Abstract
Background
Uveal melanoma (UM) arises from malignant transformation of melanocytes in the uveal tract of the eye. This rare tumor has a poor outcome with frequent chemo-resistant liver metastases. BAP1 is the only known predisposing gene for UM. UMs are generally characterized by low tumor mutation burden, but some UMs display a high level of CpG>TpG mutations associated with MBD4 inactivation. Here, we explored the incidence of germline MBD4 variants in a consecutive series of 1093 primary UM case patients and a series of 192 UM tumors with monosomy 3 (M3).
Methods
We performed MBD4 targeted sequencing on pooled germline (n = 1093) and tumor (n = 192) DNA samples of UM patients. MBD4 variants (n = 28) were validated by Sanger sequencing. We performed whole-exome sequencing on available tumor samples harboring MBD4 variants (n = 9). Variants of unknown pathogenicity were further functionally assessed.
Results
We identified 8 deleterious MBD4 mutations in the consecutive UM series, a 9.15-fold (95% confidence interval = 4.24-fold to 19.73-fold) increased incidence compared with the general population (Fisher exact test, P = 2.00 × 10–5, 2-sided), and 4 additional deleterious MBD4 mutations in the M3 cohort, including 3 germline and 1 somatic mutations. Tumors carrying deleterious MBD4 mutations were all associated with high tumor mutation burden and a CpG>TpG hypermutator phenotype.
Conclusions
We demonstrate that MBD4 is a new predisposing gene for UM associated with hypermutated M3 tumors. The tumor spectrum of this predisposing condition will likely expand with the addition of MBD4 to diagnostic panels. Tumors arising in such a context should be recognized because they may respond to immunotherapy.
Uveal melanoma (UM) is the most frequent primary intraocular tumor in adults with an overall mean incidence of 5.2 per million per year in the United States (1). Metastases arise in more than 30% of case patients, almost invariably in the liver, with a dismal prognosis because of the absence of effective treatment (median survival of 10 months) (2,3). UMs with high risk of developing metastases are characterized by loss of chromosome 3 and by BAP1 (encoded in 3p21) inactivation resulting from loss-of-function (LoF) mutations and loss of the remaining wild-type copy on chromosome 3 (4). Rare familial UMs are associated with germline mutations of BAP1 (Mendelian Inheritance in Man [MIM]: 614327) (5,6), which is the only known highly penetrant UM predisposition gene. UM mainly affects individuals of European ancestry and is associated with fair skin and light iris color. However, the low tumor mutation burden (TMB) and lack of an ultraviolet-associated mutational signature argue against a role for ultraviolet radiation in UM oncogenesis (7).
Recently, the characterization of a metastatic UM patient with an exceptional response to anti-Programmed cell death protein 1 (anti-PD-1) therapy led us to identify a CpG>TpG mutator phenotype linked to germline protein truncating variants (PTV) in MBD4 (Methyl-CpG Binding Domain Protein 4) and somatic loss of the wild-type allele in tumors in 2 patients with UM and 1 with glioma (8). Another UM patient responding to immune checkpoint inhibitors was subsequently reported with a germline MBD4 PTV (9). MBD4 encodes a glycosylase involved in the base excision repair of DNA damage arising from spontaneous deamination of 5-methylcytosine to thymine (10,11), which is consistent with the mCpG>TpG transitions [mutational signature SBS1 (12)] observed in MBD4-inactivated tumors (12). MBD4, located on chromosome 3, is thought to act as a tumor suppressor gene, following Knudson’s 2-hit model with loss of the wild-type allele by monosomy 3 (M3) in UMs (8). Altogether, these germline deleterious MBD4 variants in UM prompted us to investigate the role of MBD4 as a predisposing gene for UM. Here, we performed MBD4 targeted-sequencing in germline DNA of a large consecutive series of 1093 UM patients and in tumor DNA of a second cohort of 192 UM patients with M3 and investigated the TMB and mutational signature in patients harboring MBD4 mutations.
Methods
Study Patients
The 1099 individuals with UM were diagnosed at Institut Curie, France, from 2013 to 2018. The sex proportion of female to male was 52.2 to 47.8 ± 3.0% (95% confidence interval [CI]) and the median age at diagnosis was 64 years old (Q1-Q3 quartile interval = 54-73). All patients provided written informed consent to perform germline genetic analyses and somatic genetic analyses of tumor samples. Six patients were subsequently removed from the study: the UM diagnosis was not confirmed for 5 patients, and the sixth patient had undergone a bone marrow transplantation and his blood sample corresponded to his donor’s (Supplementary Figure 1A, available online). The study was conducted in accordance with the declaration of Helsinki and was approved by the ethical committee and institutional review board of the Institut Curie. Germline DNA was extracted from the blood of all patients (DNeasy Blood and Tissue kit, Qiagen). When available, tumor genomic status as part of the prognostication assessment was extracted from the medical record and classified as M3, including isodisomy 3) or disomy 3 (D3). Tumor samples were collected from primary eye tumors. A second series of 192 UM tumor samples with M3 was also accrued at Institut Curie, of which 120 patients were independent from the consecutive germline cohort.
MBD4 Targeted Sequencing
Germline DNA of 1099 UM patients from the UM consecutive series (before removal of the 6 a forementioned patients) and DNA of a series of 192 M3 UM tumors were screened for MBD4 variants by pooled MBD4 targeted sequencing. Details on the sequencing strategy and bioinformatics pipeline are described in the Supplementary Methods (available online). Deconvolution of the identified pooled DNA samples with an MBD4 variant was carried out by Sanger sequencing.
Identified MBD4 variants (Supplementary Table 1, available online) were defined following the recommendations of the Human Genome Variation Society (http://varnomen.hgvs.org/) and numbered based on the MBD4 (MIM: 603574) cDNA and protein sequences (GenBank accession numbers NM_003925.2 and NP_003916.1, respectively).
Glycosylase Activity Assay
Wild-type and mutant MBD4 were expressed to assess their enzymatic activity by in vitro MBD4 glycosylase assay as previously described (13,14) (Supplementary Methods, available online).
Whole-Exome Sequencing (WES) and Mutation Calling
WES was performed on tumor samples from MBD4 variant carriers who consented to germline studies (Supplementary Table 2, available online). Variant calling and TMB analysis are described in Supplementary Methods (available online).
Statistical Analysis
Statistical analysis was performed using R software v3.6. Fisher exact test was used to calculate P values between our cohort and the general population. Random subsampling 1 000 000 times of 2186 alleles from the GnomAD cohort was also used as statistical “matching” strategy to calculate P values between our cohort and the general population. A Mann-Whitney U test was used to compare the median ± median absolute deviation for the mutation burden and CpG>TpG proportion between MBD4-deficient (MBD4def) and MBD4-proficient patients. To compare age of UM onset, a Wilcoxon Rank-Sum test was used. For survival analysis using Kaplan-Meier curves, statistical analysis was carried out using the log-rank test. All statistical tests were 2-sided, except for 1-sided Wilcoxon Rank-Sum test for early age of onset in MBD4def patients. Confidence intervals were carried out at 95% confidence level. Confidence intervals for relative risk (RR) measurement was calculated as previously described (15). A P value less than .05 was considered to be statistically significant.
Results
Mining MBD4 Germline Variants in Public UM Cohorts
To evaluate the potential predisposing role of MBD4 in UM, we mined all available public UM cohorts for germline MBD4 variants. We identified 1 case harboring the germline deleterious MBD4 PTV c.1443delT (p.Leu482Trpfs*9) in a first cohort containing 37 UM patients (phs001421.v1.p1) (16). A second cohort of 98 UM patients (phs000823.v1.p1) included a second case with an MBD4 c.1020delA (p.Asp341Thrfs*13) PTV. Collectively, 5 MBD4 germline deleterious variants were found in 268 analyzed UM patients (1.9%) (8,9,16–18). In contrast, such variants are exceedingly rare in an unselected population (88 of approximately 125 000 individuals in GnomAD v2.1.1). Out of these 5 UM case patients with germline MBD4 variants, 4 had available tumor profiles that all showed M3 and somatic BAP1 inactivation (8,9,16), presumably because of the localization of both MBD4 and BAP1 on chromosome 3.
Identification of MBD4 Germline Variants in the In-House Consecutive UM Series
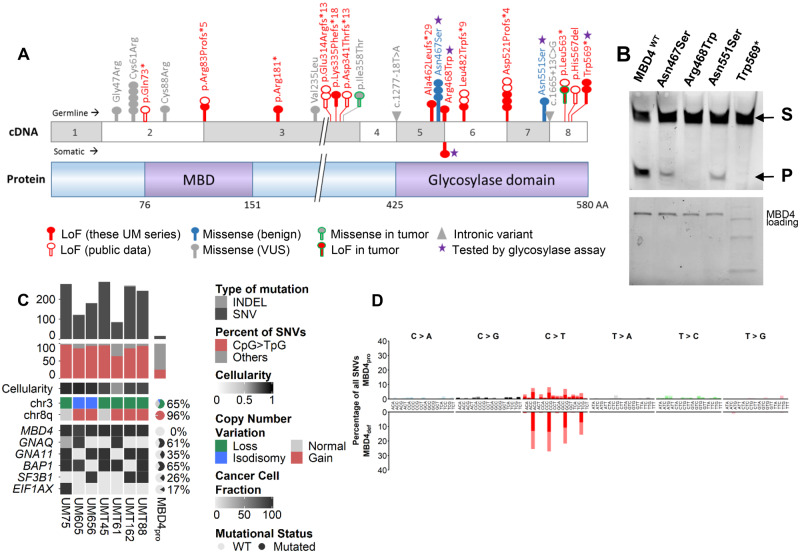
To assess the actual prevalence of MBD4 germline deleterious variants in UM, we next explored an in-house cohort of 1093 (approximately one-half the annual incidence in United States) consecutive patients diagnosed with UM at Institut Curie between 2013 and 2018. Targeted next-generation sequencing in pooled patient DNA followed by Sanger sequencing revealed germline MBD4 PTVs in 7 patients (Table 1): 2 splice site variants (c.1562-1G>T [p.Asp521Profs*4] in 2 patients and a c.335 + 1G>A [p.Arg83Profs*5] variant), 2 frameshift deletion variants (c.1443delT [p.Leu482Trpfs*9] and c.1384delG [p.Ala462Leufs*29]), and 1 stop-gain near the end of the last exon of MBD4 (c.1706G>A [p.Trp569*] in 2 patients).
Table 1.
MBD4 germline deleterious variants in UM and in other malignanciesa
| Patient series | Patient | Variant | dbSNP | Mutation type | Glycosylase assay | GnomAD allele frequency (NFEb) |
||
|---|---|---|---|---|---|---|---|---|
| Allele count | Obs. allele number | Frequency | ||||||
| UMe germline consecutive series | UM75 | p.Trp569* | rs939751619c | stop_gain | Inactived | 2 | 129 130 | 1.55 × 10−5 |
| UM1033 | ||||||||
| UM49 | p.Asp521Profs*4 | rs778697654c | splice_acceptor | ND | 5 | 113 766 | 4.39 × 10−5 | |
| UM1088 | ||||||||
| UM656 | p.Leu482Trpfs*9 | rs769076971c | frameshift_deletion | ND | 3 | 113 752 | 2.64 × 10−5 | |
| UM293 | p.Arg468Trp | rs1380952147 | nonsynonymous_SNV | Inactived | 0 | 113 630 | 0.00 | |
| UM605 | p.Ala462Leufs*29 | – | frameshift_deletion | ND | — | — | — | |
| UM436 | p.Arg83Profs*5 | rs552296498c | splice_donor | ND | 3 | 129 158 | 2.32 × 10−5 | |
| UM M3 tumor series | UMT45 | p.Asp521Profs*4 | rs778697654c | splice_acceptor | ND | 5 | 113 766 | 4.39 × 10−5 |
| UMT61 | p.Lys335Phefs*18 | rs1443006605 | frameshift_deletion | ND | 0 | 113 650 | 0,00 | |
| UMT162 | p.Arg181* | rs1270271346 | stop_gain | ND | 2 | 128 972 | 1.55 × 10−5 | |
| UM (public data) | UM (9) | p.Leu563* | rs200758755 | stop_gain | ND | 8 | 113 702 | 7.04 × 10−5 |
| TCGA_UVM_1 (8) | p.Asp521Profs*4 | rs778697654c | splice_acceptor | ND | 5 | 113 766 | 4.39 × 10−5 | |
| UMphs001421.v1.p1 (16) | p.Leu482Trpfs*9 | rs769076971c | frameshift_deletion | ND | 5 | 113 752 | 4.40 × 10−5 | |
| UVM_IC (8) | ||||||||
| UMphs000823.v1.p1 | p.Asp341Thrfs*13 | – | frameshift_deletion | ND | — | — | — | |
| Other malignancies | AMLEMC-AML-1 (13) | p.His567del | rs775848563 | inframe_deletion | ND | — | — | — |
| AMLWEHI-AML-1/2 (13) | p.Asp521Profs*4 | rs778697654c | splice_acceptor | ND | 5 | 113 766 | 4.39 × 10−5 | |
| AMLWEHI-AML-1/2 (13) | p.Glu314Argfs*13 | rs558765093c | frameshift_insertion | ND | — | — | — | |
| Spiradenocarcinoma (19) | ||||||||
| TCGA_GBM_4 (8) | p.Arg83Profs*5 | rs552296498c | splice_donor | ND | 3 | 129 158 | 2.32 × 10−5 | |
| Colorectal polyposis (20) | p.Gln73* | rs148098584 | stop_gain | ND | 0 | 113 750 | 0.00 | |
| Pilocytic astrocytoma (21) | NAg | NA | NA | ND | NA | NA | NA | |
| Gastric adenocarcinoma (21) | NA | NA | NA | ND | NA | NA | NA | |
| Pancreatic adenoK (21) | NA | NA | NA | ND | NA | NA | NA | |
| Pancreatic endocrine tumor ( 21) | NA | NA | NA | ND | NA | NA | NA | |
adenoK = adenocarcinoma; AML = acute myeloid leukemia; GBM = glioblastoma; M3 = monosomy 3; NA = not available; ND = not determined; NFE = non-Finnish European; UM or UVM = uveal melanoma; — = no value given because of the absence of the variant in dbSNP and/or in the GnomAD NFE population.
NFE population of the Genome Aggregation Database (GnomAD v2.1.1).
Variant found in more than 1 nonrelated patient.
Inactive: absence of glycosylase activity of the recombinant protein carrying the variant.
We also identified and characterized 9 rare germline MBD4 variants (frequency <1% in the general population): 7 missense variants in 13 patients and 2 intronic variants in 2 patients (Figure 1A). Out of these, 3 were predicted with possible splicing consequences: 2 missense variants (c.1652A>G [p.Asn551Ser] and c.1400A>G [p.Asn467Ser]), and 1 intronic variant (c.1277-18T>A) (Supplementary Table 1, available online).
Figure 1.
Functional consequences and phenotype associated with germline and somatic MBD4 deleterious variants. A) Schematic representation of MBD4 cDNA (top) and protein (bottom) sequences. Functional methyl-binding domain (MBD) and glycosylase domain are indicated. The position of all MBD4 variants identified in the 2 uveal melanoma (UM) series (consecutive germline UM series and tumor monosomy 3 [M3] series) is highlighted, with germline and somatic variants above and below the cDNA sequence, respectively, and the 2 variants from the tumor M3 cohort with unknown somatic or germline origin circled in green. These MBD4 variants include loss-of-function (LoF, in red), missense (either benign, in blue-filled circles, or of unknown biological significance [VUS] in gray-filled circles) and intronic (gray triangles) variants. Each circle represents 1 patient harboring the variant. Other MBD4 germline deleterious variants mined on public data are also shown (empty red circles). B) Top: Glycosylase activity assay of recombinant wild-type MBD4 (MBD4WT) and mutant proteins resulting from missense variants and 1 stop gain variant (purple star in 1A) residing in the MBD4 glycosylase domain. Substrate = S; cleaved product = P. Bottom: loading blot for MBD4 wild-type and mutant recombinant proteins corresponding to the glycosylase assay. C) Tumor characteristics of MBD4-deficient (MBD4def) patients compared with that of MBD4-proficient UM patients (MBD4pro) (18). MBD4def patients include UM75, UM605, and UM656 from the consecutive germline series and UMT45, UMT61, UMT162, and UMT88 from the M3 UM tumor series. All patients harbor germline MBD4 variants, except for UMT88 with a somatic MBD4 variant. Top: tumor mutation burden estimated by number of variants (single nucleotide variants [SNVs] in dark gray, and insertions-deletions [INDELs] in light gray) in the exome; middle: proportion of CpG>TpG transitions (red) relative to all SNVs (gray); bottom: copy number alterations in chromosomes 3 and 8q, and mutational status of MBD4, GNAQ, GNA11, BAP1, SF3B1, and EIF1AX, represented as percentage for the MBD4pro series (18). The clonality or subclonality of these key mutational events is indicated by their cancer cell fraction in black-gray gradation, taking into account the variant allele frequency (VAF), copy number change, and cellularity. A plot of the VAF distribution of all variants in the 7 exomes is available in Supplementary Figure 4 (available online). For each exome in the MBD4def group, tumor cellularity is indicated by black-gray shading (and quantified in Supplementary Table 2, available online). D) Mutational patterns of the MBD4def (top) and MBD4pro (bottom) groups based on the relative proportion (y-axis) of each of the 96 types of trinucleotide substitution (x-axis). Dark or bright colors correspond to sense or antisense strands. Individual mutational pattern for all tumor exomes assessed are available in Supplementary Figure 3 (available online).
Functional Assessment of MBD4 Variants
Exon-trapping assays performed on the 3 aforementioned variants demonstrated the use of an alternative acceptor site with c.1277-18T>A, albeit to a lesser extent than the canonical splice site, whereas c.1652A>G and c.1400A>G did not show any measurable effect (Supplementary Figure 2, available online).
All missense variants within the MBD4 glycosylase domain [aa425-580] were assessed by in vitro glycosylase assay together with p. Trp569* because of its localization near the end of the protein (Figure 1; Table 1). The assay confirmed that p. Trp569* results in a catalytically inactive protein and further demonstrated p. Arg468Trp to be a LoF variant (Figure 1). Consistent with this finding, the key role of Arg468 was previously established in binding at the G/T mismatch site, maintaining the T base in a position required for catalysis, and interacting with the orphan G base through hydrogen bonding (22).
We next characterized by WES the 3 available tumors from patients carrying germline MBD4 LoF variants (UM75: p. Trp569*, UM605: p. Ala462Leufs*29, and UM656: p. Leu482Trpfs*9) (8). We confirmed that the 3 MBD4 germline LoF mutations were associated with somatic loss of the wild-type allele in the tumors by either M3 (UM75 and UM605) or isodisomy 3 (UM656) (Figure 1C;Supplementary Table 1, available online), consistent with the 3q21.3 location of MBD4.
MBD4 Mutations in the M3 Tumor Series and Tumor Signature
To further evaluate the incidence of MBD4 alterations in UM, and assuming that MBD4def UMs are associated with M3, we accrued a series of 192 UM tumor samples with M3 (of which 120 case patients were independent from the present consecutive UM series) and screened for MBD4 mutations using the aforementioned strategy (Supplementary Figure 1B, available online). We identified 6 additional MBD4 variants, including 4 LoF mutations (UMT62: c.1688T>A [p.Leu563*], UMT45: c.1562-1G>T [p.Asp521Profs*4], UMT61: c.1002delTTTG [p.Lys335Phefs*18], UMT162: c.541C>T [p.Arg181*]) and 2 missense variants (UMT88: c.1402C>T [p.Arg468Trp] and UMT105: c.1073T>C [p.Ile358Thr]) (Table 1; Figure 1; Supplementary Table 1, available online). Of these, 4 patients (UMT45, UMT61, UMT162, and UMT88) had available germline DNA and all consented to germline studies. Characterization by WES of their tumor samples showed that the 3 tested LoF mutations were germline variants, the missense p. Arg468Trp was somatic, and all were associated with Loss Of Heterozygosity (LOH) of the wild-type allele in tumors (Figure 1, A and B; Supplementary Table 1, available online).
MBD4 inactivation has been associated with a high TMB and a CpG>TpG mutational pattern (8). We confirmed the high TMB in all 3 available MBD4def UMs from the germline consecutive cohort with 275, 122, and 181 variants per exome in UM75, UM605, and UM656, respectively, compared with 16 ± 4.0 (median ± median absolute deviation) variants in MBD4-proficient UMs (18) (Figure 1C;Supplementary Table 2, available online). CpG>TpG transitions represented 96.4%, 85.7%, and 92.8% of all single nucleotide variants (SNVs), respectively, compared with 24.3 ± 7.6% in MBD4-proficient UMs (18) (Figure 1, C and D; Supplementary Table 2, available online). In line with the glycosylase assay, TMB results and somatic chromosome 3 LOH further confirmed the deleterious effect of p. Trp569* in UM75 (Figure 1). Similarly, within the M3 UM tumor series, the 3 available UMs carrying a germline LoF MBD4 variant exhibited a high TMB (269, 288, and 86 variants per exome in UMT162, UMT45, and UMT61, respectively) and a predominance of CpG>TpG transitions (85.6%, 94.4%, and 63.9%, respectively) among all SNVs (Figure 1, C and D; Supplementary Table 2, available online). The tumor sample of patient UMT88, carrying a somatic p. Arg468Trp variant identical to that found as a germline variant in UM293, also carried a high TMB (243 variants) and the CpG>TpG mutational pattern (92.5%) (Figure 1, C and D; Supplementary Table 2, available online), thereby confirming the deleterious effect of this missense variant previously demonstrated in the glycosylase assay (Figure 1). Taken together, these 7 patients with MBD4 deleterious mutations had a 15-fold increase in number of variants per exome (MBD4def: 243 ± 66.7 variants vs MBD4pro: 16 ± 4.0, Mann-Whitney P = 8.72 × 10–5) and a statistically significantly higher CpG>TpG median proportion among SNVs (MBD4def: 92.5 ± 5.7% vs MBD4pro: 24.3 ± 7.5%, P = 9.82 × 10–7) (18).
In addition, we characterized the available tumor samples from 2 patients harboring missense variants that were not predicted to be deleterious (UM102: c.139G>A [p.Gly47Arg] and UM350: c.1652A>G [p.Asn551Ser]; Figure 1; Supplementary Table 1; Supplementary Figure 2, available online). The low TMB (33 and 40 variants per exome, respectively) and absence of CpG>TpG signature confirmed their neutral effect (Supplementary Table 2; Supplementary Figure 3, available online).
Incidence of Germline MBD4 Deleterious Mutations in UM
Taken together, we thus identified 8 LoF germline variants in MBD4 among the 1093 consecutive UM case patients, including p. Arg468Trp with deleterious effect on MBD4 glycosylase activity (Table 1). These account for a statistically significant 9.15-fold increase in deleterious variant frequency compared with the general population, even when restricting ourselves to truncating and splicing MBD4 LoFs as defined by GnomAD (7 LoFs of 2186 observed alleles in UM, representing a variant allele frequency [VAF] of 0.0032 vs 88 LoFs out of a median of 251 450 alleles in the GnomAD v2.1 general population; VAF = 3.50 × 10–4; Fisher exact test P = 2.00 × 10–5). To circumvent the imbalanced dataset, a “matching” subsampling approach was used, giving a similar P value (1.60 × 10–5). Therefore, we demonstrate that the prevalence of MBD4 germline deleterious variants in UM is approximately 0.7%, close to that of BAP1 germline mutation in UM (1.6%) (23), and that MBD4 mutations strongly predispose to UM with an RR of 9.15 (95% CI = 4.24 to 19.73). A comparison between MBD4 germline LoF frequency in this UM consecutive series and in different subsets of the GnomAD population (in the general and European populations) is further presented in Table 2.
Table 2.
Frequency of MBD4 germline deleterious variants in the UM series compared with various populations of the GnomAD databasea
| Study population | No. of LoF variants | Allele countc | Frequency | RRd(95% CIe) | Fisher test (P value) | |
|---|---|---|---|---|---|---|
| UM consecutive series | 7f | 2186 | 0.00320 | — | — | |
| GnomAD v2.1.1 | NFEb | 47 | 113 736 | 0.00041 | 7.75 (3.51 to 17.12) | 6.86 × 10−5 |
| Populationb | 88 | 251 450 | 0.00035 | 9.15 (4.24 to 19.73) | 2.00 × 10−5 | |
| GnomAD v2.1.1 (controls only) | NFE | 13 | 42 768 | 0.00030 | 10.53 (4.20 to 26.38) | 2.82 × 10−5 |
| General population | 33 | 109 404 | 0.00030 | 10.62 (4.70 to 23.97) | 1.16 × 10−5 | |
| GnomAD v2.1.1 (noncancer only) | NFE | 41 | 102 730 | 0.00040 | 8.02 (3.60 to 17.86) | 5.89 × 10−5 |
| General population | 82 | 236 912 | 0.00035 | 9.25 (4.28 to 19.99) | 1.90 × 10−5 | |
| GnomAD v3 | NFE | 20 | 64 571 | 0.00031 | 10.34 (4.38 to 24.42) | 2.00 × 10−5 |
| General population | 39 | 143 286 | 0.00027 | 11.76 (5.27 to 26.27) | 5.50 × 10−5 | |
CI = confidence interval; LoF = loss-of-function (deleterious) variants; NFE = non-Finnish European; RR = relative risk; UM = uveal melanoma; — = no value given here because the relative risk, confidence interval, and statistic tests are presented between the UM consecutive series and each GnomAD subpopulation in the rows below.
NFE population subset of the Genome Aggregation Database (GnomAD v2.1.1).
For all GnomAD populations described, refers to the median number of allele count.
RR here is calculated by dividing the LoF frequency in the UM consecutive series by the LoF frequency in the corresponding GnomAD population subset.
Confidence interval of the relative risk is calculated as previously described (15).
Seven LoF variants correspond to the 8 deleterious MBD4 variants identified in this study, with removal of the missense deleterious variant p. Arg468Trp so as to restrict the analysis to LoF variants as defined by GnomAD for accurate comparison.
Within the UM tumor cohort with M3, we identified a total of 5 MBD4 LoF variants, including at least 3 of germline origin and 1 somatic, out of 192 UM patients. These 3 germline LoF variants by themselves account for a VAF of 0.016, more than twice that found in the germline consecutive UM cohort. This was expected given the recurrence of approximately 50% of chromosome 3 loss event among all UM patients (24). This finding confirms that MBD4 deficiency in UM is mainly associated with M3 and that MBD4 germline mutations specifically predispose to hypermutated high-risk M3 UMs.
Defining the MBD4 Predisposition Syndrome
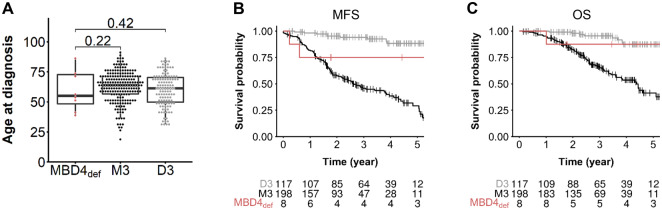
To further characterize this new cancer predisposition, we investigated the medical records of MBD4 mutation carriers. In contrast with the high RR of 9.15 (and therefore an approximately 9-fold higher risk of developing a UM) conferred by MBD4 LoF germline mutations, none of these individuals had familial or bilateral UM. With a lifetime risk of UM estimated at 7.69 × 10–5 in the general population (25), an RR of 9 would result in a lifetime risk of UM of 6.92 × 10–4. Such incidence is still too low to observe familial aggregation, which is consistent with our finding. Assuming that all MBD4def UMs are associated with M3, we compared their medical records with patients from this cohort with available tumor genomic status, that is, 198 MBD4 wild-type M3 and 117 MBD wild-type D3 UMs. Surprisingly, no early-onset UM was found in MBD4 carriers compared with noncarriers regardless of their chromosome 3 status (MBD4def: median age and Q1-Q3 quartile interval = 55.5, 95% CI = 48.4 to 72.8, N = 8; D3: 61.3, 95% CI = 49.8 to 70.3, N = 117; M3: 63.4, 95% CI = 56.6 to 71.4, N = 198; Wilcoxon test, 1-sided: MBD4def vs M3: P = .22, MBD4def vs D3: P = .42; no age difference found between D3 and M3 groups, Wilcoxon test, 2-sided P = .087; Figure 2A). Although the size of the MBD4def series prevents any definitive conclusion, we observed no difference in metastatic-free survival or overall survival between MBD4def and M3 UM patients in contrast with the better outcome in D3 compared with M3 UMs (Figure 2, B and C). Only 1 MBD4 carrier (UM49) had another cancer, a thyroid papillary carcinoma unrelated to MBD4 (low TMB without CpG>TpG signature; data not shown).
Figure 2.
Uveal melanoma (UM) clinical characteristics in an MBD4-deficient (MBD4def) context. A) Age of UM onset of MBD4def patients (n = 8) in the germline consecutive UM series compared with disomy 3 (D3, n = 117) and monosomy 3 (M3, n = 198) MBD4-proficient (MBD4pro) UMs. Wilcoxon test, 1-sided (testing early UM onset in MBD4def patients): MBD4def vs M3: P = .22, MBD4def vs D3: P = .42; no age difference found between D3 and M3 groups, Wilcoxon test, 2-sided P = .087; – not shown). B and C) Metastasis-free survival (MFS, B) and overall survival (OS, C) of MBD4def UM patients (n = 8) and MBD4pro UM patients with M3 or D3. Time zero refers to time at primary UM diagnosis. MFS was defined as the interval between the date of primary UM diagnosis and the date of distant metastasis (first imaging) or death from any cause. The number of patients in each group at each time point (year) is indicated. Survival distributions were estimated by the Kaplan-Meier method and compared using the log-rank test: log-rank test, 2-sided, M3 vs D3: P = 1.98 × 10–9 (OS), P = 1.11 × 10–16 (MFS); M3 vs MBD4def: P = .11 (OS), P = .06 (MFS); D3 vs MBD4def: P = .62 (OS), P = .10 (MFS).
Discussion
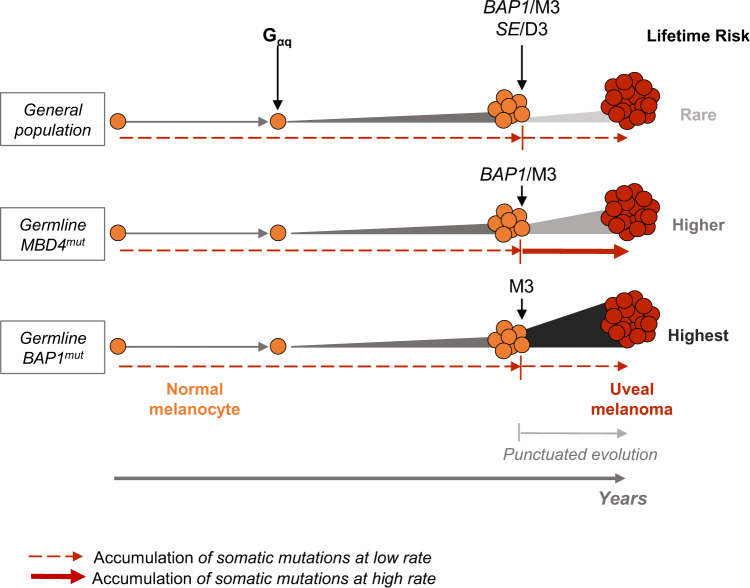
This study demonstrates that MBD4 is a predisposing gene for UM, conferring an RR of 9.15 for this dismal disease. We further demonstrated that MBD4 deficiency specifically predisposes to high-risk M3 UM. One surprising observation for this new cancer-predisposing condition is the absence of early-onset UM. Interestingly, the same is observed in germline BAP1-mutant carriers, even with the high penetrance in that context (6,23). A potential explanation for this paradox is that neither MBD4 nor BAP1 predisposing genes can act before a first step in the malignant transformation. This first step, presumably the Gαq-initiating event consisting of mutually exclusive activating mutations in GNAQ, GNA11, PLCB4, or CYSLTR2 (26–29), would be the main determinant of age of onset (Figure 3). The second step in the malignant transformation, composed of mutations in BAP1, SF3B1, or EIF1AX (“BSE” events), leads to a punctuated evolution of UM (16), which would marginally influence age of onset (Figure 3). Whether MBD4 deficiency favors malignant transformation by increasing driver mutations by modifying the methylation landscape or a distinct mechanism has yet to be determined.
Figure 3.
Working model for uveal melanoma (UM) malignant transformation process throughout time in different genetic backgrounds. Germline MBD4mut/BAP1mut: population with MBD4/BAP1 germline mutation. Following the Gαq activating mutation, secondary mutational events in each genetic background are indicated, along with their association with either disomy 3 (D3) or monosomy 3 (M3). SE = SF3B1 or EIF1AX mutation. Relative lifetime risk of UM is represented by the expansion size and color from normal melanocytes to UM. Dashed and full red arrows indicate the rate of accumulation of somatic mutations throughout time (low and high, respectively).
Importantly, germline MBD4 mutations were recently reported in other types of malignancy, including a polyposis-associated colorectal adenocarcinoma (20), a spiradenocarcinoma (19), a glioblastoma (8), a pilocytic astrocytoma, a gastric adenocarcinoma, a pancreatic adenocarcinoma, and a pancreatic endocrine tumor (21) (Table 1). Furthermore, although the above case patients were all heterozygous in the germline, biallelic germline deleterious MBD4 mutations were reported in 3 individuals who developed acute myeloid leukemias, 2 of which had additional colonic polyposis (13). It is therefore likely that the tumor spectrum associated with MBD4 germline mutations will expand when this gene becomes more systematically explored in clinical diagnosis. It is already clear that this spectrum mostly includes relatively rare tumors and some biological tumor features may underlie their association with MBD4. To be noticed, both leukemias and UMs associated with MBD4 inactivation share a consistent inactivation of the BAP1-ASXL complex (8,13).
It should be noticed that we found no other MBD4-related tumors in our UM series. However, the follow-up and cohort size of this prospective series are limited, and future studies will better characterize the medical history of MBD4 carriers. Larger cohorts will also more precisely define MBD4 mutation frequency in UM patients and the RR conferred by these mutations. Another limitation to the study is the bias for a European population in our cohort, which reflects the higher incidence of the disease in this population (30).
Interestingly, 5 recurrent MBD4 germline deleterious mutations were identified when taking together the LoF variants from our UM cohort and those found in public databases and reports of other cancer types: c.1706G>A [p.Trp569*] (2 patients), c.1562G>T [p.Asp521Profs*4] (4 patients), c.1443delT [p.Leu482Trpfs*9] (3 patients), c.335 + 1G>A [p.Arg83Profs*5] (2 patients), and c.939insA [p.Glu314Argfs*13] (2 patients) (Table 1), suggesting founder mutations. Furthermore, the observation of different tumor types associated with the same MBD4 germline mutation suggests a more global role of MBD4 in cancer predisposition. The peculiar UM proneness in MBD4-mutant carriers (13 out of 23 carriers; Table 1) remains unexplained, but the fact that the frequent M3 in UM inactivates wild-type copies of both BAP1 and MBD4 suppressor genes may at least in part explain the frequent inactivation of MBD4 in UM.
In summary, we described here a novel autosomal-dominant syndrome that is caused by germline mutations of MBD4, characterized by a high RR of developing hypermutated UM and possibly other malignancies. Tumors arising in such a context are associated with a CpG>TpG mutator phenotype and have clinical relevance because they may respond to immune-checkpoint inhibitors.
Funding
Supported by funding from the European Commission under the Horizon 2020 program and innovation program under the Marie Skłodowska-Curie grant agreement No 666003 (A-CD), the Horizon 2020 program UM Cure (LM; Project number: 667787), the INCa/ITMO/AVIESAN PhD fellowship program “Formation à la recherche translationelle” (MR), the Ligue Nationale Contre le Cancer (AE), the Institut National de la Santé et de la Recherche Médicale (INSERM), the Institut Curie, the Ligue Nationale Contre le Cancer (Labellisation), the Programme de Recherche Translationnelle en Cancérologie (PRT-K19-51) INCa-DGOS, and the Site de Recherche Intégrée sur le Cancer (SiRIC) de l’Institut Curie. The Institut Curie ICGex NGS platform is funded by the EQUIPEX “investissements d’avenir” program (ANR-10-EQPX- 03) and ANR10-INBS-09–08 from the Agence Nationale de le Recherche.
Notes
Role of the funder: The funders had no role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; and the decision to submit the manuscript for publication.
Disclosures: The authors have no conflict of interest to declare except for the following: M.R. received a grant support from Bristol-Myers Squibb and Merck.
Author contributions: A.-C.D. and M.R. conceived the study, performed experiments, interpreted the data and wrote the manuscript. A.E. and A.H. performed bioinformatics analyses. L.M., S.A., A.E.-M. and G.P. interpreted the data and provided critical advice. O.M., A.M., S.G. and N.C. provided patients specimens and critical advice. S.D., D.L. and G.P. performed biological analyses. C.C. provided clinical data and critical advice. M.-H.S. conceived and guided the study, interpreted the data and wrote the manuscript. All authors reviewed and approved the final manuscript.
Acknowledgment: The authors thank Joshua J. Waterfall for his insightful comments.
Supplementary Material
References
- 1. Aronow ME, Topham AK, Singh AD.. Uveal melanoma: 5-year update on incidence, treatment, and survival (SEER 1973-2013). Ocul Oncol Pathol. 2018;4(3):145–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Khoja L, Atenafu EG, Suciu S, et al. Meta-analysis in metastatic uveal melanoma to determine progression free and overall survival benchmarks: an international rare cancers initiative (IRCI) ocular melanoma study. Ann Oncol. 2019;30(8):1370–1380. [DOI] [PubMed] [Google Scholar]
- 3. Carvajal RD, Schwartz GK, Tezel T, et al. Metastatic disease from uveal melanoma: treatment options and future prospects. Br J Ophthalmol. 2017;101(1):38–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Harbour JW, Onken MD, Roberson EDO, . et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330(6009):1410–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wiesner T, Murali R, Fried I, et al. A distinct subset of atypical Spitz tumors is characterized by BRAF mutation and loss of BAP1 expression. Am J Surg Pathol. 2012;36(6):818–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Walpole S, Pritchard AL, Cebulla CM, et al. Comprehensive study of the clinical phenotype of germline BAP1 variant-carrying families worldwide. J Natl Cancer Inst. 2018;110(12):1328–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Furney SJ, Pedersen M, Gentien D, et al. SF3B1 mutations are associated with alternative splicing in uveal melanoma. Cancer Discov. 2013;3(10):1122–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rodrigues M, Mobuchon L, Houy A, et al. Outlier response to anti-PD1 in uveal melanoma reveals germline MBD4 mutations in hypermutated tumors. Nat Commun. 2018;9(1):1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Johansson PA, Stark A, Palmer JM, et al. Prolonged stable disease in a uveal melanoma patient with germline MBD4 nonsense mutation treated with pembrolizumab and ipilimumab. Immunogenetics. 2019;71(5–6):433–436. [DOI] [PubMed] [Google Scholar]
- 10. Hendrich B, Hardeland U, Ng HH, et al. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature. 1999;401(6750):301–304. [DOI] [PubMed] [Google Scholar]
- 11. Yoon JH, Iwai S, O'Connor TR, et al. Human thymine DNA glycosylase (TDG) and methyl-CpG-binding protein 4 (MBD4) excise thymine glycol (Tg) from a Tg: G mispair. Nucleic Acids Res. 2003;31(18):5399–5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Alexandrov LB, Kim J, Haradhvala NJ, et al. PCAWG Mutational Signatures Working Group. The repertoire of mutational signatures in human cancer. Nature. 2020;578(7793):94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sanders MA, Chew E, Flensburg C, et al. MBD4 guards against methylation damage and germ line deficiency predisposes to clonal hematopoiesis and early-onset AML. Blood. 2018;132(14):1526–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hashimoto H, Liu Y, Upadhyay AK, et al. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res. 2012;40(11):4841–4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rothman KJ. Epidemiology, an Introduction. 2nd ed New York, NY: Oxford University Press; 2002. [Google Scholar]
- 16. Field MG, Durante MA, Anbunathan H, et al. Punctuated evolution of canonical genomic aberrations in uveal melanoma. Nat Commun. 2018;9(1):116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Robertson AG, Shih J, Yau C, et al. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell. 2017;32(2):204–220.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rodrigues M, Mobuchon L, Houy A, et al. Evolutionary routes in metastatic uveal melanomas depend on MBD4 alterations. Clin Cancer Res. 2019;25(18):5513–5524. [DOI] [PubMed] [Google Scholar]
- 19. Davies HR, Hodgson K, Schwalbe E, et al. Epigenetic dysregulation underpins tumorigenesis in a cutaneous tumor syndrome. bioRxiv 2019; doi: 10.1101/687459. [DOI]
- 20. Tanakaya K, Kumamoto K, Tada Y, et al. A germline MBD4 mutation was identified in a patient with colorectal oligopolyposis and early-onset cancer: a case report. Oncol Rep. 2019;42(3):1133–1140. [DOI] [PubMed] [Google Scholar]
- 21. Waszak SM, Tiao G, Zhu B, et al. Germline determinants of the somatic mutation landscape in 2,642 cancer genomes. bioRxiv 2017; doi: 10.1101/208330. [DOI]
- 22. Morera S, Grin I, Vigouroux A, et al. Biochemical and structural characterization of the glycosylase domain of MBD4 bound to thymine and 5-hydroxymethyuracil-containing DNA. Nucleic Acids Res. 2012;40(19):9917–9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gupta MP, Lane AM, DeAngelis MM, et al. Clinical characteristics of uveal melanoma in patients with germline BAP1 mutations. JAMA Ophthalmol. 2015;133(8):881–887. [DOI] [PubMed] [Google Scholar]
- 24. Prescher G, Bornfeld N, Hirche H, et al. Prognostic implications of monosomy 3 in uveal melanoma. Lancet. 1996;347(9010):1222–1225. [DOI] [PubMed] [Google Scholar]
- 25. Singh AD, De Potter P, Fijal BA, et al. Lifetime prevalence of uveal melanoma in white patients with oculo(dermal) melanocytosis. Ophthalmology. 1998;105(1):195–198. [DOI] [PubMed] [Google Scholar]
- 26. Johansson P, Aoude LG, Wadt K, et al. Deep sequencing of uveal melanoma identifies a recurrent mutation in PLCB4. Oncotarget. 2016;7(4):4624–4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Van Raamsdonk CD, Bezrookove V, Green G, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457(7229):599–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Van Raamsdonk CD, Griewank KG, Crosby MB, et al. Mutations in GNA11 in uveal melanoma. N Engl J Med. 2010;363(23):2191–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moore AR, Ceraudo E, Sher JJ, et al. Recurrent activating mutations of G-protein-coupled receptor CYSLTR2 in uveal melanoma. Nat Genet. 2016;48(6):675–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mobuchon L, Battistella A, Bardel C, et al. A GWAS in uveal melanoma identifies risk polymorphisms in the CLPTM1L locus. NPJ Genom Med. 2017;2(1):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.