Abstract
The outbreak of the novel corona virus disease (COVID-19) in December 2019 has led to global crisis around the world. The disease was declared pandemic by World Health Organization (WHO) on 11th of March 2020. Currently, the outbreak has affected more than 200 countries with more than 37 million confirmed cases and more than 1 million death tolls as of 10 October 2020. Reverse-transcription polymerase chain reaction (RT-PCR) is the standard method for detection of COVID-19 disease, but it has many challenges such as false positives, low sensitivity, expensive, and requires experts to conduct the test. As the number of cases continue to grow, there is a high need for developing a rapid screening method that is accurate, fast, and cheap. Chest X-ray (CXR) scan images can be considered as an alternative or a confirmatory approach as they are fast to obtain and easily accessible. Though the literature reports a number of approaches to classify CXR images and detect the COVID-19 infections, the majority of these approaches can only recognize two classes (e.g., COVID-19 vs. normal). However, there is a need for well-developed models that can classify a wider range of CXR images belonging to the COVID-19 class itself such as the bacterial pneumonia, the non-COVID-19 viral pneumonia, and the normal CXR scans. The current work proposes the use of a deep learning approach based on pretrained AlexNet model for the classification of COVID-19, non-COVID-19 viral pneumonia, bacterial pneumonia, and normal CXR scans obtained from different public databases. The model was trained to perform two-way classification (i.e., COVID-19 vs. normal, bacterial pneumonia vs. normal, non-COVID-19 viral pneumonia vs. normal, and COVID-19 vs. bacterial pneumonia), three-way classification (i.e., COVID-19 vs. bacterial pneumonia vs. normal), and four-way classification (i.e., COVID-19 vs. bacterial pneumonia vs. non-COVID-19 viral pneumonia vs. normal). For non-COVID-19 viral pneumonia and normal (healthy) CXR images, the proposed model achieved 94.43% accuracy, 98.19% sensitivity, and 95.78% specificity. For bacterial pneumonia and normal CXR images, the model achieved 91.43% accuracy, 91.94% sensitivity, and 100% specificity. For COVID-19 pneumonia and normal CXR images, the model achieved 99.16% accuracy, 97.44% sensitivity, and 100% specificity. For classification CXR images of COVID-19 pneumonia and non-COVID-19 viral pneumonia, the model achieved 99.62% accuracy, 90.63% sensitivity, and 99.89% specificity. For the three-way classification, the model achieved 94.00% accuracy, 91.30% sensitivity, and 84.78%. Finally, for the four-way classification, the model achieved an accuracy of 93.42%, sensitivity of 89.18%, and specificity of 98.92%.
Keywords: COVID-19, Non-COVID-19 viral pneumonia, Bacterial pneumonia, AlexNet, Chest X-rays images (CXR)
Introduction
Pneumonia is a common disease caused by different microbial species such as bacteria, virus, and fungi. The word “Pneumonia” comes from the Greek word “Pneumon” which translates to the lungs. Thus, the word pneumonia is associated with lung disease. In medical terms, pneumonia is a disease that causes inflammation of either one or both lung parenchyma [1]. However, other causes of pneumonia include food aspiration and exposure to chemicals. Based on infection, pneumonia occurs as a result of inflammation caused by pathogens which lead the lung’s alveoli to fill up with fluid or pus and thereby leading to decrease of carbon dioxide (CO2) and oxygen (O2) exchange between blood and the lungs, making it hard for infected persons to breathe. Some of the symptoms of pneumonia include the shortness of breath, fever, cough, chest pain, etc. Moreover, the people at risk of pneumonia are elderly people (above 65 years), children (below the age of 5 years), and people with other complications such as HIV/AIDS, diabetes, chronic respiratory diseases, cardiovascular diseases, cancer, hepatic disease, etc. [2–5]. Table 1 presents a classification of pathogens that causes pneumonia.
Table 1.
Classification of pneumonia based on pathogens
| Pathogen | Specie |
|---|---|
| Bacterial | Streptococcus pneumoniae |
| Legionella pneumophila | |
| Mycoplasma pneumoniae | |
| Chlamydophila pneumoniae | |
| Viruses | Influenza virus |
| Severe acute respiratory syndrome coronavirus (SAR-CoV-1 and 2) | |
| Middle East Respiratory Syndrome (MERS) Coronavirus | |
| Respiratory Syncytial virus (RSV) | |
| Adenovirus | |
| Hantavirus | |
| Rhinovirus | |
| Varicella-zoster virus | |
| Human metapneumovirus | |
| Enteroviruses | |
| Fungi | Pneumocystis jirovecii |
| Aspergillus spp. | |
| Mucoromycetes | |
| Histoplasmosis | |
| Coccidioidomycosis | |
| Cryptococcus |
Diagnosis and Treatment of Pneumonia
There are different approaches for the diagnosis of pneumonia, some of these approaches include chest X-rays and CT scan (which form the basis of our contribution), sputum test, pulse oximetry, thoracentesis, blood gas analysis, bronchoscopy, pleural fluid culture, complete blood count, etc. Mostly, pneumonia infection is treated based on the causative pathogen. For bacterial pneumonia, antibiotics are used; for viral pneumonia such as influenzas, SARS and MERS, antiviral drugs are used while antifungal drugs are used for fungal pneumonia [6–8].
COVID-19 and Pneumonia
COVID-19 is an extremely contagious disease caused by severe acute respiratory syndrome coronavirus 2 (SAR-CoV-2) which is the recent disease that is caused by one of the family members of Coronaviridae family. In the past, two members of this family known as SARS-CoV-1 and Middle East Respiratory Syndrome Coronavirus (MERS-CoV) have caused a global epidemic. The first case of COVID-19 was reported in Wuhan, Hubei province of mainland China on the 31st December 2019. The virus spread from city to city and from one country to another leading to a global health crisis. However, it was not until March 11, 2020 that WHO declared it as pandemic [8–10].
COVID-19 can be transmitted through respiratory droplets that are exhaled or secreted by infected persons. Coronaviruses invade the lung’s alveoli (an organ responsible for exchange of O2 and CO2), thereby causing pneumonia. The symptoms of COVID-19 include dry cough, fatigue, fever, septic shock, organ failure, anorexia, dyspnea, myalgias, sputum secretion severe pneumonia, acute respiratory distress syndrome (ARDS), etc. [11–14]. The pandemic caused by SAR-CoV-2 is alarming due to the fact that there is no approved drug or vaccine [15].
In order to curb further spread of the virus, governments of various countries and states impose city lockdowns [56, 57], flight cancellations, border restrictions, closure of workplaces, restaurants, postponement of sport, religious, cultural, and entertainment event and activities, wearing of face mask, social distancing of 1–2 m, and creating awareness on hygiene. Many countries are facing challenges regarding the number of reported cases of COVID-19 as a result of the lack of RT-PCR test kit and delay in the test kit. This delay is detrimental as it leads to more cases due to interaction between the infected patients waiting for result with a healthy population [16, 17].
SARS-CoV1 and MERS-CoV
The first pandemic of the coronavirus family started in China’s South Eastern part and Hong Kong in the year 2002 due to the discovery of pathogenic virus known as SARS-CoV-1. The disease spread globally due to air travel of patients from one country to another leading to SARS pandemic. According to WHO 2004, more than 8000 confirmed cases of SARS-CoV and 774 associated deaths were recorded globally. The outbreak has caused both public and economic burdens with estimated losses of billions of dollars [18]. The second pandemic of coronavirus is caused by β-CoVs in 2012 in Saudi Arabia; unlike SARS-CoV, MERS is caused by MERS-CoV which was firstly identified in a patient sputum sample of a 60-year-old Saudi Arabian man. According to WHO reports, there were more than 1850 confirmed cases of MERS-CoV with 30% mortality rate [19]. And up until November 2019, there are closed to 2500 cases of MERS-CoV resulting in 858 deaths.
Identification of Drugs, Development of Vaccines, and Prediction of COVID-19 using AI-Driven Models
Currently, there is no exact cure for COVID-19 disease. However, scientists are sprinting to develop a vaccine which is the most effective way to contain and prevent the spread of the virus. There are multiple technologies which are under evaluation such as inactivated virus, protein subunit, nucleic acid, and replicating and non-replicating viral vector. To speed up the process, scientist must overcome so many challenges which include developing a viable immunogen, development of successful animal model, clinical trials, viral mutations, time constraints, and funding [20, 21]. Recently, Russia has approved a vaccine based on adenovirus vector developed by the Gamaleya National Center of Epidemiology and Microbiology Moscow. However, scientists around the world have criticized the efficiency of the vaccine, the phases it undergoes, and the lack of published results related to the earlier trial stages [22].
Apart from vaccines, there is ongoing research by different companies and laboratories on repurposing existing antiviral drugs such as remdesivir, ritonavir, and lopinavir. To generate potential drugs against COVID-19, scientist turned to artificial intelligence and machine learning approach for efficient identification of potent drugs. A study carried out by Beck et al. [23] using pretrained deep learning model identified “Atazanavir” an antiretroviral drug that is used for the prevention of HIV as the potential potent COVID-19 drug among all the commercially available antiviral drugs. On the other hand, Li et al. [24] identified Nelfinavir, Bictegravir, Tegobuv, and Prulifloxacin as potential drugs for COVID-19 after screening 8000 approved and experimental molecules and compounds obtained from Drugbank.
Medical experts (e.g., epidemiologists) are facing challenges such as the need for accurate data collection, analysis and precise estimation, and forecasting of the number of cases related to pandemics and epidemics. Recently, AI-driven models have proven to be valuable in terms of prediction and classification of medical data. Since the outbreak of COVID-19, data analyst and medical practitioners adopted the use of AI models for prediction and forecasting of COVID-19 cases [25]. The first company to report the use AI for prediction of COVID-19 and sent warning alerts to its customers prior to 2020 is the BlueDot AI-based company. However, other scientists have reported the use of AI models such as the use of model-free methods by Zeng et al. [26] for the prediction of inter-provincial transmissions of COVID-19 in mainland China. Hu et al. [27] utilized AI-driven methods for real-time forecasting of COVID-19 outbreak within China based on lengths, size estimations, and ending time of the outbreak based on data obtained from WHO (collected between 19 January 2020 and 16 February 2020).
Deep Learning and Transfer Learning
Deep learning (DL) is a branch of machine learning (ML), a subset of artificial intelligence (AI) inspired by the make-up of the human brain. It is termed as a sub-field of ML that works similar to the biology of human brains by taking data and processing the data through neural networks. Many biomedical health issues such as cancer (brain tumor and breast cancer) detections are using computer-aided diagnosis based on AI models. Precisely, DL models can detect hidden features in images which are not apparent or cannot be detected by medical experts. In terms of DL, convolutional neural network (CNN) is the leading DL tool that is popularly used in different sub-field of healthcare system due to their ability to extract features and learn to distinguish between different classes (i.e., positive and negative, infected and healthy, cancer and non-cancer, etc.). Transfer learning (TL) has provided easier approach to quickly retrain neural networks on selected datasets with high accuracy [28, 29].
AlexNet
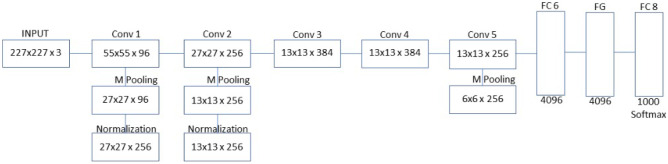
AlexNet model is a DL model proposed by Alex Krizhevsky which utilize rectified linear unit (ReLu) in place of Sigmoid function which is used in traditional neural networks [58]. The model achieved 84% accuracy in 2012 ImageNet Large Scale Visual Recognition Challenge (ILSVRC). It contains five convolution (CONV) blocks or layer with convolutional filter size 3 × 3 without padding and 2 × 2 window size of max pooling operation. The last three layers are two fully connected layers (FCL) and the output layer as shown in Fig. 1. Other terms include batch normalization (BN) and feature map (FM). SoftMax activation function is utilized in the output layer for classification [30, 31].
Fig. 1.
AlexNet architecture
The standard input size for AlexNet model is 227 × 227 × 3. The model consists of overall eight layers, five are convolutional layers, and three fully connected layers. The first two convolutional layers are made of three operations, which include convolution, pooling, and normalization. AlexNet uses ReLU as an activation function, unlike Tanh and sigmoid functions that are used in traditional machine learning. ReLU converts negative numbers to zeros and helps models learn non-linear functions [30].
Max pooling is the most common pooling methods in which its main function is to down sample or to reduce image size by pooling most important feature or by pooling out the number with a highest pixel value. The next two layers are mainly convolution layers without pooling and normalization and the final convolution layer consists of only convolution and pooling without normalization. The first two fully connected layers are dropout layers in which its main function is to reduce overfitting by reducing the number of neurons. The final fully connected layer is known as classification layer [32].
Challenges
As the number of COVID-19 patients grows exponentially, there is a high need for massive detection, which is critical for prevention and control. Medical practitioners all over the world require a sophisticated system to accurately diagnose COVID-19 disease. Different approaches are currently in use for detection of different types of pneumonia. However, detection of different strains of pathogens using molecular testing is still not up to standard of point of care diagnostics. Instead, specimens are collected from site of infections and transfer to a fully equipped or specialized laboratories for diagnosis using RT-PCR sequencing approach which is the current gold standard [33]. This method is deemed expensive and often lead to false results. Moreover, underdeveloped countries and remote areas with limited testing kit and unequipped hospitals with few or zero number of ventilators have become the epicenter of the disease. Thus, there is a high need for developing an alternative approach which is fast, cheap, simple, and reliable. The use of X-ray has proven to be an alternative; however, this method is sometimes tedious for qualified radiologist [34]. These challenges can be addressed by computer-aided detection method using the DL approach, which is accurate, fast, and precise.
The majority of research in the literature only classified two classes (such as COVID-19 vs normal). However, few researches addressed multi classes and classified each class from another. Moreover, the current articles in the state-of-the-art only addressed three-way classification. In this article, we present four-way classification (three types of pneumonia and normal CXR images). We also compare each type with another which is also missing in the majority of the literature.
Contribution
Accordingly, our contributions have been summed up as follows.
We suggested the use of Pretrained (transfer learning) AlexNet model to detect COVID-19 pneumonia non-COVID-19 viral pneumonia, bacterial pneumonia and normal/healthy patients using CXR image.
- We trained the models separately to discriminate between:
- COVID-19 pneumonia vs normal/healthy patients
- Non-COVID-19 Viral pneumonia vs normal/healthy patients
- Bacterial pneumonia vs normal/healthy patients
- COVID-19 pneumonia vs non-COVID-19 viral pneumonia patients
- COVID-19 pneumonia vs bacterial pneumonia vs normal/healthy patients
- COVID-19 pneumonia vs non-COVID-19 viral pneumonia, bacterial pneumonia, and normal/healthy patients
We assessed the performance of the network based on accuracy, sensitivity and specificity.
Related Work
The last decade has seen an exponential rise in the application of DL in the healthcare system. Different studies have shown that DL models can be used for classification of pathological cancer images, diabetic retinopathy, CT scans of pneumonia, and tuberculosis as well as microbial slide images. In the field of pathology, pathologist, computer scientist, and radiologist have been working together to detect diseases such as cancer, pneumonia, and tuberculosis using computer-aided diagnosis [35–37].
In terms of application of DL models for detection of pneumonia using CT scan and X-ray images, we presented a literature review based on studies that:
Classified/distinguished between COVID-19, non-COVID-19 viral pneumonia, and healthy CXR images or between COVID-19, bacterial pneumonia, and healthy images (i.e., four-way and three-way classifications).
Classified/distinguished between COVID-19 and non-COVID-19 viral pneumonia, COVID-19 and healthy CXR images, non-COVID-19 viral pneumonia, and healthy CXR images, bacterial pneumonia, and healthy CXR images (i.e., binary classification).
Chest scans based on chest X-ray or CT scan are an approach radiologist used to distinguish between patient suffering from pneumonia and healthy person. The difference is based on the presence of white hazy patches known as “Ground-glass opacity” in infected patient which is absent in healthy persons. However, as a result of scarcity of test for diagnosing COVID-19 as well as the high cost (120–130 USD), time consuming, low sensitivity, laborious of RT-PCR method, scientist turns to chest scan such as CT scans and X-rays as an alternative approach for diagnosis of severe pneumonia caused by SAR-CoV-2 and bacterial pneumonia [38]. Moreover, this approach has its own challenges such as shortage of expert (i.e., radiologist) that can interpret the result and the tediousness of interpreting thousands of CT scan and CXR images. These challenges are addressed by AI-driven models which have shown high efficiency in assisting medical expert in classification and prediction of disease [39, 40].
Many studies have reported the use of CXR and CT scans along with DL models in order to achieve automated detection of COVID-19 pneumonia and other types of pneumonia such as non-COVID-19 viral pneumonia and bacterial pneumonia. Moreover, many studies have shown the viability of using TL models which are deep networks pretrained on the ImageNet database for classification of pneumonia from healthy CT scans [41–43].
Classification of Multiclass Pneumonia Using AI-Driven Models
The approach of TL in DL is utilized by Chowdhury et al. [17] to differentiate between COVID-19 and viral pneumonia based on a dataset acquired from a public database. The models were trained using 423 COVID-19, 1458 viral pneumonia, and 1579 normal chest X-ray images based on (i) augmentation and (ii) without augmentation. The models achieved higher accuracies, sensitivities, and specificities. A multi-dilation CNN is utilized by Mahmud et al. [44] to classify COVID-19 and other types of pneumonia. The study utilized a deep CNN as COVXNet with modifications based on varying dilation rates for feature extraction, optimization, stacking algorithms, and gradient-based discriminative localization to train dataset containing 1493 non-COVID-19 viral pneumonia, 305 COVID-19 pneumonia, and 2780 bacterial pneumonia. The model achieved 97.4% accuracy for COVID-19 vs normal, 96.9% for COVID-19 vs non-COVID-19 viral pneumonia, 94.7% for COVID-19 vs bacterial pneumonia, and 90% for multi-class.
In order to show the difference between COVID-19 and community-acquired pneumonia (CAP), Li et al. [45] utilized a three-dimensional DL framework known as COVID-19 detection neural network (COVNet) using 4352 CT scans (1292 of COVID-19, 1735 of CAP, and 1325 normal CT scans). The models achieved 90% sensitivity and 96% specificity for detection of COVID-19 and 87% sensitivity and 92% specificity for detection of CAP. Apostolopoulos et al. [41] utilized TL approach on datasets that contain 1427 X-ray images (504 normal X-ray images, 700 bacterial pneumonia, and 224 COVID-19 X-ray images). The model was able to achieve 96.78% accuracy, 96.46% specificity, and 98.66% sensitivity.
Classification of COVID-19 and Normal X-ray Images Using AI-Driven Models
Abbas et al. [46] utilized a CNN model know as Decompose, Transfer, and Compose (DeTraC) for the classification of COVID-19 using chest X-ray images. The study utilized multiple datasets acquired from different hospitals around the world. The model achieved 95.12% accuracy, 97.91% sensitivity, and 91.87% specificity. Two deeper models are utilized by Narin et al. [42] for the classification of COVID-19 and normal CXR images obtained from public domain. The study utilized 100 images (50:50) and the models achieved 97% accuracy for InceptionV3 and 87% accuracy for Inception-ResNetV2). Moreover, an inception migration-learning model is used by Wang et al. [47] for the classification of 453 confirmed cases of COVID-19 with previously diagnosed typical pneumonia. The model achieved external testing accuracy of 73.1%, sensitivity of 74%, and specificity of 67%.
The summary of the application of AI for detection of pneumonia such as COVID-19, bacterial pneumonia, non-COVID-19 viral pneumonia, and normal X-ray images is presented in Tables 2 and 3 based on binary classifications and multiclass, respectively (see Table 4).
Table 2.
Detection of different types of pneumonia (multiclass) using AI-driven tools
| Reference | Type of pneumonia | Dataset | Result |
|---|---|---|---|
| [45] | COVID-19 and community-acquired pneumonia (CAP) | 4352 CT scans (1292 of COVID-19, 1735 of CAP, and 1325 normal CT scans) | The models achieved 90% SV and 96% SF for detection of COVID-19 and 87% SV and 92% SF for detection of CAP |
| [17] | COVID-19 and non-COVID-19 VP | 423 COVID-19, 1458 viral pneumonia, and 1579 normal chest X-ray images | The models achieved higher accuracies, sensitivities and specificities |
| [44] | COVID-19, non-COVID-19 VP, BP | 1493 non-COVID-19 viral pneumonia, 305 COVID-19 pneumonia, 2780 bacterial pneumonia | The model achieved 97.4% AC for COVID-19 vs normal, 96.9% for COVID-19 Vs non-COVID-19 VP, 94.7% for COVID-19 vs BP, and 90% for multi-class |
| [48] | Non-COVID-19 VP and BP (strains not specified) | 5856 chest X-ray | The model achieved Ac of 96.2% accuracy for BP and 93.6% for non-COVID-19 VP |
Ac accuracy, BP bacterial pneumonia, Sv sensitivity, Sf specificity, VP viral pneumonia
Table 3.
Classification of CXR images (two classes) using AI-driven models
| Reference | Type of pneumonia | Dataset | Result |
|---|---|---|---|
| [42] | COVID-19 | 50 COVID-19 and 50 normal CXR images | The models achieved 97% accuracy for InceptionV3 and 87% accuracy for Inception-ResNetV2 |
| [49] | Non-COVID-19 VP (strain not specified) | 5856 X-ray images | The model achieved average Ac of 94.81% for training and 93.01% for validation |
| [50] | Non-COVID-19 VP | 453 CT scan images | The models achieved validation AC of 82.9%, SV of 84% and SF of 80.5%, testing AC of 73.1%, SV of 74%, and SF of 67% |
| [51] | Non-COVID-19 VP | 5863 Chest X-ray images | The model achieved AC of 95.30% |
| [52] | VP (COVID-19, Influenza-A) | 618 CT scan images | The model achieved AC of 86.7% |
| [46] | COVID-19 | 185 normal CXR images and 11 COVID-19 | The model achieved 95.12% accuracy, 97.91% sensitivity, and 91.87% specificity |
| [47] | COVID-19 | 453 COVID-19 CXR images | The model achieved external testing accuracy of 73.1%, sensitivity of 74%, and specificity of 67% |
Ac accuracy, Sv sensitivity, Sf specificity, BP bacterial pneumonia, VP viral pneumonia
Table 4.
Dataset description
| Type of dataset | Number of dataset |
|---|---|
| COVID-19 pneumonia | 371 |
| Non-COVID-19 viral pneumonia | 4237 |
| Bacterial pneumonia | 4078 |
| Healthy (i.e., normal) | 2882 |
Methodology
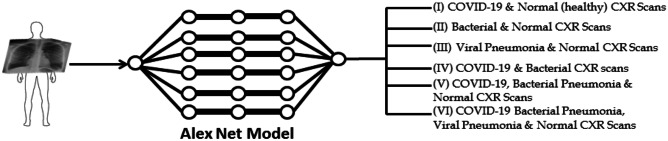
In this section, we presented the proposed approach procedures and its main assumptions. The workflow of the proposed approach is schematically shown in Fig. 2. CXR images are used to train the network using pretrained AlexNet model for classification of (i) COVID-19 and normal (healthy) CXR scans, (ii) bacterial and normal CXR scans, (iii) viral pneumonia and normal CXR scan, (iv) COVID-19 and bacterial CXR scans, (v) COVID-19, bacterial pneumonia, and normal CXR scans, and (vi) COVID-19, bacterial pneumonia, viral pneumonia, and normal CXR scans. TL on DL models has shown to perform efficiently even with small amount of dataset compared with DL models built from scratch which requires large amounts of dataset to reach optimum performance [53].
Fig. 2.
The complete workflow of the proposed method
Dataset
Even though there are more than 30 million confirmed cases of COVID-19 globally, the amount of CT scan images that are available online are very few and limited. As shown in Fig. 3, we obtained COVID-19 pneumonia, non-COVID-19 viral pneumonia, bacterial pneumonia, and normal CXR images from the following website:
153 images from GitHub (https://github.com/ieee8023/covid-chestxray-dataset)
219 images from Kaggle (https://www.kaggle.com/tawsifurrahman/covid19-radiography-database). We removed 1 image due to low contrast, making the total number of images 371. We also obtained 1341 normal X-ray images, 1345 non-COVID-19 viral pneumonia
1341 normal, non-COVID-19 viral pneumonia, 4274 bacterial pneumonia from https://www.kaggle.com/sudalairajkumar/novel-corona-virus-2019-dataset
We obtained CXR images made available by Kermany et al. [54]. The dataset contains three folders (training, validation, and testing with a total number of 5856 positive and negative cases). In each folder, there is a subfolder with names pneumonia and normal folders. The dataset description is based on X-ray images collected from retrospective pediatric patients between the age of 1 and 5. The number of each CXR images used is presented in Table 4.
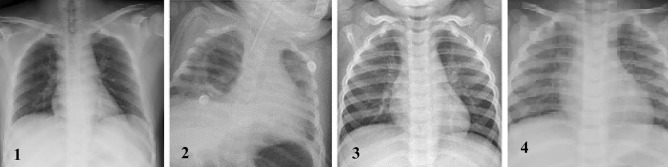
Fig. 3.
CXR scans. 1 COVID-19, 2 non-COVID 19 viral pneumonia, 3 normal CXR scan, 4 bacterial pneumonia
Model Training
For training of datasets, we employed Matlab installed on personal computer with 64-bit window, 8 GB Random Access Memory (RAM), with an intel ® Core i7-3537U and Graphical Processing Unit (GPU). Seventy percent of the datasets are used for training and 30% for testing (evaluating the model performance) as shown in Table 5. Pretrained AlexNet model is employed due to its high accuracy in carrying out feature extraction and image classification. For default settings based on the stochastic gradient descent with momentum, the epoch count has been set to be 20 at most with an initial learning rate of 0.0001 according to [59].
Table 5.
The dataset split
| Model | Training 70% | Testing (30%) | ||||||
|---|---|---|---|---|---|---|---|---|
| Non-COVID-19 VP and healthy | Non-COVID 19 VP | Healthy | Non-COVID 19 VP | Healthy | ||||
| 2966 | 2017 | 1271 | 965 | |||||
| BP and healthy | Bacterial | Healthy | Bacterial | Healthy | ||||
| 2853 | 2017 | 1225 | 965 | |||||
| COVID-19 and healthy | COVID-19 | Healthy | COVID-19 | Healthy | ||||
| 260 | 2017 | 111 | 965 | |||||
| COVID-19 and non-COVID-19 VP | COVID 19 | Non-COVID-19 VP | COVID-19 | Non-COVID-19 VP | ||||
| 260 | 2966 | 111 | 1271 | |||||
| COVID-19, BP and healthy | COVID-19 | BP | Healthy | COVID-19 | BP | Healthy | ||
| 260 | 2853 | 2017 | 111 | 1225 | 965 | |||
| COVID-19, non-COVID-19 VP, BP, and healthy | COVID-19 | BP | Non-COVID-19 VP | Healthy | COVID-19 | BP | Non-COVID-19 VP | Healthy |
| 260 | 2853 | 2966 | 2017 | 111 | 1225 | 1271 | 965 | |
BP bacterial pneumonia, VP viral pneumonia
Evaluation and Confusion Matrix
To evaluate the performance of the trained models, three parameters are considered: accuracy, sensitivity, and specificity. Accuracy is termed as the ratio of correctly classified images over the total number of images; it is also termed as the sum of sensitivity and specificity. For evaluating the accuracy and loss of a model, the following formulas are utilized:
| 1 |
| 2 |
Confusion matrix is the common approach used for evaluation of model performance based on true positive (TP), true negative (TN), false positive (FP), and false negative (FN). TPs are the number of samples that are correctly identified by the model as positive cases or number of cases that are actually positive (i.e., pneumonia) according to each model. TNs are the number of samples that are correctly identified by the model as negative cases or number of cases that are actually healthy (normal) and classified as negative according to each model. FPs are the number of samples that are incorrectly classified as negative by the model or number of cases that are actually negative (normal or healthy) but classified as positive (pneumonia) according to each model. FNs are the number of samples that are incorrectly classified as positive (pneumonia) by the model or number of cases that are actually positive but classified as normal or healthy according to each model as shown in Table 6.
Table 6.
Confusion matrix
| Actual | |||
|---|---|---|---|
| Predicted | True positive ( +) | False negative ( −) | |
| True positive | True + | False + | |
| False negative | False − | True − | |
True positive rate (sensitivity) is the proportion of positive image samples that are correctly identified as positive sample (i.e., it shows the percentage of positive samples that are correctly identified as positives).
| 3 |
False positive rate (FPR) also known as specificity is the proportion of positive samples that are incorrectly identified as positive samples (i.e., it shows the percentage of negative samples that are incorrectly identified as positives).
| 4 |
Performance Evaluation
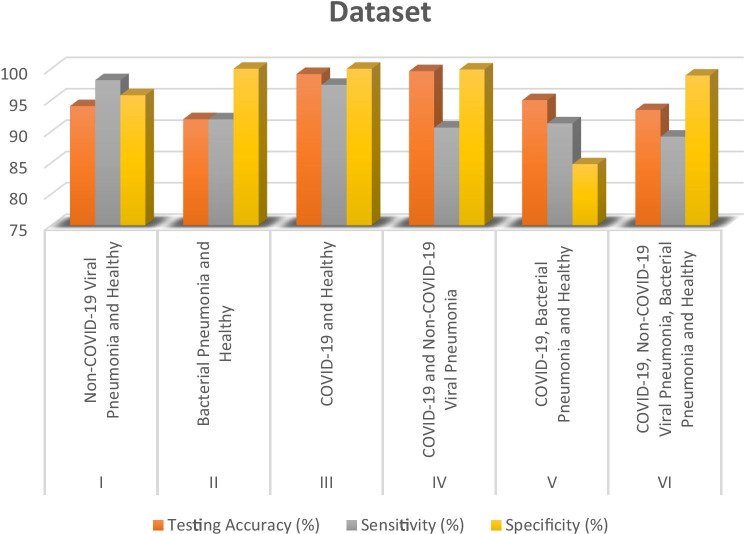
The binary classification of COVID-19 vs healthy CXR images, COVID-19 vs non-COVID-19 viral pneumonia, non-COVID-19 viral pneumonia vs healthy CXR images, bacterial pneumonia vs healthy CXR images, three-way classification (COVID-19 vs bacterial pneumonia and healthy CXR images), and four-way classification (COVID-19 viral pneumonia, bacterial pneumonia, non-COVID-19 viral pneumonia, and healthy CXR) are presented in Table 7 and Fig. 4. Moreover, comparison between some of the state-of-the-art approaches with our models is presented in Tables 8 and 9.
Table 7.
Performance evaluation
| S/N | Dataset | Training accuracy (%) | Testing accuracy (%) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|
| I | Non-COVID-19 viral pneumonia and healthy | 96.43 | 94.05 | 98.19 | 95.78 |
| II | Bacterial pneumonia and healthy | 95.28 | 91.96 | 91.94 | 100.00 |
| III | COVID-19 and healthy | 99.71 | 99.16 | 97.44 | 100.00 |
| IV | COVID-19 and non-COVID-19 viral pneumonia | 99.57 | 99.62 | 90.63 | 99.89 |
| V | COVID-19, bacterial pneumonia, and healthy | 97.40 | 95.00 | 91.30 | 84.78 |
| VI | COVID-19, non-COVID-19 viral pneumonia, bacterial pneumonia, and healthy | 94.18 | 93.42 | 89.18 | 98.92 |
Fig. 4.
Performance evaluation of the models under study based on accuracy, sensitivity, and specificity
Table 8.
Comparison between our result (based on binary classification) with the state-of-the-art
| Class | Reference | Dataset | Result | ||
|---|---|---|---|---|---|
| Ac | Sv | Sf | |||
| COVID-19 and non-COVID-19 VP | [44] | 305 COVID-19 P and 1493 non-COVID-19 VP | 96.9% | - | - |
| Our model (COVID-19 and non- COVID-19 VP) | 371 COVID-19 and 4237 non-COVID-19 VP | 99.62% | 90.63% | 99.89% | |
| Non-COVID-19 VP and healthy datasets | [49] | 5856 CXR images | 93.01% | - | - |
| [50] | 453 CXR images | 73.1%, | 74% | 67% | |
| [51] | 5863 CXR images | 95.30% | - | - | |
| [51] | 618 CXR images | 86.7% | - | - | |
| [48] | 5856 CXR | 96.2% for BP and 93.6% for Non-COVID-19 VP | - | - | |
| Our model (Non-COVID-19 VP and healthy datasets | 4237 non-COVID-19 VP and 2882 healthy datasets | 94.43% | 98.19% | 95.78% | |
| COVID-19 and healthy datasets | [42] | COVID-19 50 COVID-19 and 50 normal CXR images | 97% for InceptionV3 and 87% for Inception-ResNetV2 | - | - |
| [44] | 305 COVID-19 P | 97.4% | - | - | |
| [46] | 185 normal CXR images and 11 COVID-19 | 95.12% | 97.91% | 91.87% | |
| [47] | 453 COVID-19 CXR images | 73.1% | 74.00% | 67.00% | |
| Our model (COVID-19 and healthy datasets | 371 COVID-19 and 2882 healthy datasets | 99.16% | 97.44% | 100% | |
Ac accuracy, Sv sensitivity, Sf specificity, P pneumonia, VP viral pneumonia, BP bacterial pneumonia, CXR chest X-ray
Table 9.
Multiclass comparison between our result with the state-of-the-art
| Reference | Dataset | Result | ||
|---|---|---|---|---|
| Ac | Sv | Sf | ||
| [45] | 4352 CT scans (1292 of COVID-19, 1735 of CAP and 1325 normal CT scans) | - |
90% for COVID-19 87% for CAP |
96% for COVID-19 92% for CAP |
| [17] | 423 COVID-19, 1458 VP, and 1579 normal chest X-ray images | - | - | - |
| [44] | 1493 non-COVID-19 VP, 305 COVID-19 P, 2780 BP | 90% | - | - |
| Our model (three-way classification) | 371 COVID-19, 4078 BP, and 2882 healthy | 94.00% | 91.30% | 84.78% |
| Our model (four-way classification) | 371 COVID-19, 4237 non-COVID-19 VP, 4078 BP, and 2882 healthy | 93.42% | 89.18% | 98.92% |
Ac accuracy, Sv sensitivity, Sf specificity, P pneumonia, VP viral pneumonia, BP bacterial pneumonia, CXR chest X-ray
Results
The datasets are divided into two: 70% used in training and 30% used for testing. The performances of the models are evaluated based on testing accuracy, sensitivity, and specificity. Initially, a pilot study was carried out using 371 CXR images each for COVID-19, non-COVID-19, bacterial pneumonia, and healthy images. We obtained low accuracy, sensitivity, and specificity due to low number of datasets. We carried out this study to analyze the linearity of the dataset by using same amount training and testing dataset due to the fact that we have only 371 COVID-19 CXR images. Before we carried out a multiclass classification, we trained each type of pneumonia with healthy (non-pneumonia or non-infected) CXR images. For non-COVID-19 viral pneumonia and healthy datasets, we achieved 94.43% testing accuracy, 98.19% sensitivity, and 95.78% specificity. In terms of bacterial pneumonia and healthy datasets, we achieved 91.43% testing accuracy, 91.94% sensitivity, and 100% specificity. This shows that the model has learned to classify negative images (non-infected/healthy) accurately compared with positive CXR images (bacterial pneumonia). Moreover, the majority of the recent studies focused on COVID-19 pneumonia and non-infected CXR dataset. Our model trained based on COVID-19 pneumonia and non-infected (healthy) CXR images achieved high performance with 99.16% testing accuracy, 97.44% sensitivity, and 100% specificity. The CXR scan images of different types of viral pneumonia are similar, making it hard for radiologist to distinguish COVID-19 with other viral pneumonia. This limitation can lead to misdiagnosis and at the same time can lead to non-COVID-19 viral pneumonia misdiagnosed as COVID-19 pneumonia [17]. To address this limitation, we trained our model to distinguish between COVID-19 pneumonia and non-COVID-19 viral pneumonia. The model achieved 99.62 testing accuracy, 90.63% sensitivity, and 99.89% specificity.
For multiclass dataset, before we train the whole classes, we examine the performance of the model based on three-way classification (COVID-19, bacterial pneumonia, and healthy) to see how the model will perform before integrating non-COVID-19 viral pneumonia. The model achieved low accuracy compared with models trained to distinguish between binary classifications with 94.00% testing accuracy, 91.30% sensitivity, and 84.78% specificity. Based on this result, we hypothesized to achieve lower performance based on four-way classification (COVID-19, non-COVID-19 viral pneumonia, bacterial pneumonia, and healthy). However, the model (i.e., trained based on four-way classification) achieved a lower accuracy compared with three-way classification in terms of testing accuracy (93.42%) and sensitivity (89.18%) while it achieved a higher specificity (98.92%) compared with three-way classification as shown in Table 7 and Fig. 4.
Comparison with the State-of-the-Art
As shown in Tables 8 and 9, the performances of pretrained AlexNet models are compared with other proposed models. Compared with our work, the study carried out by Li et al. [45] grouped viral and bacterial pneumonia as community-acquired pneumonia (CAP). However, our study disputes this approach; COVID-19 as a viral disease resembles other viral pneumonia. The result we achieved when comparing COVID-19 and other viral pneumonia has shown lower sensitivity and specificity (90.63% and 99.89%, respectively) in comparison with COVID-19 and healthy which achieved 97.44% sensitivity and 100% specificity. Our claim is also supported by Chowdhury et al. [17] who stated that “models performed extremely well when used for classifying COVID-19 and normal images compared to COVID-19 and other viral pneumonia.” Both Bai et al. [55] and Narin et al. [42] have also reported high degree of similarity between COVID-19 and other viral pneumonia when considering physiological and clinical prospective.
With regards to the classification of COVID-19 and normal CXR images, it can be observed that our model provides significantly a better performance in comparison with studies that utilized a small number of datasets such as [42, 44, 46, 47] and models developed from scratch such as [29]. However, [46] achieved higher sensitivity (i.e., 97.91%) compared with our model (with sensitivity of 97.44%). The impressive performance of the model is attributed to the use of TL based on pretrained models with higher number of layers which have shown to perform efficiently with less amount of data in comparison with models designed from scratch. In terms of classification between non-COVID-19 viral pneumonia and healthy CXR images, several studies utilized same datasets made available by Kermany et al. [54]. The majority of these studies achieved higher performance of above 90% accuracy such as studies carried out by Stephen et al. [49], Saravia et al. [51], and Rajaraman et al. [48]. However, our model achieved result within the same range with 94.43% accuracy.
Conclusion
This work presents the use of deep neural network based on the TL approach (known as a pretrained AlexNet model) for automatic detection of COVID-19 pneumonia, non-COVID-19 viral pneumonia, and bacterial pneumonia. The models were trained based on two binary classes and multiclass (three and four classes). For two binary classifications, each of COVID-19, non-COVID-19 viral pneumonia, and bacterial pneumonia are classified along with healthy CXR images. For multiclass, the models are trained based on (1) three-way classification (COVID-19, bacterial pneumonia, and healthy CXR images) and (2) four-way classification (COVID-19, non-COVID-19 viral pneumonia and bacterial pneumonia and healthy CXR images). The models were evaluated based on accuracy, sensitivity, and specificity. However, the result has shown that for classification of for non-COVID-19 viral pneumonia and healthy datasets, the model achieved 94.43% testing accuracy, 98.19% sensitivity, and 95.78% specificity. For bacterial pneumonia and healthy datasets, the model achieved 91.43% testing accuracy, 91.94% sensitivity, and 100% specificity. In terms of COVID-19 pneumonia and healthy CXR images, the model achieved 99.16% testing accuracy, 97.44% sensitivity, and 100% specificity. For classification of COVID-19 pneumonia and non-COVID-19 viral pneumonia, the model achieved 99.62 testing accuracy, 90.63% sensitivity, and 99.89% specificity. For multiclass datasets, the model achieved 94.00% testing accuracy, 91.30% sensitivity, and 84.78% specificity for three-way classification (COVID-19, bacterial pneumonia, and healthy) and testing accuracy of 93.42%, sensitivity of 89.18%, and specificity of 98.92% for four-way classification (COVID-19, non-COVID-19 viral pneumonia, bacterial pneumonia, and healthy).
The higher performance achieved for classification of COVID-19 pneumonia and non-COVID-19 viral pneumonia and COVID-19 pneumonia with healthy CXR images has shown that computer-aided detection approach can be used as an alternative or a confirmatory approach against RT-PCR method which has shown to be less sensitive, time consuming, and laborious. One of the limitations of this research is the fact that we used a small dataset of COVID-19 pneumonia. This challenge makes it difficult to generalize our result. In the future, we hope to acquire more dataset and to train the images using deeper neural networks such as pretrained GoogleNet and ResNet. With the relative amount of dataset, cross validation approach can be carried out to evaluate the performance of the model. Moreover, hybrid models have shown to perform better when compared with single models. Combining CNN models with support vector machine (SVM) and support vector regression (SVR) can also improve performance of the model.
Compliance and Ethical Standards
Ethical Approval
This article does not contain any studies with human participants performed by any of the authors.
Contributor Information
Abdullahi Umar Ibrahim, Email: Abdullahi.umaribrahim@neu.edu.tr.
Mehmet Ozsoz, Email: mehmet.ozsoz@neu.edu.tr.
Sertan Serte, Email: sertan.serte@neu.edu.tr.
Fadi Al-Turjman, Email: fadi.alturjman@neu.edu.tr.
Polycarp Shizawaliyi Yakoi, Email: ypolycarp@gmail.com.
References
- 1.Obaro SK, Madhi SA. Bacterial pneumonia vaccines and childhood pneumonia: are we winning, refining, or redefining? Lancet Infect Dis. 2006;6(3):150–161. doi: 10.1016/S1473-3099(06)70411-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pound MW, Drew RH, Perfect JR. Recent advances in the epidemiology, prevention, diagnosis, and treatment of fungal pneumonia. CurrOpin Infect Dis. 2002;15(2):183–194. doi: 10.1097/00001432-200204000-00014. [DOI] [PubMed] [Google Scholar]
- 3.Virkki R, Juven T, Rikalainen H, Svedström E, Mertsola J, Ruuskanen O. Differentiation of bacterial and viral pneumonia in children. Thorax. 2002;57(5):438–441. doi: 10.1136/thorax.57.5.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones RN. Microbial etiologies of hospital-acquired bacterial pneumonia and ventilator-associated bacterial pneumonia. Clin. Infect. Dis. 2010;51(Supplement_1):S81–7. [DOI] [PubMed]
- 5.Ruuskanen O, Lahti E, Jennings LC, Murdoch DR. Viral pneumonia. The Lancet. 2011;377(9773):1264–1275. doi: 10.1016/S0140-6736(10)61459-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization . Revised WHO classification and treatment of childhood pneumonia at health facilities: evidence summaries. Geneva: World Health Organization; 2014. [PubMed] [Google Scholar]
- 7.Gao J, Tian Z, Yang X. Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Bioscience trends. 2020. [DOI] [PubMed]
- 8.Fan Y, Zhao K, Shi ZL, Zhou P. Bat coronaviruses in China Viruses. 2019;11(3):210. doi: 10.3390/v11030210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gómez P, Semmler M, Schützenberger A, Bohr C, Döllinger M. Low-light image enhancement of high-speed endoscopic videos using a convolutional neural network. Med Biol Eng Comput. 2019;57(7):1451–1463. doi: 10.1007/s11517-019-01965-4. [DOI] [PubMed] [Google Scholar]
- 10.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z. Clinical features of patients infected with 2019 novel coronavirus in Wuhan China. The lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen SG, Chen JY, Yang YP, Chien CS, Wang ML, Lin LT. Use of radiographic features in COVID-19 diagnosis: challenges and perspectives. J Chin Med Assoc. 2020;83(7):644. doi: 10.1097/JCMA.0000000000000336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KS, Lau EH, Wong JY, Xing X. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N Engl J Med. 2020. [DOI] [PMC free article] [PubMed]
- 13.Lai CC, Shih TP, Ko WC, Tang HJ, Hsueh PR. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and corona virus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrob Agents. 2020;17:105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, Xiang H, Cheng Z, Xiong Y, Zhao Y. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan China. JAMA. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rothan HA, Byrareddy SN. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J Autoimmun. 2020;26:102433. doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mueller M, Derlet PM, Mudry C, Aeppli G. Using random testing to manage a safe exit from the COVID-19 lockdown. arXiv preprint arXiv: 2004.04614. 2020.
- 17.Chowdhury ME, Rahman T, Khandakar A, Mazhar R, Kadir MA, Mahbub ZB, Islam KR, Khan MS, Iqbal A, Al-Emadi N, Reaz MB. Can AI help in screening viral and COVID-19 pneumonia?. arXiv preprint arXiv: 2003.13145. 2020.
- 18.Keogh-Brown MR, Smith RD. The economic impact of SARS: how does the reality match the predictions? Health Policy. 2008;88(1):110–120. doi: 10.1016/j.healthpol.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oboho IK, Tomczyk SM, Al-Asmari AM, Banjar AA, Al-Mugti H, Aloraini MS, Alkhaldi KZ, Almohammadi EL, Alraddadi BM, Gerber SI, Swerdlow DL. 2014 MERS-CoV outbreak in Jeddah—a link to health care facilities. N Engl J Med. 2015;372(9):846–854. doi: 10.1056/NEJMoa1408636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ella KM, Mohan VK. Coronavirus vaccine: light at the end of the tunnel. Indian Pediatr. 2020;57:407–410. doi: 10.1007/s13312-020-1812-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghebreyesus T, Lancet SS-T, 2020 U. Scientists are sprinting to outpace the novel coronavirus. thelancet.com [DOI] [PMC free article] [PubMed]
- 22.Mahase E. Covid-19: Russia approves vaccine without large scale testing or published results. BMJ: British Med J (Online). 2020:13;370. [DOI] [PubMed]
- 23.Beck BR, Shin B, Choi Y, Park S, Kang K. Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Comput Struct Biotechnol J. 2020. [DOI] [PMC free article] [PubMed]
- 24.Li Y, Zhang J, Wang N, Li H, Shi Y, Guo G, Liu K, Zeng H, Zou Q. Therapeutic drugs targeting 2019-nCoV main protease by high-throughput screening. BioRxiv. 2020.
- 25.Vaishya R, Javaid M, Khan IH, Haleem A. Artificial Intelligence (AI) applications for COVID-19 pandemic. Diabetes & Metabolic Syndrome: Clin Res Rev. 2020. [DOI] [PMC free article] [PubMed]
- 26.Zeng T, Zhang Y, Li Z, Liu X, Qiu B. Predictions of 2019-ncov transmission ending via comprehensive methods. arXiv preprint arXiv: 2002.04945. 2020.
- 27.Hu Z, Ge Q, Jin L, Xiong M. Artificial intelligence forecasting of covid-19 in china. arXiv preprint arXiv: 2002.07112. 2020.
- 28.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 29.Tan C, Sun F, Kong T, Zhang W, Yang C, Liu C. A survey on deep transfer learning. InInternational Conference on Artificial Neural Networks 2018. pp. 270–279. Springer Cham.
- 30.Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. In Advances in neural information processing systems. pp. 1097–1105. 2012.
- 31.You Y, Zhang Z, Hsieh CJ, Demmel J, Keutzer K. Imagenet training in minutes. InProceedings of the 47th International Conference on Parallel Processing. pp. 1–10. 2018.
- 32.Han X, Zhong Y, Cao L, Zhang L. Pre-trained alexnet architecture with pyramid pooling and supervision for high spatial resolution remote sensing image scene classification. Remote Sens. 2017;9(8):848. doi: 10.3390/rs9080848. [DOI] [Google Scholar]
- 33.Lippi G, Simundic AM, Plebani M. Potential preanalytical and analytical vulnerabilities in the laboratory diagnosis of coronavirus disease 2019 (COVID-19). Clinical Chemistry and Laboratory Medicine (CCLM). 2020;1(ahead-of-print). [DOI] [PubMed]
- 34.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, Yu T. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. The Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ching T, Himmelstein DS, Beaulieu-Jones BK, Kalinin AA, Do BT, Way GP, Ferrero E, Agapow PM, Zietz M, Hoffman MM, Xie W. Opportunities and obstacles for deep learning in biology and medicine. J R Soc Interface. 2018;15(141):20170387. doi: 10.1098/rsif.2017.0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kallianos K, Mongan J, Antani S, Henry T, Taylor A, Abuya J, Kohli M. How far have we come? Artificial intelligence for chest radiograph interpretation. Clin Radiol. 2019;1;74(5):338–45. [DOI] [PubMed]
- 37.Wang F, Casalino LP, Khullar D. Deep learning in medicine—promise, progress, and challenges. JAMA Intern Med. 2019;179(3):293–294. doi: 10.1001/jamainternmed.2018.7117. [DOI] [PubMed] [Google Scholar]
- 38.Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. Jama. 2020; 12:323(18):1843–4. [DOI] [PMC free article] [PubMed]
- 39.Shan F, Gao Y, Wang J, Shi W, Shi N, Han M, Xue Z, Shi Y. Lung infection quantification of covid-19 in CT images with deep learning. arXiv preprint arXiv: 2003.04655. 2020.
- 40.Yang W, Yan F. Patients with RT-PCR-confirmed COVID-19 and normal chest CT. Radiology. 2020;295(2): E3-. [DOI] [PMC free article] [PubMed]
- 41.Apostolopoulos ID, Mpesiana TA. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. 2020;3:1. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Narin A, Kaya C, Pamuk Z. Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks. arXiv preprint arXiv: 2003.10849. 2020. [DOI] [PMC free article] [PubMed]
- 43.Wang L, Wong A. COVID-Net: A Tailored Deep Convolutional Neural Network Design for Detection of COVID-19 Cases from Chest X-Ray Images. arXiv preprint arXiv: 2003.09871. 2020. [DOI] [PMC free article] [PubMed]
- 44.Mahmud T, Rahman MA, Fattah SA. CovXNet: a multi-dilation convolutional neural network for automatic COVID-19 and other pneumonia detection from chest X-ray images with transferable multi-receptive feature optimization. Comput Biol Med. 2020;1(122):103869. doi: 10.1016/j.compbiomed.2020.103869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li L, Qin L, Xu Z, Yin Y, Wang X, Kong B, Bai J, Lu Y, Fang Z, Song Q, Cao K. Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology. 2020. [DOI] [PMC free article] [PubMed]
- 46.Abbas A, Abdelsamea MM, Gaber MM. Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. arXiv preprint arXiv: 2003.13815. 2020. [DOI] [PMC free article] [PubMed]
- 47.Wang S, Kang B, Ma J, Zeng X, Xiao M, Guo J, Cai M, Yang J, Li Y, Meng X, Xu B. A deep learning algorithm using CT images to screen for Corona Virus Disease (COVID-19). MedRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 48.Rajaraman S, Candemir S, Kim I, Thoma G, Antani S. Visualization and interpretation of convolutional neural network predictions in detecting pneumonia in pediatric chest radiographs. Appl Sci. 2018;8(10):1715. doi: 10.3390/app8101715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stephen O, Sain M, Maduh UJ, Jeong DU. An efficient deep learning approach to pneumonia classification in healthcare. J healthc eng. 2019;27:2019. doi: 10.1155/2019/4180949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang S, Sun J, Mehmood I, Pan C, Chen Y, Zhang YD. Cerebral micro-bleeding identification based on a nine-layer convolutional neural network with stochastic pooling. Concurrency and Computation: Practice and Experience. 2020;32(1):5130. [Google Scholar]
- 51.Saraiva AA, Ferreira NM, de Sousa LL, Costa NJ, Sousa JV, Santos DB, Valente A, Soares S. Classification of images of childhood pneumonia using convolutional neural networks. InBIOIMAGING. 2019;1:112–119. [Google Scholar]
- 52.Rajpurkar P, Irvin J, Zhu K, Yang B, Mehta H, Duan T, Ding D, Bagul A, Langlotz C, Shpanskaya K, Lungren MP. Chexnet: Radiologist-level pneumonia detection on chest x-rays with deep learning. arXiv preprint arxiv: 1711.05225. 2017.
- 53.Tajbakhsh N, Shin JY, Gurudu SR, Hurst RT, Kendall CB, Gotway MB, Liang J. Convolutional neural networks for medical image analysis: Full training or fine tuning?. IEEE transactions on medical imaging. 2016;7;35(5):1299–312. [DOI] [PubMed]
- 54.Kermany D, Zhang K, Goldbaum M. Labeled optical coherence tomography (OCT) and Chest X-Ray images for classification. Mendeley data. 2018;2.
- 55.Bai HX, Hsieh B, Xiong Z, Halsey K, Choi JW, Tran TM, Pan I, Shi LB, Wang DC, Mei J, Jiang XL. Performance of radiologists in differentiating COVID-19 from viral pneumonia on chest CT. Radiology. 2020. [DOI] [PMC free article] [PubMed]
- 56.Kolhar M, Al-Turjman F, Alameen A, Abualhaj M. A three layered decentralized IoT biometric architecture for city lockdown during COVID-19 outbreak. IEEE Access. 2020;8(1):163608–163617. doi: 10.1109/ACCESS.2020.3021983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rahman M, Zaman N, Asyharia A, Al-Turjman F, Bhuiyan M, Zolkipli M. Data-driven dynamic clustering framework for mitigating the adverse economic impact of Covid-19 lockdown practices. Elsevier Sustainable Cities and Societies. 2020;62:102372. doi: 10.1016/j.scs.2020.102372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Waheed A, Goyal M, Gupta D, Khanna A, Al-Turjman F, Pinheiro P.R. “CovidGAN: Data Augmentation using Auxiliary Classifier GAN for Improved Covid-19 Detection”, IEEE Access, 2020. [DOI] [PMC free article] [PubMed]
- 59.Mathwork. Train neural network for deep learning https://www.mathworks.com/help/deeplearning/ref/trainnetwork.html. Accessed 12 Oct 2020.