Abstract
Background
In the PURE-01 study (NCT02736266), we aimed to evaluate the ability to predict the pathologic complete response (pT0N0) after pembrolizumab by using clinical and tumor biomarkers.
Methods
In an open-label, single-arm, phase 2 study, 3 courses of 200 mg pembrolizumab preceding radical cystectomy were administered in patients with T2-4aN0M0 muscle-invasive bladder cancer. The analyses included a comprehensive genomic profiling and programmed cell-death-ligand-1 (PD-L1)–combined positive score assessment (CPS; Dako 22C3 antibody) of pre- and posttherapy samples. Multivariable logistic regression analyses evaluated baseline clinical T stage and tumor biomarkers in association with pT0N0 response. Corresponding coefficients were used to develop a calculator of pT0N0 response based on the tumor mutational burden (TMB), CPS, and the clinical T stage. Decision-curve analysis was also performed. All statistical tests were 2-sided.
Results
From February 2017 to June 2019, 112 patients with biomarker data were enrolled (105 with complete TMB and CPS data). Increasing TMB and CPS values featured a linear association with logistic pT0N0 probabilities (P = .02 and P = .004, respectively). For low TMB values (≤11 mut/Mb, median value, n = 53), pT0N0 probability was not associated with increasing CPS. Conversely, for high TMB values (>11 mut/Mb, n = 52), pT0N0 was statistically significantly associated with higher CPS (P = .004). The C index of the pT0N0 probability calculator was 0.77. On decision-curve analysis, the net benefit of the model was higher than the “treat-all” option within the clinically meaningful threshold probabilities of 40%-50%.
Conclusions
The study presents a composite biomarker-based pT0N0 probability calculator that reveals the complex interplay between TMB and CPS, added to the clinical T stage.
Patients with muscle-invasive bladder cancer (MIBC) have curative standard therapy possibilities despite the aggressiveness of the disease (1‐3). However, the advent of immune-checkpoint inhibitors paved the way for a novel road in the treatment of bladder tumors throughout the clinical stages of the disease (4,5). In patients with MIBC, the early results from the use of single-agent neoadjuvant pembrolizumab or atezolizumab constituted the rationale for the development of several clinical trials that are currently being offered to these patients worldwide (6-8). These trials combine immunotherapy with chemotherapy or with other novel agents, including, in the near future, targeted therapy. Therefore, it is clear that the ability to accurately select those patients who are most likely to benefit from a neoadjuvant single-agent checkpoint inhibitor will become a priority of clinical research to maximize the use of resources and limit the risk of undertreating patients or delay a potentially curative surgery. Achieving this goal is particularly important in studies like the PURE-01, because of the inherent characteristics of the trial, which enrolled patients regardless of their cisplatin eligibility. From the initial results of PURE-01, a few biomarkers that may be predictive of the pathologic complete response (pT0N0) to treatment have emerged. These biomarkers were identified by employing a comprehensive molecular characterization of pretherapy transurethral resection of the bladder tumor (TURBT) samples and the determination of immunohistochemistry (IHC)-based expression of the programmed cell-death ligand-1 (PD-L1). The present study focused on these mainstream biomarker analyses that characterized the PURE-01 study.
Patients and Methods
Amended Study Design and Patient Selection Criteria
According to the amended protocol (March 2018), eligible patients had a confirmed diagnosis of MIBC, were scheduled for radical cystectomy (RC), and had a clinical (c)T2-4aN0M0 stage. The remaining inclusion and exclusion criteria have been already described, as well as the study design (3 courses of 200 mg pembrolizumab intravenously every 3 weeks, preceding RC), the endpoints, and the remaining trial procedures (6,7). All RC patients underwent extended pelvic lymph node dissection (9,10). The pT0N0 response was defined as the absence of viable tumor in examined tissue from RC and pelvic lymph node dissection. The PURE-01 study is currently recruiting participants at the time of these analyses. The primary goal of the current study was to illustrate the biomarkers and their interplay. To achieve this goal, we developed a risk-prediction tool to quantify the pT0N0 response probability based on the interim data from patients who have completed the study and had complete clinical and tumor biomarker information. All patients provided signed informed consent, and the study was approved by the Ethical Committee of the Fondazione IRCCS Istituto Nazionale dei Tumori, Milan, Italy. The study is registered in ClinicalTrials.gov, number NCT02736266.
Tumor Biomarker Analyses
The mainstream biomarker analyses that characterized the PURE-01 study were represented by a comprehensive genomic profiling that was performed in a Clinical Laboratory Improvement Amendments–certified, College of American Pathologists–accredited laboratory (Foundation Medicine, Cambridge, MA) and by PD-L1 expression that was determined by IHC (Dako 22C3 PharmDx assay; Agilent Technologies, Carpinteria, CA, USA) at the local laboratory. The details of these analyses are presented in the Supplementary Material.
For descriptive purposes, targetable genomic alterations and signatures were categorized according to the European Society for Medical Oncology Scale for Clinical Actionability of molecular Targets (ESCAT) (11). All the assays have been performed blinded to the patient’s outcomes.
Statistical Analyses
The outcome of interest in our study was the association between the baseline clinical variables and tumor biomarkers with pT0N0 (primary endpoint of the clinical trial). The secondary objective was to analyze the dynamic changes in biomarker landscape from matched pretherapy and posttherapy tumor samples. Descriptive statistics included frequencies and proportions for categorical variables. Medians and interquartile ranges were reported for continuous variables. For single gene alterations, comparisons between pathologic response subgroups were made via Fisher exact tests, with the Bonferroni adjustment for multiple hypothesis testing (based on 40 genes that resulted to have ≥3 alterations, with and without variants of unknown significance). Comparison between continuous biomarker groups were made via t tests. Logistic models were used to analyze associations between the tumor mutational burden (TMB) and pT0N0, as well as between the combined positive score (CPS) and pT0N0. Here, logarithmic probabilities of pT0N0 were plotted according to the continuously coded values of the TMB and CPS, respectively. Additionally, logarithmic probabilities of pT0N0 according to CPS values were explored in the split cohorts of patients according to the median TMB cutoff (11 mut/Mb). Univariable and multivariable logistic regression analyses were used to predict the pT0N0 response after RC. We also developed a logistic-based model for predicting the pT0N0 endpoint. The predefined-included variables were the cT stage, TMB, and CPS. The discrimination of the model was tested using the Harrel C index, corrected for overfitting using the 2000-bootstrap resamples of the model. The predictive model coefficients were used to calculate the probability to achieve a pT0N0 response for each patient and to develop the corresponding probability calculator. An interactive web‐based application was then developed to predict the percentage probability of pT0N0 on an individual patient level, derived from the prediction index of the logistic model, using the open-source software R Shiny. We tested the ability of the model to fit the outcome with or without the TMB variable, by relying on the likelihood-ratio test. Finally, decision-curve analysis was used to evaluate the net benefit of the predictive model. All statistical tests were 2-sided with a level of statistical significance set at P less than .05. Analyses were performed using the R software (version 3.5.0, R Foundation for Statistical Computing, Vienna, Austria).
Results
Patient and Tumor Characteristics
From February 2017 to June 2019, 112 patients were enrolled and treated and were evaluable for comprehensive genomic profiling and IHC analyses. Complete TMB data were available for 105 patients (Supplementary Figure 1, available online). The baseline patient characteristics are shown in Table 1. In total, there were 52 patients (46.4%) presenting with a cT3–4 stage (46.4%), and 19 (17%) presented with a predominant variant histology. Table 1 also reports the updated pathologic response outcomes of the analyzed population. A total of 42 patients (37.5%, 95% confidence intervals [CI] = 28.5 to 47.2) achieved a pT0N0 and 65 patients (58%, 95% CI = 48.3 to 67.3) a downstaging to pT no more than 1N0. Forty-seven patients (42%) were treatment failures because they showed a residual high-risk disease (pT2-4N0 and/or pN+, n = 36) or because they radiologically resulted to be nonresponders (n = 11) and received sequential standard chemotherapy.
Table 1.
Patient and tumor characteristics of the included populationa
| Clinical characteristics | No. (%) |
|---|---|
| Total | 112 (100.0) |
| Age, median (IQR), y | 66 (61-73) |
| Weeks from last pembrolizumab dose – RC, median (IQR) | 3 (3-4) |
| Female gender | 15 (13.4) |
| Smoking status | |
| No smoker | 32 (28.6) |
| Former smoker | 51 (45.5) |
| Current smoker | 29 (25.9) |
| Tumor histology | |
| UC | 78 (69.6) |
| Nonpredominant VH | 15 (13.4) |
| Predominant VH | 19 (16.9) |
| Previous NMIBC | 21 (18.8) |
| Previous BCG | 14 (12.5) |
| Clinical stage | |
| T2N0 | 60 (53.6) |
| T3N0 | 49 (43.7) |
| T4N0 | 3 (2.7) |
| Biological features on TURBT sample | |
| Median TMB, IQR (mut/Mb) | 11 (6.1-15) |
| Missing TMB value | 7 (6.2) |
| TMB ≥20 mut/Mb | 14 (12.5) |
| Median PD-L1-CPS (%, IQR) | 69 (61.6) |
| PD-L1-CPS ≥10 (%) | 69 (61.6) |
| Microsatellite instability | 0 |
| Pathologic response outcomes | |
| pT0N0 (95% CI) | 42 (37.5) (95% CI = 28.5 to 47.2) |
| pT ≤ 1N0 (95% CI) | 65 (58) (95%CI = 48.3 to 67.3) |
| pT2N0 | 9 (8) |
| pT3–4N0 | 11 (9.8) |
| pTany pN+ | 16 (14.3) |
| Nonresponse – CT postpembrolizumab | 11 (9.8) |
BCG = Bacillus Calmette-Guérin; CI = confidence interval; CPS = combined positive score; CT = chemotherapy; IQR = interquartile range; NMIBC = nonmuscle-invasive bladder carcinoma; PD-L1 = programmed cell-death ligand-1; RC = radical cystectomy; TMB = tumor mutational burden; TURBT = transurethral resection of the bladder; UC = urothelial carcinoma; VH = variant histology.
Thirty-one patients (29.5%) harbored an ESCAT tier of at least 1-2 genomic alterations suggesting benefit from approved or investigational targeted therapies (Supplementary Figure 2, available online). In particular, tier 1B class was represented by FGFR3 mutations or fusions (n = 17, 15.2%).
Biomarkers in Association With the Pathologic Response
The comparison of baseline genomic alterations between the outlier pathologic responders is presented in Supplementary Table 1 (available online). No statistically significant associations were found, because it was also observed at the single-gene alterations level (Supplementary Figure 3, available online). Univariable analyses are presented in Table 2. In brief, only TMB (P = .02), CPS (P = .004), and cT stage (P = .01) were statistically significantly associated with pT0N0. On multivariable analyses, only CPS remained statistically significant (odds ratio [OR] = 1.02, 95% CI = 1.01 to 1.04; P = .005), whereas TMB (OR = 1.04, 95% CI = 0.9 to 1.10; P = .09) and the cT stage (P = .21) were not. However, there are complexities in modeling biomarkers like TMB and CPS, mainly because their biological effect could not be fully mirrored by the multivariable models. TMB and CPS displayed a linear association with the probability of obtaining a pT0N0 (logistic probability), as shown in Supplementary Figure 4 (available online). Additionally, we found that the probability of obtaining a pT0N0 was not dependent (P = .09) from increasing values of CPS in patients with TMB below the median value (≤11 mut/Mb, n = 53). Conversely, a statistically significant increase (P = .004) of pT0N0 probability was found for increasing values of CPS in patients with TMB values of more than 11 mut/Mb (n = 52; Supplementary Figure 5, available online).
Table 2.
Logistic regression analyses evaluating the association with pT0N0 response
| Variable | Univariable analyses |
Multivariable analyses |
Logistic regression coefficients for the model predicting pT0N0 | ||
|---|---|---|---|---|---|
| OR (95% CI) | P a | OR (95% CI) | P a | Intercept: -1.95 | |
| TMB (mut/Mb) | 1.06 (1.01 to 1.11) | .02 | 1.04 (0.98 to 1.10) | .09 | .04 |
| CPS | 1.02 (1.01 to 1.03) | .004 | 1.02 (1.01 to 1.04) | .005 | .02 |
| Clinical T stage | |||||
| cT3–4 | 1.00 (Reference) | 1.00 (Reference) | |||
| cT2 | 2.71 (1.24 to 6.14) | .01 | 1.78 (0.74 to 4.33) | .21 | .58 |
| Sex | |||||
| Male | 1.00 (Reference) | ||||
| Female | 0.97 (0.32 to 3.09) | .9 | — | — | — |
| Smoking status | |||||
| Never smoker | 0.41 (0.16 to 0.98) | .05 | — | — | — |
| Current or previous smoker | 1.00 (Reference) | ||||
| No previous NMIBC | 1.79 (0.66 to 5.41) | .27 | — | — | — |
| No previous BCG | 1.19 (0.38 to 4.12) | .77 | — | — | — |
| Tumor histology | |||||
| UC | 1.00 (Reference) | ||||
| Nonpredominant VH | 1.64 (0.54 to 5.13) | .38 | — | — | — |
| Predominant VH | 0.38 (0.10 to 1.17) | .11 | — | — | — |
2-sided Wald test P value. BCG = Bacillus Calmette-Guérin; CI = confidence intervals; CPS = combined positive score; NMIBC = non muscle-invasive bladder cancer; OR = odds ratio; TMB = tumor mutational burden; UC = urothelial carcinoma; VH = variant histology.
Development of a pT0N0 Response Prediction Tool
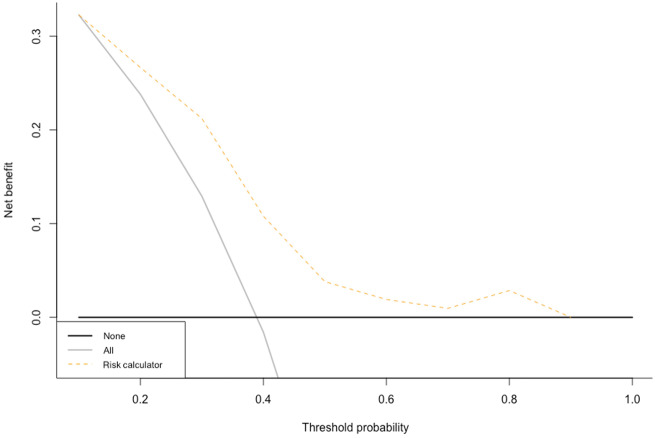
The coefficients of the multivariable model including TMB, CPS, and cT stage were used to develop a calculator for the pT0N0 response probability. This calculator is freely available as an online web resource at https://marco-bandini-md-sanraffaele.shinyapps.io/pure01/. The bootstrapped C index of the model was 0.77 (95% CI = 0.68 to 0.86). Decision-curve analysis illustrated the net benefit of the model, which was higher than the “treat-all” option within the clinically meaningful range of threshold probabilities of 40%-50% (Figure 1). The contribution of TMB and CPS toward the pT0N0 probability is visually represented in Figure 2. We also tested the ability of our calculator to fit the outcome with or without the TMB variable, by relying on the likelihood-ratio test. The model with the TMB resulted to fit the outcome statistically significantly better compared with the model without the TMB (χ2 = 4.04; P = .04).
Figure 1.
Decision curve analysis exploring the net benefit related to the use of our risk calculator in comparison with the treat-all and treat-none options. Net benefit was achieved for threshold probabilities higher than 10%.
Figure 2.
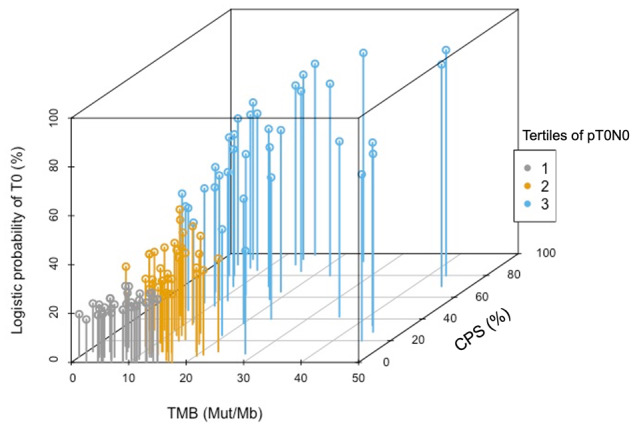
Three-dimensional plot exploiting the correlation between TMB and CPS score according to pretreatment probability of pathologic complete response (pT0N0). The plot is based on the actual biomarker data obtained from the PURE-01 study (n = 112). The x- and y-axes reported continuous values of TMB (0–45 mutations/Mb) and CPS (0%-99%), respectively. The z-axis reported the logistic probability of pT0N0 derived from interaction between TMB and CPS. Colors represent the pT0N0 probability tertile: <26% (first tertile), 26%-40% (second tertile), >40% (third tertile). CPS = combined positive score; TMB = tumor mutational burden.
Comparison of Matched Pre- and Posttherapy Tumor Samples
In total, there were 38 patients with matched pre- and posttherapy comparison who were suitable for CPS analyses. Of these case patients, 24 passed the quality control of tumor purity in posttherapy samples to allow for TMB assessment.
Supplementary Figure 6 (available online) summarizes the results of the matched TMB and CPS analyses. There was a statistically significant decrease of TMB in posttherapy lesions: the median TMB in posttherapy tumors was 10.1 mut/Mb vs 5 mut/Mb in TURBT samples (P = .005). Conversely, no statistically significant differences in CPS were observed: the median CPS value was 10% for both the TURBT and RC samples. No genomic alteration showed statistically significant differences between pretherapy and posttherapy samples.
Discussion
In this biomarker study, we developed a tool that was suitable for illustrating the complex interplay between the key biomarkers constituting the primary biomarker analyses of PURE-01 study: TMB and CPS. Similar findings are likely to emerge from neoadjuvant therapy trials using different checkpoint inhibitors. We have proposed a predictive biomarker model that quantifies the contribution of different tumor biomarkers against the pathologic response, pending validation with external datasets. The data presented here indicate that the main driver of pembrolizumab activity in the perioperative setting is undoubtedly the PD-L1 IHC expression, because it was the most robust predictor of pT0N0 response from multivariable analyses. These observations agree with the results obtained in the first-line metastatic setting where the label of pembrolizumab and atezolizumab use is restricted to PD-L1-positive patients (4).
Disappointingly, our results disagree with those reported with preoperative atezolizumab in the ABACUS study in which the authors did not find a statistically significant association between PD-L1 IHC expression and the pathologic response. In this study, the level of preexisting immunity, documented by CD8+ T-cell infiltration or by immune-related gene signatures, was associated with the pathologic response endpoint (8).
Indeed, unsolved complexities persist as regards the use and harmonization of different companion diagnostic assays. In addition, the prediction ability of PD-L1 expression could change with the use of combination neoadjuvant immunotherapies. The IHC-based CPS, which is used as a companion diagnostic test for pembrolizumab, can be used as a continuous variable, thus allowing the assessment of effects occurring across the entire spectrum of IHC expression. Despite the identification of a wide spectrum of genomic alterations in the RC specimens, when analyzed either as single-gene alterations or as genomic-pathway alterations, TMB emerged as a statistically significant predictor of tumor response to pembrolizumab, which corroborated our previous observations (6,7). Results from several studies have linked TMB with objective response, progression-free survival and overall survival with immune-checkpoint inhibitors in solid tumors (12-16). A meta-analysis across 27 tumor types showed a positive association between average response-rate and log (TMB) by linear regression analysis (16). However, there are complexities represented by the different activity profile of checkpoint inhibition in peculiar tumor entities and, most importantly, by the interaction with other biomarkers, most of which are unknown or not ready for clinical use. Owing to the complexity of tumor-immune interaction, marginal statistical association—or lack of association—between TMB and pT0N0 response in multivariable analyses, as seen in the current study, should be evaluated with extreme caution. Furthermore, when assessing the dual contribution of TMB and CPS in determining the pT0N0 probability, we found that very high TMB levels were associated with a statistically significant potential to achieve a pT0N0 response regardless of the PD-L1 expression, because this association was modeled using our calculator. As illustrated in Figure 2, there were, in fact, 26%-40% probabilities of pT0N0 (second tertile) also in a not negligible number of patients yielding TMB values in the range of 10-30 mut/Mb, despite a CPS less than 10%. We decided to incorporate the cT stage in the final model despite its lack of association with pT0N0 in multivariable models. The goal was to account for the effect of possible radical TURBT when analyzing the efficacy results, and the possibility to develop a pT0N0 prediction that relies on the clinical stage discrimination is certainly a valuable feature, despite the limitations in imaging assessment. As a practical application, when we have to decide between administering neoadjuvant pembrolizumab and providing immediate RC for a patient presenting with a cT2 stage tumor and who is cisplatin ineligible,a relatively high probability of achieving pT0N0 (eg, 30%) is warranted to justify the neoadjuvant therapy. Conversely, for a patient presenting with a cT4 tumor who cannot receive neoadjuvant chemotherapy, a 10% cutoff might be sufficient to consider a therapeutic success. Our predictive tool can also be useful to decide between using neoadjuvant pembrolizumab and standard chemotherapy, based on a consistent 30% pT0N0 rate achievable with the latter option. Therefore, our results could be used and validated to refine the design of some ongoing clinical trials, mainly those using single-agent pembrolizumab before RC. In particular, the Keynote-905 study (NCT03924895) is a randomized, open-label, phase 3 study of perioperative pembrolizumab plus RC vs RC alone in cisplatin-ineligible patients with MIBC. It allows the inclusion of all-comers regardless of any biomarker feature. However, according to our proposed calculator, a patient presenting with cT3-4 tumor, whose tumor is PD-L1–negative (CPS = 0-9), should have at least a TMB of 5 mut/Mb to have a 15% probability of pT0N0 after pembrolizumab, which may be considered close to the RC benchmark. After validation of our tool, lower predicted probabilities would not make the neoadjuvant phase ethical for this patient.
Limitations in this study should be acknowledged. First, we have observed substantial changes in posttherapy tumors regarding the TMB, and similar changes may happen in tumors progressing after a nonmuscle-invasive phase, as preliminarily suggested (17). Thus, the TMB value to use for the calculator should rely exclusively on the pretherapy TURBT samples revealing MIBC. Second, the incorporation of CPS and the heterogeneity in assessing PD-L1 expression based on the type of checkpoint inhibitor used will require validation of our findings within studies of other compounds, because several models could be generated using different checkpoint inhibitors. Third, substantial limitations in cT stage definition have already been presented from PURE-01 study (18). To date, we are unable to overcome this limitation, although the centralization of radiological imaging assessment to centers of excellence where one or more dedicated radiologists will stage the patient might substantially circumvent such unavoidable biases. Fourth, we are still waiting for mature follow-up duration to run survival analyses in our study. The notion that the pT0N0 response, or the tumor pathologic downstaging, is a surrogate endpoint of overall survival may not be necessarily valid in the postimmunotherapy context. Other surrogate features like the tumor regression grade (19) or the immune-infiltration and/or immune-gene signature features revealed within the posttherapy samples might have a more profound impact on survival postoperatively, thus making our tool suboptimal to predict patients’ survival. Last, as there are no similar data in the published literature that could serve as an external validation of this predictive model, the applicability of our findings will require a prospective multicentric collaboration with shared next-generation sequencing assays.
As an important next step, the model could be further optimized through the incorporation of gene expression data, which have recently emerged as additional biomarker findings from PURE-01. In particular, the use of immune-gene signatures allowed us to identify features characterizing the pretherapy tumor samples (eg, elevated scores of Decipher’s Immune190 signature), which were robustly associated with pathologic response to neoadjuvant pembrolizumab but not to neoadjuvant chemotherapy (20).
In summary, in the largest study conducted so far with neoadjuvant pembrolizumab in patients with MIBC, we have found that 37.5% of these patients have achieved a pT0N0 response postpembrolizumab but with critical differences between patient subgroups according to key predictive biomarkers TMB and CPS. We propose that the incorporation of the composite biomarker tool described in this study into the translational body of analyses of ongoing prospective studies has the potential in the future to optimize the selection of patients with MIBC who are best suited for neoadjuvant immune-checkpoint inhibitor therapy alone.
Funding
Merck & Co., Inc., Kenilworth, NJ, USA; Associazione Italiana per la Ricerca sul Cancro (AIRC); ClinicalTrials.gov, number NCT02736266.
Notes
Role of the funder: The funder had no role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; and the decision to submit the manuscript for publication.
Disclosures: R Madison, SM Ali, JH Chung, JS Ross are employees and stock owners of Foundation Medicine Inc. A Necchi is 1) consultant/advisor for Merck, Astra Zeneca, Janssen, Incyte, Roche, Rainier Therapeutics, Clovis Oncology, Bayer, and Astellas/Seattle Genetics, Ferring, Immunomedics; 2) grant recipient from Merck, Ipsen, and Astra Zeneca; 3) recipient of travel expenses/honoraria from Roche, Merck, Astra Zeneca, and Janssen. The other authors have no conflicts of interest to disclose.
Role of the authors: Design of the study: AN (Principal investigator). Manuscript draft: AN, MB. Manuscript revision and approval: All authors. Statistical analysis: MB. Supervision: FM.
Supplementary Material
References
- 1. Alfred Witjes J, Lebret T, Compérat EM, et al. Updated 2016 EAU guidelines on muscle-invasive and metastatic bladder cancer. Eur Urol. 2017;71(3):462–475. [DOI] [PubMed] [Google Scholar]
- 2. Witjies JA, Bruins M, Cathomas R, et al. EAU Guidelines. Edn. Presented at the EAU Annual Congress Barcelona; 2019. ISBN 978-94-92671-04-2.
- 3. Grossman HB, Natale RB, Tangen CM, et al. Neoadjuvant chemotherapy plus cystectomy compared with cystectomy alone for locally advanced bladder cancer. N Engl J Med. 2003;349(9):859–866. [DOI] [PubMed] [Google Scholar]
- 4. Feld E, Harton J, Meropol NJ, et al. Effectiveness of first-line immune checkpoint blockade versus carboplatin-based chemotherapy for metastatic urothelial cancer. Eur Urol. 2019;76(4):524–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Niglio SA, Jia R, Ji J, et al. Programmed death-1 or programmed death ligand-1 blockade in patients with platinum-resistant metastatic urothelial cancer: a systematic review and meta-analysis. Eur Urol. 2019;76:782–789. [DOI] [PubMed] [Google Scholar]
- 6. Necchi A, Anichini A, Raggi D, et al. Pembrolizumab as neoadjuvant therapy before radical cystectomy in patients with muscle-invasive urothelial bladder carcinoma (PURE-01): an open-label, single-arm, phase II study. J Clin Oncol. 2018;36(34):3353–3360. [DOI] [PubMed] [Google Scholar]
- 7. Necchi A, Raggi D, Gallina A, et al. Updated results of PURE-01 with preliminary activity of neoadjuvant pembrolizumab in patients with muscle-invasive bladder carcinoma with variant histologies. Eur Urol. 2020;77:439–446. [DOI] [PubMed] [Google Scholar]
- 8. Powles T, Kockx M, Rodriguez-Vida A, et al. Clinical efficacy and biomarker analysis of neoadjuvant atezolizumab in operable urothelial carcinoma in the ABACUS trial. Nat Med. 2019;25(11):1706–1714. [DOI] [PubMed] [Google Scholar]
- 9. Briganti A, Gandaglia G, Scuderi S, et al. Surgical safety of radical cystectomy and pelvic lymph node dissection following neoadjuvant pembrolizumab in patients with bladder cancer: prospective assessment of perioperative outcomes from the PURE-01 trial. Eur Urol. 2020;77:576–580. [DOI] [PubMed] [Google Scholar]
- 10. Moschini M, Arbelaez E, Cornelius J, et al. Pattern of node metastases in patients treated with radical cystectomy and extended or superextended pelvic lymph node dissection due to bladder cancer. Urol Oncol. 2018;36(6):307.e9–307.e14. [DOI] [PubMed] [Google Scholar]
- 11. Mateo J, Chakravarty D, Dienstmann R, et al. A framework to rank genomic alterations as targets for cancer precision medicine: the ESMO scale for clinical actionability of molecular targets. Ann Oncol. 2018;29(9):1895–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee JS, Ruppin E.. Multiomics prediction of response rates to therapies to inhibit programmed cell death 1 and programmed cell death 1 ligand 1. JAMA Oncol. 2019;5(11):1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tan TZ, Rouanne M, Tan KT, et al. Molecular subtypes of urothelial bladder cancer: results from a meta-cohort analysis of 2411 tumors. Eur Urol. 2019;75(3):423–432. [DOI] [PubMed] [Google Scholar]
- 14. Lu S, Stein JE, Rimm DL, et al. Comparison of biomarker modalities for predicting response to PD-1/PD-L1 checkpoint blockade: a systematic review and meta-analysis. JAMA Oncol. 2019;5(8):1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Simeone I, Hendrickx W, Miller L, et al. Toward a comprehensive view of cancer immune responsiveness: a synopsis from the SITC workshop. J Immunother Cancer. 2015;3(suppl 1):P1. [Google Scholar]
- 16. Zhu J, Zhang T, Li J, et al. Association between tumor mutation burden (TMB) and outcomes of cancer patients treated with PD-1/PD-L1 inhibitions: a meta-analysis. Front Pharmacol. 2019;10:673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Meeks JJ, Carneiro BA, Pai SG, et al. Genomic characterization of high-risk non-muscle invasive bladder cancer. Oncotarget. 2016;7(46):75176–75184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Necchi A, Bandini M, Calareso G, et al. Multiparametric magnetic resonance imaging as a noninvasive assessment of tumor response to neoadjuvant pembrolizumab in muscle-invasive bladder cancer: preliminary findings from the PURE-01 study. Eur Urol. 2020;77:636–643. [DOI] [PubMed] [Google Scholar]
- 19. Voskuilen CS, Oo HZ, Genitsch V, et al. Multicenter validation of histopathologic tumor regression grade after neoadjuvant chemotherapy in muscle-invasive bladder carcinoma. Am J Surg Pathol. 2019;43(12):1600–1610. [DOI] [PubMed] [Google Scholar]
- 20. Necchi A, Raggi D, Gallina A, et al. Impact of molecular subtyping and immune infiltration on pathological response and outcome following neoadjuvant pembrolizumab in muscle-invasive bladder cancer. Eur Urol. 2020;77:701–710. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.