Abstract
Aberrant MET signaling can drive tumorigenesis in several cancer types through a variety of molecular mechanisms including MET gene amplification, mutation, rearrangement, and overexpression. Improvements in biomarker discovery and testing have more recently enabled the selection of patients with MET-dependent cancers for treatment with potent, specific, and novel MET-targeting therapies. We review the known oncologic processes that activate MET, discuss therapeutic strategies for MET-dependent malignancies, and highlight emerging challenges in acquired drug resistance in these cancers.
INTRODUCTION
The MET proto-oncogene encodes the tyrosine kinase receptor of the hepatocyte growth factor (HGF) and regulates embryogenesis, wound healing, liver regeneration, angiogenesis, and immunomodulation, among other physiologic processes (1-5). In cancer, aberrant MET oncogenic signaling has been known to play a role in promoting tumor invasion, angiogenesis, and metastasis; however, the initial development of targeted therapies against MET in unselected patient populations and across different tumor types did not translate into improved clinical outcomes (1,6-8). More recently, advances in the identification of new biomarkers of MET dysregulation in cancer have renewed immense interest in identifying MET-dependent malignancies and in developing effective treatment options.
MET is a single-pass transmembrane receptor composed of an extracellular domain, transmembrane and juxtamembrane domains and a tyrosine kinase domain. The extracellular portion of MET is the binding site for its ligand, HGF, and contains a semaphorin (SEMA) domain, as well as plexin-semaphorin-integrin (PSI) and the Ig-like, plexins, transcription factors (IPT) domains (9). Upon ligand binding, MET homodimerization results in the phosphorylation of key intracellular tyrosine residues at positions Y1234/35 within the kinase domain and Y1349/56 in the docking site(10). These events directly recruit downstream effectors including the SRC proto-oncogene and signal transducer and activator of transcription 3 (STAT3), as well as adaptor proteins like the growth factor receptor-bound protein 2 (GRB2) and SHC, leading to downstream activation of the mitogen-activated protein kinase (MAPK) and phosphatidylinositol 3-kinase (PI3K)/AKT/mTOR pathways which promote cell migration, proliferation and survival (9,11). MET protein receptor stability and degradation is regulated by the intracellular juxtamembrane domain which is encoded in part by MET exon 14 and contains the tyrosine Y1003 residue that, when phosphorylated, serves as the binding site for the casitas B-lineage lymphoma (CBL) E3 ubiquitin ligase (12). CBL-mediated ubiquitination results in receptor internalization from the cell membrane to endocytic vesicles and subsequent proteasomal degradation (13,14). Exon 14 in MET is 141 nucleotides long, spanning from nucleotides c.2888 to c.3028, corresponding to amino acids p.D963 to p.D1010 (NM_000245.2, variant 2). Of note, in an alternative splice isoform of MET which is 54 nucleotides longer (NM_001127500.2 Variant 1), exon 14 spans from nucleotides c.2942 to c.3082, corresponding to amino acids p.D981 to p.D1028. This alternative numbering of nucleotides and amino acids has led to some confusion in the literature. In this review, we will primarily use the shorter isoform (variant 2) when referring to MET sequences but will indicate the corresponding sequence variant of the longer isoform (variant 1).
The oncogenic role of MET was first described with the discovery of the TPR-MET genomic rearrangement induced by the exposure of a non-tumorigenic human osteogenic sarcoma cell line to the carcinogen N-methyl-N'-nitro-N-nitrosoguanidine (MNNG) (15). Since then, across diverse tumor types, multiple biological alterations in MET have been discovered including exon 14 skipping mutations, activating mutations in the kinase domain, gene amplification, and protein overexpression. Recognition and detection of these MET biomarkers have fueled the clinical development of MET tyrosine kinase inhibitors (TKIs), antibodies, and antibody-drug conjugates (ADCs) aimed at targeting MET or its interactions with HGF or other binding partners.
MOLECULAR MECHANISMS OF MET ACTIVATION IN CANCER
MET exon 14 alterations
Approximately 3% of advanced non-small cell lung cancers (NSCLC) harbor point mutations or deletions in MET exon 14 or its flanking introns which affect splicing sequences including 5’- and 3’-splice sites, the branch-point adenosine, or the polypyrimidine tract (12,16,17). These alterations result in MET exon 14 skipping during pre-mRNA splicing, resulting in loss of the CBL-binding site and increased half-life of the MET receptor (Figure 1A) (18). As has been described in some hereditary syndromes, point mutations in the last nucleotide of an exon often result in exon skipping (19), and point mutations in the last nucleotide of MET exon 14 (c.3028 = c.3082; D1010X = D1028X in the shorter and longer isoforms of MET, respectively) also tend to cause exon 14 skipping. Additional point mutations causing amino acid substitution of the Y1003 residue (=Y1021), such as Y1003C or Y1003F, are predicted to cause a similar biologic effect through loss of the CBL binding site without causing exon 14 skipping (20).
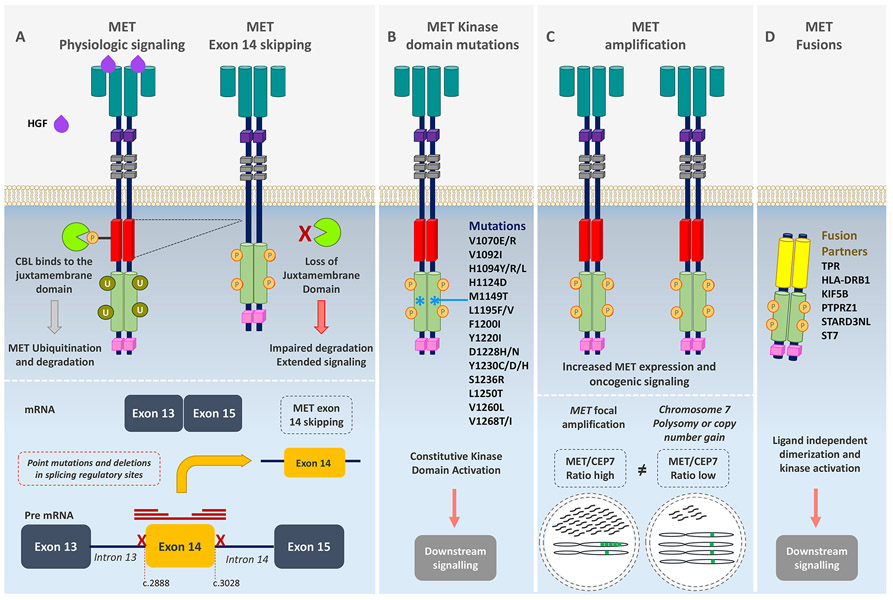
Figure 1. Mechanisms of MET oncogenic activation.
(A) MET activation is initiated after ligand binding of the hepatocyte growth factor (HGF) to the SEMA domain to the extracellular portion of the MET receptor, inducing receptor dimerization and phosphorylation of the intracellular domain leading to downstream signaling of several pathways. The E3 ubiquitin ligase CBL binds to reside Y1003 in the juxtamembrane domain (encoded in part by exon 14), resulting in receptor ubiquitination and degradation. MET exon 14 alterations including mutations or deletions in splicing regulatory sites, leading to exon skipping, deletion of a portion of the juxtamembrane domain, impaired CBL binding, decreased MET turnover, and ongoing oncogenic downstream pathway signaling. (B) Point mutations in the tyrosine kinase domain result in ligand-independent MET activation through autophosphorylation of the tyrosine kinase domain, conveying sustained oncogenic downstream pathway signaling. (C) Focal MET amplification leads to higher levels of MET transcription and MET expression. MET amplification (high MET/CEP7 ratio) differs from chromosome 7 polysomy or copy number gain in which the entire chromosome is duplicated (low MET/CEP7 ratio). Focal MET amplification and, consequently high MET expression, leads to enhanced ligand-independent oncogenic signaling by receptor auto-dimerization or oligomerization and auto-phosphorylation. (D) Gene rearrangements involving the MET tyrosine kinase domain result in fusion proteins that typically self-dimerize in a ligand-independent manner, leading to phosphorylation of the tyrosine kinase domain.
Splice site and Y1003X mutations differ from other mutations within exon 14, like T992I (=T1010I). The oncogenic potential of this variant is controversial, as some preclinical studies suggest that it imparts greater tumor growth and invasion potential whereas others report a lack of transforming capacity; this variant does not cause MET exon 14 skipping or seem to affect the CBL binding site (21-23).
MET exon 14 mutant NSCLC tends to occur in older patients (median age ~72) with a history of tobacco use, but can also be found in never smokers (24). These alterations are most commonly detected in lung adenocarcinomas, but are also enriched in pulmonary sarcomatoid (pleomorphic) carcinomas and can be found in squamous and adenosquamous NSCLC, and also rarely in other cancer types like gliomas and unknown primary tumors (16,25,26). De novo MET exon 14 alterations are almost always mutually exclusive with other oncogenic driver mutations like KRAS, EGFR, ALK, ROS1, and RET (16,24), and commonly co-occurring genomic alterations include mutations in TP53, loss of CDKN2A/B, and amplifications of MET, MDM2, and CDK4/6 (16,24,26).
The large diversity of MET genomic alterations leading to exon 14 skipping can pose a challenge for the routine detection of these mutations and deletions in clinical practice. Current methods used to identify lung cancers harboring MET exon 14 mutations include DNA next generation sequencing (NGS) platforms, Sanger sequencing of exon 14 and its flanking introns, and the detection of MET exon 14 skipping using RNA-based assays like reverse transcription polymerase chain reaction (RT-qPCR) and RNA based NGS (27). At present, there are no certified companion diagnostic tests for the detection of MET exon 14 skipping alterations that are recommended to identify patients for treatment with specific MET inhibitors; however NGS and RT-qPCR assays appear to demonstrate higher sensitivity compared to Sanger sequencing (28). With greater use of tumor- and blood-based NGS, these alterations are being identified with increased frequency in patients whose tumors undergo broad panel-based sequence analysis (29). With mounting evidence that MET exon 14 mutant NSCLC can respond to MET tyrosine kinase inhibitors (see ”Targeting MET alterations in cancer”), the inclusion of this promising biomarker in clinical sequencing assays among patients with advanced NSCLC should be strongly encouraged (30).
MET kinase domain mutations
Activating oncogenic mutations can also occur in other domains of MET, including in the tyrosine kinase domain (TKD), leading to ligand-independent receptor phosphorylation and signaling (Figure 1B) (31). Activating hereditary or sporadic point mutations in the MET TKD are found in about 13-20% of type 1 papillary renal cell carcinomas (pRCC) and result in constitutive MET receptor activation by affecting the inhibitory conformation of the activation loop, favoring kinase domain phosphorylation and prompting the downstream oncogenic signaling cascades (31-36). The variety of MET activating mutations found in pRCC include: V1092I (V1110), H1094Y/R/L (H1112), H1124D (H1142), L1195F/V (L1213), F1200I (F1218), V1188L (V1206), Y1220I (Y1238), D1228H/N (D1246), Y1230C/D/H (Y1248), M1131T (M1149), and M1250T (M1268) (31,35,37). Additionally, higher levels of MET expression are observed in pRCC due to allelic imbalances resulting from chromosome 7 polysomy, suggesting that additive oncogenic mechanisms are required to drive MET dependency in this tumor type (38).
Early trials with MET inhibitors have shown signs of clinical activity in patients with MET-dependent pRCC (see ”Targeting MET alterations in cancer”), potentially broadening the spectrum of targeted therapies in kidney cancer (39,40). Interestingly, several of these activating MET mutations can also cause resistance to MET TKIs. For example, the L1195V/F, D1228X and Y1230X mutations, which are activating MET mutations, also cause acquired resistance to MET TKIs in MET exon 14 mutant lung cancers (See “Resistance Mechanisms to MET TKI”) (41-44). A similar phenomenon has been observed with activating mutations of ALK; for example, the ALK F1174L driver mutation in neuroblastoma is a known mechanism of acquired resistance in TKI-treated ALK-rearranged lung cancer (45,46). This suggests that TKI-resistance mutations in MET may also result in increases in kinase activity.
In addition to MET kinase domain mutations, mutations affecting the Sema domain in the extracellular compartment have been described in cancer. Sema mutations may affect ligand-binding, though the functional implications of these mutations in MET signaling and their therapeutic relevance is unknown (21,47,48).
MET amplification
In addition to MET kinase domain and exon 14 mutations, another mechanism of oncogenic activation is focal genomic amplification of wild-type MET located on chromosome 7 (Figure 1C) (49-51). There is no clear consensus about the optimal molecular test and or cutoff to define MET amplification clinically. Using fluorescent in situ hybridization (FISH), MET amplification can be measured by estimating the ratio between the number of MET copies in relation to the copies of the chromosome 7 centromere (CEP7). This distinguishes focal MET amplification from MET polysomy, where the gene copy number (GCN) of MET is increased due to an increase in the number of copies of chromosome 7. In MET amplified tumors, the MET/CEP7 ratio is high, whereas in polysomy, similar MET and CEP7 signals are observed. Different cutoffs of MET/CEP7 ratios have been explored to define MET amplification (41-43); one schema classifies tumors as MET/CEP7 low (≥1.8 to ≤2.2), intermediate (>2.2 to <5), or high (≥5). Higher MET/CEP7 ratios ≥5 seem to better predict responses to treatment with MET inhibitors, and may also identify a subgroup of lung cancers that are more likely to be dependent on MET signaling, because at high levels of MET amplification, there is less overlap with other oncogenic alterations involving genes such as KRAS, EGFR, and ALK compared to lower MET levels (52,53). Most recently, this classification was modified to use a MET/CEP7 ratio ≥4 to identify high levels of MET amplification, which also correlated with responses to MET TKIs (53). To differentiate MET polysomy from amplification in treatment-naïve EGFR-mutant lung cancers, another approach defined MET amplification as the combination of both MET copies per cell ≥ 5 and a MET/CEP7 ratio ≥ 2 (49). By this method, true MET amplification was detected in 3% of cases and MET gain by polysomy was observed in 23% of cases, showing that more stringent biomarker definitions can accurately identify true MET amplification. In addition to FISH, recent advances in NGS platforms have enabled the identification of focal MET amplification, although criteria for optimal copy number cut-offs to define MET amplification by this technique are lacking (54).
Identifying tumors with high-level MET amplification is both prognostic and predictive in different tumor types, including gastric cancer and NSCLC (55,56). De novo MET amplification occurs in ~1% of NSCLC and has been associated with poor survival in patients with surgically resected early-stage disease (50,57,58). In MET exon 14 mutant lung cancer, concomitant MET amplification is found in ~15% of cases, showing that MET dependency in lung cancer can be driven by synergistic genomic events (26). In addition to lung cancer, MET copy number alterations are often present in other tumor types, including ~6% of gastroesophageal carcinomas (59). Similarly, MET amplification in this setting is associated with higher histological grade, advanced disease and an unfavorable prognosis (60).
MET amplification is a common determinant of acquired resistance to EGFR tyrosine kinase inhibitors (TKIs) in patients with lung cancers harboring EGFR sensitizing mutations (8). This has been reported in ~5% of tumors upon progression to first generation EGFR TKIs and ~10% of patients treated with osimertinib (a mutant-selective EGFR TKI) for EGFR T790M positive NSCLC (61,62). MET amplification has also recently been shown to cause resistance to anaplastic lymphoma kinase (ALK) inhibitors in ALK-rearranged NSCLC (63). In these settings, enhanced MET signaling leads to sustained bypass activation of downstream oncogenic pathways, resulting in apoptosis inhibition and enhanced tumor proliferation (8). This has guided the design of clinical trials testing the activity of EGFR and MET inhibitors in combinations such as gefitinib (a first-generation EGFR TKI) and capmatinib (a type Ib MET TKI), or osimertinib with savolitinib (a type Ib MET TKI) (see ”Targeting MET alterations in cancer”) (64,65). Given the implications of MET amplification both in cancer biology as a promising therapeutic target, clearer guidelines for identifying MET amplification consistently and reproducibly in clinical samples is warranted.
MET Fusions
Another common genomic mechanism of oncogenic activation is through chromosomal translocations, in which the functional domain (such as a kinase domain) from one protein becomes ectopically expressed through its fusion to another gene. Rearrangements involving the kinase domain of ALK, ROS1, RET, and NTRK occur in lung cancer and other tumors and are predictive biomarkers of clinical benefit with selected tyrosine kinase inhibitors (66-69). Typically, the fusion partner contains a dimerization domain, which results in the constitutive activation of the tyrosine kinase domain (Figure 1D).
Although rare, MET rearrangements have been reported across a variety of tumor types. Interestingly, the oncogenic potential of MET rearrangements have been known for decades, with the original characterization of the TPR-MET fusion protein in a human osteogenic sarcoma cell line (15). In addition to the constitutive activation of MET by homodimerization, in the TPR-MET rearrangement, the juxtamembrane domain encoded by exon 14 is lost, further contributing to the oncogenic potential of this fusion through loss of this degradation domain (70).
MET rearrangements have been identified in patients with NSCLC (~0.5%), pediatric (10%) and adult glioblastomas (3%), and as single case reports in patients with salivary secretory carcinoma and infantile spindle sarcomas (71-75). Since the discovery of the TPR-MET fusion, several fusion partners have been identified and characterized including HLA-DRB1, KIF5B, PTPRZ1, STARD3NL and ST7. Interestingly, a ST7-MET fusion was reported as an acquired resistance mechanism to the third-generation ALK TKI lorlatinib, in a patient with EML4-ALK rearranged NSCLC, which was targetable by dual ALK-MET inhibition (63). In secondary glioblastoma, PTPRZ1-MET fusions are detected in 14% of cases and are associated with detrimental survival outcomes (76). Tumors bearing MET fusions may be sensitive to treatment with MET TKIs (see ”Targeting MET alterations in cancer”).
MET overexpression
In the absence of MET amplification or other genomic alterations, MET can be also overexpressed in a variety of tumor types including gastroesophageal adenocarcinoma, cholangiocarcinoma, colon cancer, kidney cancer, glioblastomas and lung cancer (77-81). Elevated levels of MET expression have been associated with enhanced tumor invasion, metastases, and poor survival outcomes in patients with gastrointestinal malignancies (82,83). MET overexpression is also common in NSCLC, and has been found in about 20-25% of cases by immunohistochemistry (IHC); however the impact of MET overexpression as an independent prognostic factor in this disease is controversial (81,84).
The level of MET expression is usually assessed in formalin-fixed paraffin-embedded (FFPE) tissues with the SP44 antibody and is scored using a semiquantitative approach ranging from 0 to 3+. There is no agreed-upon threshold to define high MET expression, but some studies have been performed in cases with a MET staining level of moderate (2+) or strong (3+) intensity in ≥ 50% of tumor cells (85). Thus far, MET overexpression by IHC has not been demonstrated to be an effective biomarker as has not successfully predicted responses to treatment with MET targeted therapies (86). In addition, MET expression by IHC has a weak correlation with MET/CEP7 ratio (50,87). In a study of pulmonary sarcomatoid carcinomas, the reported sensitivity and specificity of MET IHC positivity for detecting MET amplification by FISH is 50% and 83%, respectively, with a positive predictive value of 21.4% and a negative predictive value of 94% (87). Therefore, MET IHC positivity is not a surrogate nor an effective screening tool for identifying MET amplification.
TARGETING MET ALTERATIONS IN CANCER
There are currently no approved drugs with an on-label indication for the treatment of patients with MET-dysregulated cancer. However, several pharmacological agents are in clinical development including: MET tyrosine kinase inhibitors, monoclonal antibodies directed against the SEMA domain or HGF, antibody-drugs conjugates, and bispecific antibodies (Table 1).
Table 1.
Preliminary activity of MET tyrosine kinase inhibitors in the treatment of MET exon 14 mutant non-small cell lung cancer, de novo and acquired MET amplification in lung cancer and MET-dysregulated papillary renal cell carcinomas in clinical trials
| Study | Phase | Drug | N | Line | MET alteration | ORR | PFS | DOR | Ref |
|---|---|---|---|---|---|---|---|---|---|
| MET exon 14 mutations and de novo MET amplification (NSCLC) | |||||||||
| PROFILE 1001 NCT00585195 | I | Crizotinib 250mg BID | 69 | 1ST 26 > 2nd: 43 | MET exon 14 skipping | 32% | 7.3 months (95% CI: 5.4, 9.1) | 9.1 months (95% CI: 6.4, 12.7) | (90) |
| GEOMETRY mono-1 NCT02414139 | II | Capmatinib 400mg BID | 28 | 1st line | MET exon 14 skipping | BIRC: 67.9% Inv: 60.7% | 9.69 months (95% CI: 5.52-13.86) | 11.14 months (95% CI : 5.55-NR) | (91) |
| 69 | 2nd/3rd line | BIRC: 40.6% Inv: 42% | 5.42 months (95% CI: 4.17-6.97) | 9.72 months (95% CI : 5.55-12.98) | |||||
| VISION NCT02864992 | II | Tepotinib 500mg/day | 87 | 1st: 33 2nd: 31 > 2nd: 23 | MET exon 14 skipping | BIRC: liquid biopsy: 50% tissue biopsy: 45.1% | BIRC: liquid biopsy : 9.5 months (95%CI: 6.7-NR) tissue biopsy : 10.8 months (95%CI: 6.9-NR) | Not reported | (92) |
| Inv: liquid biopsy: 55.3% tissue biopsy: 54.9% | Inv: liquid biopsy: 9.5 months (95% CI: 5.3-21.1) tissue biopsy : 12.2 months (95% CI: 6.3-NR) | ||||||||
| NCT02897479 | II | Savolitinib 600mg/day | 34 | 1st: 17 2nd: 13 > 2nd: 4 | MET exon 14 skipping | 12/31 (38.7%) | Not reported | 34 weeks (range: 16-96) | (93) |
| METROS NCT02499614 | II | Crizotinib 250mg BID | 26 | 2nd: 21 > 2nd: 5 | MET exon 14 skipping MET/CEP7 ratio > 2.2 | 7 (27%) | 4.4 months (95%CI: 3.0-5.8) | Not reported | (94) |
| NCT00585195 | I | Crizotinib 250mg BID | 3 | Low: ≥1.8-≤2.2 copies | 1 (33.3%) | 1.8 months (95% CI: 0.8, 14.0) | 12.1 months (95% CI:12.1–12.1) | (53) | |
| 14 | Medium: >2.2-<4 copies | 2 (14.3%) | 1.9 months (95% CI: 1.3, 5.5) | 3.7 months (95% CI: 3.7–3.7) | |||||
| 20 | 8 (40%) | 6.7 months (95% CI: 3.4, 7.4) | 5.5 months (95 CI: 3.3–25.8) | ||||||
| High: ≥ 4 copies | |||||||||
| MET amplification in EGFR TKI resistance (NSCLC) | |||||||||
| NCT01610336 | II | Gefitinib 250mg daily Capmatinib 400mg BID | 41 | Post 1st/2nd | Copy number < 4 | 5 (12%) | Copy number ≥ 6 5.49 months (95% CI: 4.21-7.29) | Not reported | (64) |
| 18 | generation | Copy number ≥ 4 < 6 | 4 (22%) | ||||||
| 36 | EGFR TKI | Copy number ≥ 6 | 17 (47%) | ||||||
| 4 | 2nd: 86 | IHC: 0 | 1 (25%) | ||||||
| 2 | IHC: +1 | 0 (0%) | IHC: +3 5.45 months (95% CI: 3.71-7.10) | ||||||
| 16 | >2nd: 75 | IHC: +2 | 3 (19%) | ||||||
| 78 | IHC: +3 | 25 (32%) | |||||||
| TATTON NCT02143466 | Ib | Savolitinib 300mg or 600mg + Osimertinib 80mg/daily | 51 | Post 1st/2nd generation EGFR TKI T790M- | EGFR mutant and MET amplification (FISH copies ≥ 5 or MET/CEP7 ratio ≥ 2, NGS 20% tumor cells and ≥ copies), or MET overexpression (3+) | 33 (65%) | 9.0 months (95% CI: 5·5–11·9) | 9.0 months (95% CI: 6·1–22·7) | (65) |
| 18 | Post 1 st/2nd generation EGFR TKI T790M+ | 12 (67%) | 11·0 months (95% CI: 4·0–NR) | 12.4 months (95% CI: 2·8–NR) | |||||
| 69 | Post 3rd-generation EGFR TKI | 21 (30%) | 5.4 months (95% CI: 4·1-8·0) | 7.9 months (95% CI: 4.0-10.5) 8.0 months (95% CI: 4·5–NR) | |||||
| Savolitinib 300mg + Osimertinib 80mg/daily | 36 | Post 1st/2nd generation EGFR TKI T790M- | 23 (64%) | 9.1 months (95% CI: 5·4–12·9) | 8.0 months (95% CI: 4·5–NR) | ||||
| MET kinase domain mutant/amplified (pRCC) | |||||||||
| NCT02127710 | II | Savolitinib 600mg/daily | 44 | 1st: 26 > 1s: 18 | MET mutation, polysomy, amplification | 4 (18%) | 6.2 months (95% CI: 4.1 - 7.0) | Range: 2.4 - 16.4 months | (40) |
| 46 | 1st: 23 >1st: 23 | MET-Independent | 0 (0%) | 1.4 months (95% CI: 1.4 - 2.7) | |||||
| 19 | 1st: 11 >1st: 8 | Unknown MET status | 0 (0%) | ||||||
| NCT00726323 | II | Foretinib | 74 | 1st: 60 ≥ 2nd: 14 | MET mutation MET amplification Chromosome 7 polysomy | 10 (13.5%) | 9.3 months (95% CI: 6.9 - 12.9) | 18.5 months | (39) |
BID, twice daily; BIRC, blinded independent review committee; DOR, duration of response; FISH, fluorescence in situ hybridization; IHC, immunohistochemistry; Inv, investigator; NSCLC, non-small cell lung cancer; NR, not reached; ORR, objective response rate; PFS, progression-free survival; pRCC, papillary renal cell carcinoma; Ref, reference.
Small molecule inhibitors of the kinase domain are typically classified into three types: type I inhibitors, which bind to the active conformation of the kinase in the ATP pocket; type II inhibitors, which bind to the inactive conformation of the kinase in the ATP pocket; and type III inhibitors, which are non-ATP-competitive allosteric inhibitors, binding outside the ATP pocket (88). Several type I MET TKIs are currently in clinical development, and are further subdivided according to their interaction with the solvent front residue G1163: type Ia inhibitors like crizotinib (Xalkori, Pfizer) interact with this residue, and type Ib inhibitors like tepotinib (MSC2156119J, Merck), capmatinib (INC280, Novartis), and savolitinib (AZD6094, AstraZeneca) and APL-101 (Apollomics) which are independent of G1163 interaction (89). In addition, type II MET inhibitors include cabozantinib (Cabometyx, Exelixis), foretinib (XL-880, GlaxoSmithKline), merestinib (LY2801653, Lilly), and glesatinib (MGCD265, Mirati Therapeutics). In addition to TKIs, a number of MET-directed therapies are currently in development which target the MET extracellular domain including: a mixture of monoclonal antibodies (Sym015), a METxMET bispecific antibody (REGN5093), an EGFR-MET bispecific antibody (JNJ-61186372) and a MET antibody-drug conjugate (ABBV-399, telisotuzumab vedotin).
Several clinical trials are studying the efficacy of MET TKIs in the treatment of patients with MET exon 14 mutant NSCLC. Results presented in studies of crizotinib (NCT00585195), capmatinib (NCT02414139), tepotinib (NCT02864992), and savolitinib (NCT02897479) have shown a response rate to type I TKIs ranging from 32% to 68% (Table 1) (90-94). Early reporting shows median progression free survival (PFS) times ranging from 5.4 months to 12.2 months depending on the drug and the line of therapy (Table 1) (90-92). Whether outcomes are improved based on line of therapy, first line versus second or subsequent lines, remains unknown and future studies will be needed to define the optimal treatment sequencing approach in the context of other treatment options like immunotherapy and chemotherapy (91,95). In addition to type I MET inhibitors, clinical trials with type II TKIs like cabozantinib (NCT01639508) and merestinib (NCT02920996) are ongoing. Although additional prospective data are needed to assess the efficacy of available MET inhibitors in the central nervous system, intracranial responses have been reported with agents such as cabozantinib, capmatinib, and PLB-1001/CBT-101/APL-101 (76,91,96).
Beyond MET exon 14 mutant NSCLC, MET TKIs have been explored in the setting of MET amplification, either de novo, or in the setting of acquired resistance to EGFR TKIs. Given the lower incidence of de novo MET amplification in the absence of other oncogenic drivers, that population has been slower to enroll, but preliminary data has shown that in patients with high levels of MET amplification, defined as a MET/CEP7 ratio ≥4, about 40% of patients achieved an objective response to crizotinib with a median PFS of 6.7 months (53) (Table 1). In the setting of acquired resistance to first- or second-generation EGFR TKIs, the combination of gefitinib with capmatinib (NCT01610336) and osimertinib with savolitinib (TATTON study) can overcome MET bypass activation (64,65). This was replicated in patients previously treated with third-generation EGFR inhibitors with concurrent or acquired MET amplification, were the combination of osimertinib and savolitinib demonstrated responses in 30% of patients with a median progression-free survival of 5.4 months (65). This combination is being further explored prospectively in the SAVANNAH (NCT03778229) and ORCHARD studies (NCT03944772), which also aim to refine the optimal method and tool for defining MET amplification in a clinically-relevant setting.
In papillary renal cell carcinomas harboring MET kinase domain mutations, there are limited data on the efficacy of MET TKIs. In a phase II study of savolitinib among patients with MET-driven pRCC, the objective response rate was 18% and the median PFS was 6.2 months (95% CI: 4.1-7.0) (40). A confirmatory trial is being conducted comparing savolitinib to sunitinib as first line treatment for patients with pRCC (SAVOIR, NCT03091192). Furthermore, a phase II study of the dual MET/ VEGFR2 inhibitor foretinib (GSK1363089, GlaxoSmithKline) in patients with MET-altered pRCC (including germline mutations, somatic MET mutations, amplifications or chromosome 7 duplications) showed a 13.5% response rate and a median PFS of 9.3 months (95% CI: 6.9-12.9)(39). Together, these early studies support the development of MET TKIs for the treatment of MET-driven papillary renal carcinomas; however, to identify which specific activating mutations are sensitive to MET TKIs, detailed genomic information among responders and non-responders should be made available from these studies. MET TKIs have also been reported to have activity against tumors that harbor MET fusions, as has been illustrated in case reports of patients with NSCLC and primary brain tumors treated with crizotinib or the MET inhibitor PLB-1001/CBT-101/APL-101, respectively (71,72,76,97).
Targeting MET with monoclonal antibodies directed against the receptor extracellular domain or the HGF ligand has been more challenging. In the randomized phase III of the monoclonal antibody onartuzumab (MetMAb, Roche) in unselected patients with NSCLC, adding onartuzumab to erlotinib in patients previously treated with chemotherapy had a detrimental effect on overall survival; this therapy also showed no benefit in an exploratory analysis of patients with MET-amplified tumors by FISH (>5 copies) (86,98). Similarly, the addition of onartuzumab to chemotherapy treatment for patients with advanced NSCLC, gastrointestinal tumors, and glioblastoma was not effective (99-102). A similar compound emibetuzumab (LY2875358, Lilly) was initially evaluated but its development was later stopped (103). Rilotumumab, a monoclonal antibody directed against soluble HGF, also resulted in deleterious outcomes when combined with chemotherapy in patients advanced gastric and esophagogastric tumors (104). These early trials lacked adequate biomarker-enrichment and patient selection, and MET expression by IHC has not proven to be a reliable indicator of MET oncogenic dependency. The etiology of the negative impact of combining MET inhibition with chemotherapy in unselected patients is unclear, but one hypothesis is that MET inhibitors may dysregulate immune-mediated cytotoxicity and factors in the tumor microenvironment (105). For instance, MET inhibition can impair interferon gamma induction of programmed cell death ligand 1 (PD-L1) expression in vitro and decrease neutrophil antitumor activity (106,107). Given these preclinical observations, the effects of MET inhibitors on tumor immunology in patients warrant further investigation.
Given the advances in the understanding of MET signaling and biomarker selection, in addition to TKIs, novel MET directed therapies are being developed. Sym015 (Symphogen A/S, Denmark) is a mixture of two IgG1 humanized monoclonal antibodies (Hu9006 and Hu9338) directed against non-overlapping epitopes in the SEMA α-domain of MET (108). This combination of antibodies confers potent in vitro and in vivo activity in MET-amplified and MET exon 14 mutant models and is currently in clinical development (NCT02648724) (109). Also, the dual MET antibody REGN5093 (Regeneron) is currently being clinically developed for patients with MET-altered NSCLC (NCT04077099). In addition, the bispecific EGFR/MET antibody JNJ-61186372 (Genmab/Jansen) appears to be active in the setting of MET-amplified, EGFR-TKI resistant EGFR-mutant NSCLC through several proposed mechanisms including by impeding ligand-receptor binding, enhancing receptor internalization, and promoting antibody-dependent cellular cytotoxicity (ADCC) (110,111). Preliminary reports of the phase I dose escalation cohort showed encouraging clinical activity for patients with EGFR mutations (112).
In addition to antibody-based therapies, antibody-drug conjugates are also being developed to target MET. The ADC telisotuzumab vedotin (ABBV-399, Abbvie) is composed of the MET monoclonal antibody ABT-700 conjugated with monomethylauristatin E (MME), a cytotoxic microtubular inhibitor, with a cleaved valine-citruline linker, and has in vitro cytotoxic activity in MET-amplified preclinical models (113). A phase I dose-escalation study of ABBV-399 in patients with advanced solid tumors and MET overexpression, showed that this drug has a tolerable safety profile with signs of activity in patients with MET-positive NSCLC (114). A phase II study (NCT03539536) is currently enrolling patients with lung cancer to test this compound in the second- or third-line setting; identification of the optimal predictive biomarkers of response to these antibodies and ADCs will be necessary to enrich for patients who will benefit from these therapies.
MECHANISMS OF RESISTANCE TO MET TYROSINE KINASE INHIBITORS
The diversity and complexity of biological mechanisms underlying the adaptation of cancer cells to kinase inhibitors is a matter of intense study. Established biological mechanisms of resistance to TKIs include: 1) acquisition of on-target kinase domain mutations affecting drug binding to the receptor or its ATP affinity, 2) bypass track activation of oncogenic signaling pathways, and 3) histological transformation such epithelial-mesenchymal transition or small cell lung cancer transformation (115), although in many cases, the molecular mechanisms of drug resistance are unknown.
In the MET-amplified or MET exon 14-alterated NSCLC, secondary MET kinase domain mutations have been clinically documented and preclinically characterized to confer resistance to type I and type II MET TKIs (41,42,116). MET mutations in codons D1228 (=D1246) and Y1230 (=Y1248) can mediate resistance to type I MET TKIs by hindering drug binding to the receptor (117-119) (Figure 2A), but do not affect the binding of type II MET TKIs (41,120). The solvent front mutation G1163R, which is analogous to the ALK G1202R and ROS1 G2023R TKI resistance solvent front mutations, confers resistance to the type Ia MET inhibitor crizotinib but not to type Ib or II MET TKIs (Figure 2B) (41,43). Multiple MET mutations can be detected at the time of progression to MET TKIs suggesting that polyclonal resistance can emerge in this setting (116). The efficacy of type II MET inhibitors in overcoming resistance to type I TKI-resistant tumors, particularly with acquiring mutations at residues D1228 and Y1230, is supported by preclinical rational and clinical reports (42,43,116,121).
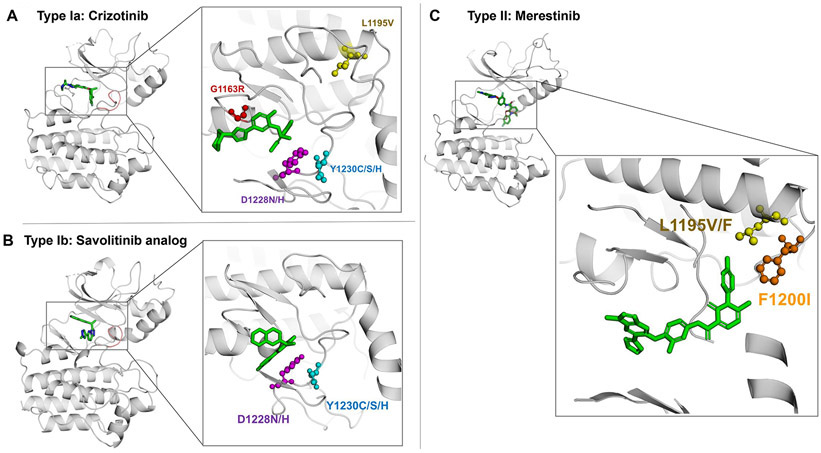
Figure 2. Mechanisms of resistance to MET tyrosine kinase inhibitors in MET exon 14 mutant lung cancer.
Crystal modelling of the MET kinase domain and binding of MET tyrosine kinase inhibitors (green). The figure displays the position of frequently mutated residues within the MET kinase domain including G1163 (red), D1228 (purple), Y1230 (blue), L1195 (yellow), and F1200 (orange) that confer acquired resistance to MET TKIs. Panel A displays interaction of the type Ia MET TKI crizotinib (PDB Ref: 2WGJ) with commonly mutated residues that confer resistance to crizotinib like G1163R, D1228X and Y1230X mutations. Panel B shows the interaction of the type Ib MET inhibitor savolitinib analog (PDB Ref: 3ZC5) with resistance mutations including D1228X and Y1230X, but the interaction with the kinase domain is not predicted to be affected by the G1163R solvent front mutation. In panel C, the type II MET TKI merestinib (PDB Ref: 4EEV) is simulated bound to the kinase domain and displays the interaction of type II MET inhibitors with key residues that can cause resistance to these compounds like L1195F/V and F1200L.
Resistance mutations may also be found infrequently at the time of initial diagnosis prior to TKI exposure. In MET exon 14 mutant NSCLC, for example, the MET Y1230C mutation was reported to be present at a low allele frequency at baseline in a patient but later emerged as a dominant mechanism of resistance to crizotinib after treatment exposure (119). Other mutations, involving residues F1200 and L1195, affect the binding of type II inhibitors to the DFG-out conformation resulting in resistance to these compounds (Figure 2C) (43). In vitro assays suggest that F1200I also confers resistance to type I MET TKI with the exception of savolitinib (43,122). The MET L1195V mutation alters the C-terminus of the alpha helix conferring resistance to crizotinib and type II MET TKI (43). Understanding the implication of these secondary kinase domain mutations in conferring sensitivity or resistance to the growing repertoire of available MET TKIs may help guide clinical strategies to sequence MET inhibitors at the time of resistance.
Bypass track activation of downstream oncogenic signaling, independent of MET inhibition, can be driven by other receptor tyrosine kinases or intracellular molecules at the time of resistance to MET TKIs. Activation of the MAPK pathway has been reported to drive acquired resistance to crizotinib through the development of wild-type KRAS amplification or KRAS mutations in patients with MET exon 14 altered tumors (123,124). Furthermore, preclinical and preliminary clinical studies suggests that baseline KRAS/NF1/RASA1 mutations are associated with primary resistance to MET TKIs, and efforts to correlate absence of MET or KRAS expression with a lack of response to MET inhibitors are underway (124,125). In addition, PI3KCA mutations and EGFR activation has been reported to drive resistance to MET inhibitors in vitro (126,127). Acquired EGFR amplification has also been detected in tumor samples from patients experiencing resistance to MET TKIs, implying that EGFR amplification can drive resistance to MET inhibitors, just as MET amplification can confer EGFR TKI resistance in EGFR-mutant NSCLC, highlighting the potential role for dual EGFR-MET inhibition in TKI-resistant NSCLC (110,116). With the expansion of MET testing in NSCLC and the improvement in access to targeted therapies with MET inhibitors, new mechanisms of resistance will be uncovered to hopefully guide the development of novel therapeutic strategies for patients.
CONCLUSION
A more comprehensive understanding of the diverse biological mechanisms driving MET dysregulation in cancer has both fueled the development of predictive biomarkers of response to MET inhibitors and enabled design of new therapies. Biomarkers like MET exon 14 skipping mutations and high levels of focal MET amplification have been effective for identifying patients who would benefit from treatment with MET TKIs. Guidelines are needed to improve and standardize diagnostic methods to identify MET-dependent cancers and inform drug development strategies. Clinical trials testing novel and selective MET inhibitors are ongoing and will lead to the approval of effective therapeutic options for patients with MET-dysregulated cancers.
STATEMENT OF SIGNIFICANCE.
Increasing evidence supports the use of MET-targeting therapies in biomarker-selected cancers that harbor molecular alterations in MET. Diverse mechanisms of resistance to MET inhibitors will require the development of novel strategies to delay and overcome drug resistance.
Acknowledgments
Sources of funding
This work was supported by the National Cancer Institute grant R01CA222823 (P.A.J. and M.M.A.), the Conquer Cancer Foundation of the American Society of Clinical Oncology grant number 6298701 (M.M.A).
Footnotes
Conflict of interest statement
G.R. Consultant/advisory board : Amgen, Pfizer, Roche. Travel grants: Pfizer, AstraZeneca.
J.C. Consultant: Soltego and Kymera Therapeutics. Shareholder: M3 bioinformatics & technology Inc.
P.A.J. Consultant: AstraZeneca, Boehringer Ingelheim, Pfizer, Merrimack Pharmaceuticals, Roche/Genentech, Chugai Pharmaceuticals, Acea Biosciences, Ignyta, LOXO Oncology, Eli Lilly pharmaceuticals, Araxes pharmaceuticals, SFJ Pharmarceuticals, Voronoi, Daiichi Sankyo, Biocartis, Novartis, Takeda Oncology. Stock Ownership: Gatekeeper Pharmaceuticals, LOXO Oncology, Sanofi. Research Funding: Astellas Pharmaceuticals, AstraZenenca, Daiichi Sankyo, PUMA, Eli Lilly pharmaceuticals, Boehringer Ingelheim, Takeda Oncology, Revolution Medicines. Patents, Royalties, Other Intellectual Property: LabCorp
M.M.A: Consultant/advisory board: Bristol-Myers Squibb, AstraZeneca, Achilles, AbbVie, Neon, Maverick, Nektar, Hegrui, Syndax, Gritstone. Institutional research funding: from Bristol-Myers Squibb, AstraZeneca, Lilly, Genentech.
REFERENCES
- 1.Bussolino F, Di Renzo MF, Ziche M, Bocchietto E, Olivero M, Naldini L, et al. Hepatocyte growth factor is a potent angiogenic factor which stimulates endothelial cell motility and growth J Cell Biol. United States; 1992;119:629–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bladt F, Riethmacher D, Isenmann S, Aguzzi A, Birchmeier C. Essential role for the c-met receptor in the migration of myogenic precursor cells into the limb bud Nature. England; 1995;376:768–71. [DOI] [PubMed] [Google Scholar]
- 3.Huh C-G, Factor VM, Sanchez A, Uchida K, Conner EA, Thorgeirsson SS. Hepatocyte growth factor/c-met signaling pathway is required for efficient liver regeneration and repair Proc Natl Acad Sci U S A. United States; 2004;101:4477–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okunishi K, Dohi M, Nakagome K, Tanaka R, Mizuno S, Matsumoto K, et al. A novel role of hepatocyte growth factor as an immune regulator through suppressing dendritic cell function J Immunol. United States; 2005;175:4745–53. [DOI] [PubMed] [Google Scholar]
- 5.Benkhoucha M, Santiago-Raber M-L, Schneiter G, Chofflon M, Funakoshi H, Nakamura T, et al. Hepatocyte growth factor inhibits CNS autoimmunity by inducing tolerogenic dendritic cells and CD25+Foxp3+ regulatory T cells Proc Natl Acad Sci U S A. United States; 2010;107:6424–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giordano S, Bardelli A, Zhen Z, Menard S, Ponzetto C, Comoglio PM. A point mutation in the MET oncogene abrogates metastasis without affecting transformation Proc Natl Acad Sci U S A. United States; 1997;94:13868–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glodde N, Bald T, van den Boorn-Konijnenberg D, Nakamura K, O’Donnell JS, Szczepanski S, et al. Reactive Neutrophil Responses Dependent on the Receptor Tyrosine Kinase c-MET Limit Cancer Immunotherapy Immunity. United States; 2017;47:789–802.e9. [DOI] [PubMed] [Google Scholar]
- 8.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling Science. United States; 2007;316:1039–43. [DOI] [PubMed] [Google Scholar]
- 9.Organ SL, Tsao M-S. An overview of the c-MET signaling pathway Ther Adv Med Oncol. England; 2011;3:S7–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ponzetto C, Bardelli A, Zhen Z, Maina F, dalla Zonca P, Giordano S, et al. A multifunctional docking site mediates signaling and transformation by the hepatocyte growth factor/scatter factor receptor family Cell. United States; 1994;77:261–71. [DOI] [PubMed] [Google Scholar]
- 11.Boccaccio C, Ando M, Tamagnone L, Bardelli A, Michieli P, Battistini C, et al. Induction of epithelial tubules by growth factor HGF depends on the STAT pathway Nature. England; 1998;391:285–8. [DOI] [PubMed] [Google Scholar]
- 12.Kong-Beltran M, Seshagiri S, Zha J, Zhu W, Bhawe K, Mendoza N, et al. Somatic mutations lead to an oncogenic deletion of met in lung cancer Cancer Res. United States; 2006;66:283–9. [DOI] [PubMed] [Google Scholar]
- 13.Petrelli A, Gilestro GF, Lanzardo S, Comoglio PM, Migone N, Giordano S. The endophilin-CIN85-Cbl complex mediates ligand-dependent downregulation of c-Met Nature. England; 2002;416:187–90. [DOI] [PubMed] [Google Scholar]
- 14.Mohapatra B, Ahmad G, Nadeau S, Zutshi N, An W, Scheffe S, et al. Protein tyrosine kinase regulation by ubiquitination: critical roles of Cbl-family ubiquitin ligases Biochim Biophys Acta. Netherlands; 2013;1833:122–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park M, Dean M, Cooper CS, Schmidt M, O’Brien SJ, Blair DG, et al. Mechanism of met oncogene activation Cell. United States; 1986;45:895–904. [DOI] [PubMed] [Google Scholar]
- 16.Frampton GM, Ali SM, Rosenzweig M, Chmielecki J, Lu X, Bauer TM, et al. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors Cancer Discov. United States; 2015;5:850–9. [DOI] [PubMed] [Google Scholar]
- 17.Lee J-H, Gao CF, Lee CC, Kim MD, Vande Woude GF. An alternatively spliced form of Met receptor is tumorigenic Exp Mol Med. United States; 2006;38:565–73. [DOI] [PubMed] [Google Scholar]
- 18.Abella J V, Peschard P, Naujokas MA, Lin T, Saucier C, Urbe S, et al. Met/Hepatocyte growth factor receptor ubiquitination suppresses transformation and is required for Hrs phosphorylation Mol Cell Biol. United States; 2005;25:9632–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anna A, Monika G. Splicing mutations in human genetic disorders: examples, detection, and confirmation J Appl Genet. England; 2018;59:253–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peschard P, Fournier TM, Lamorte L, Naujokas MA, Band H, Langdon WY, et al. Mutation of the c-Cbl TKB domain binding site on the Met receptor tyrosine kinase converts it into a transforming protein Mol Cell. United States; 2001;8:995–1004. [DOI] [PubMed] [Google Scholar]
- 21.Ma PC, Kijima T, Maulik G, Fox EA, Sattler M, Griffin JD, et al. c-MET mutational analysis in small cell lung cancer: novel juxtamembrane domain mutations regulating cytoskeletal functions Cancer Res. United States; 2003;63:6272–81. [PubMed] [Google Scholar]
- 22.Liu S, Meric-Bernstam F, Parinyanitikul N, Wang B, Eterovic AK, Zheng X, et al. Functional consequence of the MET-T1010I polymorphism in breast cancer Oncotarget. United States; 2015;6:2604–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tyner JW, Fletcher LB, Wang EQ, Yang WF, Rutenberg-Schoenberg ML, Beadling C, et al. MET receptor sequence variants R970C and T992I lack transforming capacity Cancer Res. United States; 2010;70:6233–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Awad MM, Oxnard GR, Jackman DM, Savukoski DO, Hall D, Shivdasani P, et al. MET Exon 14 Mutations in Non-Small-Cell Lung Cancer Are Associated With Advanced Age and Stage-Dependent MET Genomic Amplification and c-Met Overexpression J Clin Oncol. United States; 2016;34:721–30. [DOI] [PubMed] [Google Scholar]
- 25.Liu X, Jia Y, Stoopler MB, Shen Y, Cheng H, Chen J, et al. Next-Generation Sequencing of Pulmonary Sarcomatoid Carcinoma Reveals High Frequency of Actionable MET Gene Mutations J Clin Oncol. United States; 2016;34:794–802. [DOI] [PubMed] [Google Scholar]
- 26.Schrock AB, Frampton GM, Suh J, Chalmers ZR, Rosenzweig M, Erlich RL, et al. Characterization of 298 Patients with Lung Cancer Harboring MET Exon 14 Skipping Alterations J Thorac Oncol. United States; 2016;11:1493–502. [DOI] [PubMed] [Google Scholar]
- 27.Descarpentries C, Lepretre F, Escande F, Kherrouche Z, Figeac M, Sebda S, et al. Optimization of Routine Testing for MET Exon 14 Splice Site Mutations in NSCLC Patients J Thorac Oncol. United States; 2018;13:1873–83. [DOI] [PubMed] [Google Scholar]
- 28.Kim EK, Kim KA, Lee CY, Kim S, Chang S, Cho BC, et al. Molecular Diagnostic Assays and Clinicopathologic Implications of MET Exon 14 Skipping Mutation in Non-small-cell Lung Cancer Clin Lung Cancer. United States; 2019;20:e123–32. [DOI] [PubMed] [Google Scholar]
- 29.Aggarwal C, Thompson JC, Black TA, Katz SI, Fan R, Yee SS, et al. Clinical Implications of Plasma-Based Genotyping With the Delivery of Personalized Therapy in Metastatic Non-Small Cell Lung Cancer JAMA Oncol. United States; 2019;5:173–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalemkerian GP, Narula N, Kennedy EB, Biermann WA, Donington J, Leighl NB, et al. Molecular Testing Guideline for the Selection of Patients With Lung Cancer for Treatment With Targeted Tyrosine Kinase Inhibitors: American Society of Clinical Oncology Endorsement of the College of American Pathologists/International Association for the . J Clin Oncol. United States; 2018;36:911–9. [DOI] [PubMed] [Google Scholar]
- 31.Jeffers M, Schmidt L, Nakaigawa N, Webb CP, Weirich G, Kishida T, et al. Activating mutations for the met tyrosine kinase receptor in human cancer Proc Natl Acad Sci U S A. United States; 1997;94:11445–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Albiges L, Guegan J, Le Formal A, Verkarre V, Rioux-Leclercq N, Sibony M, et al. MET is a potential target across all papillary renal cell carcinomas: result from a large molecular study of pRCC with CGH array and matching gene expression array Clin Cancer Res. United States; 2014;20:3411–21. [DOI] [PubMed] [Google Scholar]
- 33.Lubensky IA, Schmidt L, Zhuang Z, Weirich G, Pack S, Zambrano N, et al. Hereditary and sporadic papillary renal carcinomas with c-met mutations share a distinct morphological phenotype Am J Pathol. United States; 1999;155:517–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidt L, Duh FM, Chen F, Kishida T, Glenn G, Choyke P, et al. Germline and somatic mutations in the tyrosine kinase domain of the MET proto-oncogene in papillary renal carcinomas Nat Genet. United States; 1997;16:68–73. [DOI] [PubMed] [Google Scholar]
- 35.Schmidt L, Junker K, Nakaigawa N, Kinjerski T, Weirich G, Miller M, et al. Novel mutations of the MET proto-oncogene in papillary renal carcinomas Oncogene. England; 1999;18:2343–50. [DOI] [PubMed] [Google Scholar]
- 36.Linehan WM, Spellman PT, Ricketts CJ, Creighton CJ, Fei SS, Davis C, et al. Comprehensive Molecular Characterization of Papillary Renal-Cell Carcinoma N Engl J Med. United States; 2016;374:135–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Graveel C, Su Y, Koeman J, Wang L-M, Tessarollo L, Fiscella M, et al. Activating Met mutations produce unique tumor profiles in mice with selective duplication of the mutant allele Proc Natl Acad Sci U S A. United States; 2004;101:17198–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fischer J, Palmedo G, von Knobloch R, Bugert P, Prayer-Galetti T, Pagano F, et al. Duplication and overexpression of the mutant allele of the MET proto-oncogene in multiple hereditary papillary renal cell tumours Oncogene. England; 1998;17:733–9. [DOI] [PubMed] [Google Scholar]
- 39.Choueiri TK, Vaishampayan U, Rosenberg JE, Logan TF, Harzstark AL, Bukowski RM, et al. Phase II and biomarker study of the dual MET/VEGFR2 inhibitor foretinib in patients with papillary renal cell carcinoma J Clin Oncol. United States; 2013;31:181–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choueiri TK, Plimack E, Arkenau H-T, Jonasch E, Heng DYC, Powles T, et al. Biomarker-Based Phase II Trial of Savolitinib in Patients With Advanced Papillary Renal Cell Cancer J Clin Oncol. United States; 2017;35:2993–3001. [DOI] [PubMed] [Google Scholar]
- 41.Fujino T, Kobayashi Y, Suda K, Koga T, Nishino M, Ohara S, et al. Sensitivity and Resistance of MET Exon 14 Mutations in Lung Cancer to Eight MET Tyrosine Kinase Inhibitors In Vitro J Thorac Oncol. United States; 2019; [DOI] [PubMed] [Google Scholar]
- 42.Bahcall M, Sim T, Paweletz CP, Patel JD, Alden RS, Kuang Y, et al. Acquired METD1228V Mutation and Resistance to MET Inhibition in Lung Cancer Cancer Discov. United States; 2016;6:1334–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Engstrom LD, Aranda R, Lee M, Tovar EA, Essenburg CJ, Madaj Z, et al. Glesatinib Exhibits Antitumor Activity in Lung Cancer Models and Patients Harboring MET Exon 14 Mutations and Overcomes Mutation-mediated Resistance to Type I MET Inhibitors in Nonclinical Models Clin Cancer Res. United States; 2017;23:6661–72. [DOI] [PubMed] [Google Scholar]
- 44.Rotow JK, Gui P, Wu W, Raymond VM, Lanman RB, Kaye FJ, et al. Co-occurring Alterations in the RAS-MAPK Pathway Limit Response to MET Inhibitor Treatment in MET Exon 14 Skipping Mutation-Positive Lung Cancer. Clin cancer Res an Off J Am Assoc Cancer Res. 2020;26:439–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen Y, Takita J, Choi YL, Kato M, Ohira M, Sanada M, et al. Oncogenic mutations of ALK kinase in neuroblastoma Nature. England; 2008;455:971–4. [DOI] [PubMed] [Google Scholar]
- 46.Sasaki T, Okuda K, Zheng W, Butrynski J, Capelletti M, Wang L, et al. The neuroblastoma-associated F1174L ALK mutation causes resistance to an ALK kinase inhibitor in ALK-translocated cancers Cancer Res. United States; 2010;70:10038–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kong-Beltran M, Stamos J, Wickramasinghe D. The Sema domain of Met is necessary for receptor dimerization and activation Cancer Cell. United States; 2004;6:75–84. [DOI] [PubMed] [Google Scholar]
- 48.Miao W, Sakai K, Sato H, Imamura R, Jangphattananont N, Takagi J, et al. Impaired ligand-dependent MET activation caused by an extracellular SEMA domain missense mutation in lung cancer Cancer Sci. England; 2019;110:3340–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lai GGY, Lim TH, Lim J, Liew PJR, Kwang XL, Nahar R, et al. Clonal MET Amplification as a Determinant of Tyrosine Kinase Inhibitor Resistance in Epidermal Growth Factor Receptor-Mutant Non-Small-Cell Lung Cancer J Clin Oncol. United States; 2019;JCO1800177. [DOI] [PubMed] [Google Scholar]
- 50.Guo R, Berry LD, Aisner DL, Sheren J, Boyle T, Bunn PAJ, et al. MET IHC Is a Poor Screen for MET Amplification or MET Exon 14 Mutations in Lung Adenocarcinomas: Data from a Tri-Institutional Cohort of the Lung Cancer Mutation Consortium J Thorac Oncol. United States; 2019;14:1666–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tong JH, Yeung SF, Chan AWH, Chung LY, Chau SL, Lung RWM, et al. MET Amplification and Exon 14 Splice Site Mutation Define Unique Molecular Subgroups of Non-Small Cell Lung Carcinoma with Poor Prognosis Clin Cancer Res. United States; 2016;22:3048–56. [DOI] [PubMed] [Google Scholar]
- 52.Noonan SA, Berry L, Lu X, Gao D, Baron AE, Chesnut P, et al. Identifying the Appropriate FISH Criteria for Defining MET Copy Number-Driven Lung Adenocarcinoma through Oncogene Overlap Analysis J Thorac Oncol. United States; 2016;11:1293–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Camidge DR, Otterson GA, Clark JW, Ou S-HI, Weiss J, Ades S, et al. Crizotinib in patients (pts) with MET-amplified non-small cell lung cancer (NSCLC): Updated safety and efficacy findings from a phase 1 trial. J Clin Oncol [Internet]. American Society of Clinical Oncology; 2018;36:9062 Available from: 10.1200/JCO.2018.36.15_suppl.9062 [DOI] [Google Scholar]
- 54.Jordan EJ, Kim HR, Arcila ME, Barron D, Chakravarty D, Gao J, et al. Prospective Comprehensive Molecular Characterization of Lung Adenocarcinomas for Efficient Patient Matching to Approved and Emerging Therapies Cancer Discov. United States; 2017;7:596–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lutterbach B, Zeng Q, Davis LJ, Hatch H, Hang G, Kohl NE, et al. Lung cancer cell lines harboring MET gene amplification are dependent on Met for growth and survival Cancer Res. United States; 2007;67:2081–8. [DOI] [PubMed] [Google Scholar]
- 56.Smolen GA, Sordella R, Muir B, Mohapatra G, Barmettler A, Archibald H, et al. Amplification of MET may identify a subset of cancers with extreme sensitivity to the selective tyrosine kinase inhibitor PHA-665752 Proc Natl Acad Sci U S A. United States; 2006;103:2316–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Collisson E a., Campbell JD, Brooks AN, Berger AH, Lee W, Chmielecki J, et al. Comprehensive molecular profiling of lung adenocarcinoma Nature. England; 2014;511:543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cappuzzo F, Marchetti A, Skokan M, Rossi E, Gajapathy S, Felicioni L, et al. Increased MET gene copy number negatively affects survival of surgically resected non-small-cell lung cancer patients J Clin Oncol. United States; 2009;27:1667–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jardim DLF, Tang C, Gagliato DDM, Falchook GS, Hess K, Janku F, et al. Analysis of 1,115 patients tested for MET amplification and therapy response in the MD Anderson Phase I Clinic Clin Cancer Res. United States; 2014;20:6336–45. [DOI] [PubMed] [Google Scholar]
- 60.Lennerz JK, Kwak EL, Ackerman A, Michael M, Fox SB, Bergethon K, et al. MET amplification identifies a small and aggressive subgroup of esophagogastric adenocarcinoma with evidence of responsiveness to crizotinib J Clin Oncol. United States; 2011;29:4803–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu HA, Arcila ME, Rekhtman N, Sima CS, Zakowski MF, Pao W, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers Clin Cancer Res. United States; 2013;19:2240–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oxnard GR, Hu Y, Mileham KF, Husain H, Costa DB, Tracy P, et al. Assessment of Resistance Mechanisms and Clinical Implications in Patients With EGFR T790M-Positive Lung Cancer and Acquired Resistance to Osimertinib JAMA Oncol. United States; 2018;4:1527–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dagogo-Jack I, Yoda S, Lennerz JK, Langenbucher A, Lin JJ, Rooney MM, et al. MET Alterations are a Recurring and Actionable Resistance Mechanism in ALK-Positive Lung Cancer Clin Cancer Res. United States; 2020. [published online ahead of print, 2020 February 21] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu Y-L, Zhang L, Kim D-W, Liu X, Lee DH, Yang JC-H, et al. Phase Ib/II Study of Capmatinib (INC280) Plus Gefitinib After Failure of Epidermal Growth Factor Receptor (EGFR) Inhibitor Therapy in Patients With EGFR-Mutated, MET Factor-Dysregulated Non-Small-Cell Lung Cancer J Clin Oncol. United States; 2018;JCO2018777326. [DOI] [PubMed] [Google Scholar]
- 65.Sequist LV, Han J-Y, Ahn M-J, Cho BC, Yu H, Kim S-W, et al. Osimertinib plus savolitinib in patients with EGFR mutation-positive, MET-amplified, non-small-cell lung cancer after progression on EGFR tyrosine kinase inhibitors: interim results from a multicentre, open-label, phase 1b study Lancet Oncol. England; 2020;21:373–86. [DOI] [PubMed] [Google Scholar]
- 66.Solomon BJ, Kim D-W, Wu Y-L, Nakagawa K, Mekhail T, Felip E, et al. Final Overall Survival Analysis From a Study Comparing First-Line Crizotinib Versus Chemotherapy in ALK-Mutation-Positive Non-Small-Cell Lung Cancer J Clin Oncol. United States; 2018;JCO2017774794. [DOI] [PubMed] [Google Scholar]
- 67.Shaw AT, Ou S-HI, Bang Y-J, Camidge DR, Solomon BJ, Salgia R, et al. Crizotinib in ROS1-Rearranged Non–Small-Cell Lung Cancer N Engl J Med, United States: 2014;371:1963–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Subbiah V, Gainor JF, Rahal R, Brubaker JD, Kim JL, Maynard M, et al. Precision Targeted Therapy with BLU-667 for RET-Driven Cancers Cancer Discov. United States; 2018;8:836–49. [DOI] [PubMed] [Google Scholar]
- 69.Laetsch TW, DuBois SG, Mascarenhas L, Turpin B, Federman N, Albert CM, et al. Larotrectinib for paediatric solid tumours harbouring NTRK gene fusions: phase 1 results from a multicentre, open-label, phase 1/2 study Lancet Oncol. England; 2018;19:705–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vigna E, Gramaglia D, Longati P, Bardelli A, Comoglio PM. Loss of the exon encoding the juxtamembrane domain is essential for the oncogenic activation of TPR-MET Oncogene. England; 1999;18:4275–81. [DOI] [PubMed] [Google Scholar]
- 71.Plenker D, Bertrand M, de Langen AJ, Riedel R, Lorenz C, Scheel AH, et al. Structural Alterations of MET Trigger Response to MET Kinase Inhibition in Lung Adenocarcinoma Patients Clin Cancer Res. United States; 2018;24:1337–43. [DOI] [PubMed] [Google Scholar]
- 72.Recurrent MET fusion genes represent a drug target in pediatric glioblastoma Nat Med. United States; 2016;22:1314–20. [DOI] [PubMed] [Google Scholar]
- 73.Ferguson SD, Zhou S, Huse JT, de Groot JF, Xiu J, Subramaniam DS, et al. Targetable Gene Fusions Associate With the IDH Wild-Type Astrocytic Lineage in Adult Gliomas J Neuropathol Exp Neurol. England; 2018;77:437–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rooper LM, Karantanos T, Ning Y, Bishop JA, Gordon SW, Kang H. Salivary Secretory Carcinoma With a Novel ETV6-MET Fusion: Expanding the Molecular Spectrum of a Recently Described Entity Am J Surg Pathol. United States; 2018;42:1121–6. [DOI] [PubMed] [Google Scholar]
- 75.Flucke U, van Noesel MM, Wijnen M, Zhang L, Chen C-L, Sung Y-S, et al. TFG-MET fusion in an infantile spindle cell sarcoma with neural features Genes Chromosomes Cancer. United States; 2017;56:663–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hu H, Mu Q, Bao Z, Chen Y, Liu Y, Chen J, et al. Mutational Landscape of Secondary Glioblastoma Guides MET-Targeted Trial in Brain Tumor Cell. United States; 2018;175:1665–1678.e18. [DOI] [PubMed] [Google Scholar]
- 77.Catenacci DVT, Ang A, Liao W-L, Shen J, O’Day E, Loberg RD, et al. MET tyrosine kinase receptor expression and amplification as prognostic biomarkers of survival in gastroesophageal adenocarcinoma Cancer. United States; 2017;123:1061–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Heo MH, Kim HK, Lee H, Kim K-M, Lee J, Park SH, et al. The Clinical Impact of c-MET Over-Expression in Advanced Biliary Tract Cancer (BTC) J Cancer. Australia; 2017;8:1395–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gayyed MF, Abd El-Maqsoud NMR, El-Hameed El-Heeny AA, Mohammed MF. c-MET expression in colorectal adenomas and primary carcinomas with its corresponding metastases J Gastrointest Oncol. China; 2015;6:618–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Peltola KJ, Penttila P, Rautiola J, Joensuu H, Hanninen E, Ristimaki A, et al. Correlation of c-Met Expression and Outcome in Patients With Renal Cell Carcinoma Treated With Sunitinib Clin Genitourin Cancer. United States; 2017;15:487–94. [DOI] [PubMed] [Google Scholar]
- 81.Reis H, Metzenmacher M, Goetz M, Savvidou N, Darwiche K, Aigner C, et al. MET Expression in Advanced Non-Small-Cell Lung Cancer: Effect on Clinical Outcomes of Chemotherapy, Targeted Therapy, and Immunotherapy Clin Lung Cancer. United States; 2018;19:e441–63. [DOI] [PubMed] [Google Scholar]
- 82.Wang A, Wang HY, Liu Y, Zhao MC, Zhang HJ, Lu ZY, et al. The prognostic value of PD-L1 expression for non-small cell lung cancer patients: a meta-analysis Eur J Surg Oncol. 2015;41:450–6. [DOI] [PubMed] [Google Scholar]
- 83.Sun Y, Liu W, Ma G, Gao D, Jiang Y, Liu Q, et al. Expression of HGF and Met in human tissues of colorectal cancers: biological and clinical implications for synchronous liver metastasis Int J Med Sci. Australia; 2013;10:548–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Watermann I, Schmitt B, Stellmacher F, Muller J, Gaber R, Kugler C, et al. Improved diagnostics targeting c-MET in non-small cell lung cancer: expression, amplification and activation? Diagn Pathol. England; 2015;10:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Koeppen H, Yu W, Zha J, Pandita A, Penuel E, Rangell L, et al. Biomarker analyses from a placebo-controlled phase II study evaluating erlotinib+/−onartuzumab in advanced non-small cell lung cancer: MET expression levels are predictive of patient benefit Clin Cancer Res. United States; 2014;20:4488–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Spigel DR, Edelman MJ, O’Byrne K, Paz-Ares L, Mocci S, Phan S, et al. Results From the Phase III Randomized Trial of Onartuzumab Plus Erlotinib Versus Erlotinib in Previously Treated Stage IIIB or IV Non-Small-Cell Lung Cancer: METLung J Clin Oncol. United States; 2017;35:412–20. [DOI] [PubMed] [Google Scholar]
- 87.Mignard X, Ruppert A-M, Antoine M, Vasseur J, Girard N, Mazieres J, et al. c-MET Overexpression as a Poor Predictor of MET Amplifications or Exon 14 Mutations in Lung Sarcomatoid Carcinomas J Thorac Oncol. United States; 2018;13:1962–7. [DOI] [PubMed] [Google Scholar]
- 88.Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors Nat Rev Cancer. England; 2009;9:28–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yamaoka T, Kusumoto S, Ando K, Ohba M, Ohmori T. Receptor Tyrosine Kinase-Targeted Cancer Therapy Int J Mol Sci. Switzerland; 2018;19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Drilon A, Clark JW, Weiss J, Ou S-HI, Camidge DR, Solomon BJ, et al. Antitumor activity of crizotinib in lung cancers harboring a MET exon 14 alteration Nat Med. United States; 2020;26:47–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wolf J, Seto T, Han J-Y, Reguart N, Garon EB, Groen HJM, et al. Capmatinib (INC280) in METΔex14-mutated advanced non-small cell lung cancer (NSCLC): Efficacy data from the phase II GEOMETRY mono-1 study. J Clin Oncol. American Society of Clinical Oncology; 2019;37:9004. [Google Scholar]
- 92.Paik PK, Veillon R, Cortot AB, Felip E, Sakai H, Mazieres J, et al. Phase II study of tepotinib in NSCLC patients with METex14 mutations. J Clin Oncol. American Society of Clinical Oncology; 2019;37:9005. [Google Scholar]
- 93.Lu S, Fang J, Cao L, Li X, Guo Q, Zhou J, et al. Abstract CT031: Preliminary efficacy and safety results of savolitinib treating patients with pulmonary sarcomatoid carcinoma (PSC) and other types of non-small cell lung cancer (NSCLC) harboring MET exon 14 skipping mutations Cancer Res. 2019;79:CT031 LP-CT031. [Google Scholar]
- 94.Landi L, Chiari R, Tiseo M, DIncà F, Dazzi C, Chella A, et al. Crizotinib in MET deregulated or ROS1 rearranged pretreated non-small-cell lung cancer (METROS): a phase II, prospective, multicentre, two-arms trial Clin Cancer Res. United States; 2019;25(24):7312–7319. [DOI] [PubMed] [Google Scholar]
- 95.Sabari JK, Leonardi GC, Shu CA, Umeton R, Montecalvo J, Ni A, et al. PD-L1 expression, tumor mutational burden, and response to immunotherapy in patients with MET exon 14 altered lung cancers Ann Oncol Off J Eur Soc Med Oncol. England; 2018;29:2085–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Klempner SJ, Borghei A, Hakimian B, Ali SM, Ou S-HI. Intracranial Activity of Cabozantinib in MET Exon 14-Positive NSCLC with Brain Metastases J Thorac Oncol. United States; 2017;12:152–6. [DOI] [PubMed] [Google Scholar]
- 97.Davies KD, Ng TL, Estrada-Bernal A, Le AT, Ennever PR, Camidge DR, et al. Dramatic Response to Crizotinib in a Patient with Lung Cancer Positive for an HLA-DRB1-MET Gene Fusion JCO Precis Oncol. United States; 2017:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Spigel DR, Ervin TJ, Ramlau RA, Daniel DB, Goldschmidt JHJ, Blumenschein GRJ, et al. Randomized phase II trial of Onartuzumab in combination with erlotinib in patients with advanced non-small-cell lung cancer J Clin Oncol. United States; 2013;31:4105–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bendell JC, Hochster H, Hart LL, Firdaus I, Mace JR, McFarlane JJ, et al. A Phase II Randomized Trial (GO27827) of First-Line FOLFOX Plus Bevacizumab with or Without the MET Inhibitor Onartuzumab in Patients with Metastatic Colorectal Cancer Oncologist. United States; 2017;22:264–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shah MA, Bang Y-J, Lordick F, Alsina M, Chen M, Hack SP, et al. Effect of Fluorouracil, Leucovorin, and Oxaliplatin With or Without Onartuzumab in HER2-Negative, MET-Positive Gastroesophageal Adenocarcinoma: The METGastric Randomized Clinical Trial JAMA Oncol. United States; 2017;3:620–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cloughesy T, Finocchiaro G, Belda-Iniesta C, Recht L, Brandes AA, Pineda E, et al. Randomized, Double-Blind, Placebo-Controlled, Multicenter Phase II Study of Onartuzumab Plus Bevacizumab Versus Placebo Plus Bevacizumab in Patients With Recurrent Glioblastoma: Efficacy, Safety, and Hepatocyte Growth Factor and O(6)-Methylguanine-DNA Met J Clin Oncol. United States; 2017;35:343–51. [DOI] [PubMed] [Google Scholar]
- 102.Wakelee H, Zvirbule Z, De Braud F, Kingsley CD, Mekhail T, Lowe T, et al. Efficacy and Safety of Onartuzumab in Combination With First-Line Bevacizumab- or Pemetrexed-Based Chemotherapy Regimens in Advanced Non-Squamous Non-Small-Cell Lung Cancer Clin Lung Cancer. United States; 2017;18:50–9. [DOI] [PubMed] [Google Scholar]
- 103.Rosen LS, Goldman JW, Algazi AP, Turner PK, Moser B, Hu T, et al. A First-in-Human Phase I Study of a Bivalent MET Antibody, Emibetuzumab (LY2875358), as Monotherapy and in Combination with Erlotinib in Advanced Cancer Clin Cancer Res. United States; 2017;23:1910–9. [DOI] [PubMed] [Google Scholar]
- 104.Catenacci DVT, Tebbutt NC, Davidenko I, Murad AM, Al-Batran S-E, Ilson DH, et al. Rilotumumab plus epirubicin, cisplatin, and capecitabine as first-line therapy in advanced MET-positive gastric or gastro-oesophageal junction cancer (RILOMET-1): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. England; 2017;18:1467–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Finisguerra V, Prenen H, Mazzone M. Preclinical and clinical evaluation of MET functions in cancer cells and in the tumor stroma Oncogene. England; 2016;35:5457–67. [DOI] [PubMed] [Google Scholar]
- 106.Martin V, Chiriaco C, Modica C, Acquadro A, Cortese M, Galimi F, et al. Met inhibition revokes IFNgamma-induction of PD-1 ligands in MET-amplified tumours Br J Cancer. England; 2019;120:527–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Finisguerra V, Di Conza G, Di Matteo M, Serneels J, Costa S, Thompson AAR, et al. MET is required for the recruitment of anti-tumoural neutrophils Nature. England; 2015;522:349–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Grandal MM, Havrylov S, Poulsen TT, Koefoed K, Dahlman A, Galler GR, et al. Simultaneous Targeting of Two Distinct Epitopes on MET Effectively Inhibits MET- and HGF-Driven Tumor Growth by Multiple Mechanisms Mol Cancer Ther. United States; 2017;16:2780–91. [DOI] [PubMed] [Google Scholar]
- 109.Poulsen TT, Grandal MM, Skartved NJO, Hald R, Alifrangis L, Koefoed K, et al. Sym015: A Highly Efficacious Antibody Mixture against MET-Amplified Tumors Clin Cancer Res. United States; 2017;23:5923–35. [DOI] [PubMed] [Google Scholar]
- 110.Moores SL, Chiu ML, Bushey BS, Chevalier K, Luistro L, Dorn K, et al. A Novel Bispecific Antibody Targeting EGFR and cMet Is Effective against EGFR Inhibitor-Resistant Lung Tumors Cancer Res. United States; 2016;76:3942–53. [DOI] [PubMed] [Google Scholar]
- 111.Grugan KD, Dorn K, Jarantow SW, Bushey BS, Pardinas JR, Laquerre S, et al. Fc-mediated activity of EGFR x c-Met bispecific antibody JNJ-61186372 enhanced killing of lung cancer cells MAbs. United States; 2017;9:114–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cho BC, Lee J-S, Han J-Y, Cho EK, Haura E, Lee KH, et al. 1497PJNJ-61186372 (JNJ-372), an EGFR-cMET bispecific antibody, in advanced non-small cell lung cancer (NSCLC): An update on phase I results. Ann Oncol [Internet]. 2018;29:mdy292.118-mdy292.118. Available from: 10.1093/annonc/mdy292.118 [DOI] [Google Scholar]
- 113.Wang J, Anderson MG, Oleksijew A, Vaidya KS, Boghaert ER, Tucker L, et al. ABBV-399, a c-Met Antibody-Drug Conjugate that Targets Both MET-Amplified and c-Met-Overexpressing Tumors, Irrespective of MET Pathway Dependence Clin Cancer Res. United States; 2017;23:992–1000. [DOI] [PubMed] [Google Scholar]
- 114.Strickler JH, Weekes CD, Nemunaitis J, Ramanathan RK, Heist RS, Morgensztern D, et al. First-in-Human Phase I, Dose-Escalation and -Expansion Study of Telisotuzumab Vedotin, an Antibody-Drug Conjugate Targeting c-Met, in Patients With Advanced Solid Tumors J Clin Oncol. United States; 2018;JCO2018787697. [DOI] [PubMed] [Google Scholar]
- 115.Rotow J, Bivona TG. Understanding and targeting resistance mechanisms in NSCLC Nat Rev Cancer. England; 2017;17:637–58. [DOI] [PubMed] [Google Scholar]
- 116.Recondo G, Bahcall M, Spurr LF, Che J, Ricciuti B, Leonardi GC, et al. Molecular mechanisms of acquired resistance to MET tyrosine kinase inhibitors in patients with MET exon 14 mutant NSCLC Clin Cancer Res. United States; 2020. [published online ahead of print, 2020 February 7] [DOI] [PubMed] [Google Scholar]
- 117.Qi J, McTigue MA, Rogers A, Lifshits E, Christensen JG, Janne PA, et al. Multiple mutations and bypass mechanisms can contribute to development of acquired resistance to MET inhibitors Cancer Res. United States; 2011;71:1081–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Heist RS, Sequist L V, Borger D, Gainor JF, Arellano RS, Le LP, et al. Acquired Resistance to Crizotinib in NSCLC with MET Exon 14 Skipping J Thorac Oncol. United States; 2016;11:1242–5. [DOI] [PubMed] [Google Scholar]
- 119.Ou S-HI, Young L, Schrock AB, Johnson A, Klempner SJ, Zhu VW, et al. Emergence of Preexisting MET Y1230C Mutation as a Resistance Mechanism to Crizotinib in NSCLC with MET Exon 14 Skipping J Thorac Oncol. United States; 2017;12:137–40. [DOI] [PubMed] [Google Scholar]
- 120.Yu Z, Ma Y, Ai J, Chen D, Zhao D, Wang X, et al. Energetic factors determining the binding of type I inhibitors to c-Met kinase: experimental studies and quantum mechanical calculations Acta Pharmacol Sin. United States; 2013;34:1475–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Li A, Yang J-J, Zhang X-C, Zhang Z, Su J, Gou L-Y, et al. Acquired MET Y1248H and D1246N Mutations Mediate Resistance to MET Inhibitors in Non-Small Cell Lung Cancer Clin Cancer Res. United States; 2017;23:4929–37. [DOI] [PubMed] [Google Scholar]
- 122.Tiedt R, Degenkolbe E, Furet P, Appleton BA, Wagner S, Schoepfer J, et al. A drug resistance screen using a selective MET inhibitor reveals a spectrum of mutations that partially overlap with activating mutations found in cancer patients Cancer Res. United States; 2011;71:5255–64. [DOI] [PubMed] [Google Scholar]
- 123.Bahcall M, Awad MM, Sholl LM, Wilson FH, Xu M, Wang S, et al. Amplification of Wild-type KRAS Imparts Resistance to Crizotinib in MET Exon 14 Mutant Non-Small Cell Lung Cancer Clin Cancer Res. United States; 2018;24:5963–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Suzawa K, Offin M, Lu D, Kurzatkowski C, Vojnic M, Smith RS, et al. Activation of KRAS Mediates Resistance to Targeted Therapy in MET Exon 14-mutant Non-small Cell Lung Cancer. Clin cancer Res an Off J Am Assoc Cancer Res. 2019;25:1248–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Guo R, Offin M, Brannon AR, Chow A, Delasos L, Somwar R, et al. MET inhibitor resistance in patients with MET exon 14-altered lung cancers. J Clin Oncol [Internet]. American Society of Clinical Oncology; 2019;37:9006 Available from: https://ascopubs.org/doi/abs/10.1200/JCO.2019.37.15_suppl.9006 [Google Scholar]
- 126.McDermott U, Pusapati R V, Christensen JG, Gray NS, Settleman J. Acquired resistance of non-small cell lung cancer cells to MET kinase inhibition is mediated by a switch to epidermal growth factor receptor dependency Cancer Res. United States; 2010;70:1625–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Nisa L, Hafliger P, Poliakova M, Giger R, Francica P, Aebersold DM, et al. PIK3CA hotspot mutations differentially impact responses to MET targeting in MET-driven and non-driven preclinical cancer models Mol Cancer. England; 2017;16:93. [DOI] [PMC free article] [PubMed] [Google Scholar]