Abstract
The clinical translation of messengerRNA (mRNA) drugs has been slowed by a shortage of delivery vehicles that potently and safely shuttle mRNA into target cells. Here, we describe the properties of a particularly potent branched-tail lipid nanoparticle that delivers mRNA to >80% of three major liver cell types. We characterize mRNA delivery spatially, temporally, and as a function of injection type. Following intravenous delivery, our lipid nanoparticle induced greater protein expression than two benchmark lipids, C12-200 and DLin-MC3-DMA, at an mRNA dose of 0.5 mg/kg. Lipid nanoparticles were sufficiently potent to codeliver three distinct mRNAs (firefly luciferase, mCherry, and erythropoietin) and, separately, Cas9 mRNA and single guide RNA (sgRNA) for proof-of-concept nonviral gene editing in mice. Furthermore, our branched-tail lipid nanoparticle was neither immunogenic nor toxic to the liver. Together, these results demonstrate the unique potential of this lipid material to improve the management of diseases rooted in liver dysfunction.
Keywords: mRNA delivery, lipidoid, lipid nanoparticles, gene editing, protein expression, liver delivery
Graphical Abstract
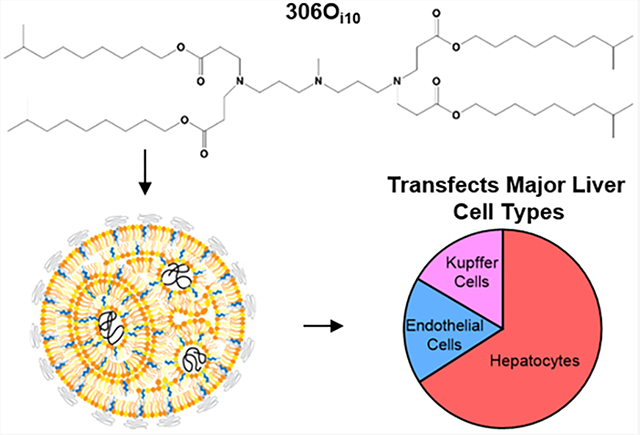
1. INTRODUCTION
In recent years, messengerRNA (mRNA) has emerged as an exciting strategy to express therapeutic proteins for vaccines, cancer immunotherapies, protein replacement therapies, and gene editing. Although there are dozens of ongoing clinical trials, many mRNA therapies that require in vivo delivery make use of the same or similar lipid nanoparticle delivery formulations. Given the considerable commercial interest in this space, the identification of additional potent and safe delivery materials is expected to accelerate clinical translation.
Many effective mRNA delivery vehicles discovered to date were identified by high-throughput screening of material libraries for positive “hits”.1–4 Although this strategy has been fruitful, its emphasis on potency end points can limit characterization of top materials. To advance the field of mRNA therapeutics, better understanding is needed of when, where, and how top delivery materials are effective in vivo. Additionally, thorough characterization of potent mRNA delivery materials, including assessment of toxicity and immunogenicity, will accelerate clinical translation.5
In this study, we characterize lipid nanoparticles (LNPs) formulated with the next-generation, branched-tail, ionizable lipid-like (lipidoid) material 306Oi10, which is one of the most promising mRNA delivery materials identified to date.1,4,6–10 When administered systemically, 306Oi10 LNPs outperformed two gold-standard lipids, codelivered multiple RNAs within the same formulation, and enabled CRISPR-mediated gene editing in mouse livers. Interestingly, 306Oi10 LNPs facilitated protein expression in all major cell types of the liver, unlike many hepatocyte-limited delivery systems.3,11,12 Finally, we show that intravenous delivery of 306Oi10 LNPs results in negligible increases in serum cytokine and IgG levels and no observable liver toxicity. Together, these results demonstrate that 306Oi10 is a versatile mRNA delivery material with high potency and low immunogenicity/toxicity, rendering it promising for future applications.
2. MATERIALS AND METHODS
2.1. Materials.
The amine 3,3′-diamino-N-methyldipropylamine (306), the tail 1,2-epoxyhexadecane (C12), cholesterol, and 2-(p-toluidinyl)naphthalene-6-sulfonic acid (TNS) were purchased from Sigma-Aldrich (St. Louis. MO). The amine 2[4-2(2-aminoethyl) amino)ethylpiperazine-1-YL)ethan-1-amine (200) was purchased from Enamine (Princeton, NJ). The tail isodecyl acrylate (Oi10) was purchased from Sartomer (Colombes, France). The lipid DLin-MC3-DMA was purchased from Biorbyt (San Francisco, CA). The phospholipid 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) and C14-PEG2000 were purchased from Avanti Polar Lipids (Alabaster, AL). All mRNAs were purchased from Trilink Biotechnologies (San Diego, CA) and included the 5-methoxyuridine (5moU) base modification. XenoLight D-Luciferin Potassium Salt was purchased from PerkinElmer (Waltham, MA). Alexa Fluor 488 Phalloidin, Hoeschst 33342, IL-6, and TNFα mouse ELISA kits were purchased from Thermo Fisher (Waltham, MA). Mouse IgG ELISA kit was purchased from Abcam (Cambridge, MA).
2.2. Lipidoid Synthesis.
Lipidoids were synthesized as previously described.13 Briefly, the amine 3,3′-diamino-N-methyldipropylamine (306) was reacted with the tail isodecyl acrylate (Oi10) at a molar ratio of 1:4 to form the lipidoid 306Oi10. The amine 2[4-2( 2-aminoethyl)amino)-ethylpiperazine-1-YL)ethan-1-amine (200) was reacted with the tail 1,2-epoxyhexadecane (C12) at a molar ratio of 1:5 to form the lipidoid C12–200. Amines and tails were combined in glass scintillation vials and stirred at 90 °C for 3 days without solvent. The lipidoids were purified using a Teledyne ISCO Chromatography system (Thousand Oaks, CA) to isolate the fully substituted lipidoid product. The structures of the final products are shown in Figure S1.
2.3. Formulation of LNPs.
The lipidoids 306Oi10 and C12-200 were formulated into LNPs as previously described,6,11 by combining lipidoid, DOPE, cholesterol, and C14-PEG2000 at a molar ratio of 35:16:46.5:2.5 in 90% (v/v) ethanol and 10% (v/v) 10 mM sodium citrate. mRNA was diluted in 10 mM sodium citrate buffer and mixed with an equal volume of lipid solution by vortexing. The final weight ratio of lipidoid:mRNA was 10:1. DLin-MC3-DMA was formulated into LNPs as previously described14 by combining ionizable lipid, cholesterol, DSPC, and DMG-PEG2000 at a molar ratio of 50:10:38.5:1.5 in 100% ethanol. mRNA was dissolved in 5 mM citric acid buffer. The lipid and mRNA solutions were combined in a microfluidic device (Precision Nanosystems) at a flow ratio of 1:3 (ethanol:aqueous phase) with a total flow rate of 12 mL/min. All LNPs were dialyzed against PBS for 60–90 min in 3500 g/mol molecular weight cutoff cassettes (Thermo Fisher). Lipid structures and N/P ratios for all formulations are shown in Figure S1.
2.4. LNP Characterization.
Particle size (number-average diameter) and zeta potential were determined using a Malvern Zetasizer Nano ZSP (Malvern, UK). mRNA entrapment was determined using the Quant-iT Ribogreen RNA assay (Thermo Fisher). LNP surface pKa was determined using a TNS assay.15
2.5. In Vivo Biodistribution Studies.
All animal experiments were conducted using institutionally approved protocols (IACUC). Female C57BL/6 mice (Charles River Laboratories, Wilmington, MA) received injections of 306Oi10 LNPs containing 0.5 mg/kg Cy5-labeled luciferase mRNA. Mice were injected by intravenous (tail vein), intraperitoneal, subcutaneous (scruff of the neck), and intramuscular (hind flanks) injections. Mice were sacrificed 1 h post-injection, and organs/tissue were harvested and imaged for Cy5 fluorescence using an IVIS imaging system (PerkinElmer) at a excitation/emission = 649/670 nm. Total radiant efficiency was determined using Living Image software (PerkinElmer).
2.6. in Vivo Luciferase mRNA Delivery.
Female C57BL/6 mice received intravenous, intraperitoneal, subcutaneous, or intramuscular injections of LNPs containing 0.5 mg/kg luciferase mRNA. Fifteen minutes prior to imaging, mice received an intraperitoneal injection of 130 μL 30 mg/mL d-luciferin. Organs/tissue were harvested and imaged for bioluminescence using the IVIS. For temporal studies, mice were anesthetized with isoflurane in oxygen, and ventral whole-body bioluminescence imaging was performed using the IVIS. Total luminescent flux was determined using Living Image software.
2.7. Confocal Microscopy.
Female C57BL/6 mice received a tail vein injection of either 306Oi10 LNPs carrying 0.5 mg/kg mCherry mRNA or naked mCherry mRNA. Mice were sacrificed 6 h post-injection, and the livers were harvested and fixed in 4% formaldehyde. Tissues were sectioned, stained with Alexa Fluor 488 Phalloidin and Hoeschst 33342, and imaged on a Zeiss LSM 700 confocal microscope (Oberkochen, Germany).
2.8. Spatiotemporal Analysis.
Female C57BL/6 mice received tail vein injections of 306Oi10 LNPs carrying 0.5 mg/ kg luciferase mRNA. Expression was analyzed 1, 3, 6, 12, 24, and 48 h post-injection (n = 3 mice/time point). At each time point, mice received an intraperitoneal injection of 130 μL 30 mg/mL D-luciferin. After 15 min, mice were sacrificed and major organs were harvested and imaged for bioluminescence using the IVIS. Total luminescent flux was determined using the Living Image software.
2.9. Multiplexed In Vivo mRNA Delivery.
Female C57BL/6 mice received tail vein injections of 306Oi10 LNPs carrying luciferase, mCherry, and erythropoietin mRNAs at a total dose of 1 mg/kg (0.33 mg/kg each mRNA). Six hours post-injection, mice received an intraperitoneal injection of 130 μL 30 mg/mL d-luciferin. Mice were sacrificed after 15 min, blood was drawn via cardiac puncture, and organs were harvested. Organs were imaged for mCherry fluorescence and bioluminescence using the IVIS. Serum was isolated by centrifuging blood samples in BD Microtainer Serum Separator Tubes at 14,000 rpm for 10 min. Serum erythropoietin concentrations were determined using the Human Erythropoietin Quantikine IVD ELISA Kit (R&D Systems, Minneapolis, MN).
2.10. In Vivo Gene Editing Studies.
Single guide RNAs (sgRNAs) targeting the LoxP locus (CGTATAGCATACATTATACG) were synthesized using the GeneArt Precision gRNA Synthesis Kit (Thermo Fisher). Female Ai9 mice (The Jackson Laboratory, Bar Harbor, ME) received tail vein injections of 306Oi10 LNPs carrying 1.6 mg/kg Cas9 mRNA + 0.4 mg/kg sgRNA. 72 h later, mice were sacrificed and organs were harvested and imaged for tdTomato fluorescence using the IVIS.
2.11. Flow Cytometry.
Female mT/mG mice (The Jackson Laboratory) received tail vein injections of 306Oi10 LNPs carrying 2.0 mg/kg Cre Recombinase mRNA. Twenty-four hours later, mice were sacrificed and the livers harvested in HBSS (Thermo Fisher). Livers were digested with a liver dissociation kit and a gentleMACS Octo Dissociator (Miltenyi Biotec). Red blood cells were lysed, and 2 × 106 cells were suspended in blocking buffer (HBSS + 1% FBS + 1:1000 Fc block; BioLegend) and stained with antibodies against F4/80 (Kupffer cells, BioLegend clone BM8), CD31 (endothelial cells, BioLegend clone 390), and ASGR1 (hepatocytes, ProteinTech) for 20 min at 4 °C. Cells were counterstained with 7AAD (Thermo Fisher Scientific) and analyzed by flow cytometry using a NovoCyte 3000 (ACEA Biosciences). Flow cytometry data were analyzed using NovoExpress software (ACEA Biosciences).
2.12. Immunogenicity and Histology Analysis.
Female C57BL/6 mice received tail vein injections of 306Oi10, C12-200, or DLin-MC3-DMA LNPs carrying 0.5 mg/kg luciferase mRNA. Blood was drawn via the submandibular vein at 2, 4, 6, 72, and 144 h post-injection and serum isolated. For TNFα and IL-6 analysis, serum was diluted 1:20. For IgG, serum was diluted 1:200,000. ELISAs were performed using the manufacturer’s instruction. For histology, mice were sacrificed 2 weeks after injection, and the livers were fixed overnight in 4% formaldehyde and transferred to 70% ethanol. Samples were embedded in paraffin, sectioned, and stained with hematoxylin and eosin.
3. RESULTS AND DISCUSSION
Given the potency of lipid nanoparticles (LNPs) formulated with the ionizable lipidoid 306Oi10 (Figure S1A), we sought a thorough understanding of their molecular properties and delivery capabilities. Our standard mRNA-optimized LNP formulation yielded an N/P ratio of 6.3 (Figure S1D); resultant nanoparticles were 124 nm by dynamic light scattering (Figure S2A). Using the TNS assay, we found the surface pKa of this LNP was 6.4 (Figure S2B), consistent with other efficacious LNPs.13,16 This material had a neutral zeta potential at physiological pH (0.43 mV) and an mRNA entrapment efficiency of 91% (Figure S2C).
Next, we compared the in vivo efficacy of 306Oi10 to two benchmark RNA delivery vehicles. The first, DLin-MC3-DMA (MC3, Figure S1B), is the ionizable lipid in the first LNP formulation to be FDA-approved for siRNA therapy17 and has been investigated extensively for mRNA delivery.14,18,19 The second material, C12-200 (Figure S1C), is another ionizable lipid commonly used for mRNA delivery.12,20 We formulated each lipid into LNPs carrying 0.5 mg/kg mLuc and injected them in mice via the tail vein. All three LNPs induced significant luciferase expression almost entirely in the liver (Figure 1A). However, 306Oi10 LNPs produced over 3-fold higher total organ expression than MC3 and over 20-fold higher expression than C12–200 (Figure 1B). The especially high potency of 306Oi10 is likely due to its ability to take on a strong positive charge under the conditions of the late endosome (pH 5).6
Figure 1.
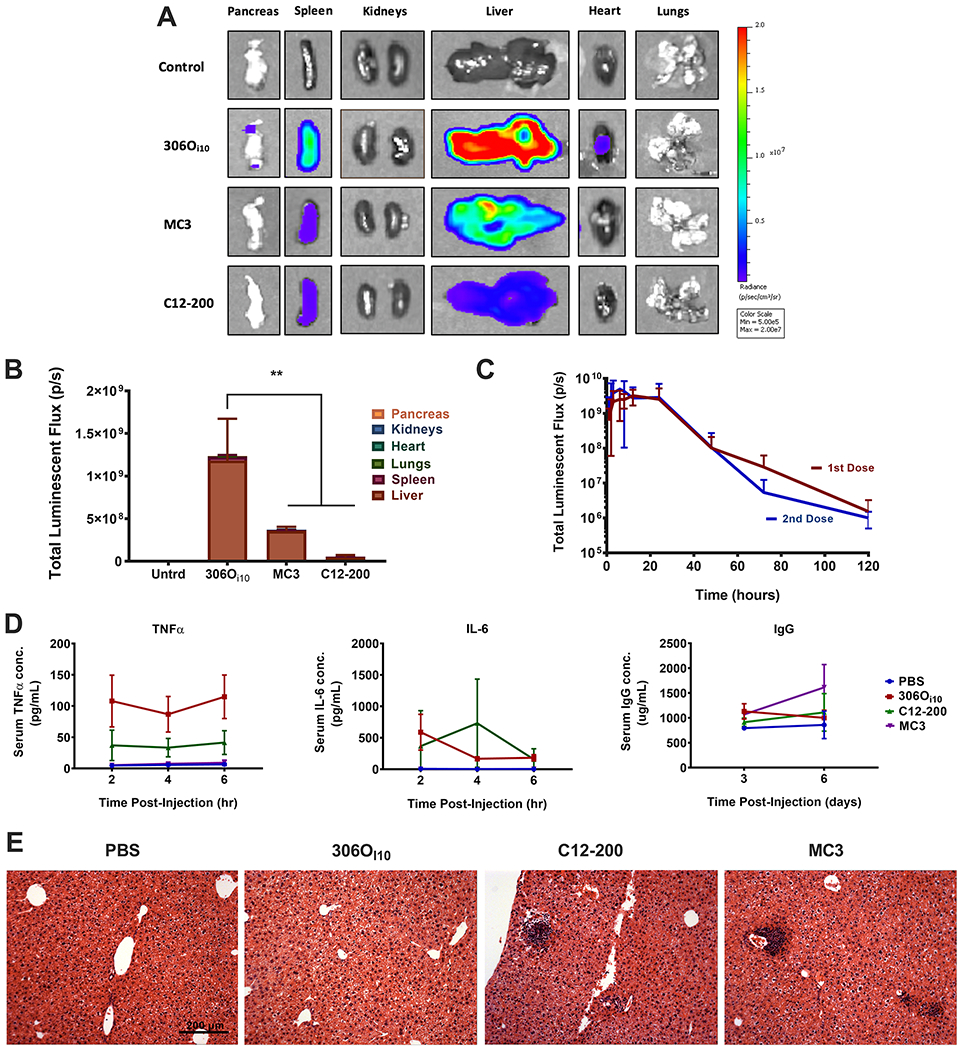
306Oi10 outperforms the benchmark ionizable lipids DLin-MC3-DMA (MC3) and C12-200. Mice were injected via tail vein with LNPs carrying 0.5 mg/kg firefly luciferase mRNA. (A) Organs were harvested and imaged for luminescence 6 h after injection. (B) Quantification of the total luciferase signal. (C) 306Oi10 LNPs did not lose potency upon repeat dosing one month after initial IV injection of a 0.5 mg/kg dose of mLuc. (D) Serum cytokine levels (TNFα and IL-6) obtained 3–6 h after injection and antibody titer (IgG) obtained 3–6 days after injection were measured by ELISA to assess acute inflammatory and humoral immune response. None of the lipids induced clinically significant elevations in signal. (E) Histological analysis (hematoxylin and eosin staining) of liver sections; scale bars = 200 μm. **p < 0.01, as determined by an unpaired Student’s t test. Error bars represent s.d. (n = 3).
Next, we assessed the performance of 306Oi10 upon repeat dosing to ensure that an adaptive immune response would not abrogate LNP efficacy over time. We injected mice with two doses of LNPs containing mLuc (0.5 mg/kg) spaced 30 days apart. After each dose, we tracked luciferase expression between 1 and 120 h using whole-body IVIS imaging. As shown in Figure 1C, luciferase expression was nearly identical after both doses, indicating that 306Oi10 LNPs maintained their efficacy upon repeat dosing. This finding suggests that antibodies are not formed in response to 306Oi10 LNPs, a phenomenon that has prevented the repeat dosing of other potent materials identified by our lab and others (data not shown).
We were further interested in comparing the safety of 306Oi10 with benchmark materials. To examine immunogenicity, we measured serum levels of the cytokines TNFα and IL-6 between 2-6 hours after administration and the antibody IgG 3-6 days after LNP administration (0.5 mg/kg). While TNFα and IL-6 increased slightly with 306Oi10 and C12-200 LNPs (Figure 1D), their values did not indicate an acute immune response.21 None of the three LNPs produced significant increases in serum IgG (Figure 1D).
We also performed histological analysis on mouse livers harvested 2 weeks after injection. In livers exposed to 306Oi10 LNPs, there were no signs of infiltrating immune cells, nor evidence of necrosis (Figure 1E). However, we observed dark clusters of cells, which can indicate necrosis and acute toxicity, in two of three samples treated with C12-200 and MC3. MC3 is the main component of the FDA-approved siRNA formulation, Patisiran (MC3), which is given clinically at substantially lower doses than described here. These results demonstrate that 306Oi10 LNPs are well-tolerated upon intravenous delivery at a dose of 0.5 mg/kg mRNA. The decreased immune infiltration of 306Oi10 relative to MC3 and C12-200 may be related to its degradability. 306Oi10 contains four ester groups that may confer biodegradability in vivo upon exposure to liver enzymes. In contrast, C12-200 contains no degradable groups, and MC3 contains only one. Enhanced degradation may improve biocompatibility and accelerate lipid clearance relative to MC3 and C12-200.22
Having confirmed that 306Oi10 LNPs compare favorably to “gold standards”, we investigated the route versatility of 306Oi10 for delivering mRNA. Specifically, we quantified biodistribution and protein expression for four routes of injection: intravenous (IV, tail vein), intraperitoneal (IP), subcutaneous (SC, scruff of neck), and intramuscular (IM, hind flank). These injection routes were chosen for their relevance to clinical payload delivery. IV administration is used to deliver molecules to the liver, spleen, and tumor sites, while IP injection provides access to organs within the peritoneal space.23 SC injection is relevant for small protein delivery (e.g., insulin),24 and IM injection is the most common route of vaccine administration.25 To assess biodistribution, mice were treated with 306Oi10 LNPs containing Cy5-labeled mRNA, while unlabeled mLuc was used to assess protein expression. Following treatment, organs were excised and imaged for Cy5 fluorescence (Figures 2A and S3) or luminescence (Figures 2B and S4), respectively. IV injection resulted in distribution (81%) and protein expression (96%) predominantly in the liver, while IP injection facilitated higher levels of LNP accumulation in the pancreas (11%), kidneys (12%), and lungs (15%) and protein expression in the liver (67%), pancreas (17%), and spleen (13%). SC and IM injections create drug depots that drain through capillaries and the lymphatic system.26,27 These injections distributed similarly, with half of the fluorescent signal remaining at the injection site. SC injections produced expression exclusively at the injection site (99%), while IM injections yielded expression at the injection site (68%), liver (12%), and kidneys (11%). Notably, the fractional liver expression resulting from SC and IM injections was less than the fractional distribution to the liver. This was in contrast to IV delivery, in which the fractional liver expression was higher than fractional liver distribution. These differences may be due to the relative ease of LNP extravasation (IV) compared to the exit from the lymphatic system (SC/IM). In other words, the larger pore size of the blood vessels compared to lymphatic vessels allows enhanced accumulation in and transfection of liver cells.
Figure 2.
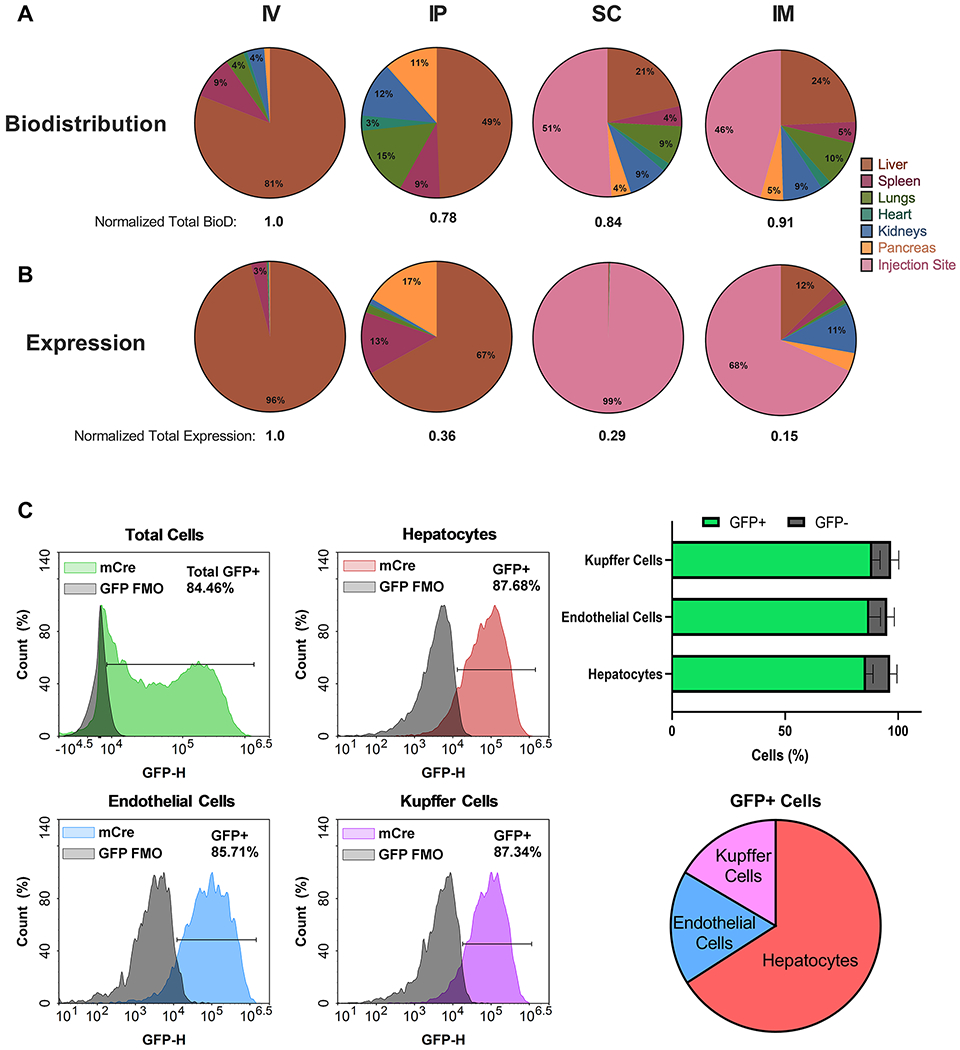
306Oi10 LNP biodistribution and protein expression varied with injection type, producing maximal expression in liver cells. (A) Biodistribution was determined 1 h postinjection of 306Oi10 LNPs carrying Cy5-labeled mRNA at a dose of 0.5 mg/kg. (B) Luciferase expression was determined 6 h post-injection of 306Oi10 LNPs carrying 0.5 mg/kg mLuc. (C) Flow cytometric liver analysis shows the cellular distribution of protein expression 24 h post-injection of 306Oi10 LNPs. Approximately 85% of all three cell types examined were transfected. Error bars represent s.d. (n = 3).
Because the liver is the predominant site of protein expression, we determined which liver cell types were transfected with 306Oi10 LNPs. We injected mT/mG mice, which express GFP upon Cre-mediated recombination, intravenously with 306Oi10 LNPs carrying Cre mRNA (2.0 mg/kg). After 24 h, mice were sacrificed and liver cells were isolated and analyzed by flow cytometry to identify GFP+ hepatocytes, Kupffer cells, and endothelial cells (Figure 2C). 306Oi10 LNPs transfected hepatocytes, Kupffer cells, and endothelial cells with 86–88% efficiency. This contrasts with other reported lipid nanoparticles. For example, one LNP transfected 100% of Kupffer cells, but only 20% of hepatocytes.28 Further, cKK-E12 LNPs robustly deliver siRNA to hepatocytes but not to liver endothelial cells or immune cells.29 Consequently, most mRNA therapies have been evaluated for diseases rooted in hepatocyte dysfunction, including hemophilia,12,18 urea cycle disorder,30,31 and glycogen storage disorders.32 Because 306Oi10 transfects the major liver cell types with equal potency, its potential therapeutic applications extend to liver diseases that affect multiple cell types, such as hepatitis, fatty liver disease, cancer, and inherited diseases.33–36 For example, an mRNA therapy for hepatocellular carcinoma could target malignant hepatocytes, endothelial cells, and Kupffer cells simultaneously.
A key aspect of mRNA delivery is the rate at which translation occurs in different organs. Although 306Oi10-mediated mRNA delivery predominantly occurs in the liver, there was also detectable expression in the spleen, pancreas, kidneys, heart, and lungs. To perform a spatiotemporal analysis of protein expression, per-organ luminescence was quantified at 1, 3, 6, 12, 24, and 48 h post-injection with 306Oi10 LNPs by IVIS imaging. Luciferase expression was observed within the first hour, and high levels of expression were maintained for at least 48 h (Figure 3A,B).
Figure 3.
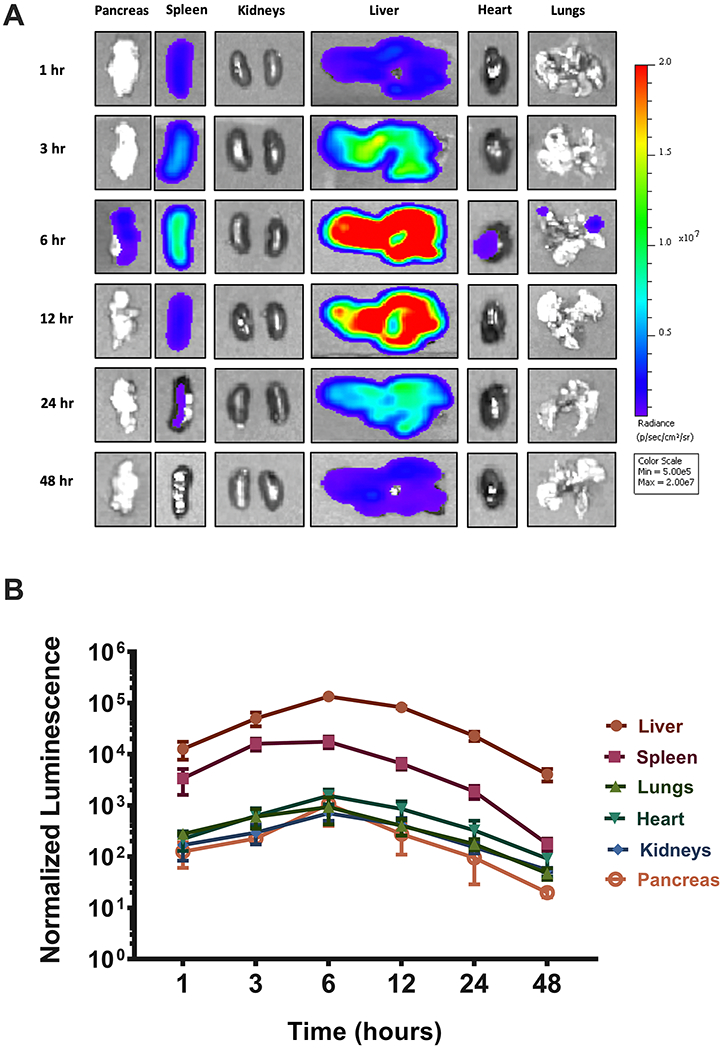
LNP-induced protein expression kinetics did not vary by organ. Mice were injected via tail vein with 306Oi10 LNPs carrying 0.5 mg/kg mLuc. (A) Mice were sacrificed, and organs were harvested and imaged by IVIS for luminescence at six times between 1 and 48 h. (B) Quantification of luminescence as a function of time and organ. Normalized luminescence was calculated by dividing the signal in each organ to the luminescence values in that organ for untreated mice. Error bars represent s.d. (n = 3).
Interestingly, we found that expression peaked in all organs at 6 h post-injection. Therefore, despite the diversity of cells being transfected, LNPs reach distinct tissues and release their cargo within cells at similar rates. This was surprising, given that different cell types exert translational control using tissue-dependent mechanisms.37 This may occur due to the lack of complex structural elements within the mLuc sequence.
Given the excellent in vivo efficacy of 306Oi10, we asked if it was possible to deliver multiple mRNAs within the same LNP. To achieve this, a single LNP formulation must condense and encapsulate mRNAs of considerably different sizes. We formulated 306Oi10 LNPs with mRNAs encoding firefly luciferase (1921 nucleotides), mCherry (996 nucleotides), and erythropoietin (EPO, 858 nucleotides). LNPs were delivered to mice at a total mRNA dose of 1 mg/kg (0.33 mg/kg of each mRNA). We sacrificed the mice 6 h after injection and harvested the major organs and blood. Both luciferase and mCherry expression were detected in the liver and spleen (Figures 4AB and S5). EPO expression was detected in serum (Figure 4C). To the best of our knowledge, this represents the first example of nonviral delivery of three functionally distinct mRNAs within the same formulation in vivo.
Figure 4.
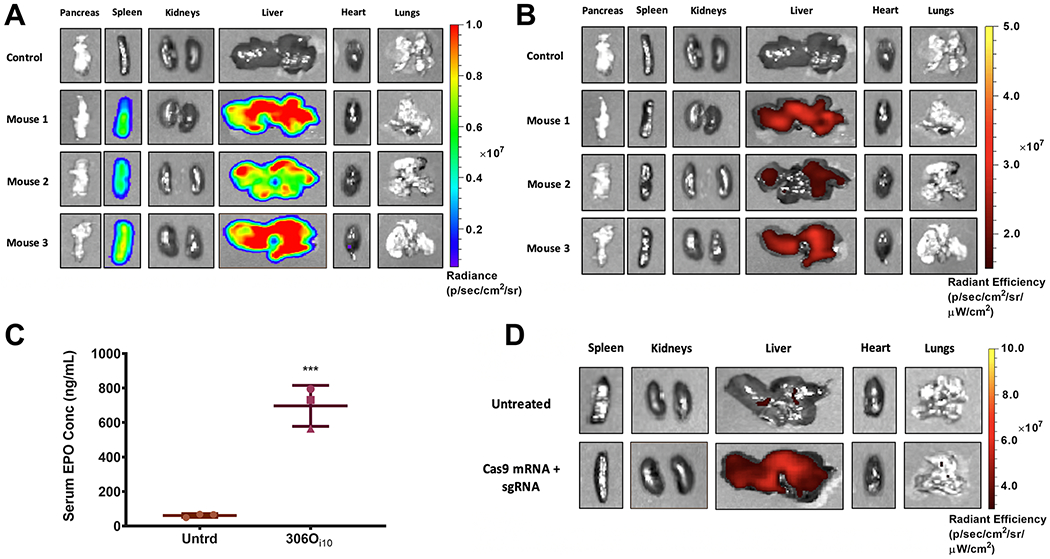
306Oi10 LNPs enabled potent codelivery of three mRNAs and gene editing in the liver. Mice were injected with LNPs containing 1 mg/ mL total mRNA (0.33 mg/mL each of firefly luciferase, mCherry, and erythropoietin (EPO) mRNA). (A) Luciferase and (B) mCherry expression were observed in the liver 6 h after injection, and (C) serum EPO levels reached >10-fold baseline levels in mice. The expression of all three proteins was consistent for each mouse. For example, mouse 2 (pink triangle) had the lowest expression for all three proteins. (D) Ai9 mice received 306Oi10 LNPs carrying 1.6 mg/kg Cas9 mRNA + 0.4 mg/kg sgRNA targeting LoxP. The successful codelivery of both RNA components resulted in expression of tdTomato fluorescence in the liver, indicating that nonviral gene editing had occurred (n = 3). Mice were sacrificed and organs were imaged for tdTomato fluorescence 72 h after IV injection. ***p < 0.001, as determined by an unpaired Student’s t test. Error bars represent s.d. (n = 3).
The delivery of multiple mRNAs within the same formulation has potential for a variety of therapeutic applications. For example, simultaneously upregulating vascular endothelial growth factor (VEGF), fibroblast growth factor (FGF), and hepatocyte growth factor (HGF) can reverse disease symptoms in a mouse myocardial infarction model.38 Additionally, the tumor suppressor genes GNMT, CEL, and SERPINI2 are downregulated in patients with pancreatic cancer, so upregulating them with mRNA may slow pancreatic cancer progression.39 Further, delivering multiple mRNAs can direct cell differentiation or reprogram somatic cells into induced pluripotent stem cells.40
Gene editing has recently emerged as an exciting strategy to treat genetic diseases. In a proof-of-concept experiment, we codelivered mRNA encoding the Cas9 nuclease (mCas9) and single guide RNAs (sgRNAs) for CRISPR-mediated gene editing. We used Ai9 mice, which ubiquitously express a cassette containing a floxed STOP codon preventing the expression of tdTomato (LoxP-Stop-LoxP-tdTomato).41 Codelivery of Cas9 mRNA and sgRNA targeting the LoxP sites (sgLoxP) to Ai9 mice results in deletion of the stop cassette and expression of tdTomato. This is a challenging in vivo gene editing model, as two cuts must be made on the same allele for tdTomato expression to occur.
For these experiments, we injected Ai9 mice with 306Oi10 LNPs carrying 1.6 mg/kg mCas9 + 0.4 mg/kg sgLoxP. Seventy-two hours later, we sacrificed the mice, harvested the major organs, and imaged them for tdTomato fluorescence. Co-delivery of mCas9 and sgLoxP produced significant tdTomato expression in the liver, indicating that gene editing had occurred (Figure 4D). These proof-of-concept data show that 306Oi10 LNPs are one of few delivery systems capable of nonviral gene editing in vivo.7,42
CONCLUSIONS
The ionizable lipidoid 306Oi10 is a potent and versatile nonviral vector for in vivo mRNA delivery. Unlike most other established lipid nanoparticles, this material facilitates the simultaneous transfection of hepatocytes, Kupffer cells, and endothelial cells in the liver. Spatiotemporal analysis revealed that 306Oi10 induces similar protein expression kinetics in all organs. Furthermore, we demonstrated for the first time that an ionizable lipid nanoparticle can encapsulate and deliver a cocktail of functionally distinct mRNAs within the same formulation, potentially enabling the treatment of diseases caused by multiple faulty proteins. The lipidoid 306Oi10 is also one of few nonviral materials capable of in vivo gene editing. Together, these results suggest that 306Oi10 is particularly promising for the nontoxic, non-immunogenic, and potent delivery of mRNA for therapeutic applications.
Supplementary Material
Acknowledgments
Funding
Funding was provided by the Defense Advanced Research Projects Agency (DARPA) (Grant No. D16AP00143) and The Shurl and Kay Curci Foundation. J.R.M. was supported by an NIH F32 fellowship (award number 1F32EB029345). The authors thank K. Fein for her feedback on the manuscript.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.nanolett.0c00596.
Structures for all lipidoids used for this work, characterization of 306Oi10 LNPs, additional IVIS images representing the quantification shown for biodistribution and efficacy studies of 306Oi10 LNPs delivered by various routes, and confocal microscopy showing mCherry expression in mouse liver cells (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.nanolett.0c00596.
The authors declare the following competing financial interest(s): K.A.W. is an inventor on U.S. Patents 9,439,968 and 9,227,917, which are related to the material described here.
Contributor Information
Khalid A. Hajj, Department of Chemical Engineering, Carnegie Mellon University, Pittsburgh, Pennsylvania 15213, United States.
Jilian R. Melamed, Department of Chemical Engineering, Carnegie Mellon University, Pittsburgh, Pennsylvania 15213, United States.
Namit Chaudhary, Department of Chemical Engineering, Carnegie Mellon University, Pittsburgh, Pennsylvania 15213, United States.
Nicholas G. Lamson, Department of Chemical Engineering, Carnegie Mellon University, Pittsburgh, Pennsylvania 15213, United States
Rebecca L. Ball, Department of Chemical Engineering, Carnegie Mellon University, Pittsburgh, Pennsylvania 15213, United States
Saigopalakrishna S. Yerneni, Department of Biomedical Engineering, Carnegie Mellon University, Pittsburgh, Pennsylvania 15213, United States
Kathryn A. Whitehead, Department of Chemical Engineering and Department of Biomedical Engineering, Carnegie Mellon University, Pittsburgh, Pennsylvania 15213, United States.
REFERENCES
- (1).Li B; Luo X; Deng B; Wang J; McComb DW; Shi Y; Gaensler KML; Tan X; Dunn AL; Kerlin BA; et al. An Orthogonal Array Optimization of Lipid-Like Nanoparticles for mRNA Delivery in Vivo. Nano Lett. 2015, 15, 8099–8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Dong Y; Dorkin JR; Wang W; Chang PH; Webber MJ; Tang BC; Yang J; Abutbul-Ionita I; Danino D; DeRosa F; et al. Poly(Glycoamidoamine) Brushes Formulated Nanomaterials for Systemic siRNA and mRNA Delivery in Vivo. Nano Lett. 2016, 16, 842–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Fenton OS; Kauffman KJ; McClellan RL; Appel EA; Dorkin JR; Tibbitt MW; Heartlein MW; DeRosa F; Langer R; Anderson DG Bioinspired Alkenyl Amino Alcohol Ionizable Lipid Materials for Highly Potent in Vivo mRNA Delivery. Adv. Mater 2016, 28, 2939–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Jarzębińska A; Pasewald T; Lambrecht J; Mykhaylyk O; Kümmerling L; Beck P; Hasenpusch G; Rudolph C; Plank C; Dohmen C A Single Methylene Group in Oligoalkylamine-Based Cationic Polymers and Lipids Promotes Enhanced mRNA Delivery. Angew. Chem., Int. Ed 2016, 55, 9591–9595. [DOI] [PubMed] [Google Scholar]
- (5).Ilinskaya AN; Dobrovolskaia MA Understanding the Immunogenicity and Antigenicity of Nanomaterials: Past, Present and Future. Toxicol. Appl. Pharmacol 2016, 299, 70–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Hajj KA; Ball RL; Deluty SB; Singh SR; Strelkova D; Knapp CM; Whitehead KA Branched-Tail Lipid Nanoparticles Potently Deliver mRNA in Vivo Due to Enhanced Ionization at Endosomal pH. Small 2019, 15, 1805097. [DOI] [PubMed] [Google Scholar]
- (7).Miller JB; Zhang S; Kos P; Xiong H; Zhou K; Perelman SS; Zhu H; Siegwart DJ Non-Viral CRISPR/Cas Gene Editing in Vitro and in Vivo Enabled by Synthetic Nanoparticle Co-Delivery of Cas9 mRNA and sgRNA. Angew. Chem., Int. Ed 2017, 56, 1059–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Kaczmarek JC; Patel AK; Kauffman KJ; Fenton OS; Webber MJ; Heartlein MW; DeRosa F; Anderson DG Polymer-Lipid Nanoparticles for Systemic Delivery of mRNA to the Lungs. Angew. Chem 2016, 128, 14012–14016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Li J; He Y; Wang W; Wu C; Hong C; Hammond PT Polyamine-Mediated Stoichiometric Assembly of Ribonucleoproteins for Enhanced mRNA Delivery. Angew. Chem 2017, 129, 13897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).McKinlay CJ; Vargas JR; Blake TR; Hardy JW; Kanada M; Contag CH; Wender PA; Waymouth RM Charge-Altering Releasable Transporters (CARTs) for the Delivery and Release of mRNA in Living Animals. Proc. Natl. Acad. Sci. U. S. A 2017, 114, E448–E456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Kauffman KJ; Dorkin JR; Yang JH; Heartlein MW; DeRosa F; Mir FF; Fenton OS; Anderson DG Optimization of Lipid Nanoparticle Formulations for mRNA Delivery in Vivo with Fractional Factorial and Definitive Screening Designs. Nano Lett. 2015, 15, 7300–7306. [DOI] [PubMed] [Google Scholar]
- (12).DeRosa F; Guild B; Karve S; Smith L; Love K; Dorkin JR; Kauffman KJ; Zhang J; Yahalom B; Anderson DG; et al. Therapeutic Efficacy in a Hemophilia B Model Using a Biosynthetic mRNA Liver Depot System. Gene Ther. 2016, 23, 699–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Whitehead KA; Dorkin JR; Vegas AJ; Chang PH; Veiseh O; Matthews J; Fenton OS; Zhang Y; Olejnik KT; Yesilyurt V; et al. Degradable Lipid Nanoparticles with Predictable in Vivo siRNA Delivery Activity. Nat. Commun 2014, 5, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Veiga N; Goldsmith M; Granot Y; Rosenblum D; Dammes N; Kedmi R; Ramishetti S; Peer D Cell Specific Delivery of Modified mRNA Expressing Therapeutic Proteins to Leukocytes. Nat. Commun 2018, 9, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Zhang J; Fan H; Levorse DA; Crocker LS Ionization Behavior of Amino Lipids for siRNA Delivery: Determination of Ionization Constants, SAR, and the Impact of Lipid pKa on Cationic Lipid–Biomembrane Interactions. Langmuir 2011, 27, 1907–1914. [DOI] [PubMed] [Google Scholar]
- (16).Jayaraman M; Ansell SM; Mui BL; Tam YK; Chen J; Du X; Butler D; Eltepu L; Matsuda S; Narayanannair Jayaprakash K; et al. Maximizing the Potency of siRNA Lipid Nanoparticles for Hepatic Gene Silencing in Vivo. Angew. Chem., Int. Ed 2012, 51, 8529–8533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Sheridan C With Alnylam’s Amyloidosis Success, RNAi Approval Hopes Soar. Nat. Biotechnol 2017, 35, 995–997. [DOI] [PubMed] [Google Scholar]
- (18).Ramaswamy S; Tonnu N; Tachikawa K; Limphong P; Vega JB; Karmali PP; Chivukula P; Verma IM Systemic Delivery of Factor IX Messenger RNA for Protein Replacement Therapy. Proc. Natl. Acad. Sci. U. S. A 2017, 114, E1941–E1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Yanez Arteta M; Kjellman T; Bartesaghi S; Wallin S; Wu X; Kvist AJ; Dabkowska A; Székely N; Radulescu A; Bergenholtz J; et al. Successful Reprogramming of Cellular Protein Production Through mRNA Delivered by Functionalized Lipid Nanoparticles. Proc. Natl. Acad. Sci. U. S. A 2018, 115, E3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Yin H; Song C-Q; Dorkin JR; Zhu LJ; Li Y; Wu Q; Park A; Yang J; Suresh S; Bizhanova A; et al. Therapeutic Genome Editing by Combined Viral and Non-Viral Delivery of CRISPR System Components in Vivo. Nat. Biotechnol 2016, 34, 328–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Copeland S; Warren HS; Lowry SF; Calvano SE; Remick D the Inflammation and the Host Response to Injury Investigators. Acute Inflammatory Response to Endotoxin in Mice and Humans. Clin. Diagn. Lab. Immunol 2005, 12, 60–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Maier MA; Jayaraman M; Matsuda S; Liu J; Barros S; Querbes W; Tam YK; Ansell SM; Kumar V; Qin J; et al. Biodegradable Lipids Enabling Rapidly Eliminated Lipid Nanoparticles for Systemic Delivery of RNAi Therapeutics. Mol. Ther 2013, 21, 1570–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Goodman M; McPartlan S; Detelich D; Saif M Chemotherapy for Intraperitoneal Use: a Review of Hyperthermic Intraperitoneal Chemotherapy and Early Post-Operative Intraperitoneal Chemotherapy. J. Gastrointest. Oncol 2016, 7 (1), 45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Gradel AKJ; Porsgaard T; Lykkesfeldt J; Seested T; Gram-Nielsen S; Kristensen NR; Refsgaard HHF Review Article Factors Affecting the Absorption of Subcutaneously Administered Insulin: Effect on Variability. J. Diabetes Res 2018, 2018, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Richner JM; Himansu S; Dowd KA; Butler SL; Salazar V; Fox JM; Julander JG; Tang WW; Shresta S; Pierson TC; et al. Modified mRNA Vaccines Protect Against Zika Virus Infection. Cell 2017, 168, 1114–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Turner P; Brabb T; Pekow C; Vasbinder MA Administration of Substances to Laboratory Animals: Routes of Administration and Factors to Consider. Journal of the American Association for Laboratory Animal Science 2011, 50, 600–613. [PMC free article] [PubMed] [Google Scholar]
- (27).Richter WF; Bhansali SG; Morris ME Mechanistic Determinants of Biotherapeutics Absorption Following SC Administration. AAPS J. 2012, 14, 559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Paunovska K; Loughrey D; Sago CD; Langer R; Dahlman JE Using Large Datasets to Understand Nanotechnology. Adv. Mater 2019, 31, 1902798–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Dong Y; Love KT; Dorkin JR; Sirirungruang S; Zhang Y; Chen D; Bogorad RL; Yin H; Chen Y; Vegas AJ; et al. Lipopeptide Nanoparticles for Potent and Selective siRNA Delivery in Rodents and Nonhuman Primates. Proc. Natl. Acad. Sci. U. S. A 2014, 111, 3955–3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Prieve MG; Harvie P; Monahan SD; Roy D; Li AG; Blevins TL; Paschal AE; Waldheim M; Bell EC; Galperin A; et al. Targeted mRNA Therapy for Ornithine Transcarbamylase Deficiency. Mol. Ther 2018, 26, 801–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Heartlein M; DeRosa F; Smith L mRNA Therapy for Argininosuccinate Synthetase Deficiency. WO 20150110859, April 23, 2015. [Google Scholar]
- (32).Roseman DS; Khan T; Rajas F; Jun LS; Asrani KH; Isaacs C; Farelli JD; Subramanian RR G6PC mRNA Therapy Positively Regulates Fasting Blood Glucose and Decreases Liver Abnormalities in a Mouse Model of Glycogen Storage Disease 1a. Mol. Ther 2018, 26, 814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Baiocchini A; Del Nonno F; Taibi C; Visco-Comandini U; D’Offizi G; Piacentini M; Falasca L Liver Sinusoidal Endothelial Cells (LSECs) Modifications in Patients with Chronic Hepatitis C. Sci. Rep 2019, 9 (8760), 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Ni Y; Li J-M; Liu M-K; Zhang T-T; Wang D-P; Zhou W-H; Hu L-Z; Lv W-L Pathological Process of Liver Sinusoidal Endothelial Cells in Liver Disease. World J. Gastroenterol 2017, 23, 7666–7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Feng T; Yu H; Xia Q; Ma Y; Yin H; Shen Y; Liu X Cross-Talk Mechanism Between Endothelial Cells and Hepatocellular Carcinoma Cells via Growth Factors and Integrin Pathway Promotes Tumor Angiogenesis and Cell Migration. Oncotarget 2017, 8, 69577–69593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Dixon LJ; Barnes M; Tang H; Pritchard MT; Nagy LE Kupffer Cells in the Liver. Comprehensive Physiology 2013, 3, 785–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Dimas A; Deutsch S; Stranger B; Montgomery S; Borel C; Attar-Cohen H; Ingle C; Beazley C; Antonarakis S Common Regulatory Variation Impacts Gene Expression in a Cell Type-Dependent Manner. Science 2009, 325, 1246–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Scimia MC; Gumpert AM; Koch WJ Cardiovascular Gene Therapy for Myocardial Infarction. Expert Opin. Biol. Ther 2014, 14, 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Goonesekere NCW Identification of Genes Highly Downregulated in Pancreatic Cancer Through a Meta-Analysis of Microarray Datasets: Implications for Discovery of Novel Tumor-Suppressor Genes and Therapeutic Targets. J. Cancer Res. Clin. Oncol 2018, 144 (2), 309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Warren L; Manos PD; Ahfeldt T; Loh Y-H; Li H; Lau F; Ebina W; Mandal PK; Smith ZD; Meissner A; et al. Highly Efficient Reprogramming to Pluripotency and Directed Differentiation of Human Cells with Synthetic Modified mRNA. Cell stem cell 2010, 7, 618–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Madisen L; Zwingman TA; Sunkin SM; Oh SW; Zariwala HA; Gu H; Ng LL; Palmiter RD; Hawrylycz MJ; Jones AR; et al. A Robust and High-Throughput Cre Reporting and Characterization System for the Whole Mouse Brain. Nat. Neurosci 2010, 13, 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Jiang C; Mei M; Li B; Zhu X; Zu W; Tian Y; Wang Q; qiann; Guo Y; Dong Y; et al. A Non-Viral CRISPR/Cas9 Delivery System for Therapeutically Targeting HBV DNA and Pcsk9 in vivo. Cell Res. 2017, 27, 440–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


