GH, Wert SE Ontology Subcommittee; NHLBI Molecular Atlas of Lung Development Program Consortium. Comprehensive anatomic ontologies for lung development: a comparison of alveolar formation and maturation within mouse and human lung. J Biomed Semantics. 2019;10:18. doi: 10.1186/s13326-019-0209-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du Y, Ouyang W, Kitzmiller JA, Guo M, Zhao S, Whitsett JA, et al. doi: 10.1101/2020.06.19.161851. Lung at a glance: an integrative web toolset of lung ontology, imaging and single cell omics [preprint]. bioRxiv; 2020 [accessed 2020 Jun 20]. Available from: [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.