Abstract
MagellanMapper is a software suite designed for visual inspection and end-to-end automated processing of large volume, 3D brain imaging datasets in a memory efficient manner. The rapidly growing number of large volume, high resolution datasets necessitates visualization of raw data at both macro- and microscopic levels to assess the quality of data and automated processing to quantify data in an unbiased manner for comparison across a large number of samples. To facilitate these analyses, MagellanMapper provides both a graphical user interface for manual inspection and a command line interface for automated image processing. At the macroscopic level, the graphical interface allows researchers to view full volumetric images simultaneously in each dimension and to annotate anatomical label placements. At the microscopic level, researchers can inspect regions of interest at high resolution to build ground truth data of cellular locations such as nuclei positions. Using the command line interface, researchers can automate cell detection across volumetric images, refine anatomical atlas labels to fit underlying histology, register these atlases to sample images, and perform statistical analyses by anatomical region. MagellanMapper leverages established open source computer vision libraries and is itself open source and freely available for download and extension.
Keywords: Image processing, 3D atlas, graphical interface, tissue clearing, microscopy images
INTRODUCTION:
The growing number of large volume, high resolution 3D imaging datasets has required the concurrent development of new tools and pipelines to analyze this wealth of data in an efficient and verifiable manner (Meijering et al., 2016; Renier et al., 2016). Previously, manual counts of cells and subcellular structures have been sufficient for 2D slices, but the advent of 3D microscopy tissue processing techniques such as serial two-photon tomography (STPT) (Kim et al., 2015; Ragan et al., 2012) and tissue clearing techniques (Mano et al., 2018) such as CLARITY (Chung et al., 2013), 3DISCO (Ertürk et al., 2012), and CUBIC (Susaki et al., 2014) combined with lightsheet microscopy (Hillman et al., 2019) for whole-organ imaging, generating thousands of slices per sample, has rendered manual approaches obsolete and required automated approaches to image processing and quantification. Additional imaging modalities such as high resolution magnetic resonance imaging (MRI) (Pallast et al., 2019) and electron microscopy (EM) (Zheng et al., 2018) have further compounded the complexity and volume of 3D imaging requiring automated analysis.
3D microscopy poses several problems for standard image processing workflows. First, images generated by these approaches are typically hundreds of gigabytes (GB) to terabytes (TB) in size, requiring efficient memory usage to view images on standard computers. Second, the vast scope of detail within these high resolution images render manual quantification unfeasible, requiring automated approaches alongside the ability to visually verify these results. Finally, images of whole organs typically need to be examined at both macro- and microscopic levels. While high resolution imaging affords unprecedented opportunities to explore the 3D organization of cellular structures, researchers often need to view these structures in the context of the whole organ to understand regional organization and quantification by anatomical region. Development of 3D atlases and registration of these atlases to samples allows individual cells to be placed within their anatomical surroundings.
To facilitate the analysis of 3D microscopy images in their anatomical frame of reference, we have developed the MagellanMapper software suite. The design philosophy behind MagellanMapper is to combine end-to-end, high-throughput, automated processing of large volume images with the ability for manual inspection and annotation of unfiltered, raw 2D images in their 3D and anatomical contexts. Inspired by the first circumnavigation of the Earth by Portuguese explorer Ferdinand Magellan, we developed the MagellanMapper suite with the vision of providing a tool to assist the circumnavigation of the brain or any other organ.
These protocols describe usage of MagellanMapper through both its graphical user interface (GUI) and command line interface (CLI) (Fig. 1). To visualize a specimen at the macroscopic level, the graphical interface includes a simultaneous orthogonal viewer, which allows navigation through the specimen in its 3D context by viewing intersecting planes from all three dimensions concurrently. At the microscopic level, a region of interest (ROI) viewer shows serial 2D planes of a chosen smaller volume from within the full volumetric image. Surface renderings of individual anatomical structures can also be viewed. For automated processing, researchers can access MagellanMapper through its command-line interface using the provided pipeline scripts or their own customized versions. Automated tasks include whole organ nuclei detection, atlas refinement in 3D, image registration through the SimpleElastix registration library (Marstal, Berendsen, Staring, & Klein, 2016), and basic statistical analysis of nuclei by anatomical region. To train and verify these detections, the graphical interface provides annotation tools such as object location marking within an ROI to build ground truth sets and anatomical label painting to refine registered atlas boundaries or to enhance the atlases themselves.
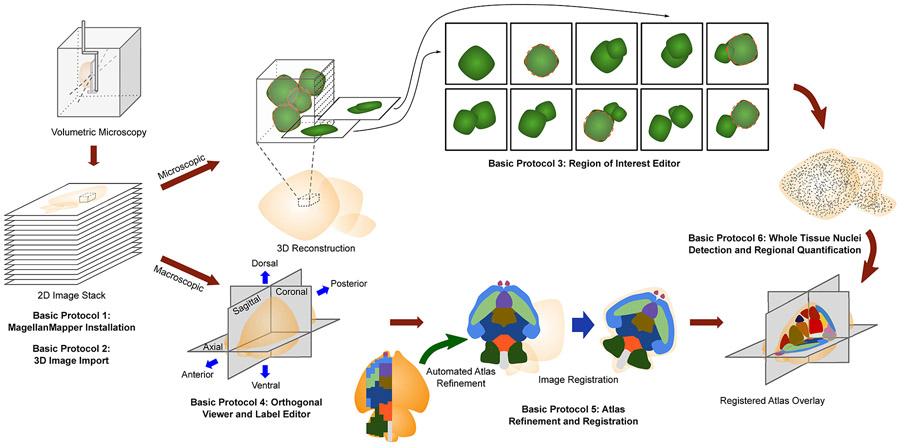
Figure 1.
Overview of MagellanMapper features and workflow for image processing of volumetric imaging. Volumetric microscopy sources include serial two-photon tomography (STPT), lightsheet imaging of tissue cleared specimen, or in vivo imaging such as MRI typically produce stacks of inherently aligned serial 2D sections that can be merged into a 3D reconstruction. MagellanMapper facilitates visualization and annotation of images at the microscopic and macroscopic scale. Basic Protocol 1 describes installation of MagellanMapper, while Basic Protocol 2 provides instruction on importing a variety of image formats into MagellanMapper. In Basic Protocol 3, we describe the Region of Interest (ROI) Editor for viewing small windows of high resolution data in a 3D context through each plane in the ROI. Automated 3D nuclei detection places circles indicating object location, and annotation tools in the editor allow repositioning, modifying, and adding or subtracting circles. Basic Protocol 4 describes the Atlas Editor used to view images at a macroscopic scale by providing simultaneous views of planes along each axis and painting tools to markup structures. Basic Protocol 5 gives instruction on atlas 3D reconstruction and registration, including tools that can take partially labeled, 2D-derived atlases and automatically complete and generate 3D atlases based on the underlying anatomical signal. This registration provides the anatomical demarcations necessary for Basic Protocol 6, which describes whole tissue 3D image processing of nuclei for quantification by anatomical region.
To facilitate interoperability with existing image software packages and pipelines, MagellanMapper has been built on open source tools to leverage established methods and implementations of computer vision techniques. In some cases, the suite accesses alternate image analysis platforms such as the BigStitcher plugin from the ImageJ/FIJI ecosystem. The software uses standard file formats to maximize cross-compatibility with other software platforms. Subpackages within MagellanMapper have been written as Python libraries to allow reuse and extension within other software packages, and the entire suite is available freely as open source software.
Basic Protocol 1 describes the installation of MagellanMapper. Basic Protocol 2 provides instructions on importing image files into MagellanMapper. Basic Protocol 3 walks the researcher through visualization and annotation of a region of interest within a high resolution image (Fig. 1). Basic Protocol 4 describes viewing a 3D image in the simultaneous orthogonal viewer. Basic Protocol 5 provides details on automated 3D atlas generation from atlases originally generated in 2D or even partially incomplete atlases, as well as registration of these atlases to a sample image and quantification of nuclei by anatomical region. Basic Protocol 6 discusses steps for whole organ nuclei detection and brings the protocols together in an end-to-end pipeline that takes a user from a raw microscopy image stack to whole organ detections. Two supporting protocols describe alternate installation pathways and an automated pipeline to import raw, tiled microscopy image files into a single 3D file for use in MagellanMapper or other Python-based software.
As MagellanMapper undergoes ongoing development, we refer the reader to the software website and README for the latest instructions, including any of which may supersede steps contained in these protocols for new features and improvements in future software versions: https://github.com/sanderslab/magellanmapper
STRATEGIC PLANNING (optional)
Using MagellanMapper begins with downloading and installing the software. MagellanMapper is freely available as open source software, and we describe several mechanisms to install it locally or in the cloud in Basic Protocol 1.
For data input, the software accepts many standard 3D formats using established open source libraries. Researchers can convert standard 2D image formats such as JPG or TIFF files, multi-page 3D TIFF files, or proprietary microscopy formats such as Zeiss CZI files into standard Numpy formats used through the suite, with the import process described in Basic Protocol 2. Other standard 3D formats such as NIfTI, MHA/MHD, and NRRD are supported through the SimpleElastix library derived from the SimpleITK library (Lowekamp, Chen, Ibanez, & Blezek, 2013; Marstal et al., 2016) and can be loaded directly, without requiring import.
Data output of 3D images is typically in the same 3D file formats. Image annotations are exported directly to labeled images for atlas labels or to an SQLite database for interest points such as nuclei locations. For figures, most windows can be exported directly to 2D image formats such as PNG, JPG, and PDF through the Matplotlib library (Hunter, 2007). The command-line interface also allows export of specified image planes and plots directly to file for automated figure generation. Statistical functions typically output data using the Pandas library (McKinney, 2010) into CSV or Excel formats.
Minimum recommended hardware
MagellanMapper is designed to run on lower-end hardware while scaling up to take advantage of any available computational resources. Based on our testing on a variety of hardware platforms, we recommend the minimum specifications:
Windows 10, MacOS 10.13, or Ubuntu 18.04
64-bit Intel- or AMD-based processor
4GB RAM
2 GB disk space
Internet connection for software download
While these protocols will assume graphical capability, the software can run in non-graphical environments to allow automated processing in cloud-based or other server environments. Please visit our software website for more information on these alternative installation and processing pathways.
BASIC PROTOCOL 1
MagellanMapper installation
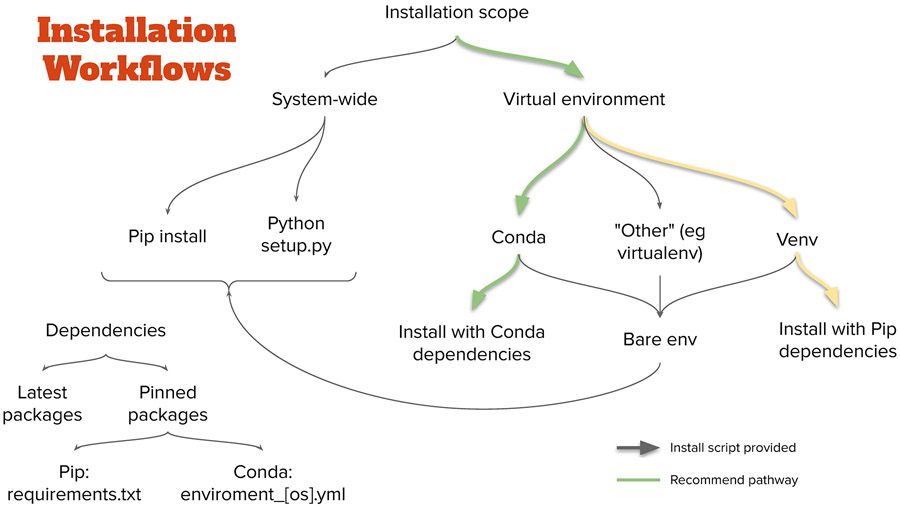
As Python based software, MagellanMapper is supported on all major operating systems (OSes), including Windows, macOS, and Linux. Installation involves downloading the software (freely available) and installing it along with its supporting packages (dependencies). Since many researchers have specific preferences for how to install Python packages, MagellanMapper supports a variety of installation pathways (Fig. 2). This protocol describes a recommended pathway requiring the least number of commands for installation. Alternate Protocol 1 offers instructions for additional installation pathways meeting other user preferences or requirements.
Figure 2.
Installation pathways. As with many Python applications, MagellanMapper can be installed through several pathways. The recommended pathway is through Conda, which handles all required dependencies and keeps them separate from any other installed Python packages in a virtual environment. For those who prefer a more “pure Python” approach, Venv will also keep packages separate in an environment while installing dependencies through Pip. Installation scripts are provided to simplify the Conda pathway. Various other pathways are also available. Typically, the latest dependencies are installed, but specifications that pin each dependency version are available for reproducibility.
Conda (https://conda.io) is an open source, cross-platform package manager that handles all of the dependencies required to run many Python and R packages such as MagellanMapper. Conda is freely available as two distributions, Miniconda and Anaconda, both of which work with MagellanMapper. MagellanMapper provides install scripts to download and install Miniconda (if Conda is not already installed), create a new Conda environment for MagellanMapper, and install MagellanMapper and all its supporting packages into the environment (Fig. 3).
Figure 3.
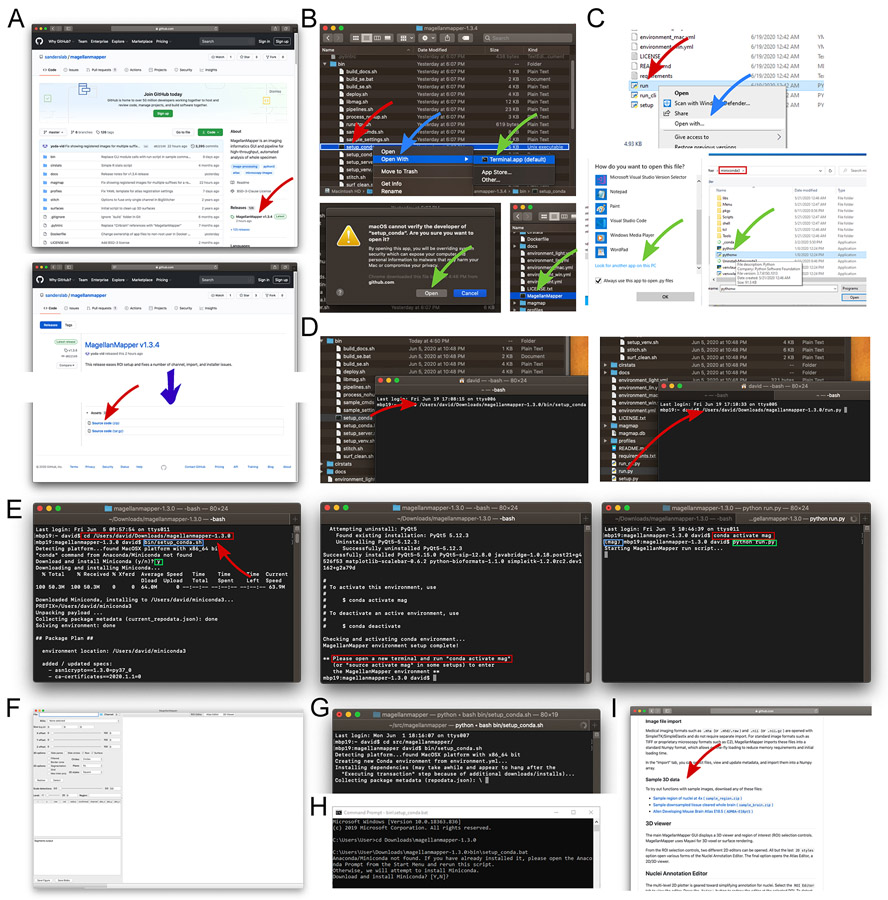
cd” (space included), the folder can be typed in directly, or dragged into the terminal similarly as before (left, red rectangle). Pressing Enter enters that folder. Typing the path to the install script (blue rectangle, here shown for Mac and Linux) and pressing Enter starts the installation process. In this case, Conda is not found and thus downloaded and installed after confirmation from the user (green rectangle). Since Conda was just installed, a new terminal will need to be opened after installation completes (middle, red rectangle). After opening a new terminal, the Conda activation command will start the newly created MagellanMapper environment (left, red rectangle). The “(mag)” at the start of the line indicates that the environment has been successfully activated (blue rectangle). The MagellanMapper launch command can now be entered (green rectangle) to (F) open MagellanMapper. (G) If Conda is already installed, the MagellanMapper install script skips Conda installation and proceeds directly to creating a new environment. (H) Equivalent install script for Windows systems. (KI) The website home page also hosts sample 3D data for use in these Protocols.
Materials:
MagellanMapper software
Computer system
Sample Data (incorporated into the protocol steps)
-
Download MagellanMapper. The software is freely available from its website, https://github.com/sanderslab/magellanmapper. Click on the “releases” link to find the latest source code release (https://github.com/sanderslab/magellanmapper/releases; Fig. 3A). In the latest release’s “Assets” section, download a “Source code” file. After downloading the file, unzip it to any desired file location.
This latest release is a stable version that has undergone more testing. For those who wish for the freshest, unreleased code, Git can be used to access and update the entire source code repository. Use this command to access the repository using Git:
Open a file browser and access the MagellanMapper download. Navigate to the downloaded file in a file browser such as Finder on macOS, File Explorer in Windows, or Files in Linux (Fig. 3B). If you downloaded the zip file, unzip it to any desired location and navigate into the extracted folder.
-
Run the installer to set up MagellanMapper in a Conda environment, which will automatically install Conda as well if necessary. The installer files are located in the
bin
folder. Enter this folder and locate the appropriate setup file for your platform:Mac or Linux: setup_conda
Windows: setup_conda.bat
We describe how to run the setup script directly through the file browser or through a terminal (Fig. 3B-D). This script will download and install Conda if not found before setting up a Conda environment for MagellanMapper.
-
Mac file browser: Right-click on
setup_conda
. Select “Open with…” and choose “Terminal.” In the security warning, press “Okay” (Fig. 3B).Mac provides a security warning as a standard reminder that the file was downloaded from the Internet.
-
Linux file browser: Double click on
setup_conda
.It may be required to first change the file browser preferences for launching text files. In the Files app on GNOME, for example, enter “Preferences,” select the “Behavior” tab, and under “Execute Text Files,” choose to either run them or ask.
-
Windows file browser: Double-click on
setup_conda.bat
. In the security warning, press “More info,” then “Run anyway.”Window also provides a standard security warning for files downloaded from the Internet.
Terminal (cross-platform): Common terminals are Terminal on macOS and Linux or Command Prompt on Windows. Drag the setup file into the terminal, which should cause the file’s path to appear in the terminal, then press Enter to start the script (Fig. 3D, left).
-
Follow the instructions in the script, including any prompt to download and install Conda and to open a new terminal after installation if Conda was installed. This entire process can take up to 5 minutes or longer, depending on the web connection for downloading supporting packages.
- Launch
MagellanMapper
. We again describe running the program through the file browser or a terminal. Note that there may be a delay of up to several minutes prior to launch by anti-malware software checks, which typically only occurs on first launch. Launching MagellanMapper will open its graphical user interface (Fig. 3F).- Mac or Linux file browser: Navigate back up to the main folder and double-click on the MagellanMapper file (Fig. 3B, bottom right).
-
Windows file browser: Navigate back up to the main folder and locate the
run.py
file. Assuming that.py
files have been associated with Python, double-click on this file to launch MagellanMapper.To associate.py
files, right-click on the file, select “Open with,” then “More apps,” and choose to look for another app on your PC. In the file dialog, navigate to the Conda installation, typically C:\Users\<your-account>\miniconda3, and selectpythonw.exe
(Fig. 3C). Once.py
files are associated with Python, you can simply double-click on the file to re-launch it. -
Terminal (cross-platform): Open a terminal, type “
python
” (note space at end), drag therun.py
file into the terminal, and press Enter.On Mac and Linux, typing “python
“ is optional. This run script takes care of the Conda activation required before launching MagellanMapper (Fig. 3D, right).When running command-line (ie non-graphical) MagellanMapper tasks as described in later protocols, we recommend running an environment activation step once in each new terminal session before the run commands to avoid reactivating the environment each time and to view full command-line output. To activate and run MagellanMapper in a terminal (Fig. 3E, right):conda activate mag python <path-to-magellanmapper>/run.py
The activation command only needs to be run once in each terminal. This run approach can also be helpful for troubleshooting any of the above launch approaches, including both command-line and graphical modes. For commands given in the rest of these protocols, we will assume that this activation step has been run and that the terminal directory has been changed to the software directory (egcd Downloads/magellanmapper-1.3.0
).
ALTERNATE PROTOCOL 1 (optional)
Alternative methods for MagellanMapper installation
Materials:
MagellanMapper software
Computer system
Sample Data (incorporated into the protocol steps)
Several other methods of installation are supported to meet platform requirements and personal preferences. We describe the most common methods here and depict a more comprehensive range of methods in Fig. 2. As these and other more advanced functions require use of the command-line, we also introduce typing (or copy-pasting) commands in a terminal.
-
Open a terminal and enter the MagellanMapper folder. For convenience, all of the commands are assumed to be run from within this folder. To enter this folder, type this command into the Terminal or Command Prompt, replacing
<magellanmapper-location>
with the location of the MagellanMapper folder, and press Enter (Fig. 3E):cd <magellanmapper-location>
For example, if the software directory is atDownloads/magellanmapper-1.3.0
in the user’s home directory:cd Downloads/magellanmapper-1.3.0
You can also simply type, “cd
“ (“cd” followed by a space), and drag and drop the MagellanMapper folder from a file browser into the terminal to add this path. -
Option 1: Install MagellanMapper using a terminal. We will use the same scripts as in Basic Protocol 1 but without using a file browser.
Type or copy-paste these commands into the terminal and press Enter (Fig. 3E).
Mac or Linux:bin/setup_conda
Windows:bin\setup_conda.bat
Follow the instructions in the script, including any prompt to download and install Conda and to open a new terminal after installation if Conda was installed.
If you have closed the terminal, reopen it and change to the MagellanMapper folder as described above.
You can open MagellanMapper for the first time or to restart it as follows (Fig. 3E, right). Again, there may be a delay of up to several minutes prior to launch by anti-malware software checks, typically only on first launch.
-
Enter the MagellanMapper folder (assuming the same example location here):
cd Downloads/magellanmapper-1.3.0
-
Activate the “mag” Conda environment:
conda activat e mag
-
e.
Start MagellanMapper:
python run.py
On Mac or Linux, the command can be simplified to./run.py
.Many parameters can be added to this command to customize the session as described in subsequent Basic Protocols. To view a complete list of command-line parameters available in MagellanMapper:
python run.py --help
-
-
Option 2: Install MagellanMapper using a Conda command. For those who already have Anaconda or Miniconda installed and feel comfortable with Conda commands, a direct Conda command can be used to create a new environment and install MagellanMapper into it:
conda env create -n mag -f environment.yml
Python packages including MagellanMapper typically have many dependent packages, each of which must match certain version numbers. Python packages are typically installed either system wide (globally) or in an isolated environment. System-wide installations typically place all packages in a common location. While often the simplest approach, this type of installation may overwrite other versions of packages that may have been installed previously, preventing other Python packages from working properly, or a future installation may prevent MagellanMapper from working properly. System-wide installations may work well for systems that are themselves isolated such as cloud-based servers.
Isolated environments such as those generated by Conda provide a mechanism to keep multiple versions of the same package concurrently. For example, one environment may contain package A with dependencies B and C, while another environment may contain package D with dependencies B and E. If package A requires package B at version 2, while package D requires package B at version 3, these installations may conflict in a system-wide install. By installing into isolated environments, however, the packages can co-exist without overwriting or conflicting with one another. Conda provides one system for creating these virtual environments to keep packages isolated from one another and handles package compatibility management as well.
-
Option 3: Install MagellanMapper using Python’s own environment manager, Venv. For those who prefer not to use Conda, we have provided a script to install MagellanMapper using Python’s built-in environment manager, Venv.
-
Install Python 3.6, which can be downloaded freely at https://www.python.org/downloads/ or accessed through sources such as Homebrew, Conda, or Pyenv.
We currently recommend Python 3.6 for MagellanMapper since we have used this version to precompile a number of supporting packages that are challenging to build and would otherwise require a compiler. Later versions of Python can be used but currently require compiling these supporting packages oneself. Please see our website for the latest updates as we add support for newer versions.
Optional: for support for importing proprietary and multi-plane TIFF image formats, install a Java Development Kit (https://openjdk.java.net/). Please see our website for more details on Java configuration.
-
Install MagellanMapper in a Venv environment. Run this script to create a new Venv environment and install MagellanMapper and its dependencies in it:
bin/setup_venv.sh
To activate the environment:
source activate ../venvs/mag
-
- Option 4: Install MagellanMapper within another environment manager or system-wide. For those who prefer to use another environment manager or none at all, MagellanMapper can be installed directly using Python’s built-in Pip tool.
- Install Python as above.
-
Use Pip to install MagellanMapper and all of its supporting packages.
pip install .[all] --extra-index-url https://pypi.fury.io/dd8/
The extra URL provides access to the supporting packages not included on PyPI that we have precompiled to simplify installation. The other install methods also access these packages.
BASIC PROTOCOL 2 (optional)
Import image files into MagellanMapper
Materials:
MagellanMapper software
sample_region.zip
image file available on the software website
Sample Data (incorporated into the protocol steps)
MagellanMapper opens a number of standard medical 3D imaging formats such as NIfTI, MHA, and NRRD directly through the SimpleITK/SimpleElastix library, without requiring file conversion. For other standard image formats such as TIFF and RAW, MagellanMapper imports these files into Numpy format (Stefan van der Walt, Colbert, & Varoquaux, 2011), an open format accessible across many scientific software platforms that allows for efficient image processing computations and memory usage. MagellanMapper can import files from four types of sources: 1) A directory of 2D image files, each of the same dimensions, 2) Multi-page TIFF files, where each file consists of a 3D stack of 2D planes for one channel of the image, 3) Proprietary microscopy 3D formats such as Zeiss CZI supported in the Bioformats library (Linkert et al., 2010), or 4) RAW data formats from microscopes and cameras.
To facilitate gaining experience with the interface, we have provided a small image stack in multiple formats to practice in MagellanMapper using this protocol. The image can be accessed in the GitHub repository home page (
sample_region.zip
, Fig. 3I). Example imports will use this sample, but the paths can be replaced to use one’s own image files.
- Option 1: Import 2D images in a directory.
- In the controls panel on the left side of MagellanMapper, open the “Import” tab.
-
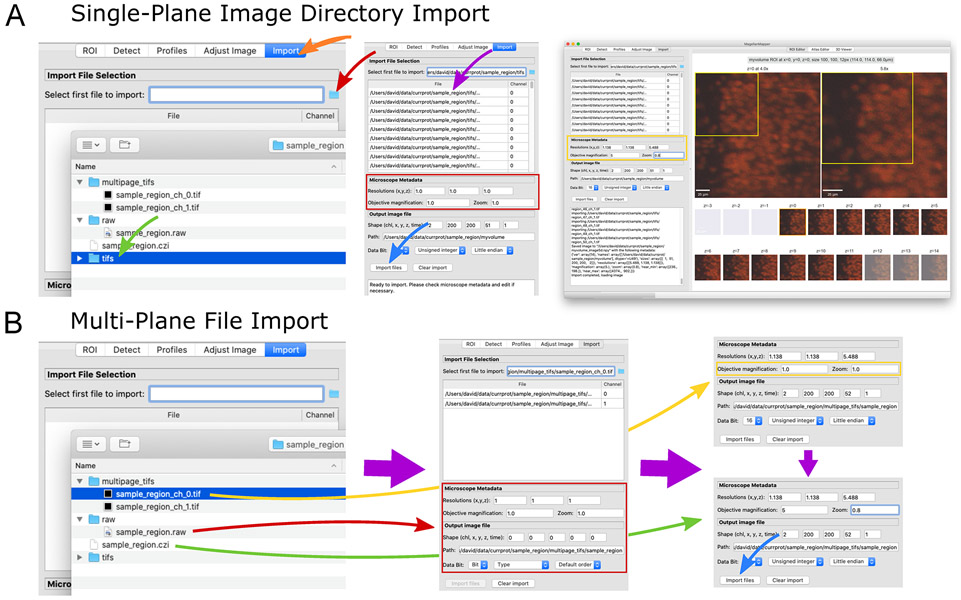
In the “Import File Selection” area, click on the file directory icon to open a file chooser, navigate to the
sample_region
directory, single-click on thetifs
directory (select the directory itself rather than entering it), and press Open (Fig. 4A, left). The files in the directory should appear in the files table with the appropriate channel based on the channel number in each file’s name.Each file in this sample folder is loaded as a successive 2D plane in the resulting 3D image file. To import multi-channel files, the importer looks for files that end with_ch_<n>
, wheren
is the channel number. Each file will be loaded as a plane within the corresponding channel. If no files have the channel identifier, all files will be assumed to belong to a single channel. -
In the “Microscope Metadata” area, enter the microscope objective information (Fig. 4A, middle). For this particular image, the resolutions are x = 1.138, y = 1.138, and z = 5.488. The magnification is 5.0, and zoom is 0.8. You can adjust the output path to the desired location and name. The resulting filenames will be based on this name.The output image shape and data type are automatically determined and can be left as-is.
-
Press “Import files” to initiate the import. The bottom text box will display progress on the import. When the import completes, the image metadata will be shown, and the image itself will appear in the right figure panel (Fig. 4A, right). The rest of the protocols will show examples from this imported file.During import, the volumetric image in Numpy format and a metadata file are generated. Either file can be selected in MagellanMapper to reopen the imported image.
-
Alternatively, you can import this directory through a single command. Launch MagellanMapper with the image path set to the directory to import. All files in the directory will be imported in alphabetical order. To import the files in the
sample_region/tifs
folder, assuming that it has been moved to the mainmagellanmapper
folder:python run.py --img sample_region/tifs --proc import_only \ --set_meta resolutions=1.138,1.138,5.488 \ magnification=5.0 zoom=0.8
As before, we assume that the MagellanMapper environment has been activated, and the command is run from the software folder. The--set_meta
option specifies additional metadata related to the microscope objective used to record this image. Resolutions are given in x,y,z order. An optional--prefix
parameter can be used to specify the output base path.
- Option 2: Import multi-page TIFF files. Multi-page TIFF files store 3D images by containing multiple individual TIFF images.
-
In the Import panel file chooser, select the first file in the
sample_regions/multipage_tifs
folder (Fig. 4B, left). Files named in a similar format to this file will be loaded into the table.As in Option 1, any files named according to the channel format will be treated as a separate channel of the resulting single image. This import initialization typically takes longer while loading the Bioformats library (Fig. 4B, middle), which automatically retrieves metadata embedded in the image files if present and also loads large images in a memory-efficient manner. If a file is added that you do not want, click on the file’s row and press the Backspace or Delete key to remove the entry. - Check the automatically entered metadata. In this case, the “Objective magnification” should be changed to “5” and “Zoom” to “0.8” since this information was not embedded in the file (Fig. 4B, top right).
- Press the “Import files” button to import the image (Fig. 4B, bottom right.
-
Alternatively, you can import these files using this command, which specifies the file excluding any channel information:
python run.py --img \ sample_region/multipage_tifs/sample_region.tif \ --proc import_only --set_meta magnification=5.0 zoom=0.8
-
- Option 3: Import a proprietary microscopy image file. MagellanMapper will import image file formats supported by the Bioformats library such as the Zeiss CZI format. Since these formats typically embed metadata such as objective resolutions, they do not need to be entered manually.
- In the Import panel, select the
sample_region/sample_region.czi
file (Fig. 4B, left). This single file containing multiple channels will appear in the table. - Check the microscope metadata extracted directly from the image and adjust the output path if desired. In this case, the file contains all necessary metadata, and no further adjustments are required.
- Press “Import files” to import the image (Fig. 4C, bottom right).
-
Alternatively, from the command-line:
python run.py --img sample_region/sample_region.czi \ --proc import_only --prefix sample_region/my_region
We demonstrated the optional--prefix
parameter to specify the output base path. If omitted, the output name is based on the original image’s filename.
- Option 4: Import a raw data file. Microscopy and camera software frequently output images in a RAW data format. To import these files into MagellanMapper, the image shape and data type will need to be known beforehand, in addition to other microscopy metadata such as resolutions and zoom.
- In the Import panel file chooser, select the
sample_regions/raw/sample_region.raw
file (Fig. 4B, left). - Enter all metadata (Fig. 4B, middle). This information will need to be known beforehand based on microscopy parameters since they are not embedded in the file. For this file, first enter the same metadata as in Option 1. Next, enter the following “Output image file” data:
- Shape: channel = 2, x = 200, y = 200, z = 51, time = 1
- Data: 16 bit, Unsigned integer type, Little Endian order
- Press the “Import files” button to import the image.
-
Alternatively, you can import these files using this command:
python run.py --img sample_region/raw/sample_region.raw \ --proc import_only --set_meta shape=2,200,200,51,1 \ dtype=uint16 resolutions=1.138,1.138,5.488 \ magnification=5.0 zoom=0.8
This command adds additional metadata specifying the image shape and data type.
Figure 4.
Import image files into a Numpy volumetric format. (A) The “Import” panel (left, orange arrow) displays selectors for choosing a file (red arrow), such as a directory of images (green arrow). The files are loaded into the table (middle, purple arrow) with channel determined based on the filename. Microscopy metadata can be entered (red box) and pressing the “Import files” button loads (blue arrow) each image file into a separate plane of a single volumetric image file (right). (B) Several types of multi-plane images can be loaded (left). Raw image formats typically contain no metadata (red arrow), and all metadata should be entered manually. For multi-plane, multichannel TIFF files, selecting the first file (yellow arrow) will cause related files to be loaded into the table (middle). In the metadata area (middle, red box), image parameters will be loaded based on available metadata embedded in the file. In this case, partial metadata is loaded, and the rest needs to be entered manually (right, yellow box). Some proprietary formats (green arrow) contain all channels and metadata in a single file, allowing immediate import (right, blue arrow).
BASIC PROTOCOL 3
Region of interest visualization and annotation
Here we introduce the graphical user interface of MagellanMapper, including the controls panel and the ROI Editor for annotating objects in high resolution data. This protocol assumes that the researcher has installed the software as described in Basic Protocol 1 and imported an image as in Basic Protocol 2. UNIX-style (including Mac and Linux) commands and syntax are also assumed, such as slashes instead of backslashes typically seen in Windows commands but can generally be replaced with Windows-style syntax on Windows platforms. If the software was installed using a Conda pathway on Windows, the Anaconda Prompt may need to be opened to provide the tools for Conda environments.
Materials:
MagellanMapper software installed as described in Basic Protocol 1
Sample image or user-supplied images imported as described in Basic Protocol 2
Launch MagellanMapper
- Load the chosen image file in MagellanMapper.
- In the “ROI” tab in the controls panel on the left, press on the file directory icon to select a file, then choose one of the image files that you imported during Basic Protocol 2 (Fig. 5A). For example, select the
sample_region/myvolume_image5d.npy
file. - Select profiles for viewing and analyzing the image. Profiles are collections of customizable settings that influence visualization and analysis. In the controls panel on the left, select the “Profiles” tab. The “Profile” dropdown contains the different types of profiles in MagellanMapper. Select “Roi” for ROI profiles, which contain settings that mainly affect analysis of smaller regions of interest (ROI) within large, higher resolution images. Next, select “Lightsheet” from the list of profile names in the “Names” dropdown box. In this case,
lightsheet
is a collection of settings optimized for nuclei detection of images taken with a Zeiss Z.1 lightsheet microscope. Select the number of channels to apply this profile to, such as 0 for the first channel, and press on “Add profile.” The profile should appear in the table. Press on “Load profiles” to load this profile into MagellanMapper. -
Alternatively, we can use the command-line interface to specify the sample image file and other parameters. The command line interface supports many parameters to configure the software at runtime. Assuming that the sample image directory is located in the MagellanMapper directory and has been imported as per Basic Protocol 2, start the software using this command:
python run.py --img sample_region/myvolume \ --roi_profile lightsheet
As before, we assume that the MagellanMapper environment has been activated and the software folder entered for these commands. We have specified the path to the imported image using the--img
parameter. Since the image import generates a number of files based on the original filename or--prefix
parameter, we use that original name and can omit the extension. This image path can be replaced with user’s image file of choice.--roi_profile
is a parameter to specify these ROI-related profiles. Multiple profiles can be strung together using “,” to separate each profile name, where each subsequent profile takes precedence over previous settings. Templates for creating custom profiles can be found in theprofiles
folder.
-
Familiarize yourself with the ROI Editor. After loading the image, the researcher should be presented with the volumetric image in the ROI Editor (Fig. 5A). The panel on the left contains controls to select a region of interest and configure display settings. The right panel contains tabs with various ways to visualize the 3D image. The first tab shows the ROI Editor, which lays out the serial 2D planes from an ROI side-by-side to allow the researcher to view the full 3D stack in its original raw images. For context, the ROI Editor also shows overview images that each progressively zoom into the ROI.
The overview plots start with a full plane of the original image and depict the ROI with a box. The listed magnification is based on the image’s microscopy magnification value. Additional overview plots zoom toward this ROI, depicting the total virtual zoom value above the plot. While the overview plots show a single plane at the value indicated above the first plot, scrolling a mouse or pressing the arrow keys will scroll through image planes. To jump to a specific plane, right-click on the desired plane, which will shift all overview plots to the corresponding plane.
Although MagellanMapper attempts to auto-adjust intensities to an optimal level, it may be desirable to control these settings manually. In the “Adjust Image” tab on the control panel, you can adjust intensity ranges, brightness, contrast, and opacity for each channel.
-
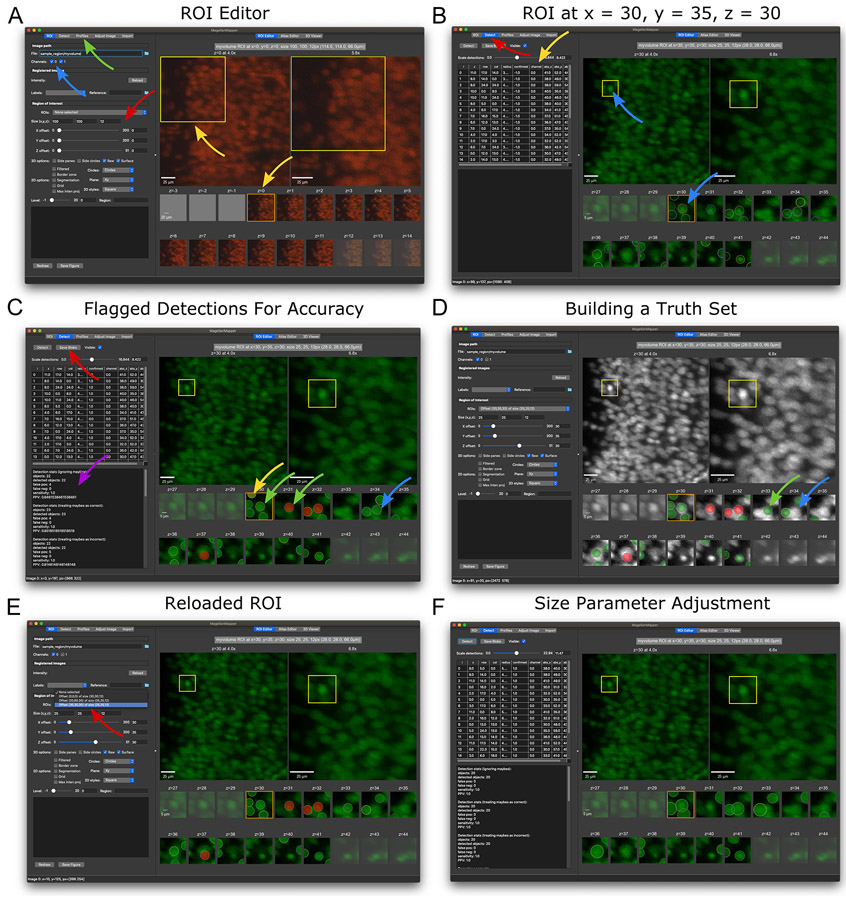
Select a new region of interest. An ROI is defined by a set of offset coordinates within the full image and a set of size measurements in each dimension. The researcher can adjust these parameters by left-clicking on the desired offset (upper left corner) position in an overview plot, which displays a preview of the new ROI position. For more precise control and z-plane selection, the ROI sliders and text boxes allow one to define the exact offset and size, respectively (Fig. 5A).
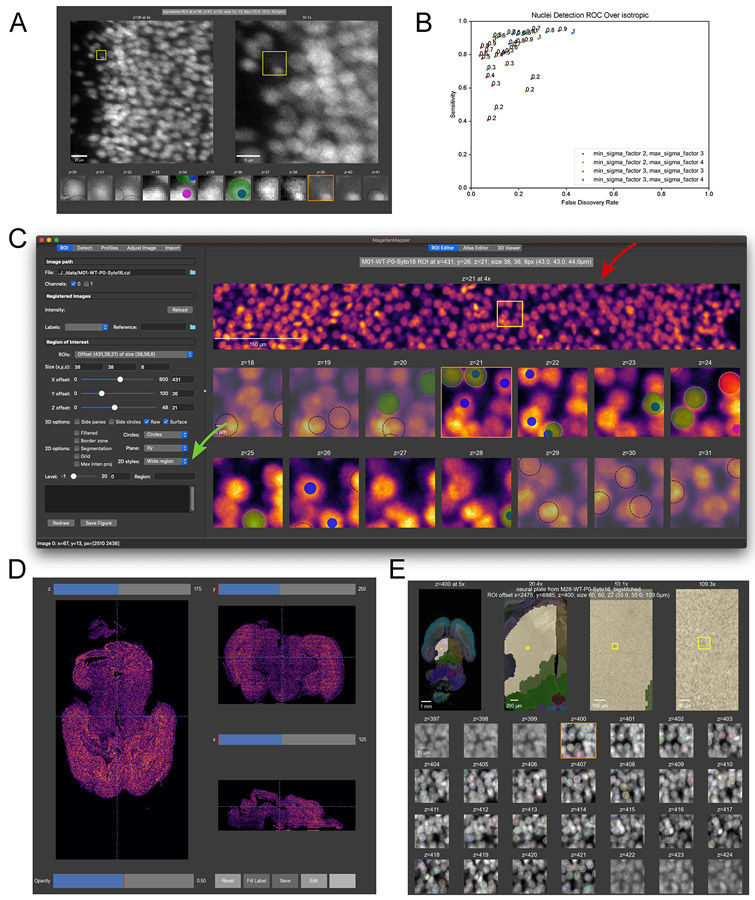
By default, the ROI Editor will overlay all channels on one another. To show only a specific channel, change the “Channel” check boxes to the desired combination of channels. To view the selected ROI, press the “Redraw” button, or double right-click in an overview plot. Practice navigating to an offset of x = 30, y = 35, and z = 30 with a size of x = 25, y = 25, and z = 12, with only channel 0 selected. The viewed ROI should be similar to that depicted in Fig. 5B.
ROI selection allows one to zoom in on a specific region within a potentially much larger volumetric image. ROI viewing can be particularly important for exploring high resolution data from whole specimens, where zooming into a smaller region may be necessary to examine fine details. When displaying an ROI, MagellanMapper will only load that portion of the image, avoiding the high memory requirements that would otherwise be incurred by loading the full image into memory. To open MagellanMapper at a given offset, add the--offset
parameter to the launch command to preselect the ROI offset in x,y,z, quickly returning to the same location with each launch. Similarly, the--size
parameter specifies the ROI size. -
Track nuclei within the ROI. Since nuclei were imaged in 3D, each nucleus typically appears in several z-planes. While following a nucleus from plane to plane, the nucleus will appear to come and go, growing larger as we approach the plane at the center of the nucleus before shrinking as we move on to planes past it. Try counting the number of nuclei in this ROI.
Visualizing an ROI in 3D can be challenging. 3D renditions may provide a very realistic, engaging depiction but typically require a degree of processing to achieve the effect (Long et al., 2012). Ideally, one could have the option to view the raw, unprocessed images through an entire stack. In the overview plots, one can scroll through the stack to see each plane sequentially, which allows one to compare the position of objects from one plane to the next but only shows one plane at a time. As a complementary approach, the serial 2D plots depict all planes from the ROI simultaneously. The researcher can follow objects from one plane to the next without flipping among them. One can link these two viewing approaches by shifting the overview plot to the currently examined serial 2D plot and scrolling up and down to see how objects shift along the z-axis.
-
Detect nuclei within the ROI. MagellanMapper incorporates a 3D blob detector to find objects such as nuclei within volumetric images. Press the “Detect” button to detect these objects. Each detection appears as a colored circle over the corresponding nucleus, with the circle size matching the detected nucleus radius (Fig. 5B). The circle’s z-plane corresponds to the detected center of the nucleus. Dotted black circles depict detections that extend into the ROI but whose centers are just outside of the ROI. Again, track nuclei across each ROI z-plane, but now assisted by the annotations depicting where the detector considered the object centered in all three dimensions.
The detector employs a Laplacian of Gaussian (LoG) blob detection technique (Lindeberg, 1993; Schmid, Schindelin, Cardona, Longair, & Heisenberg, 2010) in 3D as implemented by the scikit-image image processing library (Stéfan van der Walt et al., 2014) to find nuclei. The algorithm attempts to find all blobs matching criteria specified in the microscope profile but will inevitably make mistakes such as missing objects, detecting non-existent objects, detecting duplicates of the same object, or positioning the object incorrectly. Later, we discuss ways to optimize the detector.
-
Annotate nuclei detections. Ideally, each nucleus has only one associated circle, and each circle has an associated nucleus. To track the correctness of each detection and train the detector, these circles are editable to allow repositioning, resizing, or flagging detections. The annotation tools built into the ROI Editor allow one to edit the given annotations, typically through key-mouse combinations as documented in the software README file.
Flag detections: Clicking on a circle changes its color to flag the correctness of the detection. Click on the circle until its flag turns green. For incorrect detections, such as those that correspond to artifactual objects or duplicately detected nuclei, flag the circle as red. The yellow flag is for detections that are indeterminate, such as tightly packed cells where some nuclei may be hard to distinguish from one another. Click through all flags will to reset the flag.
Move/resize detections: To shift a circle’s position, press and hold Shift while left-clicking and dragging the circle within its plot. To alter the circle’s size, press Alt while left clicking on the circle and dragging to adjust the radius. While adjusting circles in the editor, each circle’s coordinates and size update in the table in the ROI selection window. This table shows both the coordinates relative to the ROI and the absolute coordinates within the full image.
Add/remove detections: Occasionally a detection will be missed, or extra circles may exist that do not correspond to a correctly identified object or duplicate detections of the same object. To add a circle, press Control while left-clicking on the desired location. To delete a circle, press D while left-clicking on the circle to remove.
Cut/copy/paste detection: Sometimes a circle will match an object but not in the correct plane. To shift planes, press X while left-clicking the circle to cut it, then press V while left-clicking in the desired plane to paste in the circle. Replace X with C to copy rather than cut the circle.
Save/restore detections: Press the “Save Blobs” button to save the ROI and its annotations. All ROIs are saved within themagmap.db
SQLite database in the MagellanMapper folder. If you move to another ROI or close the software, you can return to this ROI and restore your annotations by selecting the ROI tab and choosing the desired entry from the “ROIs” dropdown box. To test this function, try setting the ROI offset back to x = 0, y = 0, and z = 0, and press “Redraw.” Next, restore the ROI you previously saved using the “ROIs” dropdown box.Practice using the ROI Editor:
Flag the correctness of detections to measure detection accuracy. Check each detection in this ROI to see if it matches a corresponding nucleus, clicking on the circle to adjust its flag. Track each nucleus across z-planes to ensure that it has one and only one associated detection circle. If a detection is missing, add a circle at the appropriate position and z-plane corresponding to the center of the nucleus. If two or more circles are associated with a given nucleus, flag all but the circle closest to the nucleus’s center as incorrect. Once satisfied with the flags, save the ROI. Basic stats such as sensitivity and positive predictive value will appear in the lower left-hand text display based on your flagged detections. The annotations should appear similar to the window in (Fig. 5C). At times, some flags may be debatable depending on the resolution of the images, in which case the yellow (“maybe”) flag may come in handy.
-
Generate a ground truth set from the ROI. Going one step further from flagging the correctness of detections, we can adjust the detections for precise locations and sizes. The resulting curated set of locations will serve as ground truth to train the detector. Starting with the previously saved annotations saved in Step a, use the editor controls to adjust the circles to match each nucleus’s radius and centroid position. If necessary, cut circles from one z-plane to another z-plane to match the center plane of the nucleus. Note that the original position will remain for reference during editing but disappear after saving and reloading by selecting the ROI from the dropdown selector at the top of the ROI settings area. You can compare your truth set with our annotations (Fig. 5D).
Finding the precise location of nuclei can be difficult. First, densely packed nuclei may appear merged, where distinguishing whether an object is one, two, or even more nuclei may be unclear. Second, oddly shaped or potentially merged nuclei present a challenge in identifying the central z-plane of the nucleus, which may affect whether a detection is classified as correctly matching a given ground truth nucleus or not. Carefully following each object from plane to plane may help clarify the identity and location of each nucleus. Alternatively, browsing through planes in one location may reveal subtle nuclei shifts that highlight where one nucleus ends, and another starts. Use the overview plot scroll and jump controls to assist with this viewing. Once satisfied with the truth set, save the annotations again using the “Save Blobs” button.
The ROI selection panel contains several options to configure the ROI Editor layout. The “2D options” checkboxes allow one to overlay a grid or show a filtered view that highlights foreground, for example. The “Circles” dropdown box allows one to configure whether to show contextual circles for each detection in surrounding planes or to turn off circles completely. The “Plane” dropdown box shows the ROI from the other dimensions based on a 3D reconstructed view of the ROI. In the “2D styles” dropdown box, one can select various window layouts, such as layouts with additional overview plots or all serial 2D planes in a single row. Different layouts may be more appropriate for differently sized ROIs or for figure generation. To save the ROI Editor tab as a figure, press the “Save Figure” button.
-
Customize nuclei detection settings. The ROI profiles allow one to customize a variety of settings for the blob detector. To create your own settings, copy the blob settings template configuration file,
profiles/roi_blobs.yaml
, to a new file, such asprofiles/roi_myblobs.yaml
. Press “Load profiles” to reload the list of profile names, then select your newly added profile from the list. Add it to the profiles table and press “Load profiles” again to load your settings.Open the ROI that you saved previously using the ROI dropdown box. All circles should appear as before (Fig. 5E) since the annotations are loaded from the database rather than redetecting the ROI. Press “Redraw” and “Detect” to redetect nuclei in the ROI. Note how the circle size are now noticeably larger (Fig. 5F). A critical factor for detecting the correct number and size of nuclei are the minimum and maximum sizes, which correspond to themin_sigma_factor
andmax_sigma_factor
settings, respectively. The default settings in the file you added use larger sizes, leading to larger blob detections and fewer duplicate detections of the same nuclei.Open this file in a text editor to modify the detection settings. Adjust these settings to tweak the lower and upper size limits for detected blobs. Press “Redraw,” which will reload this profile file, and “Detect” to see detections with these new settings. Keep adjusting values until you find satisfactory circle sizes. Note that the placement or even number of detections will change depending on these size parameters. Tweaking the size limits can thus help screen for the optimal number of detections.
This profile file contains additional settings, and other ROI templates in the folder adjust further settings that affect blob detection. While determining the absolute most optimal settings may be practically impossible, often tweaking a few key settings will have large impact. In Basic Protocol 6, we will discuss automated detection verifications to guide optimizations for a small number of key settings combinations.
Alternatively, you can load the profile at startup by specifying this profile file as an additional ROI profile:
python run.py --img sample_region/myvolume \ --roi_profile lightsheet,roi_myblobs.yaml
Figure 5.
Detecting and annotating nuclei in the ROI Editor. (A) The main graphical interface for MagellanMapper shows options for defining and visualizing a region of interest (ROI) on the left, with the ROI displayed on the right. The ROI settings (red arrow) can be entered in the size boxes and offset sliders. Below the ROI selection controls are various 2D and 3D viewing options, and further down are figure redraw and save controls. Channels can be toggled (blue arrow), and profiles changed in the Profiles tab (green arrow). In the right panel, the top row shows overview plots starting with the original image on the left and progressively zooming into the ROI highlighted by a yellow box with each successive plot (top yellow arrow), with. As a volumetric image, the full image contains multiple z-planes. The z-planes within the ROI are shown as smaller plots in the bottom rows, labeled by the corresponding absolute z-plane number (starting with z = 0). Scrolling the mouse or pressing arrow keys will scroll the original image’s z-plane and shift the orange box to the corresponding ROI z-plane (bottom yellow arrow). (B) A new ROI with different offset and size parameters as shown in the left panel settings. Whereas the previous view overlaid all channels, only the first channel was selected here. Selecting the “Detect” tab (red arrow) and pressing the “Detect” button will perform 3D blob detection to identify objects such as nuclei. The detected locations appear as circles in the ROI plots, with each circle positioned at the detected center of the given blob. Notice how the bright nucleus in the middle of the yellow box corresponds to a detection shown in the z = 30 ROI plot (blue arrows). The table shows the coordinates of each circle (yellow arrow). (C) Annotating detections based on accuracy. These circles can be repositioned, added, deleted, or flagged to assess detection accuracy and annotate ground truth sets to train the detector. Clicking on circles changes their color to flag them as correct (green circle, green arrow) or incorrect (red circle, red arrow). A yellow flag (yellow circle, yellow arrow) can be used for ambiguous detections or nuclei. Missing detections can be added by Ctrl-clicking on the desired location. After saving annotations (red arrow), detection statistics are shown in the feedback panel (purple arrow). (D) An example of further annotating the ROI to build ground truth by shifting, resizing, and copying/cutting/pasting circles. An alternate colormap assists with contrast for more precise annotation. Note the circle cut from z = 33 (blue arrows in parts C and D) and pasted into the next plane (z = 34, green arrow) to match the center of the nucleus along the z-axis. (E) After saving the ROI, the ROI appears in a dropdown box (red arrow), which can be selected to restore the ROI after shifting to a different ROI or re-opening MagellanMapper. (F) Cell detection parameter adjustment. Changing the minimum and maximum detection sizes can impact detection accuracy. In this case, the detected sizes of each nuclei are somewhat large, but the number of duplicate detections of the same nuclei decreases.
Sample Data (to be incorporated directly into the steps of the protocol)
The
sample_region.zip
file with a small volumetric image has been provided to walk the user through importing and viewing this image in the ROI Editor. Screenshots show expected views when loading this image (Fig. 5).
BASIC PROTOCOL 4 (optional)
Explore an atlas along all three dimensions and register to a sample brain
In addition to focused views of high resolution data in regions of interest, higher level views of the whole volumetric image can provide a global context for the imaged specimen. Such visualization allows the researcher to understand the orientation of the specimen and its anatomical relationships while also facilitating atlas overlays to define region boundaries. Although microscopy images are typically taken as 2D planes, serial planes taken along an axis of the specimen can be reconstructed into a 3D volumetric image. MagellanMapper can display views of this 3D reconstructed image along all three major axes by showing all orthogonal planes intersecting at a given point. Multichannel images and registered atlases can be overlaid to identify correspondence among different signals. To refine atlas alignments or generate ground truth, the interface provides annotation tools to paint or interpolate labels with real-time updates in all orthogonal views.
We have provided an example whole mouse brain image at postnatal day 0 accessible from the software website to view (
sample_brain.zip
on software website, Fig. 3I). Download this archive, extract it, and move the folder into the MagellanMapper folder. To register an atlas, download the Allen Developing Mouse Brain Series (ADMBA) embryonic day 18.5 (“E18pt5”), also available on our repository home page (
ADMBA-E18pt5.zip
), which we have bundled from the Allen Institute for Brain Science website archives (Allen Institute for Brain Science, n.d.). The file names and paths can be changed, but we will assume these file locations for the rest of these protocols.
The latter part of this protocol will make use of running MagellanMapper on the command-line to perform automated processing. To run these tasks, open a terminal, activate the environment, and navigate to the MagellanMapper folder as described previously (see Alternate Protocol 1). If you are already running MagellanMapper through the terminal, you can either close MagellanMapper to run the terminal commands or open a separate terminal to activate and run the commands.
Materials:
MagellanMapper software
sample_brain.zip
(unzipped)ADMBA-E18pt5.zip
(unzipped)
-
Open the sample brain in MagellanMapper. The sample brain folder contains Numpy format image and associated metadata files imported from an image named
brain
. In the “ROI” tab and under the “Image Path” section, open the file button and select thesample_brain/brain_image5d.npy
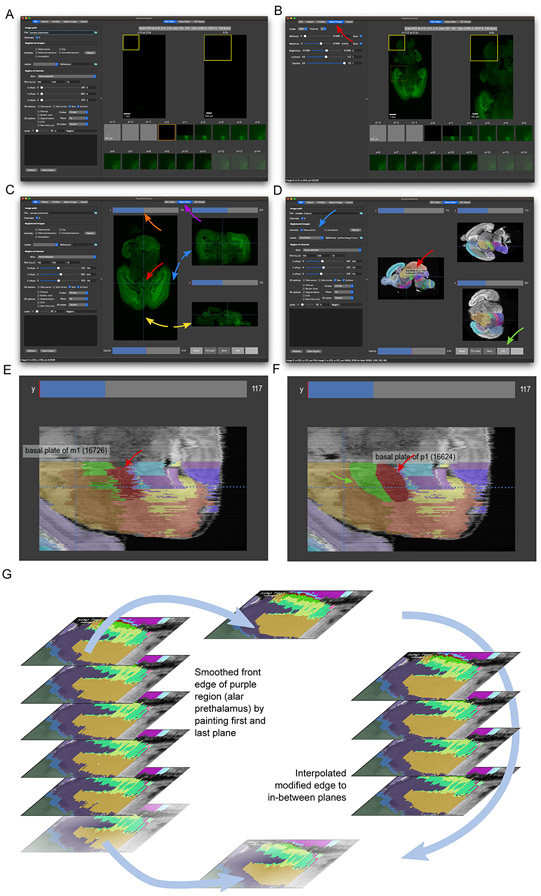
file. The opened image should appear at its bottom z-plane, showing a largely empty image (Fig. 6A).Try scrolling through image planes as you did in Basic Protocol 3. It may be necessary to adjust the image intensities in the “Adjust Image” tab (Fig. 6B).
If you are loading the file from the command-line at startup, open the sample brain image using the original (brain
) name. Assuming that this folder has been moved to the MagellanMapper directory:python run.py --img sample_brain/brain
-
Open the Atlas Editor tab to show the sample brain along each axis. In the right panel, press on the “Atlas Editor” tab to view all three orthogonal views of the brain (Fig. 6C). Again, the initial view will show empty planes from the border of the volumetric image.
To navigate through the image, scroll a mouse wheel or trackpad while hovering over a given view to navigate through planes along that axis, or click or drag on the scroll bars above each view to jump to a specific plane. Notice how the lines in the other views move during this scrolling. To center all views as a desired location, click to move the crosshairs there, which will shift the other views to the corresponding planes intersecting at that spot. Try navigating to a position similar to that shown in Fig. 6C.
To zoom in, drag the mouse up while right-clicking (or press Ctrl while left-clicking), or down to zoom out. To pan through the image, drag the mouse while middle-clicking (or press Shift while left-clicking). The image intensity adjustment controls in the “Adjust” tab will alter the displayed intensities of the currently selected viewer such as this Atlas Editor.
-
Open a brain atlas in MagellanMapper. Now that we have seen a single sample image in MagellanMapper, we will open an atlas of a similarly aged brain. The atlas consists of two images: 1) an intensity image and 2) its associated labeled image, which we will overlay to show their relationship to one another. In the ROI tab of MagellanMapper, press the File browser folder button and navigate through the folders to select the
ADMBA-E18pt5/atlasVolume.mhd
file from the unzippedADMBA-E18pt5.zip
file you downloaded. Explore the brain by selecting the “Atlas Editor” tab (Fig. 6D). Scroll planes and move the crosshairs to navigate through the brain in all dimensions.To use the colormap shown in Fig. 6D, we will load a profile with a grayscale map. In the “Profiles” tab, select the “ROI” profile type, the “Atlas” profile, and press the “Add profile” button. The profile should appear for each selected channel. Press “Load profiles” to load them.
Once you have explored the brain atlas, overlay its labels by selecting them in the “ROI” tab. In the “Registered Images” section, select the “Labels” dropdown and choose “Annotation.” Press the “Reload” button to reload the images including the labels. Try navigating to a location similar to that in Fig. 6D. Notice how two images appear overlaid on one another, with the translucent labels partially occluding the histological intensity image.
To see the identity of each label, we will load the labels reference file. Press on the “Reference” folder button and select theADMBA-E18p/ontology17.json
file, then press “Reload” again. Navigate to a desired area and readjust the image intensities if necessary. Hover the mouse over various labels to display a popup of their identity, including the label’s Allen ontological ID and corresponding anatomical structure. To adjust the labels’ level of opacity, move the opacity slider in the Atlas Editor to the desired level. You can also select “Labels” from the “Image” dropdown in the “Adjust Image” tab to adjust the opacity with the slider there.Alternatively, you can load the atlas with its associated labels files using a single command. We introduce launch options to specify the images to overlay and profiles to customize the colormaps. Assuming that the ADMBA E18.5 mouse brain atlas folder has been extracted to the MagellanMapper folder, use this command:
python run.py --img ADMBA-E18pt5/ \ --labels ADMBA-E18pt5/ontology17.json \ --reg_suffixes atlasVolume.mhd annotation.mhd \ --roi_profile atlas
The--labels
option specifies the ontology file that maps atlas labels to their appropriate structure. The first argument to--reg_suffixes
sets the intensity image to display, such as a microscopy image, and the second argument sets the labels image, such as the annotated atlas image whose pixel values are integers that correspond to the label IDs in the ontology file. We used the--roi_profile
option to specify a profile with colormaps suitable for atlas display, and this parameter could be omitted to use the default colormaps. -
Edit the atlas. By adjusting the labels’ opacity to view label edges in relation to anatomical boundaries, discrepancies between label and anatomical edges may become apparent, especially when viewed in planes from all three dimensions. For example, try zooming into a region of the y-planes to see the jagged borders arising from slight plane-to-plane annotation misalignments as shown in Fig. 6E. MagellanMapper provides an editing interface to curate these labels by painting using the controls described here. See the website or software README for controls updates.
Basic editing: To enter editing mode, press on the “Edit” button. Now, instead of navigating when clicking in an image, clicking picks up the label at that location to paint into neighboring regions. Drag while clicking to paint those pixels, such as smoothing the edge of a label or correcting an artifact. While painting in one plane, the orthogonal planes will update in real-time. Try painting the red and green labels in the same zoomed y-plane to smooth their borders as seen in Fig. 6F.
New labels: Occasionally one might need to paint a label into non-neighboring pixels. In this case, click on the desired label, then press Alt while clicking on a new location to use the last selected label. To paint any label, including one not represented in the image, click in the box next to the “Editing” button and enter the desired integer label. The next click and drag will paint using this label value. Since the colormap for each atlas is generated when first loading the image, any completely new label may not have a unique color until after reloading the image.
Brush size: To change the size of the brush, use the bracket (“[ ]”) keys, which will adjust the brush radius as seen in size of the circle/ellipse underneath the mouse pointer (Fig. 6F).
Labels opacity: The Opacity slider adjusts the visibility of the labels. Try reducing the opacity to see the underlying intensity image more prominently through the labels. Aligning labels with anatomical borders may require frequent toggling between label opacities to see the underlying intensity image. To speed toggling, press the “a” key to make labels completely invisible, and press again to restore the prior opacity level. Press Shift+a to halve the opacity, which will continue to halve the opacity with each repeated press.
Interpolate edges between planes: Editing labels in 3D can be especially challenging and time-consuming. Often borders that appear smooth along one axis will appear jagged in planes along the other two axes because borders may not line up with one another from plane to plane. To facilitate creating smooth borders between planes and to minimize repeated editing, we have provided an interpolation function to fill in spaces between edges in two separated, edited planes. By automatically connecting these planes in 3D, this approach provides a smooth transition across the filled space (Fig. 6G).
First, edit the edge of a label in one plane. Next, scroll several planes and edit the same edge of the label in that plane. The “Fill z” button should now be enabled with numbers indicating the edited plane indices and the label ID. Press this button to fill the edited edge in the intervening planes. Scroll through these planes to check whether the edited edge now transitions smoothly between the originally edited planes. Note that editing any other label will reset this button, but you can click anywhere in the given label in each boundary plane to set up the fill button again.
This method works ideally for label edges that transition linearly between planes. For complex structures with edges that transition at different rates in different places, it may help to edit the edges in multiple parts. For example, one can take a small section of an edge, edit it in one plane, jump several planes, edit that portion of the edge, and use the fill button, then repeat the process in an adjacent section of the edge. One can also “iterate” interpolations by interpolating between two relatively close planes and using the last edited plane as the first plane of the next interpolation, filling a larger volume by making several small jumps at a time.
-
Register the atlas to the sample brain. To identify the anatomical regions within the sample brain, the atlas can be registered to the sample brain by shifting and warping it to fit the shape and landmarks of the sample brain. The registered atlas labels will then demarcate the anatomical boundaries within the sample brain. To register images, MagellanMapper uses the SimpleElastix library (Marstal et al., 2016), which combines the programmatic access and image processing methods of the SimpleITK library (Lowekamp et al., 2013) with the established image registration algorithms in the Elastix toolkit (Klein, Staring, Murphy, Viergever, & Pluim, 2010). MagellanMapper will register the atlas to the sample brain using this command:
python run.py --img sample_brain/brain ADMBA-E18pt5 \ --register single --plane xz
As earlier, we assume that the MagellanMapper environment has been activated, and this command is run from the software folder. We now use the--img
option to specify two paths: 1) The fixed image, in this case the sample brain and 2) The moving image directory, in this case the atlas that will move to register with the sample brain. The--register
option sets the type of registration, such assingle
to register an atlas to a single image, orgroup
for groupwise registration of multiple sample brains to one another. The--plane
option specifies an image transposition, with the original orientation taken as “xy,” and “xz” or “yz” as orthogonal orientations, which is necessary to transpose the atlas to the same orientation as that of the sample brain.Registration will produce several files with “reg suffixes” added just before the filename extension to denote the type of registered image. The “atlasVolume” image is the registered atlas intensity image, while the “annotation” image is the corresponding registered labels image. Since these images are cropped to the fixed image after registration, the “Precur” versions show the images before this curation. The “exp” image is a copy of the original fixed image in the same file format as that of the registered images.
By default, three types of registrations will be performed: 1) Translation, a form of rigid registration that shifts and rotates the moving image, 2) Affine, which shears and scales the moving image, and 3) Non-rigid registration such as B-spline, which performs local deformations to fine-tune the alignments once the images are in a similar space. Registration may require considerable tuning to optimize for the given images and application (Nazib, Galloway, Fookes, & Perrin, 2018). The--atlas_profile
option provides access to registration profiles for configuring many of these settings in SimpleElastix. Similarly to the ROI profiles, these atlas profiles can be customized following examples in theprofiles
folder. -
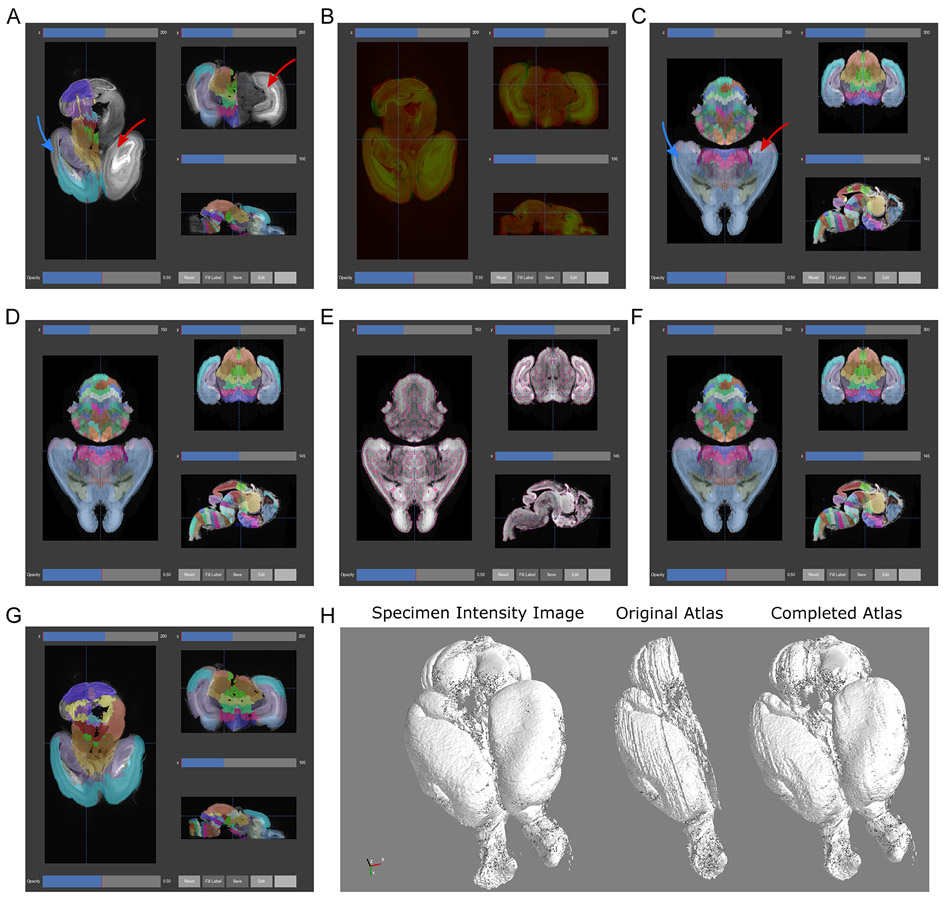
Overlay the registered atlas labels on the sample image (Fig. 7A). We will again overlay labels on an intensity image, but this time using the registered atlas labels with the sample image. Load the sample brain image as in Step 1. As in Step 3, select the registered “Annotation” file in the “Labels” dropdown box of the “Registered Images” section and press “Reload” to overlay registered atlas labels. To change the colormap of the main image to grayscale, select the “Atlas” ROI profile in the “ROI” tab. “Redraw” the image or start scrolling through planes.
Now instead of opening the atlas with its labels, the sample brain will open along with the registered annotations. Open the Atlas Editor and scroll through the image to view the overlay, where labels should conform to the landmarks of the sample brain. Again, hovering over labels will show the name and ID of the corresponding structures based on the label ontology file.
Alternatively, use this command to open the images at once:
python run.py --img sample_brain/brain \ --reg_suffixes annotation=annotation.mhd \ --labels ADMBA-E18pt5/ontology17.json --roi_profile atlas
-
Overlay the registered atlas intensity image on the sample image (Fig. 7B). One might also wish to view the alignment of the atlas intensity image and the sample brain to assess the quality of the tissue registration. To overlay these images, select the “Atlasvolumeprecur” and “Exp” check boxes for the intensity image in the registered images section. Reset the labels image (choose the empty entry), reload the images, and scroll through planes. The intensities and opacities of the overlaid images may need to be adjusted to see both images at once.
We have shown the pre-curated registered atlas volume, which has not undergone any post-processing after registration and shows the raw alignment. Careful inspection of registrations will often show poor alignment in a few places, which can be improved through customization of the atlas profiles as described earlier. While optimizing settings may improve these settings, at times they may worsen other areas of the same brain or may not be universally applicable to a wider number of brains. Sometimes it may be practical to manually edit these misaligned labels using the annotation tools. One may also use the annotation tools to curate the labels for a few structures in registered atlases to serve as ground truth for quantitative evaluation of registration. In some cases, misalignments may be related to the atlas itself, in which case automated tools for atlas refinement can help improve the underlying source of anatomical segmentations as described in Basic Protocol 5.
To overlay these files with a single command, reopen MagellanMapper using this command:
python run.py --img sample_brain/brain \ --reg_suffixes exp.mhd,atlasVolumePrecur.mhd \ --roi_profile norm --vmax 0.6,0.5 --vmin 0,0 \ --plot_labels alphas_chl=1,0.3
Here the two intensity images are rescaled to a similar range to allow display of images from different scales. The images are specified together as a single, comma-separated argument to the--reg_suffixes
parameter to join them into this common scale, where “atlasVolumePrecur” image is the registered atlas intensity image before any curation. By putting these images on a common intensity scale, we can clip each image’s intensities using the--vmax
and--vmin
parameters to make both images visible when overlaid. The--plot_labels
parameter specifies multiple subparameters that control plot characteristics, such as the relative alpha (opacity) of each image.
Figure 6.
Annotating labels in the Atlas Editor. (A) Opening the sample brain in MagellanMapper initially shows the bottom z-plane in the ROI Editor. (B) Scrolling a mouse moves the overview plots’ z-plane, while clicking on the plot moves the ROI offset controls to that position and shows a preview of the ROI (dashed box). Opening the image adjustment tab (red arrow) allows brightness and contrast control. (C) Opening the Atlas Editor tab (purple arrow) shows a simultaneous orthogonal viewer of the sample brain. Scrolling through the horizontal (axial, z, or xy-plane) plane or clicking/dragging its slider (orange arrow) moves through these planes. Clicking within the plane moves the crosshairs (red arrow) and shifts the coronal (y, or xz-plane; blue arrows) and sagittal (x, or yz-plane; yellow arrows) views to the planes corresponding to these crosshair lines. (D) The Allen Developing Mouse Brain E18.5 atlas is shown with the specified labels (blue arrow) overlaid on the microscopy intensity image. Each distinct label color corresponds to a separate label in the Allen ontology. Hovering over a given label identifies its name and Allen ID (red arrow). The tools at the bottom of the editor (green arrow) allow label display adjustment as well as edit controls for painting labels in each 2D plane. (E) Example y-plane before editing, showing jagged label edges. The white bordered ellipse shows the paint brush (red arrow), whose size can be adjusted using the bracket keys. (F) The red (red arrow) and green (green arrow) regions have been smoothed by simply dragging the brush along the label borders. (G) Example edge interpolation to smooth a label edge. Painting successive planes in 2D can lead to jagged edges when viewed in the third dimension. To paint smoothly in 3D, the label fill tool (green arrow section in part D) interpolates label edits between two separate planes. The alar prethalamus (purple label) shows jagged artifacts in each plane, including irregularity between each plane (left). The anterior edge of the label has been edited in the top and bottom planes to smooth the jaggedness (middle), without touching the middle four planes. Applying the edge interpolation applies the edits from the two manually edited planes smoothly along these unedited planes. After edits, the image can be saved (green arrow section in part D).
Figure 7.
Atlas registration and automated refinement. (A) Registration of the original Allen Developing Mouse Brain Atlas E18.5 atlas to the sample P0 mouse brain. After registration, the existing labels approximate the position, orientation, size, and shape of the corresponding parts of the sample brain. Note the missing labels in one entire hemisphere (red arrow) and the lateral planes of the labeled hemisphere (blue arrow). (B) Overlay of the sample brain (green) and registered atlas (red) intensity images as a qualitative assessment of registration. Misalignments can be optimized by adjusting settings in the software’s atlas profiles. (C) The automated 3D atlas generation pipeline performs extension of lateral labels into the unlabeled areas (blue arrow) by iteratively growing the existing labels to fit the anatomical contours (part F). After extending the lateral labels, the unlabeled hemisphere is replaced by mirroring the labeled hemisphere (red arrow), resulting in a fully labeled atlas in 3D. (D) Label smoothing removed many of the jagged artifacts most visible in the axial and coronal planes. (E) To further smooth and refine labels, edge detection (pink lines) of gross anatomical boundaries (high grayscale contrast) in the microscopy intensity image provides a 3D guide for aligning label edges. (F) Starting with the atlas from part C, each label is eroded to its core and regrown by a watershed algorithm guided by the anatomical map from part E, followed by smoothing as in part D to remove small artifacts from the watershed. (G) The same registration as performed in part A is performed but with the generated 3D atlas labels. (H) The 3D Viewer provides a macroscopic view of the specimen through 3D surface rendering of the sample brain (left). Surface rendering of the registered labels (center) before and (right) after 3D atlas generation demonstrate label completion and smoothing.
Sample Data (incorporated into the protocol steps)
The
sample_brain.zip
file with a downsampled whole brain image and
ADMBA-E18pt5.zip
file with an atlas for the corresponding age have been provided to walk the user through importing and viewing this image in the Atlas Editor. Screenshots show expected views when loading this image (Fig. 6).
BASIC PROTOCOL 5 (optional)
Automated 3D anatomical atlas construction
Many 3D atlases have been derived from 2D annotation of successive planes taken from a given specimen. This approach often leads to artifacts apparent in 3D or when planes are viewed along axes other than that of the original annotation, such as the axial and sagittal planes of an atlas annotated coronally (Ng et al., 2007; Niedworok et al., 2016; Wang et al., 2020). Annotation by hand, while typically necessary to generate atlases and ground truth, is also a time-consuming and laborious process. The Allen Developing Mouse Brain Atlas (ADMBA) series of atlases provides detailed annotations of many key structures for mouse brains at 8 different timepoints from embryonic (E11.5) to postnatal development (P56) but understandably omits the lateral portions of each hemisphere on one hemisphere and the entire hemisphere on the other side for most of its atlases (Thompson et al., 2014).
To extend and refine these labels automatically, we have developed and integrated a method to extend these lateral labels into unlabeled regions based on the underlying anatomical edges in 3D. The method also smooths and refines existing labels based on this anatomical map. Here we describe how to apply this approach to each atlas in the ADMBA, or to create custom profiles to refine other 2D-derived datasets in 3D. This protocol will make more use of running automated command-line tasks as described in the introduction to Basic Protocol 4.
Materials:
MagellanMapper software
sample_brain.zip
(unzipped) or user-supplied whole organ imageADMBA-E18pt5.zip
(unzipped) or user-supplied atlas
- Gather an atlas to refine. For this protocol, we will continue to use the ADMBA E18.5 atlas accessed in Basic Protocol 4, but any atlas organized in the following format should work with our refinement pipeline:
- An intensity image, such as the microscopy image
-
An annotated image with labels indicated by unique integer valuesBoth images should be in a 3D format, such as NIfTI, MHA/MHD, or NRRD medical imaging formats, placed in a single folder.
-
Generate a complete, 3D reconstructed atlas from a partial, 2D derived atlas. As shown previously, the ADMBA E18.5 atlas is missing labels in the lateral planes of the labeled side and the entire opposite hemisphere. MagellanMapper provides a method to fill in these labels based on the anatomical data from the atlas intensity image. Use this command to input the original atlas, extend its labels into incompletely labeled areas such as the lateral hemisphere, and mirror labels onto the unlabeled hemisphere:
python run.py --img ADMBA-E18pt5 --register import_atlas \ --plane xz --atlas_profile abae18pt5,nosmooth
As before, we assume an activated environment and running this command from the software folder. To target the range of potential atlases for refinement, MagellanMapper provides a number of customizable atlas and registration refinement settings, grouped in profiles specified by the--atlas_profile
parameter, similar to the ROI profiles. As each atlas in the ADMBA brings unique challenges, we have provided profiles for each atlas, such as the “abae18pt5” profile here for the ADMBA E18.5 atlas. Additional profiles can be layered on the atlas profile to modify steps in the pipeline, such as “raw” to turn off all refinement, “nosmooth” to apply all refinement steps except smoothing, or the default profile to apply all steps including smoothing. -
View the generated atlas. Open the new atlas using the same process as in Basic Protocol 4, Step 3, except selecting the atlas volume in the new folder,
ADMBA-E18pt5_import/atlasVolume.mhd
.After navigating to the center area of the resulting image in the Atlas Editor tab, the image should appear as seen in Fig. 7C. Compare the label coverage with the original atlas to identify where labels have been computationally inferred based on underlying anatomical boundaries. The atlas is now complete, with labels covering all major areas of the brain, although label boundary artifacts persist.
-
Generate a completed, smooth atlas. Starting with the same command as before, we simply use the E18.5 atlas profile directly:
python run.py --img ADMBA-E18pt5 --register import_atlas \ --plane xz --atlas_profile abae18pt5
The resulting atlas can be viewed using the same process as in Step 3 (Fig. 7D), or reload the registered images if the graphical interface is still open. Compare the label edges, particularly in the z-plane, with the original atlas. Notice how the previously jagged label edges in the axial and coronal views appear much smoother.
-
Generate an anatomical edge map to guide further refinement. While smoothing refines labels based on their inherent shape, we can further guide refinement by the underlying microscopy image. To do so, generate a map of gross anatomical edges from it using an edge detection algorithm:
python run.py --img ADMBA-E18pt5_import/ADMBA-E18pt5 \ --register make_edge_images --atlas_profile abae18pt5
We have loaded the imported atlas to use its symmetric microscopy image for the edge map. To view the resulting edge map, follow the same process as in Step 3 except loading theatlasEdge.mhd
file as the registered labels image.Compare the identified anatomical boundaries with the underlying intensity image to assess whether the boundaries capture discrete anatomical regions. This edge map should capture gross boundaries between structures by demarcating areas of high contrast (Fig. 7D). Scrolling among planes should reveal that the edge map generally follows these boundaries in 3D.
- Perform an “edge-aware” refinement using this anatomical map. We are now ready to use this edge map to curate label edges based on the underlying anatomical edges, which we term “edge-aware” refinement.
-
First, re-generate the completed but unsmoothed atlas as described previously since the refinement will have its own smoothing step:
python run.py --img ADMBA-E18pt5 --register import_atlas \ --plane xz --atlas_profile abae18pt5,nosmooth
-
Next, we will erode the current labels to their core, presumably the regions of highest anatomical accuracy, and regrow them to fit these anatomical edges, followed by smoothing:
python run.py --img ADMBA-E18pt5_import/ADMBA-E18pt5 \ --register merge_atlas_segs --atlas_profile abae18pt5
Again, the process described in Step 3 can be used to view the resulting refined atlas (Fig. 7E). Compare the new label edges with the original edges and the identified anatomical boundaries to determine whether the labels have conformed to these boundaries. Try adjusting the Opacity slider to find an optimal balance between the visibility of the labels and the microscopy image to assess the alignment of the label and anatomical boundaries.
-
-
Register this refined atlas to the sample brain. Now that we have extended, mirrored, and refined the atlas in 3D, we can perform whole-brain segmentation of the sample brain by registering this new atlas:
python run.py --img sample_brain/brain ADMBA-E18pt5_import \ --register single --prefix sample_brain/brain_fullatlas
The--prefix
option changes the output path to avoid overwriting previously saved images such as the prior registration. View the resulting atlas using the same process as in Basic Protocol 4, Step 7, but loading the new set of images fromsample_brain/brain_fullatlas_exp.mhd
. The registration is identical to the prior registration since the settings and microscopy images are the same, but the registration now pulls in labels with greater coverage and anatomical fidelity. - Show a 3D surface rendering of the brain and registered atlas. The 3D viewer renders surface views of the images. We can use these renderings to visualize the overall shape of the volumetric images and compare this shape between the sample and registered atlas labels.
-
View the sample brain in 3D. Select the “3D Viewer” tab. A small section corresponding to the current ROI will be shown. To view the whole sample, change the ROI settings to include the entire image. Move all three offset sliders to 0 and change the size boxes to the maximum size of each axis as shown on the right side of the offset sliders. Press “Redraw” in the “ROI” control panel to re-render the surface for this whole-brain ROI (Fig. 7H).Note that this rendering can take several minutes and requires 2-3GB of RAM available to complete for this sample, during which time the software will be non-responsive. Once rendering completes, click and drag on the visualization to rotate it, middle-click to pan, and right-click while moving the mouse up or down to zoom. If you have navigated away from the image, it may be necessary to reorient the view using the isometric view button (3D cube) in the viewer’s toolbar.
-
View the labels in 3D. Uncheck the “Raw” boxes in the “3D options.” Enter “+/-15565” (the Allen ID for the whole brain, with “+/-” added since MagellanMapper distinguishes hemispheres by these signs) in the “Region” box, then press Enter. This surface rendition is based on the atlas labels rather than the sample brain intensity image. If the registration aligned the atlas well, the two surface renditions should look similar.To compare this registered atlas with the original atlas, try reloading the originally registered atlas using the command in Basic Protocol 5, step 6. Render its labels in 3D as described here and compare label surface renderings.The “Region” box can also be used to center the ROI at any other atlas label ID. The
region_ids.csv
file included in the ADMBA-E18pt5 folder contains IDs mapped to their region name.
-
Sample Data (incorporated into the protocol steps)
The
sample_brain.zip
file and
ADMBA-E18pt5.zip
file are again used age to walk the user through 3D atlas generation and registration. Screenshots show expected views when performing these tasks (Fig. 7).
BASIC PROTOCOL 6 (optional)
Whole tissue cell detection and quantification by anatomical label
A key application for 3D atlas generation and registration is to automatically demarcate anatomical structures for cellular quantification within each structure. While traditionally such counts have been measured by hand, the scale of whole tissue imaging has rendered this manual task impracticable. Doing so instead requires whole tissue automated image processing for quantification such as nuclei or cell counts across these large, high resolution datasets. MagellanMapper provides a pipeline for whole tissue nuclei detection to measure the volume, nuclei count, and nuclei density per anatomical structure.
Materials:
-
Detect nuclei in a whole volumetric image. MagellanMapper will perform whole image processing to detect the 3D position of nuclei. To maximize performance within the capabilities of the underlying hardware, the image will be processed in parallel in smaller blocks. The number of parallel processes is determined by the number of computing cores available and memory requirements based on the block size. To demonstrate whole image nuclei detection in a small tissue volume, we will again use the sample nuclei-resolution image in
sample_region
, where the folder is assumed to be in the MagellanMapper folder. This command will process the full volume automatically:python run.py --img sample_region/myvolume --proc detect \ --channel 0 --roi_profile lightsheet,roi_myblobs.yaml
Again, we assume that the environment has been activated in the terminal and the software folder entered. The--proc
option specifies the type of processing, in this case “detect” for parallel processing of the whole image. The--channel
option limits processing to only channel 0 since the two channels contained in this image are simply the views from opposite lightsheet illuminators, which should show roughly the same image. The output prints the number of detected blobs and saves these blobs in a Numpy archive. -
Use the ROI Editor to assess whole image detections. In Basic Protocol 3, we used the ROI Editor to view on-the-fly nuclei detection within the selected region of interest. We now view detections performed beforehand on the whole image, which incorporates more contextual information and typically reduces artifacts near the ROI edge. To load the detections acquired during Step 1, open the image with this command:
python run.py --img sample_region/myvolume --proc load \ --roi_profile lightsheet,roi_myblobs.yaml --offset 30,35,30 \ --size 25,25,12
This time, we use the “load” setting to the--proc
parameter to load the detections archive associated with the image name. The--offset
and--size
parameters automatically set the ROI for viewing to the given position and shape in x, y, z ordering. Press on the “Detect” button to show the loaded nuclei detections within the ROI. Note how detections tend to clump less at borders such as the top and bottom z-planes of the ROI. -
Automatically verify the accuracy of detections. Using the ground truth you built in Basic Protocol 3 for this ROI, we can automatically assess the correspondence between detections and truth with this command:
python run.py --img sample_region/myvolume --proc detect \ --channel 0 --roi_profile lightsheet,roi_myblobs.yaml \ --truth_db verify magmap.db --grid_search gridtest
The--truth_db
option causes a ground truth database to be loaded. The “verify” argument tells MagellanMapper to perform verifications on the automated detections by comparing them with the ground truth you annotated previously. The “magmap.db” argument specifies the name of the database to use for ground truth, which is the default database name where we saved ground truth in Basic Protocol 3. The--grid_search
option performs a Grid Search algorithm with the specified profile along with automated detection verification as described below.The automated detection verification evaluates the detection accuracy based on ground truth. This verification uses the Hungarian method of optimization to assign the appropriate detection to ground truth, with missed or duplicate detections counted as false negatives or false positives, respectively. The resulting statistics of sensitivity (or recall) versus false discovery rate (FDR, or 1 - positive predictive value (PPV), the precision) are output in a CSV file. The optimal combination of chosen parameter settings will typically have a balance of high sensitivity and PPV.
-
View automated detection verifications. Although the statistics such as recall and precision can be helpful to evaluate detection accuracy, it can be helpful to view where the automated detector correctly or incorrectly identified objects such as nuclei. MagellanMapper can display these verifications of automated detections within each ground truth ROI by using this command:
python run.py --img sample_region/myvolume --proc load \ --roi_profile atlas --truth_db verified
The “atlas” profile is used for greater image contrast. The “verified” mode loads the automated verifications showing matches between detections and ground truth. To load verifications from a different database, specify the database path after the “verified” parameter.
In the ROI Selector window, the “ROIs” dropdown should show the verified ROI dimensions, which is slightly smaller than the corresponding ground truth is to account for indeterminate matches at ROI borders. Select an ROI to load its verifications (Fig. 8A). Automated detections are shown in the same green and red translucent circles as before. Green circles are correct detections (true positive), meaning that a ground truth location is within a given tolerance as set in the profile, while red circles are detections without a corresponding ground truth (false positive). For duplicate detections, the closest match is green, while any other duplicates are red. Ground truth annotations appear as solid blue or purple circles. Blue circles are nuclei that have been correctly detected (true positive), whereas purple circles are nuclei that were missed (false negative).
While one could attempt to find the optimal detector settings by repeatedly using different settings, the number of parameters available can make this process tedious and time-consuming to assess each parameter by hand. Moreover, adjustments in each parameter may affect the result from other parameters, meaning that each combination of parameter may need to be investigated. The Grid Search function can be expanded to test a range of values for multiple parameters. The researcher can create a new Grid Search profile with desired settings or combinations of settings to find more optimal detections for one’s own datasets by following example Grid Search profiles in the “profiles” folder (files starting with “grid”). These files can be copied to make one’s own search parameters, and “gridtest” replaced with the filename. Sensitivity and FDR statistics for each combination of settings are depicted in a scatter plot called a Receiver Operating Characteristic (ROC) curve to assess the optimal combination.
Often a single ROI will be insufficient to generate the optimal settings because settings appropriate for that ROI may not generalize to other ROIs. Generating additional ground truth sets will increase the generalizability of any settings obtained from them. An example of an ROC from a Grid Search with three parameters applied over 10 ROIs is shown in Fig. 8B and automated verifications in one of those ROIs in Fig. 8C.
For large images, re-detecting the entire image may take an unnecessary amount of time. To focus detections on just the area of interest, use the--subimage_offset
and--subimage_size
parameters to save a small subset of the image from which to build truth sets. For example, extract a 100 x 100 x 30 (x, y, z) subset of the sample image from an offset of 10, 10, 20 (x, y, z) with this command:python run.py --img sample_region/myvolume --proc detect \ --channel 0 --subimg_offset 10,10,20 \ --subimg_size 100,100,30 --roi_profile lightsheet \ --save_subimg
The same sub-image parameters can be used to open the sub-image, build truth sets, and run automated detections and verifications (Fig. 8C). Image subsets also allows one to share small chunks of images to collaborators for viewing and building truth sets without having to send the original, often prohibitively large, image file.
- Combine raw microscopy import, stitching, whole tissue processing, and image registration into an automated pipeline for quantification by anatomical structure. Using the previous Basic Protocols and the whole tissue processing described in this Basic Protocol, we can create an end-to-end automated pipeline. To demonstrate this pipeline, we will use the hypothetical image file, “large_brain.czi”, which can be replaced with your microscopy image in original format. We will also use the pipelines script described in more detail in Supporting Protocol 1.
-
Start a full pipeline that includes import from proprietary format, tile stitching, whole image nuclei detection, and downsampling for image registration using this command, which runs the pipelines script:
bin/pipelines.sh -i large_brain.czi -p full -g 5.0 -u 1.0
Replace the-g
with the magnification and-u
with the zoom of the objective. If necessary, add the-e
option with the resolutions of the given image inx,y,z
. Depending on the size of the image and the hardware specifications, this process may take a while, up to several days. Supporting Protocol 1 describes the various pipelines available for automated processing. - Explore the imported image in the ROI Editor and Atlas Editor. Follow Basic Protocol 3 to view selected ROI in the newly imported image to verify that the microscopy image file imported correctly at the microscopic scale. At the macroscopic level, view the downsampled image in the Atlas Editor as described in Basic Protocol 4.
- Refine an atlas of the closest matching age. For a sample of age P30, for example, the ADMBA P28 brain would be the closest available atlas from that atlas series. Follow Basic Protocol 3 to complete and refine the original atlas in preparation for registering the atlas to your sample. If another atlas or partial atlas is available, one could create a new profile to optimize settings for import since the methods integrated in MagellanMapper are purposed to be applicable to any 2D-derived volumetric atlas.
-
Register the atlas to the downsampled image. Follow Basic Protocol 5 to register the refined atlas to the downsampled version of the sample image. This registration will segment the sample into anatomical regions for quantifying the detected nuclei by structure. For example, register the brain with this command:
python run.py --img large_brain_resized(863,480,418) \ ABA-P28_import --prefix large_brain --register single
-
Generate a heat map of nuclei densities. The whole tissue nuclei coordinates detected earlier can be converted into a heat map of nuclei per voxel. Generate this heat map with this command:
python run.py --img large_brain \ --register make_density_images
The output will be an image of the same size as that of the registered atlas (Fig. 8D). To view the heat map, load it with this command:python run.py --img large_brain_heat.mhd \ --roi_profile contrast --vmin 0 --vmax 10
The--roi_profile
specifies a high-contrast colormap for the nuclei density heat map. The--vmin
and--vmax
options set the minimum and maximum intensity values, which can be titrated to the range of nuclei per voxel. -
Quantify volumes and nuclei by anatomical structure. Armed with whole tissue nuclei counts and anatomical segmentation via atlas based registration (Fig. 8E), we can now use the basic statistics subpackage included with MagellanMapper to generate statistics by anatomical label:
python run.py large_brain --register vol_stats \ --labels ABA-P28/ontology17.json --atlas_profile abap28
The output is a comma separated values (CSV) spreadsheet with measurements by each atlas label. Each row contains a separate label, including the end labels shown in the labels image and the superstructures that comprise multiple end labels or other superstructures. -
Generate a movie of the image stack. A movie can serve as a portable way to capture the full volume and share it with others.
python run.py --img large_brain_resized(863,480,418) \ --proc animated --slice none,none,1 --roi_profile atlas \ --savefig mp4 --labels ABA-P28/ontology17.json \ --reg_suffixes annotation=annotation.mhd --alphas 1,0.7
This command typically requires a library to have been installed to support output to the MP4 format, such as FFMpeg. Replace the image name to match the path to the downsampled image. The --slice option sets the start, stop, and step size for z-planes to include in the animation, where “none” will use default values. The--alphas
option keeps full opacity of the intensity image but lowers the opacity of the registered atlas to make the underlying intensity image more visible.
-
Figure 8.
Whole brain nuclei detection and assignment to anatomical labels. (A) Automated verification within the depicted sub-ROI shows truth blobs as small, opaque dots and detections as larger, translucent circles. Blue dots are correctly detected truth blobs, while purple dots are misses. Green circles are correct detections, and red circles are false. Correct detections and corresponding truth blobs may be separated by up to a few z-planes if the detector correctly identified a nucleus but slightly off its center in the z-axis. The interface allows inspection to determine where the detector may have worked or failed to guide further optimization. (B) A receiver operating characteristic (ROC) curve from a Grid Search test of several combinations of detection setting parameters with more ROIs of larger size. The minimum and maximum object size parameters and an isotropic scaling factor were varied, with the false discovery rate (FDR, x-axis) and sensitivity (y-axis) shown for each combination. The point labels show the isotropic factor (1 = isotropic, 0.5 = z-scale is half that of x and y), and the color and point symbol correspond to the legend values denoting min/max size parameters. (C) Example of automated verifications in one of these larger ROIs. A sub-image (red arrow) was extracted from the original whole-brain image and 10 ROIs annotated as ground truth, and a wide layout was selected (green arrow). (D) Density heat map of nuclei detected across a full-resolution sample brain using the pipelines script. (E) High resolution serial 2D plots from the ROI Editor show the individual detected nuclei. The ROI coordinates were found by specifying a region ID (below the green arrow in part C), Allen ID 16382 (the alar thalamus, showing the left side), which positioned the ROI to the center of this label. Overview plots show the ROI in context with anatomical labels overlaid and the left alar thalamus highlighted in light gold.
Sample Data (incorporated into the protocol steps)
SUPPORT PROTOCOL 1 (optional)
Import a tiled microscopy image in proprietary format into MagellanMapper
Materials:
MagellanMapper software
Microscopy images
ImageJ/FIJI
BigStitcher ImageJ/FIJI plugin
Sample Data (incorporated into the protocol steps)
Many microscopes store images in their manufacturers’ own proprietary formats, making direct access challenging. To access these images, MagellanMapper uses the Bioformats library (Linkert et al., 2010) to read these formats as described in Basic Protocol 2. Another challenge is that whole tissue microscopy techniques often require microscopy images to be taken and stored as multiple tiles. Each tile consists of a stack of image planes from a small part of the whole specimen, with size typically limited by the field of view of the microscope objective. Multiple tiles are taken in a grid pattern to cover the entire specimen. Afterward, the tiles must be stitched back together to generate a single volumetric image comprising the whole sample. While some microscope system software performs this stitching automatically, the software is not always present or may require additional customization not available in the software.
To stitch these tiled files automatically, MagellanMapper leverages the ImageJ/FIJI software ecosystem to access the BigStitcher plugin (Hörl et al., 2019). This plugin aligns and stitches together these tiles from large images in a memory efficient way, typically through a graphical, interactive interface, and generates a single volumetric image of the whole specimen. By using the ImageJ command-line interface and scripting capabilities, MagellanMapper fully automates the stitching task except for an optional step to verify tile alignments. Afterward, the pipeline can be extended to whole image object detection, segmentation by atlas registration, and regional quantification as described in Basic Protocol 6. The MagellanMapper import pipeline can thus automate file import from raw, tiled microscopy images to stitched images ready for whole image automated processing.
Download and install ImageJ/FIJI. We recommend FIJI as a user-friendly distribution of ImageJ with many plugins already installed. FIJI is freely available at https://fiji.sc/.
-
Launch ImageJ/FIJI. This software requires Java, which is installed during the Conda setup pathway. If the software does not launch or launches with errors, launch it from a terminal where the Conda environment has been activated using this command:
<path-to-ImageJ> --java-home “$JAVA_HOME”
Replace<path-to-ImageJ>
with the path to the ImageJ executable. The JAVA_HOME variable is specified in the Conda environment. Add the BigStitcher plugin. To add a plugin download source, run the menu item, “Help > Update…”. Press the “Manage update sites” button and select the check box next to “BigStitcher.” Press the “Add update site” and “Close” buttons. Back in the Updater window, press the “Apply changes” button to install all updates, including the new plugin.
-
Launch an image stitching pipeline using the MagellanMapper pipelines script. The
bin/pipelines.sh
script in MagellanMapper defines various automated image processing pipelines, includingstitching
to stitch and import raw microscopy image files. The file is a Bash script that runs in standard terminals on Mac and Linux but not in default Windows terminals. Although the sample image only contains one tile, we will use it to demonstrate the pipeline including launching ImageJ. To start the pipeline, use this command:bin/pipelines.sh -i sample_region/sample_region.czi \ -p stitching -g 5 -u 0.8
We assume that the MagellanMapper environment has been activated and folder entered. Option-i
specifies the image path;-p
is the pipeline, which can be one of any options in thePIPELINES
script variable; and-g
and-u
are the magnification and zoom, respectively, which should also be changed to the objective settings.-e
can also be given to specify the microscope objective resolution asx,y,z
if the information was not extracted during stitching. The objective settings will be stored in the image metadata, used for later image processing tasks, and can be ignored if the values are unknown.To view the imported image, loadsample_region/sample_region_bigstitched_image5d.npy
through the graphical interface, or use this command:python run.py --img sample_region/sample_region_bigstitched
-
Wait for the image to complete its import into BigStitcher’s custom format. Depending on the size of the microscopy image file and the computer hardware, this process may take many hours to several days.
In our experience, 500GB images typically take about 5h to import on an AWS m5.4xlarge instance with a 2TB SSD volume.
-
Explore alignments. After importing the image and aligning tiles, the pipeline will pause with the ImageJ/FIJI window open to allow verifying tile alignments. The sample image has only a single tile, but one can open the BigStitcher plugin to explore the image. Once satisfied with alignments, close ImageJ/FIJI to continue the pipeline.
Usage of BigStitcher is beyond the scope of this protocol. We refer the reader to the original methods paper by Horl, et. al. (Hörl et al., 2019) for background and instructions on using the plugin.
-
Wait for fusion and import to Numpy format. BigStitcher will first fuse the file and export it to a multipage TIFF file for each image channel. Once all channels have been exported, they will be imported into a combined Numpy archive file. The resulting image format is ready to use in MagellanMapper image processing tasks or in any other Numpy-based software.
In our experience, fusion requires approximately twice the time of initial import. The BigStitcher import format is not used further by MagellanMapper and can be discarded unless used within the ImageJ ecosystem.
-
Optional: Another pipeline available in the script downsamples large images, which allows for more efficient viewing and processing at the expense of resolution loss. Basic Protocol 2 views a whole brain at the macroscopic level using a brain image downsampled using this pipeline. The
transformation
pipeline will downsample (or upsample, if desired) images in chunks to minimize memory usage and maximize parallel processing during the operation. We will again use the hypothetical image file, “large_brain.czi”, to demonstrate the command:bin/pipelines.sh -i large_brain -p transformation \ -z 278,581,370 -- --atlas_profile abae18pt5
Option-z
specifies the output size in x, y, z dimensions. Other options include transforming the image to a different orientation, such asyx
orxz
instead of the defaultxy
orientation.To view the imported image, loadlarge_brain_resized(278,581,370)_image5d.npy
through the graphical interface, or use this command:python run.py --img “large_brain_resized(278,581,370)”
- Several options can be added during the pipeline launch in Step 2 for further customization. These options include specifying a path on AWS S3 from which the original image will be downloaded and to which a compressed archive of the output image file will be uploaded. A URL can be specified for notifications, such as a Slack bot, to notify the researcher when the pipeline is awaiting review of tile alignments or has completed. A cleanup option shuts down the computer once the pipeline completes, which may save server computing costs in cloud environments. Run
bin/pipelines.sh -h
for the full list of options.
REAGENTS AND SOLUTIONS:
None
COMMENTARY
BACKGROUND INFORMATION:
Historical background on the development of the technique(s) and/or comparison of the described technique with other methods that may be used for similar analyses.
The emergence of high resolution, 3D images from microscopy as well as medical imaging such as MRI at ever increasing scale necessitates tools for efficient visualization, annotation, and high throughput analysis (Cai et al., 2019; E. Meijering, 2012; Erik Meijering, Carpenter, Peng, Hamprecht, & Olivo-Marin, 2016). Ideally, analysis software would handle data at the full scale of resolutions, from microscopic analyses of cellular detail to meso- and macroscopic detail of anatomical structures delineated by atlases to provide global contextual information. For many analyses, the bottleneck is the availability of ground truth to guide these analyses, whether the breadth of annotated cellular structures, availability of age-specific atlases, or accuracy of atlas labels. While several open source and commercial software packages have emerged to address these aims, we have developed and provided MagellanMapper as an open source software suite specifically designed to cover analyses across this full range of resolution scales through whole tissue image processing, graphical tools for visualization and ground truth annotation, and 3D atlas refinement for registration and regional quantification. Moreover, we have designed the suite to scale with the underlying hardware to enable usage on computers ranging from a personal laptop to cloud computing.
Similar software suite include ClearMap, an integrated suite designed for analysis of 3D image data obtained from iDISCO cleared tissue including whole mouse brains (Renier et al., 2016). More recently, the suite underwent a complete rewrite as ClearMap2, which is designed for fast processing of cleared tissue 3D data (Kirst et al., 2020) but not currently available at the time of writing. The suite also supports image registration through the Elastix library and incorporates deep learning for vessel segmentation and a wobbly stitching toolkit for aligning tiles with non-rigid registration. While many of its functions are complementary to MagellanMapper, MagellanMapper also provides atlas refinement tools applicable to any 2D-derived volumetric dataset, a focus on annotation tools, and designed support for a range of computing platforms. WholeBrain is another open source software suite focused on whole brain mapping efforts, including atlas registration and conversion to vector-based, scale invariant atlases (Fürth et al., 2018). While WholeBrain is written in R, MagellanMapper is written in Python to leverage the broad Pythonic ecosystem of scientific and image processing libraries and the efficient parallel computing capabilities built into Python. Ramirez et al. (Poinsatte et al., 2019; Ramirez, Ajay, Goldberg, & Meeks, 2019) describes a pipeline for registering mouse whole brains imaged with serial two photon tomography as well as feature classification using the Ilastik machine learning toolkit, although to the best of our knowledge, the pipeline is not open source. Brainpatterns is a software suite that integrates atlas registration with regional quantification of fluorescent signal (Salinas et al., 2018). It is available as open source but uses the commercial software Imaris and Excel to control the software.
MagellanMapper seeks to take an integrated approach to whole tissue image processing and more generally to any 3D dataset. The combination of a graphical user interface and command line interface allow the software to be controlled in an interactive manner or in automated high performance computing environments. The ROI Editor and block image processing facilitate visualization and quantification of microscopic, high resolution data, while the Atlas Editor and atlas registration (via SimpleElastix) provide macroscopic context. The object and atlas annotation tools and the automated 3D atlas refinement algorithms facilitate ground truth generation across all of these scales.
CRITICAL PARAMETERS:
MagellanMapper provides several mechanisms to set parameters for analysis and visualization. For transient settings that may change with each run, the command line interface supports many parameters to quickly adjust settings. For frequently used settings or grouped settings, MagellanMapper includes profiles with collections of related settings, such as the configuration for refining a particular atlas. At times the distinction between these two types of parameters may become blurred. To use the same command line parameters repeatedly, one could use a Bash or Batch script to save frequently used commands. To use profile settings not included within MagellanMapper, one can design a new profile.
To view the available command line parameters, open the help documentation using this command:
python run.py --help
Example profiles and their specific parameter settings are stored in the “profiles” folder. These files are in YAML format for simple configuration editing.
For nuclei detection, the most critical parameters are
min_sigma_factor
and
max_sigma_factor
, which control the lower and upper size limits, respectively. The
overlap
value sets the threshold at which two potentially separate detections are counted as the same nucleus, which may be critical for separating densely packed nuclei. The detector performs image preprocessing before detection. Checking the “Filtered” box in the “2D options” group will show this preprocessed image to assist with adjusting settings in the profile preprocessing group.
For image registration, two critical parameters are the maximum number of iterations (
iter_max
) for each type of registration and the grid space voxel size (
grid_space_voxels
) for the b-spline deformable registration. The maximum iterations set an upper bound on how many times the registration will be optimized, where lower settings may prevent the registration from reaching an optimal amount, while higher settings may overshoot the registration and lead to excess deformation. The voxel size influences the granularity for finding matching structures, where small values may match smaller structures but at the potential expense of finding incorrect matches.
During atlas refinement, the
labels_edge
parameter group guides the automated lateral labels extension, including where the extension will start and how the extension incorporates the underlying anatomy. The various profiles starting with “aba” can be compared to see the range of settings used for these parameters. The
log_sigma
parameter is critical for determining the level of detail incorporated in the gross anatomical edge map. Higher sigmas use a larger Gaussian blurring filter kernel when generating the Laplacian of Gaussian (LoG) image, leading to a less detailed edge map, whereas lower sigmas will preserve more detail but may lead to more arbitrary boundaries in areas of low contrast.
TROUBLESHOOTING:
Although we have attempted to install MagellanMapper on a variety of platforms, new or different configurations may cause installation challenges. In particular, different distributions of Linux have different preinstalled packages, which may cause conflicts or may require additional package installations on distributions that we have not tested. We have prebuilt several dependencies in an effort to streamline the installation process as much as possible and avoid the need for a compiler, but these dependencies have known cross-compatibility challenges that may require recompilation on the user’s own system.
We have provided a running list of installation solutions and workaround for problems that have arisen during testing on our repository website and will continue to keep this list updated as researchers encounter new issues. We have also provided additional scripts to assist with compilation or follow alternative installation pathways when necessary, found in the
bin
folder. As an open source, scientific software suite, we welcome the feedback of researchers and other users as well as any software contributions, whether documentation to improve clarity or code updates. The software is licensed under the 3-Clause BSD License as a permissive open-source license to ensure that the code and updates remain available to the scientific community.
STATISTICAL ANALYSIS: (optional)
MagellanMapper includes two sub-packages to assist with statistical calculation and plots. These sub-packages use Python based tools such as the Scipy (Virtanen et al., 2019) and Matplotlib (Hunter, 2007) libraries to measure volume and density related statistics, especially to view results by anatomical label. The R package included in MagellanMapper is a more general-purpose package that performs and plots statistical comparisons between different atlases or subject groups. While each sub-package is tailored toward MagellanMapper, the goal is to make these statistical functions and pipelines relevant to a wider range of basic statistical applications.
UNDERSTANDING RESULTS:
To understand microscopy images fully, one must often visualize and compare images from multiple perspectives. The ROI Editor and Atlas Editor provide graphical interfaces to explore raw microscopy images in a 3D context optimized for either high resolution or macroscopic scale, respectively, but each interface also attempts to provide visualization at different scales. In the ROI Editor for example, the overview images also provide a global context, and scrolling through these planes can provide a complementary view to serial planes. In the Atlas Editor, zoom and pan functions allow one to home in on critical areas for inspection and annotation. Surface rendering in the 3D viewer provides a way to view anatomical labels from multiple angles in all dimensions. Together, these views can complement one another to provide a more holistic understanding of the image. Screenshots have been provided in the figures to depict expected output from a variety of sample commands given in the Basic Protocols.
TIME CONSIDERATIONS:
Processing times may vary greatly depending on the size of image and hardware specifications. MagellanMapper was specifically designed for analysis of whole tissue microscopy imaging on a personal laptop with the ability to scale to the cloud. As a reference, 500GB tiled microscopy images typically take about 2 days to stitch and import, followed by another 2 days for whole tissue nuclei detection on an Amazon Web Services (AWS) EC2 m5.4xlarge instance. Atlas refinement and registration on a 2014 MacBook Pro each take approximately 5-10 minutes, and statistical analysis by anatomical structure typically takes 2-5 minutes.
ACKNOWLEDGEMENTS:
This work was supported by a NARSAD Young Investigator Grant by the Brain & Behavior Research Foundation (SJS), NIMH U01 MH122681 (SJS), and NIMH R01 MH109901 to (SJS).
Footnotes
KEY REFERENCES: (optional)
References that are significant for understanding the method but not necessarily cited in the article. Each Key Reference should be accompanied by a short annotation explaining the significance of the reference.
INTERNET RESOURCES: (optional)
URLs for important sites relevant to the method. Each should be accompanied by a short description of the subject of the site.
TABLES: (optional)
Tables should be added to this document using Microsoft Word’s insert table feature.
LITERATURE CITED:
- Allen Institute for Brain Science. (n.d.). API - Allen Developing Mouse Brain Atlas. Retrieved July 3, 2019, from http://help.brain-map.org/display/devmouse/API [Google Scholar]
- Cai R, Pan C, Ghasemigharagoz A, Todorov MI, Förstera B, Zhao S, … Ertürk A (2019). Panoptic imaging of transparent mice reveals whole-body neuronal projections and skull–meninges connections. Nature Neuroscience, 22(2), 317 10.1038/s41593-018-0301-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürth D, Vaissière T, Tzortzi O, Xuan Y, Märtin A, Lazaridis I, … Meletis K (2018). An interactive framework for whole-brain maps at cellular resolution. Nature Neuroscience, 21(1), 139 10.1038/s41593-017-0027-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörl D, Rusak FR, Preusser F, Tillberg P, Randel N, Chhetri RK, … Preibisch S (2019). BigStitcher: Reconstructing high-resolution image datasets of cleared and expanded samples. Nature Methods, 16(9), 870–874. 10.1038/s41592-019-0501-0 [DOI] [PubMed] [Google Scholar]
- Hunter JD (2007). Matplotlib: A 2D Graphics Environment. Computing in Science Engineering, 9(3), 90–95. 10.1109/MCSE.2007.55 [DOI] [Google Scholar]
- Kirst C, Skriabine S, Vieites-Prado A, Topilko T, Bertin P, Gerschenfeld G, … Renier N (2020). Mapping the Fine-Scale Organization and Plasticity of the Brain Vasculature. Cell. 10.1016/j.cell.2020.01.028 [DOI] [PubMed] [Google Scholar]
- Klein S, Staring M, Murphy K, Viergever MA, & Pluim JPW (2010). elastix: A Toolbox for Intensity-Based Medical Image Registration. IEEE Transactions on Medical Imaging, 29(1), 196–205. 10.1109/TMI.2009.2035616 [DOI] [PubMed] [Google Scholar]
- Lindeberg T (1993). Detecting salient blob-like image structures and their scales with a scale-space primal sketch: A method for focus-of-attention. International Journal of Computer Vision, 11(3), 283–318. 10.1007/BF01469346 [DOI] [Google Scholar]
- Linkert M, Rueden CT, Allan C, Burel J-M, Moore W, Patterson A, … Swedlow JR (2010). Metadata matters: Access to image data in the real world. The Journal of Cell Biology, 189(5), 777–782. 10.1083/jcb.201004104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowekamp BC, Chen DT, Ibanez L, & Blezek D (2013). The Design of SimpleITK. Frontiers in Neuroinformatics, 7 10.3389/fninf.2013.00045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marstal K, Berendsen F, Staring M, & Klein S (2016). SimpleElastix: A User-Friendly, Multi-lingual Library for Medical Image Registration. In 2016 IEEE Conference on Computer Vision and Pattern Recognition Workshops (CVPRW) (pp. 574–582). 10.1109/CVPRW.2016.78 [DOI] [Google Scholar]
- McKinney W (2010). Data Structures for Statistical Computing in Python (pp. 51–56). Presented at the Proceedings of the 9th Python in Science Conference Retrieved from http://conference.scipy.org/proceedings/scipy2010/mckinney.html [Google Scholar]
- Meijering E (2012). Cell Segmentation: 50 Years Down the Road [Life Sciences]. IEEE Signal Processing Magazine, 29(5), 140–145. 10.1109/MSP.2012.2204190 [DOI] [Google Scholar]
- Meijering Erik, Carpenter AE, Peng H, Hamprecht FA, & Olivo-Marin J-C (2016). Imagining the future of bioimage analysis. Nature Biotechnology, 34(12), 1250–1255. 10.1038/nbt.3722 [DOI] [PubMed] [Google Scholar]
- Nazib A, Galloway J, Fookes C, & Perrin D (2018). Performance of Registration Tools on High-Resolution 3D Brain Images. In 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) (pp. 566–569). 10.1109/EMBC.2018.8512403 [DOI] [PubMed] [Google Scholar]
- Ng L, Pathak S, Kuan C, Lau C, Dong H, Sodt A, … Hawrylycz M (2007). Neuroinformatics for Genome-Wide 3-D Gene Expression Mapping in the Mouse Brain. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 4(3), 382–393. 10.1109/tcbb.2007.1035 [DOI] [PubMed] [Google Scholar]
- Niedworok CJ, Brown APY, Cardoso MJ, Osten P, Ourselin S, Modat M, & Margrie TW (2016). AMAP is a validated pipeline for registration and segmentation of high-resolution mouse brain data. Nature Communications, 7, 11879 10.1038/ncomms11879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallast N, Diedenhofen M, Blaschke S, Wieters F, Wiedermann D, Hoehn M, … Aswendt M (2019). Processing Pipeline for Atlas-Based Imaging Data Analysis of Structural and Functional Mouse Brain MRI (AIDAmri). Frontiers in Neuroinformatics, 13 10.3389/fninf.2019.00042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poinsatte K, Betz D, Torres VO, Ajay AD, Mirza S, Selvaraj UM, … Stowe AM (2019). Visualization and Quantification of Post-stroke Neural Connectivity and Neuroinflammation Using Serial Two-Photon Tomography in the Whole Mouse Brain. Frontiers in Neuroscience, 13 10.3389/fnins.2019.01055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez DMO, Ajay AD, Goldberg MP, & Meeks JP (2019). Serial Multiphoton Tomography and Analysis of Volumetric Images of the Mouse Brain In Hartveit E (Ed.), Multiphoton Microscopy (pp. 195–224). New York, NY: Springer; 10.1007/978-1-4939-9702-2_9 [DOI] [Google Scholar]
- Renier N, Adams EL, Kirst C, Wu Z, Azevedo R, Kohl J, … Tessier-Lavigne M (2016). Mapping of Brain Activity by Automated Volume Analysis of Immediate Early Genes. Cell, 165(7), 1789–1802. 10.1016/j.cell.2016.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas CBG, Lu TT-H, Gabery S, Marstal K, Alanentalo T, Mercer AJ, … Secher A (2018). Integrated Brain Atlas for Unbiased Mapping of Nervous System Effects Following Liraglutide Treatment. Scientific Reports, 8(1), 1–12. 10.1038/s41598-018-28496-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid B, Schindelin J, Cardona A, Longair M, & Heisenberg M (2010). A high-level 3D visualization API for Java and ImageJ. BMC Bioinformatics, 11, 274 10.1186/1471-2105-11-274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CL, Ng L, Menon V, Martinez S, Lee C-K, Glattfelder K, … Jones AR (2014). A High-Resolution Spatiotemporal Atlas of Gene Expression of the Developing Mouse Brain. Neuron, 83(2), 309–323. 10.1016/j.neuron.2014.05.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Walt Stefan, Colbert SC, & Varoquaux G (2011). The NumPy Array: A Structure for Efficient Numerical Computation. Computing in Science Engineering, 13(2), 22–30. 10.1109/MCSE.2011.37 [DOI] [Google Scholar]
- van der Walt Stéfan, Schönberger JL, Nunez-Iglesias J, Boulogne F, Warner JD, Yager N, … Yu T (2014). scikit-image: Image processing in Python. PeerJ, 2 10.7717/peerj.453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, … Contributors, S. 1 0. (2019). SciPy 1.0—Fundamental Algorithms for Scientific Computing in Python. Retrieved from https://arxiv.org/abs/1907.10121v1 [Google Scholar]
- Wang Q, Ding S-L, Li Y, Royall J, Feng D, Lesnar P, … Ng L (2020). The Allen Mouse Brain Common Coordinate Framework: A 3D Reference Atlas. Cell. 10.1016/j.cell.2020.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Lauritzen JS, Perlman E, Robinson CG, Nichols M, Milkie D, … Bock DD (2018). A Complete Electron Microscopy Volume of the Brain of Adult Drosophila melanogaster. Cell, 174(3), 730–743.e22. 10.1016/j.cell.2018.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]