Abstract
Background
With the advent of primary human papillomavirus testing followed by cytology for cervical cancer screening, visual interpretation of cytology slides remains the last subjective analysis step and suffers from low sensitivity and reproducibility.
Methods
We developed a cloud-based whole-slide imaging platform with a deep-learning classifier for p16/Ki-67 dual-stained (DS) slides trained on biopsy-based gold standards. We compared it with conventional Pap and manual DS in 3 epidemiological studies of cervical and anal precancers from Kaiser Permanente Northern California and the University of Oklahoma comprising 4253 patients. All statistical tests were 2-sided.
Results
In independent validation at Kaiser Permanente Northern California, artificial intelligence (AI)-based DS had lower positivity than cytology (P < .001) and manual DS (P < .001) with equal sensitivity and substantially higher specificity compared with both Pap (P < .001) and manual DS (P < .001), respectively. Compared with Pap, AI-based DS reduced referral to colposcopy by one-third (41.9% vs 60.1%, P < .001). At a higher cutoff, AI-based DS had similar performance to high-grade squamous intraepithelial lesions cytology, indicating a risk high enough to allow for immediate treatment. The classifier was robust, showing comparable performance in 2 cytology systems and in anal cytology.
Conclusions
Automated DS evaluation removes the remaining subjective component from cervical cancer screening and delivers consistent quality for providers and patients. Moving from Pap to automated DS substantially reduces the number of colposcopies and also achieves excellent performance in a simulated fully vaccinated population. Through cloud-based implementation, this approach is globally accessible. Our results demonstrate that AI not only provides automation and objectivity but also delivers a substantial benefit for women by reduction of unnecessary colposcopies.
Advances in digital imaging and machine learning can revolutionize cancer screening, diagnosis, and treatment by improving accuracy and reproducibility of image assessment and streamlining clinical workflow (1–4). With its requirement for high throughput and fast turnaround and its dependence on microscopic and visual technologies, automation can play a major role in improving the efficiency of cervical cancer screening. Many countries are currently switching from Pap cytology to high-risk human papillomavirus (HPV) screening (5–7). Although a negative HPV test provides great reassurance of low cervical cancer risk over the next decade (8–10), only a small subset of women with a positive HPV test require further evaluation. To avoid overburdening the system with HPV-positive women, additional triage is required for colposcopy referral (11, 12). Current triage strategies include partial HPV genotyping and Pap cytology (7, 13). The limited sensitivity and reproducibility of cytology require laborious quality control procedures and frequent retesting (14, 15). Improving the efficiency of cervical cancer screening is particularly important for vaccinated populations due to lower disease prevalence and higher demands for screening test performance.
A promising triage strategy is concomitant detection of p16 and Ki-67 in the same cell (p16/Ki-67 dual stain [DS]), 2 markers that are closely linked to cervical carcinogenesis and HPV oncoprotein actions. The HPV oncoprotein E7 interrupts cell cycle control by releasing E2F, activating p16 expression. The coexpression of p16 and Ki-67, a cell proliferation marker, in the same cell is specific to HPV-related carcinogenesis. DS has shown greater accuracy for detection of HPV-related precancers compared with cytology (16–21). Currently, artificial intelligence (AI) algorithms mostly try to match manual reading accuracy to improve automation but do not offer a substantial improvement for patients. Automated scanning and deep-learning evaluation of DS slides can improve throughput, reproducibility, and accuracy of the assay for better risk stratification and a direct benefit to women (8, 22, 23). To achieve this, we developed the CYTOREADER system that combines whole-slide scanning with automated evaluation of DS cytology slides. Cloud-based evaluation provides ample computational capacity and storage space and can provide diagnostic procedures where sufficient personnel, expertise, or infrastructure is lacking. We evaluated the clinical performance of CTYOREADER in 4253 slides from 3 epidemiological studies of HPV-positive cervical and anal precancers.
Methods
General Approach
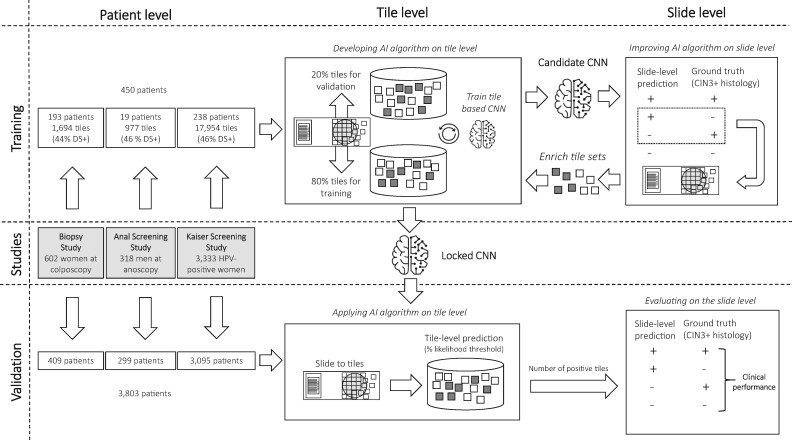
CYTOREADER uses whole-slide scanners (Hamamatsu Nanozoomers HT, XR, and S360) for imaging of ThinPrep (Hologic) or SurePath (Becton Dickinson, BD) slides, 2 widely used liquid-based cytology technologies. CYTOREADER is a cloud-based system (Google Cloud Platform) that can also run as a local installation. Training of deep-learning algorithms for automated DS evaluation was performed using small areas (tiles) from whole slides containing individual or small numbers of epithelial cells. For training of the deep-learning algorithms, tiles from training slides were manually evaluated for DS-positive cells by 3 observers (Supplementary Figure 1, available online).
Deep Learning
Two deep-learning approaches (Convolutional Neural Network with 4 layers [CNN4] and Inception-v3 with 48 layers [IncV3]) were developed sequentially as shown in Figure 1 and described in Supplementary Methods (available online). The algorithms determine the number of DS-positive cells on a slide by detecting the number of tiles above a certain likelihood threshold. A slide is considered positive if the number of DS-positive cells on a slide exceeds a certain cutoff. Training and validation were conducted on the tile level and the slide level. First, a training set from 450 patients was selected for which the clinical endpoint cervical intraepithelial neoplasia grade 3 or greater (CIN3+) was unblinded. Tiles were selected for initial training (80%) and validation (20%) of the algorithm. The deep-learning network provides a likelihood for each tile above which it is considered positive (0.5 for CNN4 and 0.4 for IncV3). The resulting candidate CNN was applied on the slide level on training slides. A cutoff of positive tiles is used to determine slide positivity (≥3 tiles per cell for CNN4 and ≥2 tiles per cell for IncV3). From misclassified slides, false-positive or false-negative tiles were extracted and fed back into the original CNN training to optimize classification accuracy of the CNN. A final locked CNN was applied on the patient level on the blinded validation set comprising 3803 slides. CNN4 showed good performance in Thinprep slides but not in Surepath slides. Subsequently, a second algorithm (IncV3) was trained specifically for Surepath slides (Supplementary Methods, available online). We published a GitHub repository and created a web page at https://github.com/stcmedhub/dual_stain_dl with a source code description of the models and the installation instructions.
Figure 1.
Study design. AI = artificial intelligence; CNN = convolutional neural network; CIN3+ = cervical intraepithelial neoplasia grade 3 or worse; DS = dual stain.
Study Populations
The Biopsy Study is a population-based study of women aged 18 years or older referred to colposcopy at the University of Oklahoma Health Sciences Center between 2009 and 2011 (24). We included DS slides from 602 women as previously described (19). The study population was split into a representative training set (193 slides with 741 DS-positive and 953 DS-negative tiles) and a validation set of 409 slides (Figure 1). This study was approved by the University of Oklahoma and National Cancer Institute (NCI) institutional review boards (IRB); written informed consent was obtained from all participants before study enrollment.
The Anal Cancer Screening Study (ACSS) was based at the San Francisco Kaiser Permanente Northern California (KPNC) Anal Cancer Screening Clinic. HIV-positive men who have sex with men 18 years or older were enrolled at KPNC between 2009 and 2010. DS slides from 318 men were generated as previously described (25). From 19 training slides, 445 DS-positive and 532 DS-negative tiles were used for training (Figure 1). This study was approved by the KPNC and NCI IRBs; written informed consent was obtained from all participants before study enrollment.
At KPNC, DS was evaluated for triage of HPV-positive women between 2012 and 2015 in a population of women aged 25 years and older who were undergoing routine cervical cancer screening (16). From a screening population of more than 300 000 women in a year, 3333 slides from HPV-positive women were included. From 238 training slides, 8215 DS-positive and 9739 DS-negative tiles were used for training (Figure 1). The study was approved by the KPNC IRB and was exempted from institutional review at the NCI by the Office of Human Subjects Research. Patient consent was waived because deidentified discarded specimens were used in this study.
Clinical Endpoints
All studies followed routine clinical practice at the respective institutions. Cytology was classified by the Bethesda System: negative for intraepithelial lesions or malignancy, atypical squamous cells of undetermined significance, low-grade squamous intraepithelial lesions, and high-grade squamous intraepithelial lesions (HSIL) (26). Final diagnosis was established by histopathology classified according to the cervical intraepithelial neoplasia (CIN) scale for cervical endpoints, which indicates the extent of dysplastic cells in the cervical epithelium: no indication for biopsy, normal CIN, grade 1 (CIN1), grade 2 (CIN2), grade 3 (CIN3), and cancer. We grouped adenocarcinoma in situ with CIN3. For anal disease endpoints, the comparable anal intraepithelial neoplasia nomenclature (AIN) was used.
p16/Ki-67 Staining and Evaluation
For the Biopsy Study and ACSS, slides were prepared from residual PreservCyt material using a T2000 processor (Hologic, Bedford, MA). For the KPNC study, slides were prepared from residual SurePath tubes according to the manufacturer’s instructions (BD, Sparks, MD). Immunostaining of cervical cytology slides for p16/Ki-67 was performed using the CINtec Plus Kit (Roche, Tucson, AZ) according to the manufacturer’s instructions. DS-trained cytotechnologists reviewed all slides; a slide was considered positive if 1 or more cervical epithelial cell(s) stained both with a brown cytoplasmic stain (p16) and a red nuclear (Ki-67) irrespective of morphologic abnormalities. Slides from the Biopsy Study and ACSS were stained and evaluated at Roche mtm laboratories AG, Heidelberg, Germany, whereas slides from the Kaiser DS study were stained and evaluated at KPNC. HPV testing with partial genotyping (HPV16 and HPV18) at KPNC was based on the cobas assay (Roche, Pleasanton, CA).
Statistical Analysis
We created boxplots and calculated medians to show the distribution of DS-positive cells in cytology and histology categories. We compared differences in DS cell counts in ordinal cytology and histology categories using 1-way analysis of variance. The primary endpoint for the Biopsy Study and the Kaiser Study was CIN3 or greater (CIN3+). For ACSS, the primary endpoint was AIN2 or AIN3 (AIN2+). Receiver operator characteristics curve analysis was conducted for the number of DS-positive cells against the primary endpoints, and the area under the curve (AUC) was calculated. Sensitivity and specificity coordinates for manual DS evaluation and cytology were plotted on the receiver operator characteristics curve for comparison. We calculated percentage positivity, sensitivity, specificity, and Youden’s index in the Biopsy Study and ACSS for the cutoff determined by CNN4 and for manual DS evaluation. In the Kaiser Study, with a representative population of women who underwent routine screening, we calculated percentage positivity, sensitivity, specificity, and positive and negative predictive values for automated and manual DS. Differences in positivity, sensitivity, and specificity were evaluated using an exact McNemar’s χ2, and differences in predictive values were evaluated using the R package DTComPair, using the generalized score statistic (27). To evaluate clinical efficiency of each strategy, we estimated the number of CIN3+ detected for different cutoffs of DS-positive cells and the ratio of the number of tests and colposcopies per case of CIN3+ detected. We also evaluated the theoretical performance of automated DS in a fully vaccinated population by excluding all women who were positive for HPV16 and/or HPV18 from the analysis. Analyses were performed in SPSS, Stata, and R. All statistical tests were 2-sided and P less than .05 was considered statistically significant.
Results
Automated Detection of DS-Positive Cells in Colposcopy and Anoscopy Populations
We developed a deep-learning algorithm for automated detection of DS-positive cells on ThinPrep slides from 2 referral populations (Biopsy Study and ACSS), including 212 training slides with 1186 DS-positive and 1485 DS-negative tiles (Figure 1). We evaluated the algorithm in independent validation slides from both studies (Figure 1). In both studies, we observed an increase in the number of DS-positive cells by increasing severity of cytology and histology, with higher absolute DS-positive cell numbers in ACSS (P < .001 for all comparison; Supplementary Figure 2, available online).
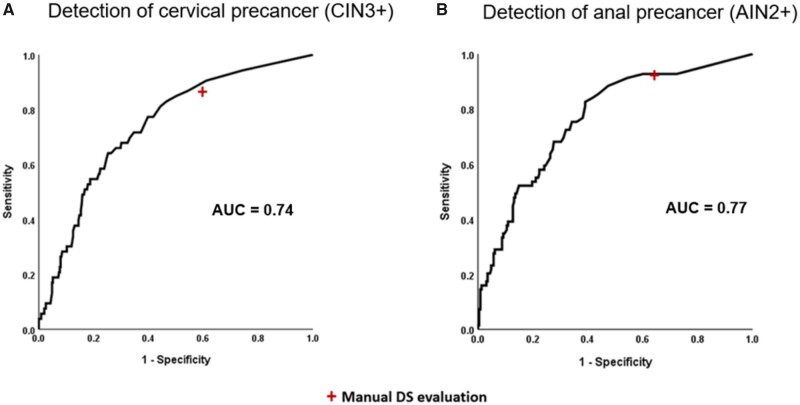
In the Biopsy Study validation set with 53 CIN3+, the AUC for detecting CIN3+ based on automated DS using CNN4 was 0.74 (Figure 2). At a cutoff of 3 DS-positive cells, the CNN4 algorithm had marginally lower positivity (58% vs 63%, respectively, P = .06) with comparable sensitivity (P = 1.0) and marginally higher specificity compared with manual DS (40.6% vs 45.7%, respectively, P = .07) (Table 1).
Figure 2.
Receiver operating curve characteristics analysis of number of dual stain (DS)-positive cells detected by CYTOREADER for detection of cervical precancer in the Biopsy Study and anal precancer in the Anal Cancer Screening Study. AUC = area under the curve; AIN2+ = anal intraepithelial neoplasia grade 2 or worse; CIN3+ = cervical intraepithelial neoplasia grade 3 or worse.
Table 1.
Accuracy for cervical and anal precancer based on manual and automated detection of DS-positive cells on ThinPrep slides in a colposcopy population (Biopsy Study, N = 409) and an anoscopy population (ACSS, N = 299)
| Evaluation | Positive |
AUC | Sensitivity |
Specificity |
Youden’s index | |||
|---|---|---|---|---|---|---|---|---|
| % | P a | % (95% CI) | P a | % (95% CI) | P a | |||
| Biopsy Study validation set (CIN3+) | ||||||||
| Manual | 63.1 | Ref | 87.0 (75.6 to 93.6) | Ref | 40.5 (35.6 to 45.7) | Ref | 0.27 | |
| CNN4 | 57.9 | .06 | 0.74 | 87.0 (75.6 to 93.6) | 1.0 | 45.6 (40.5 to 50.8) | .07 | 0.33 |
| ACSS validation set (AIN2+) | ||||||||
| Manual | 71.0 | Ref | 92.8 (82.2 to 96.5) | Ref | 36.1 (30.3 to 42.4) | Ref | 0.29 | |
| CNN4b | 62.9 | <.001 | 0.77 | 91.3 (80.2 to 95.4) | 1.0 | 46.1 (40.0 to 52.6) | <.001 | 0.37 |
Two-sided McNemar’s test. ACCSS = Anal Cancer Screening Study; AIN2+ = anal intraepithelial neoplasia grade 2 or worse; AUC = area under the curve; CIN3+ = cervical intraepithelial neoplasia grade 3 or worse; DS = dual stain; CNN = convolutional neural network; Ref = referent.
CNN4 cutoff for Biopsy: 3 or more cells; CNN4 cutoff for Anal: 3 or more cells.
In the ACSS validation set with 69 AIN2+, the AUC for detecting AIN2+ based on automated evaluation of DS slides with CNN4 was 0.77 (Figure 2). At a cutoff of 3 DS-positive cells, the positivity of the CNN4 algorithm was lower (63% vs 71%, respectively, P = .001) with comparable sensitivity (P = 1.0) and higher specificity compared with manual DS (36.1% vs 46.1%, respectively, P = .001) (Table 1).
Automated Detection of DS-Positive Cells in an HPV Screening Population
We developed the deep-learning algorithm for SurePath slides using a training set of 238 slides from the Kaiser study with 8215 DS-positive and 9739 DS-negative tiles and applied it in an independent validation set of slides from 3095 women. We observed an increase of DS-positive cells with increasing severity of cytology and histology (Supplementary Figure 3, available online).
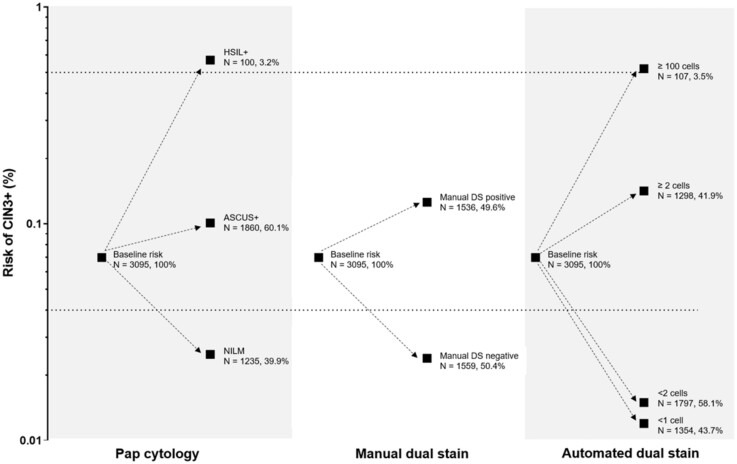
In the Kaiser validation study including 218 CIN3+, the AUC for detecting CIN3+ based on automated evaluation of DS slides was 0.82 (Figure 3). At a cutoff of 2 cells, the positivity of the algorithm was statistically significantly lower (42% vs 50%, respectively, P < .001) with equal sensitivity (P = .4) but statistically significantly higher specificity (61.5% vs 52.6%, respectively, P < .001) compared with the manual DS. At a cutoff of 100 cells, accuracy approached HSIL cytology that allows for immediate treatment according to current management guidelines (Figure 3). Automated DS provided better risk stratification compared with Pap cytology and manual DS (Figure 4): more women were reassured of a lower risk compared with the other strategies (58% for automated DS vs 50% for manual DS and 40% for cytology), and risk among positives was higher.
Figure 3.
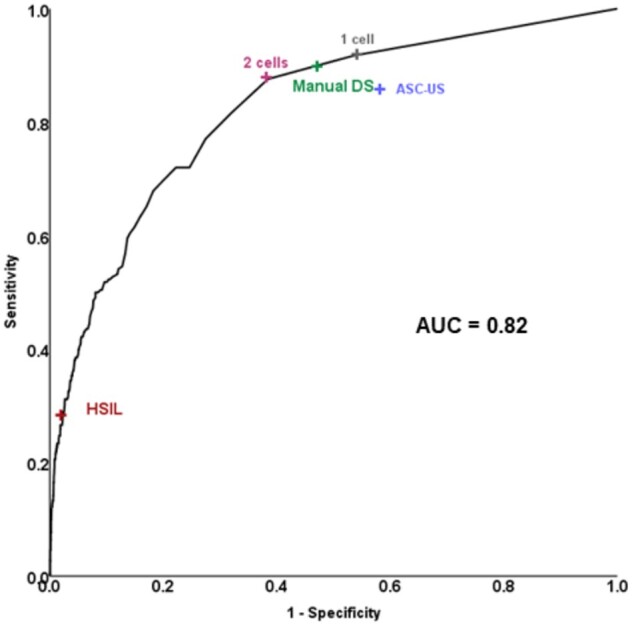
Receiver operating curve characteristics analysis of number of dual stain (DS)-positive cells detected by CYTOREADER for detection of cervical precancer in a human papillomavirus screening population in Kaiser Permanente Northern California. ASC-US = Atypical Squamous Cells of Undetermined Significance; AUC = area under the curve; HSIL = high-grade squamous intraepithelial lesions.
Figure 4.
Absolute risk of precancer for Pap cytology, manual dual stain (DS), and automated DS. ASCUS+= positive for Atypical Squamous Cells of Undetermined Significance or greater cytology results. The dotted lines show clinical action risk thresholds for colposcopy referral (4% risk) and immediate treatment (50% risk).
Clinical Efficiency of Automated DS Evaluation
We compared the clinical efficiency of Pap cytology, manual DS, and automated DS at 2 cutoffs (2 or more cells and 1 or more cells) for triage of HPV-positive women (Table 2). All DS strategies achieved equal or better sensitivity for detection of CIN3+ compared with Pap cytology while reducing unnecessary colposcopic referrals. Automated DS reduced overall referral to colposcopy by one-third for the primary automated cutoff of 2 cells (41.9% for automated DS vs 60.1% for cytology, P < .001). Automated DS at a cutoff of 2 or more cells had similar sensitivity but statistically significantly higher specificity compared with manual DS evaluation (61.5% vs 52.6%, P < .001). Automated DS detection at a cutoff of 1 or more cells achieved the highest sensitivity of all strategies, with statistically significantly higher specificity and lower colposcopy referral compared with Pap cytology. Automated DS at a cutoff of 2 or more cells had the most favorable ratio of colposcopies per CIN3+ detected compared with the least favorable ratio for the current standard, Pap cytology (6.8 vs 9.9, respectively). Extrapolating this to the full Kaiser screening population, out of 300 000 women screened annually, approximately 30 000 would test HPV-positive and more than 18 000 would be referred to colposcopy using the current approach with Pap cytology, while only 12 570 would be referred to colposcopy using automated DS. We also estimated the performance of automated DS in a fully vaccinated population. Similar to the overall evaluation, automated DS showed equal sensitivity and lower colposcopy referral compared with Pap cytology with even higher specificity (Supplementary Table 1, available online).
Table 2.
Accuracy for cervical precancer based on Pap cytology and manual and automated detection of DS-positive cells on SurePath slides in the Kaiser Validation Study (N = 3095)
| Evaluation | Colposcopy referral, No. (%) | P a (cytology/manual DS) | Sensitivity, % (95% CI) | P a (cytology/manual DS) | Specificity, % (95% CI) | P a (cytology/manual DS) | PPV, % (95% CI) | P b (cytology/ manual DS) | NPV, % (95% CI) | P b (cytology/manual DS) | Colposcopies per CIN3+ detected, No. (95% CI) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Pap cytology (188 CIN3+) | 1860 (60.1) | Ref | 85.8 (81.2 to 90.5) | Ref | 41.9 (40.1 to 43.7) | Ref | 10.1 (8.7 to 11.5) | Ref | 97.5 (96.6 to 98.3) | Ref | 9.9 (8.7 to 11.3) |
| Manual DS (197 CIN3+) | 1536 (49.6) | <.001/ Ref | 90.0 (86.0 to 93.9) | .2/Ref | 52.6 (50.8 to 54.5) | <.001/ Ref | 12.6 (11.0 to 14.3) | <.001/ Ref | 98.6 (98.0 to 99.2) | .02/Ref | 7.8 (6.8 to 8.9) |
| Automated DS (2 cells) (192 CIN3+) | 1298 (41.9) | <.001/ <.001 | 88.1 (82.5 to 91.7) | .6/.4 | 61.5 (59.7 to 63.3) | <.001/ <.001 | 14.8 (12.9 to 16.8) | <.001/ <.001 | 98.5 (97.8 to 99.0) | .03/.8 | 6.8 (5.9 to 7.7) |
| Automated DS (1 cell) (201 CIN3+) | 1741 (56.3) | .007/ <.001 | 91.8 (87.3 to 95.1) | .05/.5 | 46.5 (44.6 to 48.3) | .06/ <.001 | 11.5 (10.1 to 13.1) | .002/ <.001 | 98.7 (97.9 to 99.2) | .01/.5 | 8.7 (7.6 to 9.9) |
Two-sided McNemar’s test. CIN3+ = cervical intraepithelial neoplasia grade 3 or worse; DS = dual stain; NPV = negative predictive value; PPV = positive predictive value; Ref = referent.
Two-sided generalized score statistic.
Discussion
Using a rigorous study design, we developed a novel deep-learning–based image analysis platform for automated evaluation of DS cytology. In a large population of women undergoing HPV-based cervical cancer screening, we show that automated evaluation of DS slides dramatically increases the efficiency of cervical cancer screening by substantially reducing unnecessary colposcopies compared with current standards and similarly achieves excellent performance in a simulated fully vaccinated population. Thus, CYTOREADER exceeds human diagnostic accuracy and serves as an example of AI achieving improvements beyond the automation of a human standard.
Our results demonstrate how automation and machine learning can transform cervical cancer screening that is currently undergoing major changes. HPV testing for cervical cancer screening is an objective and reliable approach directly linked to the carcinogenic process (28). HPV-negative women are at very low risk of developing precancer or cancer over the next decade and screening intervals can be extended (8–10). Yet most HPV infections are transient, and women require additional tests to decide who needs further evaluation or treatment (11, 12). Pap cytology is recommended and approved for triage of HPV-positive women but suffers from subjectivity, lack of reproducibility, and relatively low sensitivity (14). Our previous study comparing manual DS to cytology together with the current results demonstrates that automated DS evaluation can supplant and improve the role of Pap cytology for triage of HPV-positive women and should also be evaluated for postcolposcopy and posttreatment surveillance (16). Compared with Pap cytology, manual DS has higher accuracy and can provide longer reassurance against disease when a test is negative, while the risks to patients do not differ from Pap cytology, because the same sample type is used (17, 21). We previously showed that the few DS-negative CIN3s are more likely to have no HPV16/18 and no high-grade cytology, suggesting that these cases are less likely to progress (16). Automated DS evaluation can provide a completely objective cervical cancer–screening approach, improving efficiency and reducing harms and cost related to false-positive screening results. Furthermore, by demonstrating that AI-based DS detection works for anal cytology, we show the robustness of the imaging and analysis platform. Importantly, our approach is also suited for vaccinated populations, where it may achieve even higher specificity and counterbalance the lower disease prevalence in vaccinated women (29).
Automated DS evaluation immediately quantifies the number of DS-positive cells on a slide, allowing tailoring positivity cutoffs for specific clinical decisions. Current guidelines give an option for immediate treatment in women with HSIL cytology, who have a very high probability of having underlying CIN3+ (30). A higher cutoff of DS-positive cells could be used to guide treatment decisions. Moving forward, additional criteria can be developed to expand slide assessment; for example, the presence of abnormal glandular cells to identify adenocarcinoma precursors, which is a particular challenge for Pap cytology (31).
Digitization of glass slides paired with automated evaluation in the cloud can provide high-throughput triage of HPV-positive women with inherent objectivity. Furthermore, the functionality of CYTOREADER can provide an assisted diagnostics mode for evaluating DS slides. The automatic algorithm can be used for presenting all DS-positive cells found on a slide ranked by the likelihood that a cell is DS-positive to accelerate slide evaluation. Similarly, CYTOREADER can be used for quality control of a program that is based on manual DS evaluation.
Successful implementation of CYTOREADER requires an infrastructure for high-quality staining, full-slide scanning, and running the machine-learning algorithm. However, slide preparation, scanning, and slide evaluation can be geographically separated, providing high-quality cervical cancer screening and triage in locations that currently do not have infrastructure and training to achieve reliable DS evaluation given a reliable courier system is available. Compared with manual evaluation of DS slides, the automated evaluation requires access to scanning infrastructure but may require a smaller cytotechnology workforce. Scanners are increasingly available in pathology laboratories and can process large batches of slides with limited need for a skilled operator (22, 23). Studies are warranted to evaluate if DS is amenable to self-collected specimens, a sampling strategy that is important for low-resource settings. Future efforts also need to evaluate how long a negative automated DS result provides reassurance against precancer and how automated DS can be used in women undergoing surveillance.
We conducted a large, well-powered study to evaluate performance of automated DS for triage of HPV-positive women. However, some limitations should be noted. In contrast to the large KPNC study on HPV triage based on SurePath slides, 2 studies using ThinPrep slides were comparably small, and they were conducted in colposcopy/anoscopy populations. Future studies need to evaluate automated DS in larger HPV screening populations using ThinPrep slides. Also, the positivity and sensitivity of cytology at KPNC is much higher compared with other settings, which may affect the comparison of clinical efficiency estimates.
Our approach to train and validate both on the tile level and the slide level with ground truth disease endpoints sets our work apart from other deep-learning approaches in digital pathology that focus on replicating a subjective evaluation. We recognize that there is substantial subjectivity underlying histologic endpoints of cervical disease (15). In our study, we minimized the impact by relying on the most reproducible correlate of cervical precancer, CIN3, as our primary endpoint for evaluation of triage of HPV-positive women. Our work also emphasizes the importance of integrating epidemiology and AI with the availability of population bases studies to improve medical diagnostics beyond automation. It has been proposed for a long time that “digital pathology” will become an important cornerstone of future health care. Despite this vision, image analysis currently does not contribute substantially to routine clinical practice and to the benefit of the patient. The automated evaluation of DS cytology slides has substantially improved accuracy and efficiency compared with Pap cytology and serves as an important example for introducing digital pathology and deep learning into clinical practice. This approach has the potential to substantially improve screening program performance, potentially affecting millions of women testing HPV-positive in cervical cancer screening each year.
Funding
This work was supported by the Intramural Research Program of the US National Cancer Institute, National Institutes of Health, Department of Health and Human Services.
Notes
Role of the funder: The funder had no role in the design, of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; and the decision to submit the manuscript for publication.
Conflict of interest: Drs. Wentzensen and Schiffman are employed by the National Cancer Institute (NCI), which has received cervical cancer screening assays in-kind or at reduced cost from BD and Roche for studies that Drs. Wentzensen and Schiffman are conducting. Dr Goldhoff reported receiving grants from the National Institutes of Health (NIH)/NCI during the conduct of the study. Dr Castle reported receiving cervical screening tests and diagnostics from Roche, Becton Dickinson, Cepheid, and Arbor Vita Corp at a reduced cost or no cost for research. Dr Kingery reported receiving grants from the NIH and the NCI during the conduct of the study and receiving grants from the NIH and the NCI outside the submitted work. Dr Grewal reported receiving grants from the NIH and the NCI during the conduct of the study and grants from the NIH and the NCI outside the submitted work. Dr Lorey reported receiving grants from the NIH and the NCI during the conduct of the study and grants from the NIH and the NCI outside the submitted work. No other disclosures were reported.
Data accessibility statement: The code is available at: https://github.com/stcmedhub/dual_stain_dl.
Author contributions: All authors contributed substantially to the conception and design of the study, the acquisition of data, or the analysis and interpretation. All authors drafted or provided critical revision of the article and provided final approval of the version to publish.
Supplementary Material
References
- 1. Bi WL, Hosny A, Schabath MB, et al. Artificial intelligence in cancer imaging: clinical challenges and applications. CA Cancer J Clin. 2019;69(2):127–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hinton G. Deep learning-a technology with the potential to transform health care. JAMA. 2018;320(11):1101–1102. [DOI] [PubMed] [Google Scholar]
- 3. Shouval R, Labopin M, Bondi O, et al. Prediction of allogeneic hematopoietic stem-cell transplantation mortality 100 days after transplantation using a machine learning algorithm: a European Group for blood and marrow transplantation acute leukemia working party retrospective data mining study. J Clin Oncol. 2015;33(28):3144–3151. [DOI] [PubMed] [Google Scholar]
- 4. Stead WW. Clinical implications and challenges of artificial intelligence and deep learning. JAMA. 2018;320(11):1107–1108. [DOI] [PubMed] [Google Scholar]
- 5. Wentzensen N, Arbyn M, Berkhof J, et al. Eurogin 2016 roadmap: how HPV knowledge is changing screening practice. Int J Cancer. 2017;140(10):2192–2200. [DOI] [PubMed] [Google Scholar]
- 6. Schiffman M, Wentzensen N, Wacholder S, et al. Human papillomavirus testing in the prevention of cervical cancer. J Natl Cancer Inst. 2011;103(5):368–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Curry SJ, Krist AH, Owens DK, et al. ; US Preventive Services Task Force. Screening for cervical cancer: US Preventive Services Task Force recommendation statement. JAMA. 2018;320(7):674–686. [DOI] [PubMed] [Google Scholar]
- 8. Gage JC, Schiffman M, Katki HA, et al. Reassurance against future risk of precancer and cancer conferred by a negative human papillomavirus test. J Natl Cancer Inst. 2014;106(8):dju153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dillner J, Rebolj M, Birembaut P, et al. Long term predictive values of cytology and human papillomavirus testing in cervical cancer screening: joint European cohort study. BMJ. 2008;337(oct13 1):a1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Katki HA, Kinney WK, Fetterman B, et al. Cervical cancer risk for women undergoing concurrent testing for human papillomavirus and cervical cytology: a population-based study in routine clinical practice. Lancet Oncol. 2011;12(7):663–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cuschieri K, Ronco G, Lorincz A, et al. Eurogin roadmap 2017: triage strategies for the management of HPV-positive women in cervical screening programs. Int J Cancer. 2018;143(4):735–745. [DOI] [PubMed] [Google Scholar]
- 12. Wentzensen N, Schiffman M, Palmer T, et al. Triage of HPV positive women in cervical cancer screening. J Clin Virol. 2016;76(Suppl 1):S49–S55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huh WK, Ault KA, Chelmow D, et al. Use of primary high-risk human papillomavirus testing for cervical cancer screening: interim clinical guidance. J Low Genit Tract Dis. 2015;19(2):91–96. [DOI] [PubMed] [Google Scholar]
- 14. Wright TC Jr, Stoler MH, Behrens CM, et al. Interlaboratory variation in the performance of liquid-based cytology: insights from the ATHENA trial. Int J Cancer. 2014;134(8):1835–1843. [DOI] [PubMed] [Google Scholar]
- 15. Stoler MH, Schiffman M.. Interobserver reproducibility of cervical cytologic and histologic interpretations: realistic estimates from the ASCUS-LSIL Triage Study. JAMA. 2001;285(11):1500–1505. [DOI] [PubMed] [Google Scholar]
- 16. Wentzensen N, Clarke MA, Bremer R, et al. Clinical evaluation of HPV screening with p16/Ki-67 dual stain triage in a large organized cervical cancer screening program. JAMA Intern Med. 2019;179(7):881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Clarke MA, Cheung LC, Castle PE, et al. Five-year risk of cervical precancer following p16/Ki-67 dual-stain triage of HPV-positive women. JAMA Oncol. 2019;5(2):181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wentzensen N, Fetterman B, Castle PE, et al. p16/Ki-67 dual stain cytology for detection of cervical precancer in HPV-positive women. J Natl Cancer Inst. 2015;107(12):djv257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wentzensen N, Schwartz L, Zuna RE, et al. Performance of p16/Ki-67 immunostaining to detect cervical cancer precursors in a colposcopy referral population. Clin Cancer Res. 2012;18(15):4154–4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wright TC Jr, Behrens CM, Ranger-Moore J, et al. Triaging HPV-positive women with p16/Ki-67 dual-stained cytology: results from a sub-study nested into the ATHENA trial. Gynecol Oncol. 2017;144(1):51–56. [DOI] [PubMed] [Google Scholar]
- 21. Carozzi F, Gillio-Tos A, Confortini M, et al. Risk of high-grade cervical intraepithelial neoplasia during follow-up in HPV-positive women according to baseline p16-INK4A results: a prospective analysis of a nested substudy of the NTCC randomised controlled trial. Lancet Oncol. 2013;14(2):168–176. [DOI] [PubMed] [Google Scholar]
- 22. Lahrmann B, Valous NA, Eisenmann U, et al. Semantic focusing allows fully automated single-layer slide scanning of cervical cytology slides. PLoS One. 2013;8(4):e61441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grabe N, Lahrmann B, Pommerencke T, et al. A virtual microscopy system to scan, evaluate and archive biomarker enhanced cervical cytology slides. Cell Oncol. 2010;32(1-2):109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wentzensen N, Walker JL, Gold MA, et al. Multiple biopsies and detection of cervical cancer precursors at colposcopy. J Clin Oncol. 2015;33(1):83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wentzensen N, Follansbee S, Borgonovo S, et al. Human papillomavirus genotyping, human papillomavirus mRNA expression, and p16/Ki-67 cytology to detect anal cancer precursors in HIV-infected MSM. Aids. 2012;26(17):2185–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Solomon D, Davey D, Kurman R, et al. The 2001 Bethesda system: terminology for reporting results of cervical cytology. JAMA. 2002;287(16):2114–2119. [DOI] [PubMed] [Google Scholar]
- 27. Leisenring W, Alono T, Pepe MS.. Comparisons of predictive values of binary medical diagnostic tests for paired designs. Biometrics. 2000;56(2):345–351. [DOI] [PubMed] [Google Scholar]
- 28. Schiffman M, Doorbar J, Wentzensen N, et al. Carcinogenic human papillomavirus infection. Nat Rev Dis Primers. 2016;2(1):16086. [DOI] [PubMed] [Google Scholar]
- 29. Franco EL, Cuzick J.. Cervical cancer screening following prophylactic human papillomavirus vaccination. Vaccine. 2008;26(Suppl 1):A16–A23. [DOI] [PubMed] [Google Scholar]
- 30. Massad LS, Einstein MH, Huh WK, et al. 2012 ASCCP Consensus Guidelines Conference. 2012 updated consensus guidelines for the management of abnormal cervical cancer screening tests and cancer precursors. J Low Genit Tract Dis. 2013;17(5 Suppl 1):S1–S27. [DOI] [PubMed] [Google Scholar]
- 31. Conrad RD, Liu AH, Wentzensen N, et al. Cytologic patterns of cervical adenocarcinomas with emphasis on factors associated with underdiagnosis. Cancer Cytopathol. 2018;126(11):950–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.