Abstract
We propose a fully-automatic deep learning-based algorithm for segmentation of ocular structures and microbial keratitis (MK) biomarkers on slit-lamp photography (SLP) images. The dataset consisted of SLP images from 133 eyes with manual annotations by a physician, P1. A modified region-based convolutional neural network, SLIT-Net, was developed and trained using P1’s annotations to identify and segment four pathological regions of interest (ROIs) on diffuse white light images (stromal infiltrate (SI), hypopyon, white blood cell (WBC) border, corneal edema border), one pathological ROI on diffuse blue light images (epithelial defect (ED)), and two non-pathological ROIs on all images (corneal limbus, light reflexes). To assess inter-reader variability, 75 eyes were manually annotated for pathological ROIs by a second physician, P2. Performance was evaluated using the Dice similarity coefficient (DSC) and Hausdorff distance (HD). Using seven-fold cross-validation, the DSC of the algorithm (as compared to P1) for all ROIs was good (range: 0.62 – 0.95) on all 133 eyes. For the subset of 75 eyes with manual annotations by P2, the DSC for pathological ROIs ranged from 0.69 – 0.85 (SLIT-Net) vs. 0.37 – 0.92 (P2). DSCs for SLIT-Net were not significantly different than P2 for segmenting hypopyons (p > 0.05) and higher than P2 for WBCs (p < 0.001) and edema (p < 0.001). DSCs were higher for P2 for segmenting SIs (p < 0.001) and EDs (p < 0.001). HDs were lower for P2 for segmenting SIs (p = 0.005) and EDs (p < 0.001) and not significantly different for hypopyons (p > 0.05), WBCs (p > 0.05), and edema (p > 0.05). This prototype fully-automatic algorithm to segment MK biomarkers on SLP images performed to expectations on an exploratory dataset and holds promise for quantification of corneal physiology and pathology.
Keywords: Automatic segmentation, deep learning, microbial keratitis, slit-lamp imaging
I. Introduction
Microbial keratitis (MK) is an infectious corneal disease and one of the main causes of blindness worldwide [1–5]. Risk factors include ocular trauma and contact lens overwear, as well as geography and climate [6–10]. Ophthalmologists recommend different treatments for MK based on key morphological biomarkers, including stromal infiltrate (SI) and epithelial defect (ED) sizes [11–15]. Other biomarkers are highly indicative of severe MK, such as an intraocular hypopyon, the accumulation of white blood cells (WBCs), and corneal swelling (corneal edema). As they are tightly associated with clinical outcomes, clinicians evaluate biomarkers when diagnosing disease severity, monitoring disease progression, and making treatment decisions [15, 16]. However, the subjective use of biomarkers can be inaccurate and lead to suboptimal outcomes including treatment delays, poor vision outcomes, and even blindness [17].
Slit-lamp photography (SLP) images are a low-cost technology available in all eye clinics. SLP images are high-resolution external views of the anterior structures of the eye, including the cornea [18, 19]. Different illuminations using color filters, in conjunction with the application of topical stains to the eye such as fluorescein, lissamine green, or rose bengal, may be used to enhance the visibility of certain biomarkers, such as EDs [19–22]. However, SLP is not used on all patients. Instead, clinicians typically rely on subjective assessment of biomarkers and manually record their findings in the electronic health records (EHR). Subjective techniques include manual slit-lamp caliper measurements, drawings in the EHR, and free-text descriptions [23–27]. There are no standardized, distributed strategies to quantify specific MK biomarkers based on SLP images.
Prior research shows that (1) using a standardized method to capture information of corneal MK biomarkers and (2) employing a computer-aided strategy to quantify biomarkers improves reliability of measurements compared to the clinical exam gold standard while simultaneously creating improved data fidelity with stored SLP images [28, 29]. Prior approaches have been developed for SIs and EDs based on k-means clustering [30], superpixels [31], Gaussian mixture models [32], random forests [29], and patch-based convolutional neural networks (CNNs) [33]. However, most of these algorithms require manual user input in the form of selecting landmarks to identify the corneal surface area [30, 31, 33] or seed regions to distinguish between the background and biomarker of interest [29]. Additionally, these algorithms do not segment other important biomarkers such as hypopyons, WBCs, or edema. Building upon prior approaches, a fully-automatic computerized tool can deliver a robust and objective assessment of MK biomarkers on SLP images.
We developed and tested a fully-automatic algorithm, Slit-Lamp Imaging Technology (SLIT)-Net, for segmentation of ocular structures and multiple MK biomarkers on SLP images under two different illuminations. The algorithm is based on deep learning, a sub-field of machine learning that has demonstrated excellent performance using CNNs for many image recognition and segmentation tasks [34–43] including in ophthalmic imaging [44–54]. A CNN is made up of several layers of filters that extract features from images and map them to an output. The values of the filters, commonly referred to as the weights of the network, are learned via optimization on annotated training data. We developed and evaluated SLIT-Net using SLP images collected in the USA and India. To the best of our knowledge, this is the first fully-automatic segmentation algorithm for MK. The goal is to help clinicians quantify measurements of MK biomarkers and therefore improve medical decision-making at the time of diagnosis and over the course of disease management. A preliminary version of this work has been reported [55, 56].
II. Material and methods
We developed and trained a deep learning network, SLIT-Net, to identify and segment four MK biomarkers on diffuse white light SLP images and one biomarker on diffuse blue light SLP images. Additionally, the network segments the corneal limbus and ocular surface light reflexes. We evaluated the performance of the network using the Dice similarity coefficient (DSC) and the general Hausdorff distance (HD).
A. Dataset
The dataset consisted of SLP images of 133 eyes taken at two eye centers - the University of Michigan Kellogg Eye Center (MI, USA) and Aravind Eye Care System (Madurai, India) - from 2016 to 2018. The University of Michigan Institutional Review Board and Aravind ethics committee reviewed the proposal and granted permission for this prospective investigation.
Patients were photographed under an SLP imaging setting described in prior work [29]. At both eye centers, two types of images were taken of each eye - one with diffuse white light illumination and one with diffuse blue light illumination after topical fluorescein staining. All images had an aspect ratio of 3:2. The dimensions of the images were 3888 × 2592 pixels or 3648 × 2432 pixels at the University of Michigan and 5184 × 3456 pixels at Aravind Eye Care System. For each image, the regions of interest (ROIs) were manually annotated by a physician, P1 (MFK, MD, final year ophthalmology training), at the University of Michigan using ImageJ software (NIH, MD, USA) [57]. Pathological ROIs included the following MK biomarkers - the SI, hypopyon, WBC border, and corneal edema border on diffuse white light SLP images. EDs were annotated on diffuse blue light SLP images. Non-pathological ROIs included the corneal limbus and ocular surface light reflexes. The corneal limbus size is known and can provide a reference for the true size of the MK biomarkers. Light reflexes can indicate the health of the corneal surface and can be confused for other biomarkers (e.g. SIs) if not separately identified.
Not all biomarkers were present in each eye. Additionally, in some images the superior corneal limbus border was partially occluded by the eyelid or outside the imaging field of view. In these cases, the eyelid or image boundary were annotated to create an enclosed area of the visible corneal surface. Figure 1 shows examples of the manual annotations by P1.
Figure 1:

Examples of SLP images taken with diffuse white light illumination and diffuse blue light illumination after topical fluorescein staining. Pathological and non-pathological ROIs were manually annotated by a physician (P1).
For 75 of the 133 eyes, the pathological ROIs were manually annotated by a second physician, P2 (MMT, MD, final year ophthalmology training, for diffuse white light images; KHK, MD, corneal specialist, for diffuse blue light images), at the University of Michigan.
The manual annotations by P1 were used for the development, training, and testing of the network. The manual annotations by P2 were used only for an inter-reader variability analysis.
B. Network
The proposed SLIT-Net is based on a modified version of Mask R-CNN [41, 58]. Figure 2 shows the details of SLIT-Net. The backbone architecture is a CNN, specifically ResNet-50 [36] with a feature pyramid network (FPN) [59], which extracts multi-scale features from the image.
Figure 2:
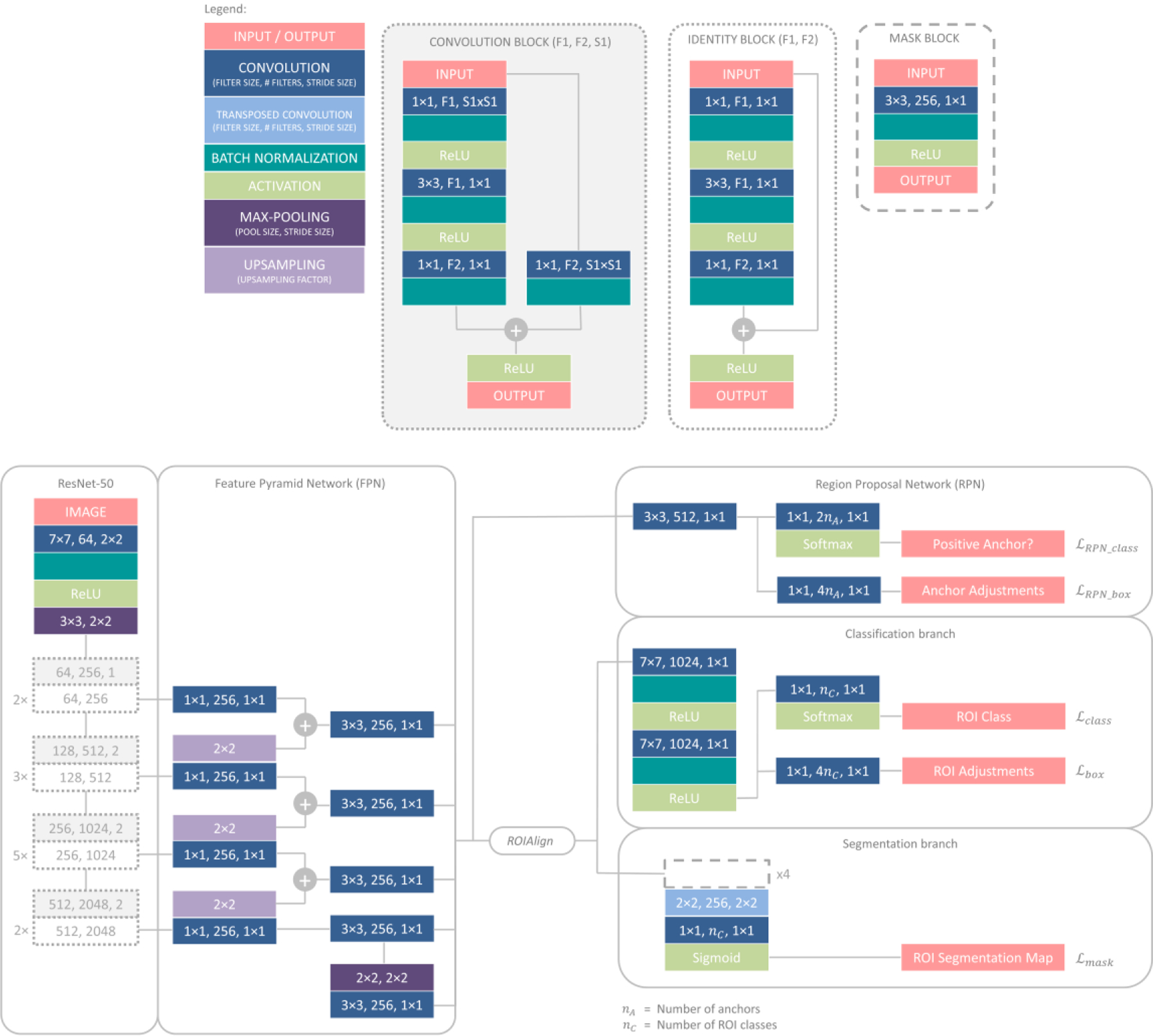
SLIT-Net architecture.
From the multi-scale features and a set of anchors, a second CNN called a regional proposal network (RPN) [40] predicts the probability of each anchor being a positive or negative class. The set of anchors correspond to the size and location of pre-determined rectangular regions on the image. We use anchors of five sizes (16, 32, 64, 128, and 256 pixels) and three aspect ratios (0.5, 1, and 2) at every pixel. An anchor is classified as positive if it overlaps with any manual annotation’s bounding box with an intersection-over-union (IoU) [60] ≥ 0.7; otherwise, it is classified as negative. The manual annotation’s bounding box is defined as the smallest rectangular region within which the manual annotation lies, by four parameters - the x-coordinate, x, and y-coordinate, y, of the top-left corner, width, w, and height, h, of the bounding box.
The RPN is trained with a focal loss [61] defined as
where nA is the number of anchors and pA is the predicted probability of the correct class of anchor A.
The RPN also predicts adjustments for the anchor to be closer to the manual annotation’s bounding box. The adjustments are defined as
where xA and yA are the x-coordinate and y-coordinate of the top-left corner and wA and hA are the width and height of anchor A. The RPN is trained with a smooth L1 loss [39] defined as
where is the predicted adjustment value for anchor A and
The anchors are sorted from highest to lowest by the probability of being a positive class, and the top 2000 anchors are retained. The predicted adjustments are made to the anchors and non-maximum suppression is applied to prune them; that is, any anchor that overlaps another with IoU > 0.7 but has a lower probability is discarded. The final set of adjusted anchors are the proposed regions.
Next, a ROIAlign layer [41] is used to extract from the multi-scale feature maps a N×N feature patch corresponding to the spatial location of each proposed region. The feature patch is used by two separate branches, each comprised of a CNN. The classification branch uses 7×7 patches to classify and predict further adjustments to the proposed region. The segmentation branch uses 14×14 patches to segment the ROI within the proposed region.
For training, any proposed region overlapping a manual annotation’s bounding box with an IoU < 0.5 is classified as a background region; otherwise, it is assigned the same class as the highest overlapping manual annotation’s bounding box and considered a positive region. All positive regions are retained, while background regions are randomly sampled such that the ratio of positive to background regions is 0.33 to avoid an overwhelming number of background regions.
Similar to the RPN, the classification branch is trained with a focal loss defined as
where nR is the number of proposed regions and pR is the predicted probability of the correct class of the proposed region R.
The adjustments with respect to the manual annotation’s bounding box are now defined as
where xR and yR are the x-coordinate and y-coordinate of the top-left corner and wR and hR are the width and height of proposed region R. The smooth L1 loss is defined as
where is the predicted adjustment value for proposed region R.
The segmentation branch predicts segmentation probability maps for the positive regions only, as background regions do not have segmentation maps. It is trained with a Hausdorff-Dice loss [62] to reduce large segmentation errors in the boundary [62] as well as balance the background and foreground pixels in the segmentation maps [63]. The Hausdorff-Dice loss is defined as
where λ is a weight factor which is calculated as the ratio of to to equalize the contribution from both terms.
[63] is defined as
where ∘ is an element-wise multiplication.
can be efficiently implemented using convolutions with circular kernels [62] and is defined as
where nI is the number of pixels in the image. r is the radius of the circular kernel Br and the elements of Br are normalized such that they sum to 1. , q and are the manual annotations, predicted segmentation probability maps, and predicted binary segmentation maps (obtained by thresholding q at 0.5), respectively. The superscript C indicates the complement while * denotes convolution. fsoft is a soft-thresholding operation defined as
where
is an estimation of the part of that does not overlap with defined as
and vice versa for .
The final loss, for training the network is the summation of the individual losses such that
C. Training
For training, the images were resized using bilinear interpolation such that the longer side was 512 pixels while retaining the aspect ratio. As the images were of different sizes, they were zero-padded to a standard size of 512×512 pixels.
Two forms of data augmentation were applied to the images on the fly during training to artificially increase the dataset size. For each image, one of four geometric transformations - horizontal flipping, cropping, applying an affine transformation, or no transformation - was randomly selected. Then, one of five intensity transformations - adding a random scalar, multiplying by a random scalar, adding Gaussian noise, applying contrast normalization, or no transformation - was randomly selected. The images were normalized by subtracting the per-channel mean across the entire dataset.
The network weights were initialized with pre-trained weights on the COCO dataset [64] except for the final layers of each branch, which were randomly initialized using Xavier initialization [65]. The weights were further optimized on our dataset using Nesterov’s accelerated stochastic gradient descent [66] with a learning rate of 0.001, momentum of 0.9, and learning rate decay of 10−6 to minimize .
The network was trained with a batch-size of five images for 100 epochs until convergence. Performance was monitored on a hold-out validation set for the last 20 epochs and the weights of the best-performing network were retained as the final weights of the network. Performance evaluation is detailed in Section II.F.
D. Testing
For testing, the images were resized and zero-padded to 512×512 pixels and normalized as described in Section II.C. No data augmentation was applied. The trained network was then used to identify and segment the ROIs on the images. The same anchors as described in Section II.C were used for the RPN and the top 1000 adjusted anchors were retained as proposed regions. Each proposed region was assigned to the class with the highest predicted probability by the classification branch. The predicted segmentation probability maps from the segmentation branch were resized using bilinear interpolation and thresholded at 0.5 to obtain predicted binary segmentation maps at the original image resolution.
E. Post-processing
Additional post-processing was applied to the output of SLIT-Net to adhere to certain pathological and anatomical constraints.
For diffuse white light images, the following constraints were applied:
For the limbus, only the ROI with the highest probability was retained as only one limbus can exist for any eye.
Any instances of pathological ROIs which occurred outside the limbus were deleted as these ROIs can only exist within the limbus.
All holes within the binary segmentation maps were filled using a morphological operation [67] to improve the quality of the segmentation maps.
For any overlapping ROIs of the same class with IoU > 0.7, only the ROI with the highest probability was retained to avoid multiple instances of the same ROI.
For the hypopyon, only the ROI with the highest probability was retained as only one hypopyon can exist for any eye.
All instances of WBCs and edema which did not co-occur or overlap with an SI were deleted as these ROIs can only exist in the presence of an SI.
For light reflexes with a high probability > 0.8, any overlapping SIs were deleted as they were most likely false positives due to the similarity of their appearances (bright white regions) on the image.
For diffuse blue light images, only constraints i – iv were applied as the hypopyon, WBCs, edema, and SIs are not detected on diffuse blue light images.
F. Quantitative analysis
To evaluate performance, the DSC [68] was calculated for each ROI class as
where TP was the number of true positive, FP was the number of false positive, and FN was the number of false negative pixels in the binary segmentation maps as compared to the manual annotations of P1 as the gold standard.
The HD [69] was calculated for each ROI class as
where
is the directed Hausdorff distance between X and Y, which are the boundaries of the predicted segmentations and the gold standard, respectively, and vice versa for hd(Y, X).
The DSC provides a measure of the proportion of overlap between the predicted segmentations and the gold standard, whereby a higher DSC indicates better performance. On the other hand, the HD provides a measure of the largest segmentation error and therefore, the scale of the error between the predicted segmentations and the gold standard, whereby a lower HD indicates better performance. Both measures are complementary and together provide a more complete performance evaluation.
As described in Section II.A, not all biomarkers were present in each eye. However, it is still important that the algorithm be able to correctly identify the absence of biomarkers. Therefore, if an absent biomarker is correctly identified as being absent by the algorithm, this is still considered a perfect overlap and results in a DSC of 1.00 and an HD of 0. However, if a biomarker is absent and the algorithm erroneously identifies its presence, this results in a DSC of 0.00. In this case, however, the HD is undefined and thus excluded following [70]. Similarly, if a biomarker is present and the algorithm erroneously identifies it as being absent, this also results in a DSC of 0.00. In this case, the HD is also undefined and thus excluded.
To assess inter-reader variability, we calculated the performance of P2. To compare the performance between SLIT-Net and P2, we used the p-value of the Wilcoxon signed-rank test to determine the statistical significance of the difference in performance in which a p-value < 0.05 was considered statistically significant.
G. Implementation
SLIT-Net was implemented in Python using the TensorFlow [71] (Version 1.5.1) and Keras [72] (Version 2.0.8) libraries on a desktop computer equipped with an Intel® Core™ i7–6850K CPU and four NVIDIA® GeForce® GTX 1080Ti GPUs.
The dataset and automatic software introduced in this study is available on GitHub at https://github.com/jessicaloohw/SLIT-Net
III. Results
We report the quantitative and qualitative results of the proposed SLIT-Net and compare it to alternative methods. As there are no fully-automatic algorithms for this particular task available for comparison, we used a U-Net [38], one of the most popular deep learning-based methods for medical segmentation that is often used as a baseline [73–87]. We also used the more recently developed nnU-Net [88] that automatically adapts the U-Net to achieve performance gains for a specific medical segmentation task by determining the ideal architecture and training strategy based on the training data. Additionally, we made comparisons with the original Mask R-CNN [41].
A. Comparison to alternative methods
We used seven-fold cross-validation to evaluate performance on all available images while ensuring independence of the training and testing sets. The 133 eyes were randomly divided into seven groups, each consisting of 19 eyes. Images from six groups were used as the training set while the remaining group was used as the testing set. From the training set, one group was used as the hold-out validation set. The groups were then rotated such that each group was used once for testing.
We trained and tested the proposed SLIT-Net as described in Sections II.C, II.D, and II.E.
For the baseline U-Net, we used the same network architecture and loss as described in the original publication [38]. For training, we used the same procedure as described in Section II.C. However, in our experiments, we noticed that 100 epochs were not sufficient for convergence; therefore, we trained the network for 1000 epochs until convergence. Besides the extended training, the pre-processing, initialization, data augmentation, optimization, and validation were otherwise kept the same. The nnU-Net [88] automatically determines the architecture, training, and validation procedures, and we followed all the provided specifications. For Mask R-CNN [41], we removed the modifications that were introduced into SLIT-Net. For testing, as described in Sections II.D and II.E, we resized the predicted segmentation probability maps using bilinear interpolation, thresholded at 0.5 to obtain predicted binary segmentation maps at the original image resolution, and applied the same post-processing constraints for all methods.
Note that separate networks were trained for diffuse white light and diffuse blue light images.
B. Quantitative analysis
Tables 1 and 2 show the average performance of the proposed SLIT-Net and alternative methods on all 133 eyes for diffuse white light and diffuse blue light images, respectively. We also report the performance for the subset of 75 eyes and the p-value of the Wilcoxon signed rank test between SLIT-Net and P2 in Table 3.
TABLE 1.
Average DSC and HD (mean ± standard deviation, median) of the baseline U-Net, nnU-Net, Mask R-CNN, and the proposed SLIT-Net on diffuse white light images
| ROIs | DSC | HD* | |||||||
|---|---|---|---|---|---|---|---|---|---|
| U-Net | nnU-Net | Mask R-CNN | SLIT-Net | U-Net | nnU-Net | Mask R-CNN | SLIT-Net | ||
| Pathological | SI | 0.34 ± 0.32, 0.29 | 0.64 ± 0.29, 0.76 | 0.68 ± 0.31, 0.80 | 0.66 ± 0.33, 0.82 | 493 ± 364, 401 | 375 ± 360, 243 | 219 ± 188, 150 | 216 ± 182, 169 |
| Hypopyon | 0.19 ± 0.36, 0.00 | 0.61 ± 0.44, 0.87 | 0.75 ± 0.40, 1.00 | 0.75 ± 0.40, 1.00 | 481 ± 498, 311 | 122 ± 237, 0 | 73 ± 180, 0 | 67 ± 151, 0 | |
| WBC | 0.40 ± 0.36, 0.43 | 0.59 ± 0.37, 0.77 | 0.64 ± 0.37, 0.79 | 0.62 ± 0.39, 0.78 | 225 ± 285, 110 | 331 ± 257, 253 | 222 ± 185, 168 | 213 ± 196, 177 | |
| Edema | 0.59 ± 0.47, 1.00 | 0.15 ± 0.30, 0.00 | 0.63 ± 0.48, 1.00 | 0.69 ± 0.46, 1.00 | 28 ± 80, 0 | 510 ± 405, 422 | 47 ± 152, 0 | 16 ± 73, 0 | |
| Non-pathological | Light reflex | 0.01 ± 0.09, 0.00 | 0.72 ± 0.18, 0.77 | 0.78 ± 0.15, 0.82 | 0.76 ± 0.17, 0.81 | 828 ± 636, 692 | 127 ± 239, 27 | 26 ± 55, 6 | 36 ± 75, 6 |
| Limbus | 0.91 ± 0.17, 0.96 | 0.96 ± 0.11, 0.98 | 0.95 ± 0.03, 0.96 | 0.95 ± 0.02, 0.96 | 294 ± 386, 177 | 287 ± 598, 103 | 334 ± 638, 152 | 342 ± 639, 161 | |
does not include undefined values
TABLE 2.
Average DSC and HD (mean ± standard deviation, median) of the baseline U-Net, nnU-Net, Mask R-CNN, and the proposed SLIT-Net on diffuse blue light images
| ROIs | DSC | HD* | |||||||
|---|---|---|---|---|---|---|---|---|---|
| U-Net | nnU-Net | Mask R-CNN | SLIT-Net | U-Net | nnU-Net | Mask R-CNN | SLIT-Net | ||
| Pathological | ED | 0.58 ± 0.41, 0.82 | 0.76 ± 0.34, 0.92 | 0.75 ± 0.33, 0.90 | 0.77 ± 0.32, 0.91 | 281 ± 363, 131 | 153 ± 218, 70 | 132 ± 138, 93 | 121 ± 143, 79 |
| Non-pathological | Light reflex | 0.56 ± 0.31, 0.68 | 0.73 ± 0.19, 0.75 | 0.74 ± 0.19, 0.80 | 0.74 ± 0.19, 0.79 | 128 ± 270, 36 | 82 ± 188, 22 | 19 ± 33, 8 | 25 ± 70, 7 |
| Limbus | 0.86 ± 0.18, 0.92 | 0.94 ± 0.15, 0.97 | 0.90 ± 0.15, 0.94 | 0.90 ± 0.17, 0.94 | 442 ± 430, 297 | 207 ± 366, 128 | 300 ± 352, 211 | 288 ± 385, 194 | |
does not include undefined values
TABLE 3.
Average DSC and HD (mean ± standard deviation, median) of the proposed SLIT-net and P2 on pathological ROIs
| ROIs | DSC | HD* | |||||
|---|---|---|---|---|---|---|---|
| SLIT-Net | P2 | p-value | SLIT-Net | P2 | p-value | ||
| Diffuse white light | SI | 0.70 ± 0.30, 0.83 | 0.81 ± 0.22, 0.90 | < 0.001 | 216 ± 182, 169 | 183 ± 169, 109 | 0.005 |
| Hypopyon | 0.73 ± 0.40, 0.94 | 0.88 ± 0.24, 1.00 | 0.052 | 67 ± 151, 0 | 59 ± 88, 0 | 0.819 | |
| WBC | 0.69 ± 0.35, 0.85 | 0.45 ± 0.44, 0.43 | < 0.001 | 213 ± 196, 177 | 270 ± 281, 181 | 0.248 | |
| Edema | 0.73 ± 0.44, 1.00 | 0.37 ± 0.46, 0.00 | < 0.001 | 16 ± 73, 0 | 112 ± 188, 0 | 0.500 | |
| Diffuse blue light | ED | 0.85 ± 0.20, 0.91 | 0.92 ± 0.13, 0.95 | < 0.001 | 121 ± 143, 79 | 89 ± 68, 64 | < 0.001 |
does not include undefined values
Overall, SLIT-Net achieved good performance with relatively high average DSCs and relatively low average HDs, although there was high variability.
Compared to U-Net, SLIT-Net achieved a higher average DSC for all ROIs and a lower average HD for all pathological ROIs on both diffuse white light and diffuse blue light images. Compared to nnU-Net, SLIT-Net achieved a higher average DSC and a lower average HD for all ROIs on both diffuse white light and diffuse blue light images, except the corneal limbus. While the average DSCs were rather similar for Mask R-CNN and SLIT-Net, the advantage of SLIT-Net was mainly reflected in the average HDs, whereby SLIT-Net achieved a lower average HD for all pathological ROIs on both diffuse white light and diffuse blue light images.
For identification and segmentation of WBCs and edema, SLIT-Net achieved significantly higher average DSCs than P2, and statistically similar average HDs. For hypopyons, the average DSC and HD were statistically similar for SLIT-Net and P2. On the other hand, P2 achieved significantly higher average DSCs and lower average HDs than SLIT-Net for both SIs and EDs. However, the average DSC for SLIT-Net in these cases was ≥ 0.70, a threshold considered highly correlated [89].
C. Qualitative analysis
Figure 3 shows the comparison between the manual annotations by P1 and the fully-automatic segmentations by the baseline U-Net, nnU-Net, Mask R-CNN, and the proposed SLIT-Net.
Figure 3:
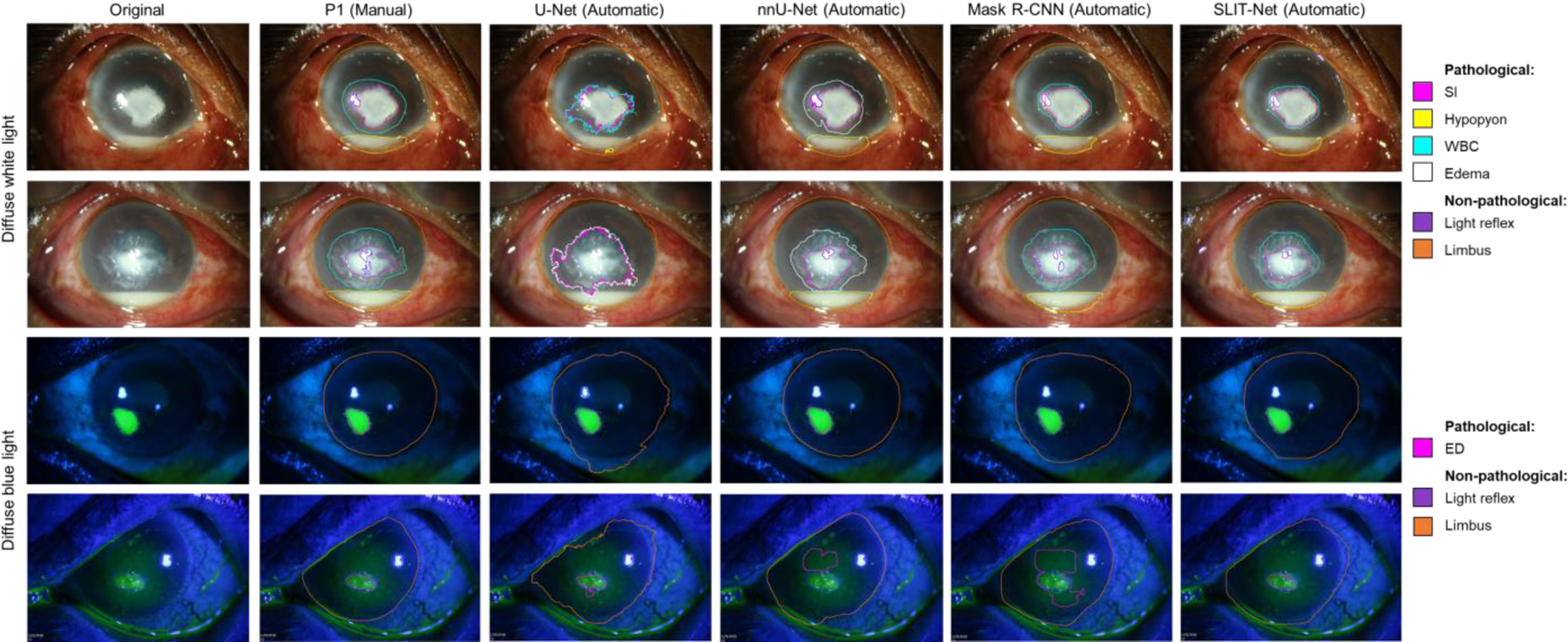
Examples of manual annotations by P1 and fully-automatic segmentations by the baseline U-Net, nnU-Net, Mask R-CNN, and the proposed SLIT-Net. SLIT-Net’s segmentations were closest to that of P1 and outperformed the alternative methods.
Figures 4–7 show the comparison between the manual annotations by P1 and P2 and the fully-automatic segmentations by SLIT-Net. Figure 4 shows examples of good agreement between the methods. Some biomarkers, such as WBCs, edema, and EDs, were difficult to identify and segment with precision due to ambiguous or “fuzzy” borders as shown in Figures 5 and 6. The heterogeneity of MK phenotypes and image quality can also worsen the ability to accurately identify and segment the ROIs as shown in Figure 7.
Figure 4:

Examples of manual annotations by P1 and P2 and fully-automatic segmentations by SLIT-Net on diffuse white light and diffuse blue light images. There was good agreement among the three methods.
Figure 7:

Examples of poor segmentation by SLIT-Net due to the heterogeneity of MK phenotype or poor image quality. In some cases, there was also low agreement between P1 and P2.
Figure 5:
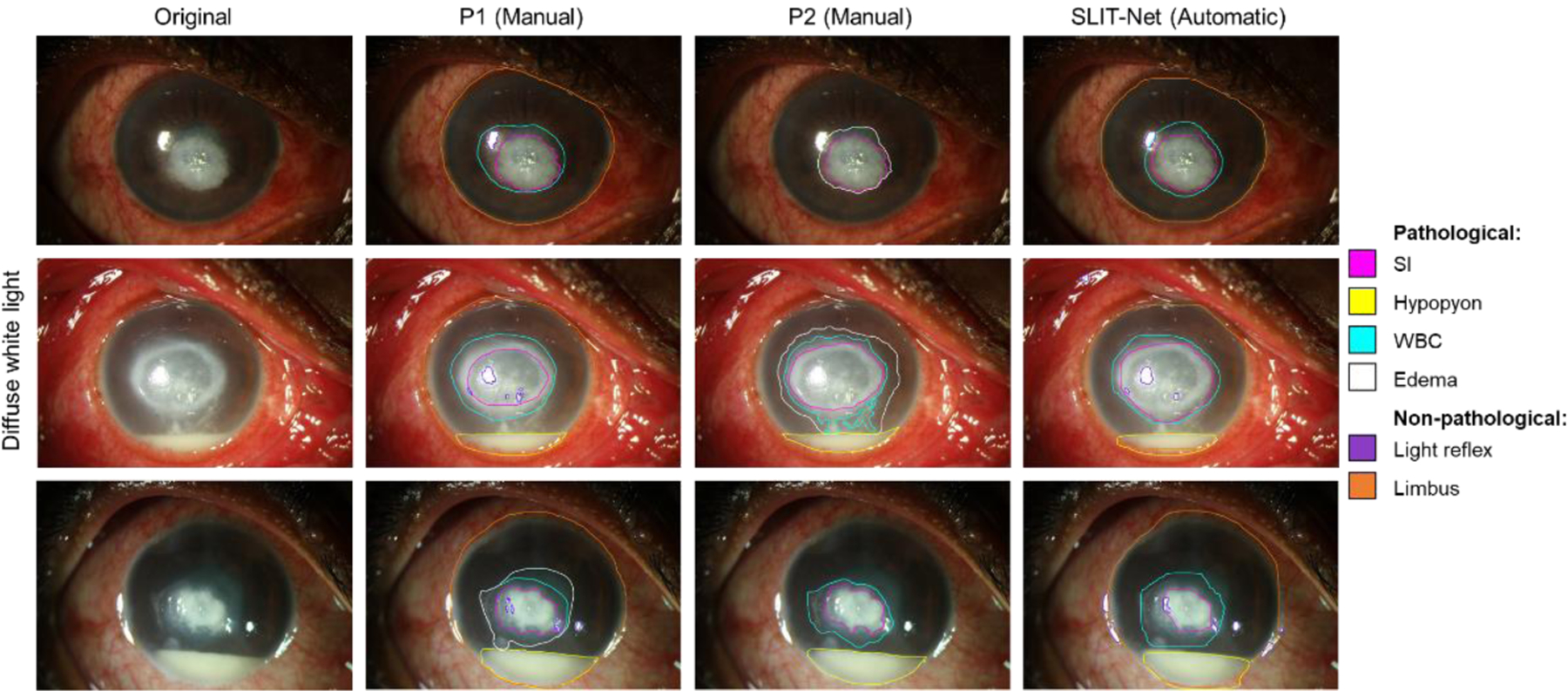
Examples of the difficulty with identification and segmentation of WBCs and edema on diffuse white light images. Top: P1 identified the region surrounding the SI as WBCs, while P2 identified the same region as edema instead. SLIT-Net’s segmentations were closer to that of P1. Middle: P1 identified the region surrounding the SI as only WBCs, while P2 identified an additional region of edema. P1 also identified a smaller SI compared to P2. SLIT-Net identified a SI that was closer to P2 but did not identify any edema. Bottom: P1 identified both WBCs and edema surrounding the SI, while P2 and SLIT-Net identified only WBCs.
Figure 6:
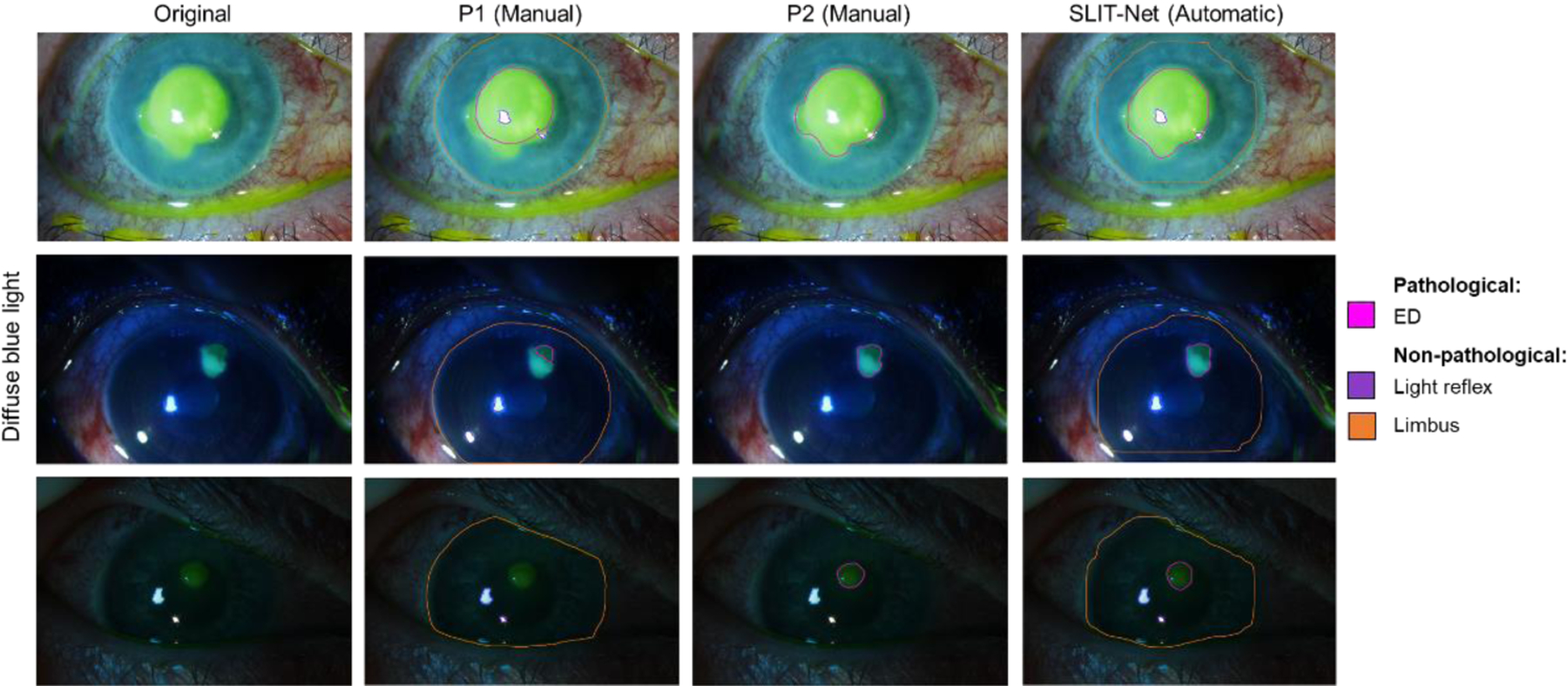
Examples of the difficulty with segmentation of EDs on diffuse blue light images. SLIT-Net’s segmentations were closer to that of P2 in these cases.
D. Ablation study
The proposed SLIT-Net uses a ResNet-50 with FPN backbone. The FPN generates multi-scale features which are ideal for identification and segmentation of the ROIs on SLP images as they are of very different scales, ranging from small light reflexes to the corneal limbus. We investigated the performance of SLIT-Net with and without the FPN in the backbone. We made the corresponding architectural changes when the FPN is removed from the backbone as described in [41]. Tables 4 and 5 show the average performance of SLIT-Net with the different backbone architectures. The overall performance of SLIT-Net drops drastically without the FPN, demonstrating that the features from ResNet-50 alone were not sufficient for identification and segmentation of the diverse range of ROIs on SLP images. Some ROIs were simply not identified, resulting in undefined HD values. Therefore, while some biomarkers such as hypopyons, WBCs, and EDs have lower average HDs, this is not necessarily an indication of better performance and the lower average DSCs need to be considered as well. An alternative HD may also be considered, using the maximum diameters of the unidentified biomarkers as a proxy for the undefined HD values. The average alternative HDs for SIs, hypopyons, WBCs, edema, and EDs were 787 ± 484, 483 ± 740, 867 ± 643, 295 ± 628, and 621 ± 517 using the ResNet-50 backbone and 297 ± 326, 273 ± 462, 422 ± 458, 422 ± 703, and 164 ± 222 using the ResNet-50 with FPN backbone. Furthermore, without the multi-scale features, distinguishing the subtle transitions between ROIs, such as SIs and WBCs, would be nearly impossible. The effectiveness of incorporating the FPN into the backbone of SLIT-Net was confirmed with the ablation study.
TABLE 4.
Average DSC and HD (mean ± standard deviation, median) of the proposed SLIT-Net with different backbone architectures on diffuse white light images
| ROIs | DSC | HD* | |||
|---|---|---|---|---|---|
| ResNet-50 | ResNet-50 with FPN | ResNet-50 | ResNet-50 with FPN | ||
| Pathological | SI | 0.04 ± 0.17, 0.00 | 0.66 ± 0.33, 0.82 | 295 ± 385, 122 | 216 ± 182, 169 |
| Hypopyon | 0.67 ± 0.48, 1.00 | 0.75 ± 0.40, 1.00 | 5 ± 50, 0 | 67 ± 151, 0 | |
| WBC | 0.22 ± 0.42, 0.00 | 0.62 ± 0.39, 0.78 | 8 ± 44, 0 | 213 ± 196, 177 | |
| Edema | 0.78 ± 0.41, 1.00 | 0.69 ± 0.46, 1.00 | 0 ± 0, 0 | 16 ± 73, 0 | |
| Non-pathological | Light reflex | 0.19 ± 0.23, 0.10 | 0.76 ± 0.17, 0.81 | 216 ± 269, 116 | 36 ± 75, 6 |
| Limbus | 0.07 ± 0.17, 0.00 | 0.95 ± 0.02, 0.96 | 2182 ± 675, 2290 | 342 ± 639, 161 | |
does not include undefined values
TABLE 5.
Average DSC and HD (mean ± standard deviation, median) of the proposed SLIT-net with different backbone architectures on diffuse blue light images
| ROIs | DSC | HD* | |||
|---|---|---|---|---|---|
| ResNet-50 | ResNet-50 with FPN | ResNet-50 | ResNet-50 with FPN | ||
| Pathological | ED | 0.18 ± 0.38, 0.00 | 0.77 ± 0.32, 0.91 | 0 ± 0, 0 | 121 ± 143, 79 |
| Non-pathological | Light reflex | 0.18 ± 0.25, 0.00 | 0.74 ± 0.19, 0.79 | 200 ± 296, 115 | 25 ± 70, 7 |
| Limbus | 0.01 ± 0.02, 0.00 | 0.90 ± 0.17, 0.94 | 2144 ± 515, 2187 | 288 ± 385, 194 | |
does not include undefined values
IV. Discussion
SLIT-Net is a fully-automatic algorithm for segmentation of ocular structures and biomarkers of MK on SLP images under two different illuminations. The performance of this first-of-a-kind automatic algorithm is considered good according to conventional standards [89]. SLIT-Net outperforms several other popular deep learning-based segmentation methods, namely U-Net, nnU-Net, and Mask R-CNN. However, there is still room for improvement until SLIT-Net matches the performance of expert clinicians. The difficulty of this task, and consequently the difficulty of automating this task, is also reflected in some relatively low agreements between clinicians on the manual annotations of certain biomarkers.
Upon further evaluation, key elements were identified. First, some biomarkers have similar characteristics. Figure 5 shows that WBCs and edema can be difficult to distinguish due to similar appearances on the diffuse white light images. Additionally, there may be a gradual transition from one to the other, lacking a definitive border. This difficulty is reflected in Table 3 by the low average DSC for P2, indicating that the physicians also had difficulty annotating these biomarkers and did not always agree on where one transitioned to the other.
Second, Figure 7 shows that the algorithm may have difficulty detecting a biomarker with high transparency, such as edema.
Third, the imaging technique and image quality may affect the ability to accurately identify and segment biomarkers. The field of view and quality of the images can vary drastically if images do not adhere to a strict protocol. In Figure 6, the intensity of fluorescein staining varies between images and the fluorescein staining can be picked up by other structures, such as the eyelid cul-de-sac. This makes it difficult to discern the true EDs and may result in different annotations by different physicians based on their judgment calls. SIs will gain a yellow tinge on diffuse white light images if the eye is imaged after the clinician stains the eye with fluorescein as shown in Figure 7, which may affect the ability of the algorithm to accurately detect the biomarkers. Besides that, the complete limbus may be visible in some images and partially occluded in others. To prevent limbus occlusion, the image often includes the photographer’s fingertips.
Evaluating an algorithm by performance metrics is the standard approach for validation, but it does not sufficiently determine reliability for clinical applications. For our next step, we will evaluate SLIT-Net on sequential clinical images to evaluate MK healing during treatment. Determination of the ability of SLIT-Net to measure the change in biomarkers on images over time generates a more clinically meaningful evaluation of the algorithm’s performance and its potential for use [90]. Measuring disease trajectories could also be used to compare different treatments and predict clinical outcomes.
The diffuse white light and diffuse blue light images of an eye were taken in one imaging session. Therefore, segmentation performance may improve if both images are provided as inputs to SLIT-Net simultaneously. We will investigate this approach as we build our dataset. We will also explore if the use of additional image types, such as slit beam images, improves performance.
Prior to clinical studies and applications, SLIT-Net has to undergo further development, improvement, and testing. Meanwhile, SLIT-Net can be used in a semi-automatic manner to provide initial annotations on SLP images. Clinicians can then evaluate and correct any algorithmic errors. This will provide clinicians with objective quantified measures of ocular structures and biomarkers of MK which can aid clinicians as they manage patients over time. For example, since SLIT-Net accurately segments the corneal limbus, this can serve as a reference point to annotate the other biomarkers. SLIT-Net can aid clinicians in identifying and segmenting ROIs more reliably, as shown with the application of other algorithms [29].
V. Conclusion
MK is an infectious corneal disease and a leading cause of blindness worldwide [1–5]. Clinicians subjectively evaluate multiple important biomarkers to inform treatment decisions. However, no fully-automatic, standardized, and distributed strategies exist. Therefore, clinicians still rely on manual and subjective assessments of these biomarkers. We have developed the first fully-automatic algorithm, SLIT-Net, for segmentation of ocular structures and biomarkers of MK on SLP images under two different illuminations. SLIT-Net identifies and segments four pathological ROIs on diffuse white light images (SIs, hypopyons, WBCs, and edema), one pathological ROI on diffuse blue light images with fluorescein staining (EDs), and two non-pathological ROIs (corneal limbus and light reflexes) on all images. Each component of our method, individually or together, can be used in many anterior eye segment research projects. To promote future advancement of automatic algorithms for diagnosis and prognosis of infectious corneal diseases, we made our dataset and algorithms freely available online as an open-source software package.
Acknowledgments
This work was supported by the National Institutes of Health under Grants R01EY031033-01 and P30EY005722.
Contributor Information
Jessica Loo, Department of Biomedical Engineering, Duke University, Durham, NC 27708 USA.
Matthias F. Kriegel, Department of Ophthalmology and Visual Sciences, Kellogg Eye Center, University of Michigan, Ann Arbor, MI 48105 USA Department of Ophthalmology, Augenzentrum am St. Franziskus Hospital Münster, 48145 Münster, NRW, Germany.
Venkatesh Prajna, Department of Cornea and Refractive Services, Aravind Eye Care System, Madurai, Tamil Nadu, India.
Maria A. Woodward, Department of Ophthalmology and Visual Sciences, Kellogg Eye Center, University of Michigan, Ann Arbor, MI 48105 USA; Institute for Healthcare Policy and Innovation, University of Michigan, Ann Arbor, MI 48109 USA.
Sina Farsiu, Department of Biomedical Engineering, Duke University, Durham, NC 27708 USA; Department of Ophthalmology, Duke University Medical Center, Durham, NC 27708 USA.
References
- [1].Bourne RR et al. , “Causes of vision loss worldwide, 1990–2010: a systematic analysis,” The lancet global health, vol. 1, no. 6, pp. e339–e349, 2013. [DOI] [PubMed] [Google Scholar]
- [2].Flaxman SR et al. , “Global causes of blindness and distance vision impairment 1990–2020: a systematic review and meta-analysis,” The Lancet Global Health, vol. 5, no. 12, pp. e1221–e1234, 2017. [DOI] [PubMed] [Google Scholar]
- [3].Resnikoff S et al. , “Global data on visual impairment in the year 2002,” Bulletin of the World Health Organization, vol. 82, pp. 844–851, 2004. [PMC free article] [PubMed] [Google Scholar]
- [4].Pascolini D and Mariotti SP, “Global estimates of visual impairment: 2010,” British Journal of Ophthalmology, vol. 96, no. 5, pp. 614–618, 2012. [DOI] [PubMed] [Google Scholar]
- [5].Whitcher JP, Srinivasan M, and Upadhyay MP, “Corneal blindness: a global perspective,” Bulletin of the world health organization, vol. 79, pp. 214–221, 2001. [PMC free article] [PubMed] [Google Scholar]
- [6].Musch DC, Sugar A, and Meyer RF, “Demographic and predisposing factors in corneal ulceration,” Archives of Ophthalmology, vol. 101, no. 10, pp. 1545–1548, 1983. [DOI] [PubMed] [Google Scholar]
- [7].Liesegang TJ, “Contact lens-related microbial keratitis: Part I: Epidemiology,” Cornea, vol. 16, no. 2, pp. 125–131, 1997. [PubMed] [Google Scholar]
- [8].Schaefer F, Bruttin O, Zografos L, and Guex-Crosier Y, “Bacterial keratitis: a prospective clinical and microbiological study,” British Journal of Ophthalmology, vol. 85, no. 7, pp. 842–847, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Cheng KH et al. , “Incidence of contact-lens-associated microbial keratitis and its related morbidity,” The Lancet, vol. 354, no. 9174, pp. 181–185, 1999. [DOI] [PubMed] [Google Scholar]
- [10].Lam D, Houang E, Fan D, Lyon D, Seal D, and Wong E, “Incidence and risk factors for microbial keratitis in Hong Kong: comparison with Europe and North America,” Eye, vol. 16, no. 5, p. 608, 2002. [DOI] [PubMed] [Google Scholar]
- [11].Krachmer J, Mannis M, and Holland E, “Cornea,” ed: Elsevier, 2011. [Google Scholar]
- [12].Vital MC, Belloso M, Prager TC, and Lanier JD, “Classifying the severity of corneal ulcers by using the “1, 2, 3” rule,” Cornea, vol. 26, no. 1, pp. 16–20, 2007. [DOI] [PubMed] [Google Scholar]
- [13].Srinivasan M et al. , “Corticosteroids for bacterial keratitis: the Steroids for Corneal Ulcers Trial (SCUT),” Archives of ophthalmology, vol. 130, no. 2, pp. 143–150, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kim RY, Cooper KL, and Kelly LD, “Predictive factors for response to medical therapy in bacterial ulcerative keratitis,” Graefe’s archive for clinical and experimental ophthalmology, vol. 234, no. 12, pp. 731–738, 1996. [DOI] [PubMed] [Google Scholar]
- [15].Lalitha P, Prajna NV, Kabra A, Mahadevan K, and Srinivasan M, “Risk factors for treatment outcome in fungal keratitis,” Ophthalmology, vol. 113, no. 4, pp. 526–530, 2006. [DOI] [PubMed] [Google Scholar]
- [16].Kent C, “Winning the Battle Against Corneal Ulcers,” 5 September. [Google Scholar]
- [17].Miedziak AI, Miller MR, Rapuano CJ, Laibson PR, and Cohen EJ, “Risk factors in microbial keratitis leading to penetrating keratoplasty,” Ophthalmology, vol. 106, no. 6, pp. 1166–1171, 1999. [DOI] [PubMed] [Google Scholar]
- [18].Martonyi CL, Bahn CF, and Meyer RF, “Slit lamp: examination and photography,” 2007. [Google Scholar]
- [19].Bennett TJ and Barry CJ, “Ophthalmic imaging today: an ophthalmic photographer’s viewpoint-a review,” Clinical & experimental ophthalmology, vol. 37, no. 1, pp. 2–13, 2009. [DOI] [PubMed] [Google Scholar]
- [20].Wilson G, Ren H, and Laurent J, “Corneal epithelial fluorescein staining,” Journal of the American Optometric Association, vol. 66, no. 7, pp. 435–441, 1995. [PubMed] [Google Scholar]
- [21].Bennett TJ and Miller G, “Lissamine green dye-an alternative to rose bengal in photo slit-lamp biomicrography,” Journal of Ophthalmic Photography, vol. 24, pp. 74–5, 2002. [Google Scholar]
- [22].Feenstra RP and Tseng SC, “Comparison of fluorescein and rose bengal staining,” Ophthalmology, vol. 99, no. 4, pp. 605–617, 1992. [DOI] [PubMed] [Google Scholar]
- [23].Mukerji N, Vajpayee RB, and Sharma N, “Technique of area measurement of epithelial defects,” Cornea, vol. 22, no. 6, pp. 549–551, 2003. [DOI] [PubMed] [Google Scholar]
- [24].VanRoekel RC, Bower KS, Burka JM, and Howard RS, “Anterior segment measurements using digital photography: a simple technique,” Optometry and Vision Science, vol. 83, no. 6, pp. 391–395, 2006. [DOI] [PubMed] [Google Scholar]
- [25].Otri AM, Fares U, Al-Aqaba MA, and Dua HS, “Corneal densitometry as an indicator of corneal health,” Ophthalmology, vol. 119, no. 3, pp. 501–508, 2012. [DOI] [PubMed] [Google Scholar]
- [26].Parikh PC et al. , “Precision of epithelial defect measurements,” Cornea, vol. 36, no. 4, p. 419, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Maganti N et al. , “Natural Language Processing to Quantify Microbial Keratitis Measurements,” Ophthalmology, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Toutain-Kidd CM et al. , “Evaluation of fungal keratitis using a newly developed computer program, Optscore, for grading digital corneal photographs,” Ophthalmic Epidemiology, vol. 21, no. 1, pp. 24–32, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Patel TP et al. , “Novel Image-Based Analysis for Reduction of Clinician-Dependent Variability in Measurement of the Corneal Ulcer Size,” Cornea, vol. 37, no. 3, pp. 331–339, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Deng L, Huang H, Yuan J, and Tang X, “Automatic segmentation of corneal ulcer area based on ocular staining images,” in Medical Imaging 2018: Biomedical Applications in Molecular, Structural, and Functional Imaging, 2018, vol. 10578, p. 105781D: International Society for Optics and Photonics. [Google Scholar]
- [31].Deng L, Huang H, Yuan J, and Tang X, “Superpixel Based Automatic Segmentation of Corneal Ulcers from Ocular Staining Images,” in 2018 IEEE 23rd International Conference on Digital Signal Processing (DSP), 2018, pp. 1–5: IEEE. [Google Scholar]
- [32].Liu Z, Shi Y, Zhan P, Zhang Y, Gong Y, and Tang X, “Automatic Corneal Ulcer Segmentation Combining Gaussian Mixture Modeling and Otsu Method,” in 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), 2019, pp. 6298–6301: IEEE. [DOI] [PubMed] [Google Scholar]
- [33].Sun Q, Deng L, Liu J, Huang H, Yuan J, and Tang X, “Patch-Based Deep Convolutional Neural Network for Corneal Ulcer Area Segmentation,” in Fetal, Infant and Ophthalmic Medical Image Analysis: Springer, 2017, pp. 101–108. [Google Scholar]
- [34].Krizhevsky A, Sutskever I, and Hinton GE, “Imagenet classification with deep convolutional neural networks,” in Advances in Neural Information Processing Systems, 2012, pp. 1097–1105. [Google Scholar]
- [35].Simonyan K and Zisserman A, “Very deep convolutional networks for large-scale image recognition,” arXiv preprint arXiv:1409.1556, 2014. [Google Scholar]
- [36].He K, Zhang X, Ren S, and Sun J, “Deep residual learning for image recognition,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2016, pp. 770–778. [Google Scholar]
- [37].Long J, Shelhamer E, and Darrell T, “Fully convolutional networks for semantic segmentation,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2015, pp. 3431–3440. [DOI] [PubMed] [Google Scholar]
- [38].Ronneberger O, Fischer P, and Brox T, “U-net: Convolutional networks for biomedical image segmentation,” in International Conference on Medical Image Computing and Computer-Assisted Intervention, 2015, pp. 234–241: Springer. [Google Scholar]
- [39].Girshick R, “Fast R-CNN,” in Proceedings of the IEEE International Conference on Computer Vision, 2015, pp. 1440–1448. [Google Scholar]
- [40].Ren S, He K, Girshick R, and Sun J, “Faster R-CNN: Towards real-time object detection with region proposal networks,” in Advances in Neural Information Processing Systems, 2015, pp. 91–99. [DOI] [PubMed] [Google Scholar]
- [41].He K, Gkioxari G, Dollár P, and Girshick R, “Mask R-CNN,” in Proceedings of the IEEE International Conference on Computer Vision, 2017, pp. 2961–2969. [Google Scholar]
- [42].Redmon J, Divvala S, Girshick R, and Farhadi A, “You Only Look Once: Unified, Real-Time Object Detection,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2016, pp. 779–788. [Google Scholar]
- [43].Liu W et al. , “SSD: Single shot multibox detector,” in European Conference on Computer Vision, 2016, pp. 21–37: Springer. [Google Scholar]
- [44].Lee CS, Baughman DM, and Lee AY, “Deep learning is effective for classifying normal versus age-related macular degeneration OCT images,” Ophthalmology Retina, vol. 1, no. 4, pp. 322–327, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].De Fauw J et al. , “Clinically applicable deep learning for diagnosis and referral in retinal disease,” Nature medicine, vol. 24, no. 9, p. 1342, 2018. [DOI] [PubMed] [Google Scholar]
- [46].Abràmoff MD, Lavin PT, Birch M, Shah N, and Folk JC, “Pivotal trial of an autonomous AI-based diagnostic system for detection of diabetic retinopathy in primary care offices,” Npj Digital Medicine, vol. 1, no. 1, p. 39, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Gargeya R and Leng T, “Automated identification of diabetic retinopathy using deep learning,” Ophthalmology, vol. 124, no. 7, pp. 962–969, 2017. [DOI] [PubMed] [Google Scholar]
- [48].Li Z, He Y, Keel S, Meng W, Chang RT, and He M, “Efficacy of a deep learning system for detecting glaucomatous optic neuropathy based on color fundus photographs,” Ophthalmology, vol. 125, no. 8, pp. 1199–1206, 2018. [DOI] [PubMed] [Google Scholar]
- [49].Fang L, Cunefare D, Wang C, Guymer RH, Li S, and Farsiu S, “Automatic segmentation of nine retinal layer boundaries in OCT images of non-exudative AMD patients using deep learning and graph search,” Biomedical optics express, vol. 8, no. 5, pp. 2732–2744, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Cunefare D, Fang L, Cooper RF, Dubra A, Carroll J, and Farsiu S, “Open source software for automatic detection of cone photoreceptors in adaptive optics ophthalmoscopy using convolutional neural networks,” Scientific Reports, vol. 7, no. 1, p. 6620, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Cunefare D et al. , “Deep learning based detection of cone photoreceptors with multimodal adaptive optics scanning light ophthalmoscope images of achromatopsia,” Biomedical Optics Express, vol. 9, no. 8, pp. 3740–3756, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Cunefare D, Huckenpahler AL, Patterson EJ, Dubra A, Carroll J, and Farsiu S, “RAC-CNN: Multimodal deep learning based automatic detection and classification of rod and cone photoreceptors in adaptive optics scanning light ophthalmoscope images,” Biomedical Optics Express, vol. 10, no. 8, pp. 3815–3832, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Loo J, Fang L, Cunefare D, Jaffe GJ, and Farsiu S, “Deep longitudinal transfer learning-based automatic segmentation of photoreceptor ellipsoid zone defects on optical coherence tomography images of macular telangiectasia type 2,” Biomedical Optics Express, vol. 9, no. 6, pp. 2681–2698, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Desai AD et al. , “Open-source, machine and deep learning-based automated algorithm for gestational age estimation through smartphone lens imaging,” Biomedical optics express, vol. 9, no. 12, pp. 6038–6052, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Farsiu S, Loo J, Kriegel MF, Tuohy M, Prajna V, and Woodward MA, “Deep learning-based automatic segmentation of stromal infiltrates and associated biomarkers on slit-lamp images of microbial keratitis,” Investigative Ophthalmology & Visual Science, vol. 60, no. 9, pp. 1480–1480, 2019. [Google Scholar]
- [56].Kriegel MF et al. , “Reliability of physicians’ measurements when manually annotating images of microbial keratitis,” Investigative Ophthalmology & Visual Science, vol. 60, no. 9, pp. 2108–2108, 2019. [Google Scholar]
- [57].Schneider CA, Rasband WS, and Eliceiri KW, “NIH Image to ImageJ: 25 years of image analysis,” Nature Methods, vol. 9, no. 7, p. 671, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Abdulla W, “Mask R-CNN for object detection and instance segmentation on Keras and TensorFlow,” Github repository, 2017. [Google Scholar]
- [59].Lin T-Y, Dollár P, Girshick R, He K, Hariharan B, and Belongie S, “Feature pyramid networks for object detection,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2017, pp. 2117–2125. [Google Scholar]
- [60].Jaccard P, “The distribution of the flora in the alpine zone,” New Phytologist, vol. 11, no. 2, pp. 37–50, 1912. [Google Scholar]
- [61].Lin T-Y, Goyal P, Girshick R, He K, and Dollár P, “Focal loss for dense object detection,” in Proceedings of the IEEE International Conference on Computer Vision, 2017, pp. 2980–2988. [Google Scholar]
- [62].Karimi D and Salcudean SE, “Reducing the Hausdorff Distance in Medical Image Segmentation with Convolutional Neural Networks,” arXiv preprint arXiv:1904.10030, 2019. [DOI] [PubMed] [Google Scholar]
- [63].Milletari F, Navab N, and Ahmadi S-A, “V-net: Fully convolutional neural networks for volumetric medical image segmentation,” in 3D Vision (3DV), 2016 Fourth International Conference on, 2016, pp. 565–571: IEEE. [Google Scholar]
- [64].Lin T-Y et al. , “Microsoft COCO: Common objects in context,” in European Conference on Computer Vision, 2014, pp. 740–755: Springer. [Google Scholar]
- [65].Glorot X and Bengio Y, “Understanding the difficulty of training deep feedforward neural networks,” in Proceedings of the Thirteenth International Conference on Artificial Intelligence and Statistics, 2010, pp. 249–256. [Google Scholar]
- [66].Sutskever I, Martens J, Dahl G, and Hinton G, “On the importance of initialization and momentum in deep learning,” in International Conference on Machine Learning, 2013, pp. 1139–1147. [Google Scholar]
- [67].Soille P, Morphological image analysis: principles and applications. Springer Science & Business Media, 2013. [Google Scholar]
- [68].Dice LR, “Measures of the amount of ecologic association between species,” Ecology, vol. 26, no. 3, pp. 297–302, 1945. [Google Scholar]
- [69].Crum WR, Camara O, and Hill DL, “Generalized overlap measures for evaluation and validation in medical image analysis,” IEEE Transactions on Medical Imaging, vol. 25, no. 11, pp. 1451–1461, 2006. [DOI] [PubMed] [Google Scholar]
- [70].Kuijf HJ et al. , “Standardized Assessment of Automatic Segmentation of White Matter Hyperintensities and Results of the WMH Segmentation Challenge,” IEEE Transactions on Medical Imaging, vol. 38, no. 11, pp. 2556–2568, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Abadi M et al. , “TensorFlow: A System for Large-Scale Machine Learning,” in OSDI, 2016, vol. 16, pp. 265–283. [Google Scholar]
- [72].Chollet F, “Keras,” ed, 2015. [Google Scholar]
- [73].Litjens G et al. , “A survey on deep learning in medical image analysis,” Medical image analysis, vol. 42, pp. 60–88, 2017. [DOI] [PubMed] [Google Scholar]
- [74].Christ PF et al. , “Automatic liver and lesion segmentation in CT using cascaded fully convolutional neural networks and 3D conditional random fields,” in International Conference on Medical Image Computing and Computer-Assisted Intervention, 2016, pp. 415–423: Springer. [Google Scholar]
- [75].Sirinukunwattana K et al. , “Gland segmentation in colon histology images: The glas challenge contest,” Medical image analysis, vol. 35, pp. 489–502, 2017. [DOI] [PubMed] [Google Scholar]
- [76].Chen H, Qi X, Yu L, Dou Q, Qin J, and Heng P-A, “DCAN: Deep contour-aware networks for object instance segmentation from histology images,” Medical image analysis, vol. 36, pp. 135–146, 2017. [DOI] [PubMed] [Google Scholar]
- [77].Roy AG et al. , “ReLayNet: Retinal layer and fluid segmentation of macular optical coherence tomography using fully convolutional networks,” Biomedical optics express, vol. 8, no. 8, pp. 3627–3642, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Li X, Chen H, Qi X, Dou Q, Fu C-W, and Heng P-A, “H-DenseUNet: Hybrid densely connected UNet for liver and tumor segmentation from CT volumes,” IEEE transactions on medical imaging, vol. 37, no. 12, pp. 2663–2674, 2018. [DOI] [PubMed] [Google Scholar]
- [79].Fu H, Cheng J, Xu Y, Wong DWK, Liu J, and Cao X, “Joint optic disc and cup segmentation based on multi-label deep network and polar transformation,” IEEE transactions on medical imaging, vol. 37, no. 7, pp. 1597–1605, 2018. [DOI] [PubMed] [Google Scholar]
- [80].Xue Y, Xu T, Zhang H, Long LR, and Huang X, “Segan: Adversarial network with multi-scale L1 loss for medical image segmentation,” Neuroinformatics, vol. 16, no. 3–4, pp. 383–392, 2018. [DOI] [PubMed] [Google Scholar]
- [81].Wang S et al. , “Central focused convolutional neural networks: Developing a data-driven model for lung nodule segmentation,” Medical image analysis, vol. 40, pp. 172–183, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Wang G et al. , “Interactive medical image segmentation using deep learning with image-specific fine tuning,” IEEE transactions on medical imaging, vol. 37, no. 7, pp. 1562–1573, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Liu F, Zhou Z, Jang H, Samsonov A, Zhao G, and Kijowski R, “Deep convolutional neural network and 3D deformable approach for tissue segmentation in musculoskeletal magnetic resonance imaging,” Magnetic Resonance in Medicine, vol. 79, no. 4, pp. 2379–2391, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Norman B, Pedoia V, and Majumdar S, “Use of 2D U-Net convolutional neural networks for automated cartilage and meniscus segmentation of knee MR imaging data to determine relaxometry and morphometry,” Radiology, vol. 288, no. 1, pp. 177–185, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Men K, Dai J, and Li Y, “Automatic segmentation of the clinical target volume and organs at risk in the planning CT for rectal cancer using deep dilated convolutional neural networks,” Medical physics, vol. 44, no. 12, pp. 6377–6389, 2017. [DOI] [PubMed] [Google Scholar]
- [86].Novikov AA, Lenis D, Major D, Hladůvka J, Wimmer M, and Bühler K, “Fully convolutional architectures for multiclass segmentation in chest radiographs,” IEEE transactions on medical imaging, vol. 37, no. 8, pp. 1865–1876, 2018. [DOI] [PubMed] [Google Scholar]
- [87].Son J, Park SJ, and Jung K-H, “Retinal vessel segmentation in fundoscopic images with generative adversarial networks,” arXiv preprint arXiv:1706.09318, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Isensee F et al. , “nnU-Net: Self-adapting framework for U-Net-based medical image segmentation,” arXiv preprint arXiv:1809.10486, 2018. [Google Scholar]
- [89].Mukaka MM, “A guide to appropriate use of correlation coefficient in medical research,” Malawi Medical Journal, vol. 24, no. 3, pp. 69–71, 2012. [PMC free article] [PubMed] [Google Scholar]
- [90].Loo J, Clemons TE, Chew EY, Friedlander M, Jaffe GJ, and Farsiu S, “Beyond Performance Metrics: Automatic Deep Learning Retinal OCT Analysis Reproduces Clinical Trial Outcome,” Ophthalmology, vol. (in press), 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]


