ABSTRACT
Recent studies indicate that microbial enterotypes may influence the beneficial effects of wholegrain enriched diets including bodyweight regulation. In a 4-week intervention trial, overweight subjects were randomized to consume either arabinoxylan-oligosaccharides (AXOS) (10.4 g/d) from wheat bran or polyunsaturated fatty acids (PUFA) (3.6 g/d). In the present study, we have stratified the subjects participating in the intervention (n = 29) according to the baseline Prevotella-to-Bacteroides (P/B) ratios through a post-hoc analysis and applied a linear mixed model analysis to identify the influence of this P/B ratio on the differences in weight changes in the intervention arms. Following AXOS consumption (n = 15), the high P/B group showed no bodyweight changes [−0.14 kg (95% CI: −0.67; 0.38, p = .59)], while the low P/B group gained 0.65 kg (95% CI: 0.16; 1.14, p = .009). Consequently, a difference of −0.79 kg was found between P/B groups (95% CI: −1.51; −0.08, p = .030). No differences were found between P/B groups following PUFA consumption (0.61 kg, 95% CI: −0.13; 1.35, p = .10). Among the Bacteroides species, B. cellulosilyticus relative abundance exhibited the highest positive rank correlation (Kendall’s tau = 0.51, FDR p = .070) with 4-week weight change on AXOS, and such association was further supported by using supervised classification methods (Random Forest). We outlined several carbohydrate-active enzyme (CAZy) genes involved in xylan-binding and degradation to be enriched in B. cellulosilyticus genomes, as well as multiple accessory genes, suggesting a supreme AXOS-derived glycan scavenging role of such species. This post-hoc analysis, ensuring species and strain demarcation at the human gut microbiota, permitted to uncover the predictive role of Bacteroides species over P/B enterotype in weight gain during a fiber-based intervention. The results of this pilot trial pave the way for future assessments on fiber fermentation outputs from Bacteroides species affecting lipid metabolism in the host and with direct impact on adiposity, thus helping to design personalized interventions.
KEYWORDS: Enterotype, arabinoxylans, Prevotella, Bacteroides, overweight, obesity, B. cellulosilyticus
Introduction
As the prevalence of overweight and obesity has reached epidemic proportions globally over the past few decades, the search for causes and management approaches continues.1 Multiple dietary interventions have been tested on weight control; however, the efficacy of a specific diet over another has not been established.2 The limited evidence for the most effective diet has given rise to conclude that there is no “one diet fits all.” Thus, the conventional view that different people will respond similarly to a specific diet might be too simplistic and instead it is more likely that the success of a diet might be predicted based on specific individual characteristics,3 including the gut microbiota.4
Identification of predictive traits for the anticipation of diet-based effects on weight loss is a matter of study, and microbial enterotypes have been suggested as promising biomarkers for such an aim.5,6 The Prevotella and Bacteroides enterotypes are characterized by different functionalities, where the Prevotella species are consistently associated with fiber-enriched diets due to their genetic ability to process complex carbohydrates of plant-origin.7 In support, Kovatcheva et al. found that subjects with a high P/B ratio specifically improved their enzymatic capacity for fiber digestion and glucose metabolism, when consuming a whole grain-rich diet.8
In four recent post-hoc analyses of studies conducted in Denmark, we have linked Prevotella abundance in the human gut microbiota to weight loss, when consuming whole grain and fiber-rich diet ad libitum.9–12 Specifically, the whole-grain fiber, arabinoxylan, is largely consumed as these are highly abundant in rye bread, a staple food item among Danish participants.13 On the other hand, Bacteroides is commonly associated with a “Western diet” low in fiber, and high in fat and refined sugars. However, the remarkable glycolytic potential of some Bacteroides species hinders the complete association of such microbes with fat-enriched diets and adiposity in humans.14,15 In our previous analyses, these subjects dominated by Bacteroides species have little weight control success when consuming diets rich in fiber and whole grain.9–12
Therefore, we investigated the influence of enterotypes (inferred as the Prevotella-to-Bacteroides [P/B] ratio16) in weight management of participants randomized to receive arabinoxylan oligosaccharides (AXOS) and polyunsaturated fatty acids (PUFA) for 4 weeks. We hypothesized that subjects with a higher P/B ratio (more abundant Prevotella content than Bacteroides) would improve body weight control on the AXOS supplemented diet (10.4 g/d) compared to the PUFA-enriched diet (3.6 g/d) that would serve as a negative control. Furthermore, as there is a large inter-individual variation in Bacteroides spp.,17 with vastly different fermentation, and short-chain fatty acid (SCFA) potentials, we further hypothesized that few species with AXOS-degrading capacity specifically would predict body weight changes.
Results
P/B-ratio predicts weight change when consuming AXOS but not PUFA
From baseline fecal samples, 29 overweight participants were stratified by the median value of the Prevotella-to-Bacteroides (P/B) ratio (−0.81) into high P/B and low P/B groups. The baseline characteristics of the two P/B groups are presented in Table S1.
Following 4-week AXOS consumption (n = 15), the high P/B group was weight stable [−0.14 kg (95% CI: −0.67; 0.38, p = .59)], whereas the low P/B group had a weight gain of 0.65 kg (95% CI: 0.16; 1.14, p = .009). Consequently, a difference of −0.79 kg was found between P/B groups (95% CI: −1.51; −0.08, p = .030). To evaluate if the weight change was an enterotype-AXOS interaction effect, we used 4-week PUFA consumption as a negative control (n = 14), and found no difference in weight changes between the P/B groups (0.61 kg, 95% CI: −0.13; 1.35, p = .10). However, we observed different trends on PUFA than on AXOS; weight gain among high P/B subjects (0.41 kg, CI: -0.11; 0.94, p = .12) and weight maintenance among the low P/B subjects (−0.20 kg, CI: −0.72; 0.32, p = .45), but these were not significant (Figure 1). Accordingly, when comparing the weight changes on AXOS to PUFA between the P/B groups, a total difference of −1.41 kg was observed (95% CI: −2.44; −0.38, p = .007).
Figure 1.
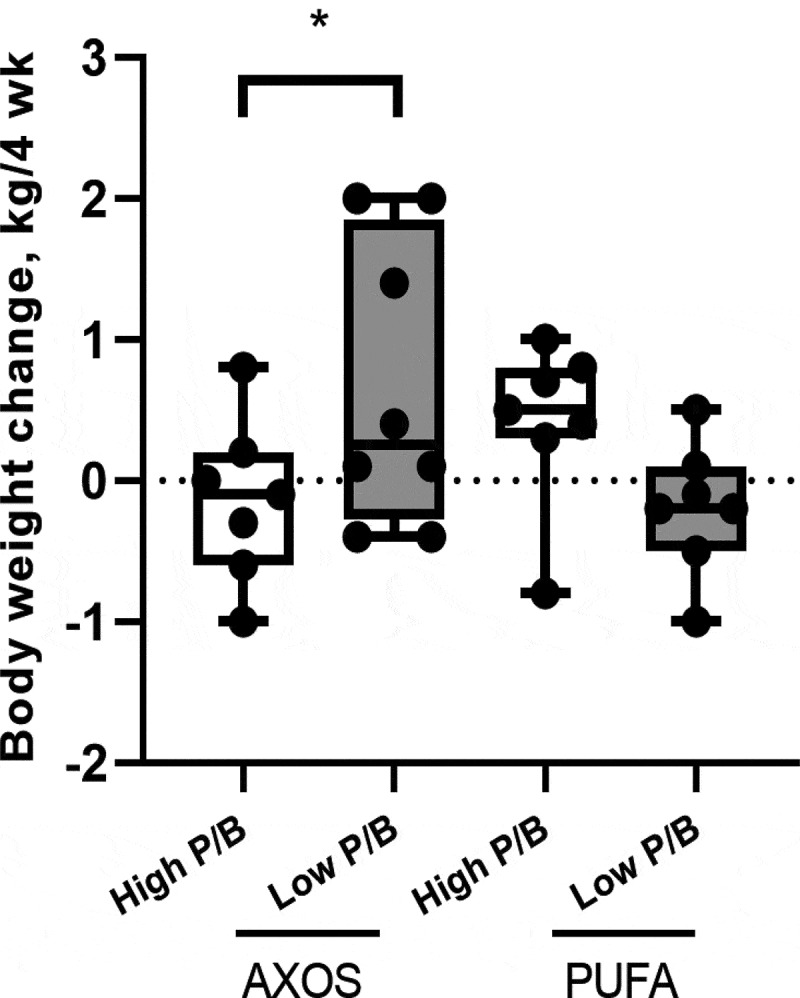
Body weight changes from weeks 0 to 4 for healthy, overweight adults (n = 29) stratified by the median baseline P/B ratio into two groups: Low P/B (n = 15) and High P/B (n = 14), when consuming AXOS and PUFA diets. *Significant difference between Low P/B and High P/B groups on each diet (P < .05) in a linear-mixed model adjusted for age, gender, and baseline BMI. AXOS, arabinoxylan oligosaccharides; P/B, Prevotella-to-Bacteroides; PUFA, polyunsaturated fatty acids
Following AXOS consumption, we found no differences in 4-week change for waist circumference, total energy intake, carbohydrate E%, or protein E% between P/B groups. However, a meaningful decrease in fat E% was observed in the high P/B group (Table S2), but this did not explain changes in body weight (Kendall’s tau = 0.18, p = .342).
Lastly, to exclude a longitudinal effect of the enterotypes, we calculated fold change of the P/B ratio and correlated it with bodyweight change after 4-week AXOS consumption, and found no relation between the two (Kendall’s tau = −0.14, p = .49). AXOS produced a notable increase of Bifidobacterium species,18 but we found no evident correlation between its abundance and body weight changes at both time-points (Kendall’s tau = −0.01 and 0.32, p = 1.000 and 0.102, respectively). Nonetheless, we did observe that changes in Bifidobacterium abundance was higher in low P/B subjects than in high P/B counterparts (414 ± 141 vs 963 ± 216 DNA reads, respectively, p = .039).
Distinct Bacteroides species predicts body weight change when consuming AXOS
Baseline P/B-ratio correlated with weight change after AXOS consumption (Kendall’s tau = −0.43, p = .029), but not after PUFA (Kendall’s tau = 0.34, p = .089) (Figure 2a-b). We then investigated whether the most prevalent Prevotella and Bacteroides species could further predict body weight following 4-week AXOS consumption (n = 15). We evaluated the abundance at the species-level, retrieved from shotgun DNA sequencing (see methods) for the 10 most abundant Bacteroides and Prevotella (metagenomic Operational Taxonomic Units) mOTUs in fecal samples (Table 1). The majority of the subjects harbored a high abundance of Bacteroides species, which is similar to previously characterized Westernized populations.5,19 Among the Bacteroides species, B. cellulosilyticus relative abundance exhibited the highest positive rank correlation with a 4-week weight change (Kendall’s tau = 0.51, p = .007, FDR p = .070) (Figure 3a and Table 2). The importance of B. cellulosilyticus as a predictor variable for weight gain during AXOS intervention was further explored by executing supervised classification with the Random Forest algorithm. B. cellulosilyticus baseline abundance was found to be the most important predictor of body weight change among the top 10 Bacteroidetes species (INP = 2.00) and the P/B ratio (Figure 3b).
Figure 2.
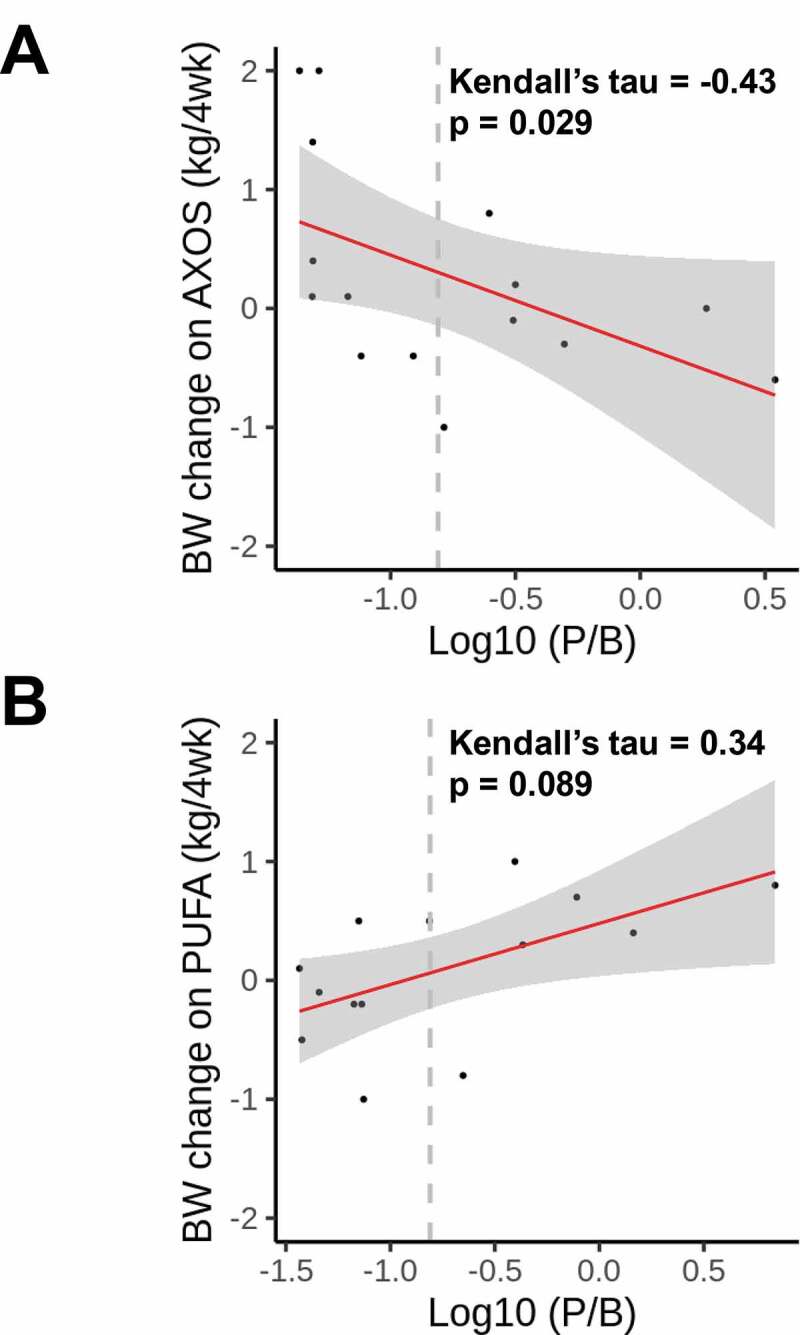
Body weight changes from weeks 0 to 4 versus baseline log10-transformed P/B ratio for healthy, overweight subjects consuming either (a) AXOS (n = 15) or (b) PUFA (n = 14). Kendall correlation coefficient (tau) and p-value is shown. Vertical dashed gray lines at x = −0.81 (P/B median) separate the enterotype groups; Low P/B and High P/B. Linear regression line is depicted in red and respective confidence interval (95%) is drawn in gray. AXOS, arabinoxylan oligosaccharides; BW, body weight; P/B, Prevotella-to-Bacteroides; PUFA, polyunsaturated fatty acids
Table 1.
Top 10 most abundant Bacteroidetes mOTUs at baseline from overweight subjects (n = 15)
| Rank | Species | mOTU | R.A1 | High P/B R.A1 | Low P/B R.A1 |
|---|---|---|---|---|---|
| 1 | Bacteroides dorei/vulgatus | ref_mOTU_v2_0898 | 5.16% (1.97–7.83) | 1.56% (1.42–4.04) | 7.78% (5.15–11.8) |
| 2 | Bacteroides uniformis | ref_mOTU_v2_0899 | 5.14% (2.28–10.2) | 3.92% (1.42–6.67) | 9.53% (4.39–10.6) |
| 3 | Bacteroides fragilis/ovatus | ref_mOTU_v2_1073 | 0.35% (0.05–1.62) | 0.07% (0.03–0.31) | 0.93% (0.34–2.82) |
| 4 | Bacteroides xylanisolvens | ref_mOTU_v2_1072 | 0.32% (0.09–0.56) | 0.28% (0.09–0.88) | 0.35% (0.21–0.47) |
| 5 | Bacteroides caccae | ref_mOTU_v2_1382 | 0.28% (0.02–0.79) | 0.28% (0.02–0.65) | 0.34% (0.02–0.99) |
| 6 | Prevotella copri | ref_mOTU_v2_4448 | 0.12% (0.09–2.81) | 2.81% (0.08–8.35) | 0.12% (0.10–0.27) |
| 7 | Bacteroides cellulosilyticus | ref_mOTU_v2_0692 | 0.03% (0.01–1.88) | 0.01% (0.01–0.03) | 1.67% (0.01–4.39) |
| 8 | Bacteroides stercoris | ref_mOTU_v2_0275 | 0.03% (0.02–0.95) | 0.03% (0.03–0.35) | 0.05% (0.02–1.50) |
| 9 | Bacteroides eggerthii | ref_mOTU_v2_1410 | 0.01% (0.01–0.23) | 0.01% (0.00–0.37) | 0.01% (0.01–0.12) |
| 10 | Bacteroides massiliensis | ref_mOTU_v2_0455 | 0.01% (0.00–0.08) | 0.02% (0.01–0.61) | 0.00% (0.00–0.02) |
1Data expressed as median with interquartile distribution (Q1-Q3). R.A ,relative abundance.
Figure 3.
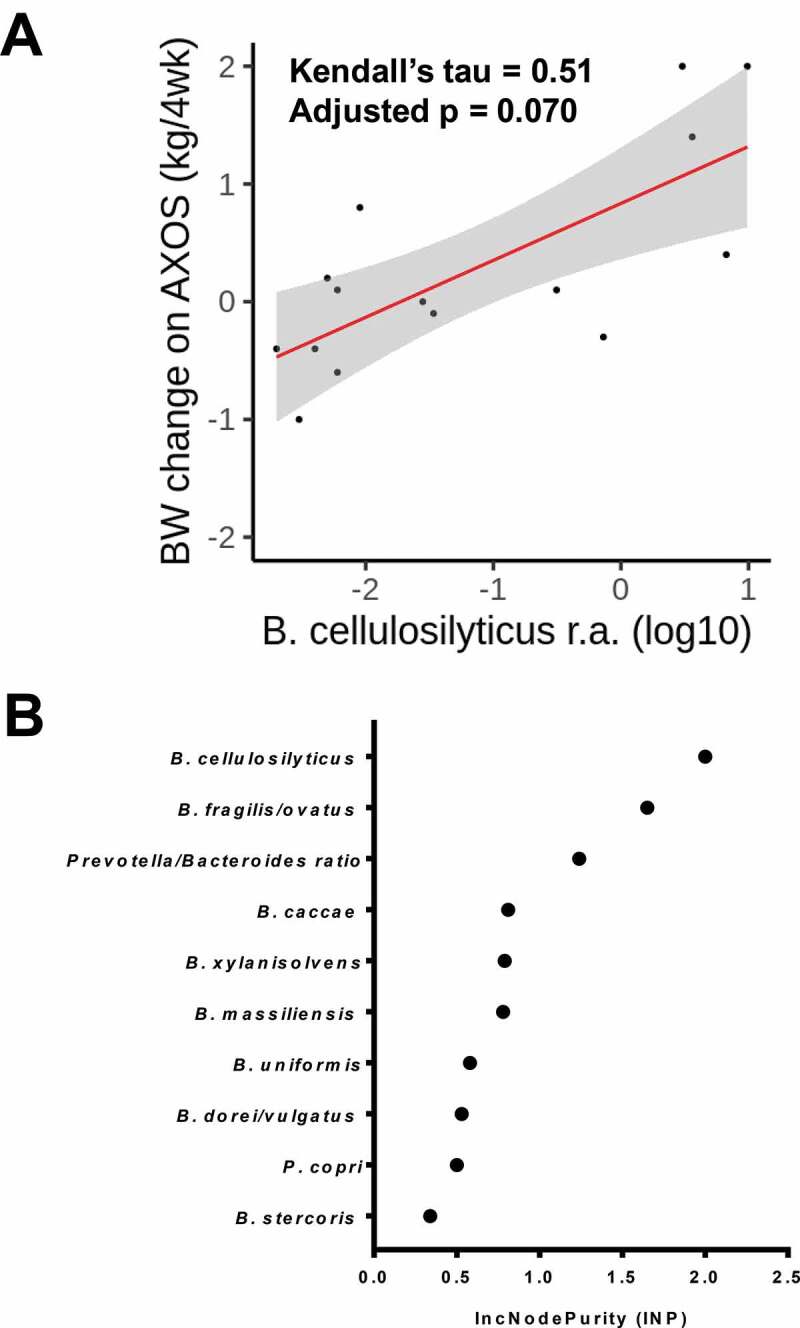
Predictive role of B. cellulosilyticus. (a) Body weight changes from weeks 0 to 4 versus baseline log10-transformed B. cellulosilyticus relative abundance (r.a.) following AXOS consumption (n = 15). Kendall correlation coefficient and adjusted p-value (FDR) is shown. Linear regression line is depicted in red and respective confidence interval (95%) is drawn in gray. AXOS, arabinoxylan oligosaccharides; BW, body weight. (b) Random Forest classification output based on the Mean Decrease Gini (IncNodePurity) (sorted decreasingly from top to bottom) of attributes as assigned by the algorithm
Table 2.
Baseline Bacteroidetes mOTU correlations with body weight change on the AXOS-enriched diet (n = 15)
| Abundant Bacteroidetes mOTUs | Kendall’s tau | p-value | FDR adjusted p-value |
|---|---|---|---|
| Bacteroides cellulosilyticus | 0.51 | 0.007** | 0.070 |
| Bacteroides fragilis/ovatus | 0.44 | 0.022* | 0.112 |
| Bacteroides caccae | 0.40 | 0.037* | 0.123 |
| Bacteroides eggerthii | 0.38 | 0.051 | 0.128 |
| Bacteroides dorei/vulgatus | 0.28 | 0.136 | 0.272 |
| Bacteroides massiliensis | −0.23 | 0.244 | 0.406 |
| Bacteroides stercoris | 0.11 | 0.584 | 0.649 |
| Bacteroides xylanisolvens | −0.14 | 0.487 | 0.609 |
| Prevotella copri | −0.17 | 0.371 | 0.530 |
| Bacteroides uniformis | 0.08 | 0.691 | 0.691 |
*significant Kendall correlation (p < 0.05), ** significant Kendall correlation (p < 0.001). FDR, False detection rate.
Lastly, we investigated whether fold changes of the 10 most abundant Bacteroides and Prevotella species differed between the low P/B and high P/B groups, but found no indications of differential effects for these species upon AXOS consumption.
Baseline co-abundance analysis among the 30 most abundant species (all phyla)
With B. cellulosilyticus pointed-out as a potential predictive biomarker of weight management on an AXOS-based diet, we wanted to identify additional and less evident predictors among other gut microbiota members. Therefore, a baseline co-abundance analysis20 using metagenomic data was applied to detect such species interacting positively or negatively with B. cellulosilyticus. B. cellulosilyticus was negatively correlated to two Clostridiales species; Clostridium.sp.CAG.138 (Kendall’s tau = −0.54, FDR p = .04), and Ruminococcus.sp.CAG.177. (Kendall’s tau = −0.54, FDR p = .04), and was positively correlated with Phascolarctobacterium.sp. (Kendall’s tau = 0.57, FDR p = .04).
There was no association between B. cellulosilyticus and P. copri; however, when introducing the strain-level information of P. copri defined as clades A, B, C, and D according to Tett and coworkers,19 we did find meaningful correlations. All four clades were detected in this westernized, overweight population, and the average proportions were for Clade A = 53.4%, B = 39%, C = 7.2%, and D = 0.4%. Clade B was positively associated with B. cellulosilyticus (Kendall’s tau = 0.61, adjusted p = .0066), while Clades A, C, and D all were inversely associated with B. cellulosilyticus (adjusted p ≤ 0.037) (Figure 4a-d).
Figure 4.
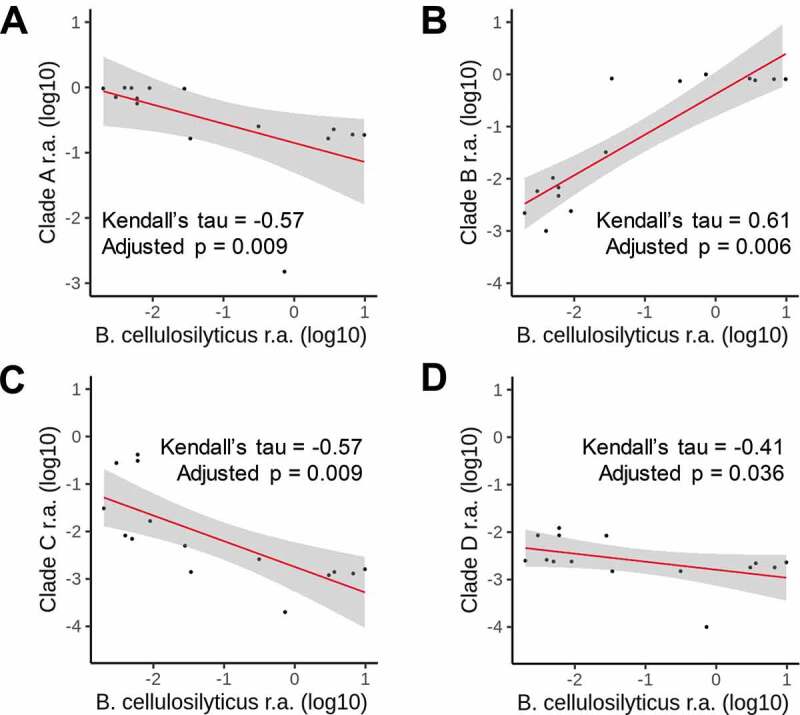
Correlations at baseline between log10-transformed B. cellulosilyticus with the four clades of the Prevotella copri complex (all log10-transformed): (a) Clade A, (b) Clade B, (c) Clade C, and (d) Clade D for subjects randomized to AXOS (n = 15). Kendall correlation coefficients and adjusted p-values (FDR) are shown. Linear regression line is depicted in red and respective confidence interval (95%) is drawn in gray
Absolute quantification to confirm B. cellulosilyticus presence when consuming AXOS
The qPCR-based approach to measure the presence of B. cellulosilyticus in baseline fecal DNA indicated that samples of subjects who controlled weight tended to have a lower abundance of this Bacteroides species (log10 rpoB molecules/ng DNA = 2.14 ± 0.81) when compared to samples derived from subjects who gained weight (3.46 ± 1.03) (p = .166). This difference was more evident in the samples after a 4-week intervention, where B. cellulosilyticus abundance increased 1.42-fold in the low P/B and weight-gain group (4.88 ± 0.76) whereas remained stable in the high P/B group (2.42 ± 0.82) (p = .023). Globally, the qPCR data fitted well with the B. cellulosilyticus mOTU relative abundance in the entire set of samples at both time-points (Kendall’s tau = 0.54, p < .001), but data correlation was higher at baseline (Kendall’s tau = 0.76, p < .001). Three samples in each P/B group produced null detection of B. cellulosilyticus, but we cannot completely discard its presence in those given the limit of detection of our qPCR approach (~600 molecules rpoB per ng DNA). The log10 average presence of B. cellulosilyticus among the three subjects with the largest weight gains (1.4, 2.0, and 2.0 kg) was 5.77 ± 0.26 rpoB molecules/ng DNA.
Distinctive genetic traits on B. cellulosiluyticus genomes
The searching for carbohydrate-active enzyme (CAZy) genes within polysaccharide utilization loci (PULs) present in more than 100 Bacteroides genomes (Table S3) permitted to evaluate of the abundance of 212 CAZy families and accessory genes. Functional enrichment analysis indicated that 10 CAZy families are more prevalent and abundant in B. cellulosilyticus genomes (Table 3). These include CBM-containing enzymes associated with xylan binding (CBM13 and CBM22) and GHs mainly dedicated to xylanase and arabinofuranosidase activities (GH5, GH8, GH10, GH43, and GH79). Two PLs were also detected to be enriched in B. cellulosilyticus genomes and they were linked to glycosaminoglycan degradation (e.g., chondroitin-(sulfate) lyase, hyaluronate lyase, and heparin-(sulfate) lyase).
Table 3.
CAZy families enriched in B. cellulosilyticus genomes
| CAZy gene | B. cellulosilyticus abundance1 | Other Bacteroides abundance2 | p-value(FDR) | Activity |
|---|---|---|---|---|
| PL8 | 6(2) | 1(0.01) | 4.37−6 | hyaluronate lyase; chondroitin lyase; xanthan lyase; heparin lyase |
| CBM22 | 4(1.33) | 0(0) | 2.25−4 | xylan binding function with affinity for mixed β-1,3/β-1,4-glucans |
| CBM13 | 5(1.67) | 6(0.06) | 0.002 | xylan binding function(e.g. Streptomyces lividans xylanase A) |
| GH43_11 | 3(1) | 0(0) | 0.002 | β-xylosidase; α-L-arabinofuranosidase; xylanase; |
| GH79 | 3(1) | 0(0) | 0.002 | β-glucuronidase; hyaluronoglucuronidase; heparanase |
| GH8 | 3(1) | 0(0) | 0.002 | chitosanase; cellulase; endo-1,4-β-xylanase; reducing-end-xylose releasing exo-oligoxylanase |
| PL37 | 3(1) | 0(0) | 0.002 | chondroitin-sulfate endolyase; heparin-sulfate lyase; ulvan lyase |
| GH5_13 | 6(2) | 19(0.18) | 0.007 | endo-β-1,4-xylanase; xyloglucan-specific endo-β-1,4-glucanase; arabinoxylan-specific endo-β-1,4-xylanase |
| GH10 | 12(4) | 99(0.91) | 0.025 | endo-1,4-β-xylanase; endo-1,3-β-xylanase; xylan endotransglycosylase |
| GH43_7 | 2(0.67) | 0(0) | 0.031 | β-xylosidase; α-L-arabinofuranosidase; xylanase |
1Sum of genes present in all B. cellulosilyticus genomes (N = 3). The density of the gene per B. cellulosilyticus genome is shown within parenthesis.
2Sum of genes present in all Bacteroides genomes explored different than B. celulosilyticus (N = 106). The density of the gene per genome is shown within parenthesis.
The functional assessment on the entire genomes using the Pfam annotation system permitted to assess the abundance of more than 3000 Pfam domains. The statistical test to determine the probable enrichment of such protein domains on B. cellulosilyticus genomes recovered 87 domain associations, and 54 of which had reliable functional annotations (Table 4). This analysis corroborated some previous observations during the CAZy gene survey. Thus, several domains associated with different GH and PL enzymes listed in Table 3, and related to xylan and glycosaminoglycan degradation were also retrieved (e.g., GH10, GH79, GH43, PL8) (Table 4). Moreover, we also observed that other domains linked to xylan binding and degradation were enriched in B. cellulosilyticus genomes (e.g., Glyco_hydro_30, CBM_6, Glyco_hydro_3, Bac_rhamnosid). Nevertheless, the glycan metabolism domains enriched in B. cellulosilyticus in comparison with other Bacteroides species is also accompanied by a higher abundance of polysaccharide degradation functions as well as of sensor and kinase subunits of several two-component systems specialized on carbohydrate uptake. Moreover, we detected an enrichment of some peptidase domains (Peptidase C25, Peptidase_M6 and Peptidase_C39), and domains of secreted proteins involved in adhesion (VCBS, fn3, Fn3-like), and flagella- and pili-independent gliding motility (SprA_N and PorP_SprF).
Table 4.
Pfam families enriched in B. cellulosilyticus genomes
| Domain | p-value (FDR) | Associated function |
|---|---|---|
| CBM26 | 2.21−14 | Starch-binding function. |
| SprA_N | 5.53−07 | Domain found the gliding motility-related SprA proteins -secretion |
| CYTH | 0.003 | Conversion of ATP to 3ʹ,5ʹ-cyclic AMP and pyrophosphate |
| Dak1 | 0.003 | Kinase domain of the dihydroxyacetone kinase family |
| Dak2 | 0.003 | Kinase domain of the dihydroxyacetone kinase family |
| Glyco_hydro_79 | 0.003 | Glycosyl hydrolase family 79 |
| Peptidase_C25 | 0.003 | Gingipains R and K type cysteine peptidases |
| Peptidase_M6 | 0.003 | Metalloendopeptidase of antibacterial humoral factors from insects |
| PorP_SprF | 0.003 | Gliding motility, cell movement without flagella |
| RHS_repeat | 0.003 | Heparing binding |
| Thi4 | 0.003 | Putative thiamine biosynthetic enzyme |
| TraN | 0.003 | Outer membrane protein involved in the mating-pair stabilization (adhesin) |
| AAA_lid_6 | 9.74−10 | ATPase domain |
| VKOR | 4.53−11 | Vitamin K epoxide reductase recycling reduced vitamin K |
| Phage_T4_gp19 | 1.76−5 | Tube protein gp19 sequences from the T4-like viruses |
| VCBS | 1.76−5 | Role for this domain in adhesion |
| Phage_sheath_1 | 1.33−4 | Domain in a variety of phage tail sheath proteins |
| PhdYeFM_antito | 7.35−4 | Toxin-antitoxin system |
| NPCBM | 3.44−9 | N-terminus of glycosyl hydrolase family 98 |
| Cthe_2159 | 6.45−9 | Cellulose and/or acid-sugar binding proteins |
| Malt_amylase_C | 0.005 | C-terminal domain of Maltogenic amylase |
| YoeB_toxin | 0.002 | Type II toxin-antitoxin system |
| fn3 | 3.44−9 | Fibronectin domain |
| Glyco_hydro_30 | 5.80−10 | endo-β-1,4-xylanase; β-glucosidase; β-glucuronidase; β-xylosidase |
| Lyase_8 | 5.12−7 | Bacterial lyase acting on hyaluronan/chondroitin in the extracellular matrix of host tissues |
| LRR_5 | 4.77−11 | BSPA-like surface antigens from Trichomonas vaginalis |
| Fucosidase_C | 1.01−4 | Alpha-L-fucosidase C-terminal domain |
| CBM_6 | 1.22−4 | Cellulose-binding function on amorphous cellulose and β-1,4-xylan |
| Peptidase_C39 | 1.43−4 | Cleavage of the ‘double-glycine’ leader peptides from bacteriocin precursors |
| Mannosidase_ig | 0.005 | Bacteroides thetaiotaomicron beta-mannosidase, BtMan2A – Mannose foraging |
| Pectate_lyase | 4.93−6 | Polygalacturonic acid lyase |
| Glyco_hydro_10 | 4.35−8 | endo-1,4-β-xylanase; endo-1,3-β-xylanase; xylan endotransglycosylase |
| Glyco_hydro_28 | 2.99−7 | Polygalacturonase; α-L-rhamnosidase; exo-polygalacturonase; rhamnogalacturonase |
| GH43_C2 | 1.91−4 | Beta xylosidase |
| Glyco_hydro_88 | 1.60−7 | d-4,5-unsaturated β-glucuronyl hydrolase |
| UpxZ | 0.001 | Family of transcription anti-terminator antagonists |
| Peptidase_S24 | 5.90−4 | Endopeptidases involved in LexA/RecA system DNA repair |
| Fn3-like | 4.35−8 | Fibronectin type III-like structure associated with GH3 |
| AAA_14 | 8.92−5 | ATPase module in search of a basic functions |
| Glyco_hydro_3 | 1.19−7 | β-glucosidase; xylan 1,4-β-xylosidase; β-glucosylceramidase; α-L-arabinofuranosidase |
| Glyco_hydro_43 | 1.19−9 | β-xylosidase; α-L-arabinofuranosidase; xylanase |
| Bac_rhamnosid | 0.002 | GH78 – α-L-rhamnosidase; rhamnogalacturonan α-L-rhamnohydrolase |
| Y_Y_Y | 3.87−10 | Periplasmic sensor domain binding unsaturated disaccharides |
| HisKA | 1.33−15 | Histidine kinase two-component system |
| HATPase_c | 3.87−10 | ATPase domains of histidine kinase |
| Glyco_hydro_2 | 3.11−5 | β-galactosidase; β-mannosidase; β-glucuronidase; α-L-arabinofuranosidase |
| HTH_18 | 1.35−8 | Helix-turn-helix (HTH) binding DNA. |
| Response_reg | 2.17−5 | Bacterial two-component systems, DNA binding effector domain |
| Phage_int_SAM | 5.78−4 | Phage integrase, N-terminal SAM-like domain |
| FecR | 5.43−4 | FecR is involved in regulation of iron dicitrate transport |
| Arm-DNA-bind_5 | 6.22−4 | DNA-binding domain found in various tyrosine recombinases |
| Glyco_hydro_20 | 2.34−5 | β-hexosaminidase; lacto-N-biosidase; β-1,6-N-acetylglucosaminidase |
| CoA_binding_3 | 0.005 | CoA-binding domain |
| STN | 5.67−4 | Secretins of the bacterial type II/III secretory system/TonB-dependent receptor proteins |
Associating B. cellulosilyticus with metabolic changes when consuming AXOS
We investigated whether baseline B. cellulosilyticus relative abundance correlated with changes in fecal SCFA concentrations, as an indication of its influence on host health and/or bacterial cross-feeding; however, B. cellulosilyticus did not correlate with changes of SCFA concentrations. Lastly, when correlating this species with clinical parameters, we found a correlation between changes in total serum cholesterol concentrations (n = 14, Kendall’s tau = 0.44, p = .028), but this correlation was not significant after correction for multiple testing (Table S4).
Discussion
We demonstrate that a 4-week intake of AXOS resulted in a bodyweight change difference of 0.79 kg when comparing the low- and high-P/B groups, which was driven by a significant 0.65 kg weight gain for the low P/B group. Distinct body weight trajectories may partially be explained by the differences in fat E% intake detected in both low- and high-P/B groups, but a direct correlation between fat intake and body weight change was not intuited. Beyond the genus-level P/B groups, we revealed that baseline B. cellulosilyticus abundance predicted weight gain with better precision than the P/B ratio. Besides body weight, we found indications of B. cellulosilyticus would affect host metabolism, as changes in total serum cholesterol levels could be associated to some extent with this Bacteroides species. Differential Bifidobacterium abundances at the end-point between high and low P/B groups may be explained by competition for substrate between Prevotella and Bifidobacterium spp.,21 as they are consistently found to be inversely correlated across different populations.22 However, we do not discard some indirect role of these species in microbiota-mediated body weight change as well, being feasible since cross-feeding interactions where Bifidobacterium species take part are well known.23
B. cellulosilyticus is a common species in the human gut and has been reported to colonize around 60% of the westerners,14 which is similar to our data [qPCR: 9/15; metagenomics (mOTU counts>100): 8/15]. B. cellulosilyticus has clearly defined xylan-degrading enzymes,14 grows especially well on wheat arabinoxylans,24 and even outcompetes other prevalent Bacteroides spp. in arabinoxylan-rich conditions.17 Our comparative analysis on CAZy gene content supports the previous experimental observations given that B. cellulosilyticus genome seems to be enriched in xylan binding and degrading enzymes, when compared to other common Bacteroides species present in the human gut, thus conferring it an advantage to uptake and metabolize this particular type of carbohydrates. Interestingly, Patnode et al. found that B. ovatus avoids arabinoxylan competition when co-residing with B. cellulosilyticus by shifting fermentation strategy during 10-day experiments, a metabolic flexibility not observed among other Bacteroides spp.17 However, this metabolic flexibility may be a temporary strategy, as B. ovatus has been shown to bloom after 10 days while abundances of B. cellulosilyticus drop concurrently with AXOS as substrate.25 This may partly be a consequence of reliance on arabinoxylans as substrate by B. cellulosilyticus, whereas B. ovatus and others may thrive equally well on a mixture of dietary fibers.24,25
Additional to the potential advantage conferred by the higher number of CAZy genes deserved to arabinoxylan degradation, we observed that the B. cellulosilyticus genome is enriched in peptidases and glycosaminoglycan degrading enzymes constituting a signal of O-glycan foraging from host gut mucins as previously inferred for this and other Bacteroides species.14 This could indicate that in absence of arabinoxylans or other complex carbohydrates from the host diet, B. cellulosilitycus would also compete with other mucin-degrading species. Mucin production in colon epithelial cells has been proven to depend on butyrate, one of the SCFAs released by gut microbiota.26–28 Therefore, this metabolic circuit seems to be pivotal in the evolutionary mutualistic relationship between the several gut microbes and the host.
However, butyrate production is mainly linked to Gram-positive bacteria from the Firmicutes phylum and alternative pathways have been stated in few species from the Bacteroidetes phylum.29,30 Consequently, an exacerbated mucin utilization by gut microbiota enriched in B. cellulosilyticus without positive feedback signals for its production (SCFAs, mainly butyrate and propionate) would weaken barrier function altering gut immune homeostasis,31,32 worsen endotoxemia, attenuate incretins production affecting satiety,33–35 and possibly impairing weight loss upon the dietary intervention. On the other hand, the presence of certain genes encoding protein domains associated with gliding motility would also confer an advantage to B. cellulosilyticus during the scavenging for nutrients, reinforcing the idea of its extreme glycan predatory role. This surface motility mechanism is widely described in non-motile (pili- or flagella-based) oral microbiota pathogens such as Porphyromonas gingivalis and Flavobacterium johnsoniae,36 together with the above-mentioned circuits for arabinoxylans and mucin O-glycan degradation, could constitute an evolutionary adaptation to outperform the carbohydrate uptake. However, the existence of this particular motility mechanism in B. cellulosilyticus should be confirmed in in vitro experiments.
While there is a notable inter-individual variation in Bacteroides species abundance, P. copri is consistently found to be the most prevalent Prevotella species in the human gut. P. copri also shows a superior ability to utilize xylans,37 and may outcompete Bacteroides spp. with overlapping glycan degrading capacity.38 In fact, intake of xylan-rich foods may be the main determinant of P. copri positive effect on human metabolism,8,19 while a diet rich in fat seems to have the opposite effect.39 However, on AXOS, we found no weight-loss effect of high baseline abundance of P. copri, which could be due to too few subjects studied or the recently discovered disparate genomic and metabolic capacity of P. copri clades.19 Interestingly, clade C has been shown to grow well on arabinoxylans, in contrast to clade B, which lacks the enzymatic capacity.37 This may help to explain why clade B as the only P. copri clade was strongly positively correlated with B. cellulosilyticus, as there may not be substrate competition. This finding further points to the necessity of considering each clade in the P. copri complex to understand the effect on host metabolism. Also, it is worth noting that the increased consumption of fat on the PUFA-enriched diet (in combination with lower fiber intake) tended to result in poorer weight regulation for the high P/B subjects than the low P/B subjects, as also observed in previous P/B-ratio studies.11,12 We hypothesize this could be a consequence of a mismatch between substrate and P. copri leading to, e.g., increased production of branch-chained amino acids and potentially insulin resistance.39
Given that the bodyweight changes may be partly due to fermentation end-products of AXOS released distally in the gastrointestinal tract, future studies should investigate potential links between B. cellulosilyticus and fecal SCFA. Acetate is the most highly produced SCFA in response to arabinoxylan fermentation38 and exclusively produced by B. cellulosilyticus upon arabinogalactan fermentation.40 While P. copri is a well-known acetate producer,41 we hypothesize that B. cellulosilyticus may not crossfeed SCFAs in a similar fashion, and thereby not contribute to create a healthy microbial community. In support, B. cellulosilyticus was negatively associated with Clostridiales spp., which may be a consequence of little acetate cross-feeding.42 Furthermore, we found that B. cellu-losilyticus tended to correlate with a change in total cholesterol concentrations. We do not discard this could be due to increased production of acetate in the upper gastrointestinal tract, where B. cellulosilyticus also thrives43 and followed by increased uptake in the liver (instead of cross-feeding) resulting in increased synthesis of cholesterol. However, such a hypothesis also needs further investigation in future studies. Interestingly, Chung et al. recently found a significant positive correlation between the relative abundance of Prevotella spp. and fecal SCFA concentrations following AXOS consumption, which was not seen for Bacteroides spp.44
A strength of this study is first and foremost the high dose of the specific whole-grain fiber, AXOS (compared to our previous post-hoc analyses with whole grain-rich diets9–12), which allow us to (further) validate the differential effects of whole-grain fiber on body weight regulation between the enterotypes,9–12 and secondly the introduction of deep-level sequencing that improves our understanding of AXOS-degrading gut microbes at species and strain level. However, it is a limitation that these species-level analyses include gut microbiomes from only 15 subjects, and thus the novel findings need to be validated in a larger sample in combination with metabolome analyses to further explore causing factors.
In conclusion, AXOS consumption promoted weight gain among subjects with a low Prevotella-to-Bacteroides ratio, and B. cellulosilyticus has been pointed out as the main predictor of the body weight gain during the intervention. This analysis paves the way for future investigations aiming at elucidating the underlying metabolic cross-talk between these species and other microbes inhabiting the human gut under AXOS administration, and how that metabolic exchange influence negatively on adiposity. Furthermore, we believe that these results underline the need to investigate enterotypes beyond the genus level and in combination with specific dietary fibers to further understand the role in human metabolism and obesity management, and to design more personalized interventions.
Methods
Study design
The assessment is a sub-study nested within a randomized cross-over trial.18 The study included two diet periods (AXOS and PUFA). To ensure body weight maintenance, the study participants had consultations (by physically present and by phone) with a dietician every week where body weight (non-fasting) and diet were evaluated. Dietary advising compliance by participants in the intervention was evaluated and results are published elsewhere.18 The present work focuses on the first period of the cross-over design, as findings from the original study found participants to be responders of the AXOS intervention only in the first period, but not in the second period.18 The participants on the AXOS intervention in the first period had a change in their microbiota composition (responders), whereas participants on the AXOS intervention in the second period did not experience this change (non-responders). This was suggested to be a consequence of a potential carry-over effect from the PUFA intervention in period 1.18 The study was registered at clinicaltrial.gov: NCT02215343 and approved by the Danish Ethical Committee. All study procedures were carried out in accordance with the Helsinki Declaration and the Danish Protection Agency. Written informed consent was obtained before the study start.
Study participants
Eligibility criteria were nonsmoking men and women between 18 and 60 years with a BMI of 25–40 kg/m2. Furthermore, participants should have a waist circumference of ≥94 cm for men and ≥80 cm for women and in addition it was required that they should have at least one criteria for metabolic syndrome; elevated fasting plasma glucose (≥5.6 mmol/L), elevated triglycerides (≥1.7 mmol/L) lowered high-density lipoprotein (HDL) (men: <1.03 mmol/L, women: <1.29 mmol/L), or elevated blood pressure (BP) (systolic BP ≥130 mmHg or diastolic BP ≥85 mmHg).
Intervention
The evaluated dietary supplements were wheat bran extract, rich in AXOS (10.4 g/d) and PUFA (3.6 g/d). For the AXOS intervention, the goal was to reach a high-fiber diet consisting of approximately 30 g fiber/day, of which 10.4 g was obtained from the AXOS supplementation. AXOS was provided as a powder (5 g/d) and biscuits (4 biscuits/d). The powder was instructed to be consumed in the morning and in the evening and should be dissolved in water. For the PUFA intervention, the goal was to reach approximately 10 energy percentage (E%) PUFA/day, whereas the participants were guided to decrease their intake of saturated fatty acids. PUFA was provided as fish oil capsules (1.32 g/d of docosahexaenoic acid (DHA) and 1.86 g/d of eicosapentaenoic acid (EPA)).
Clinical evaluation
The anthropometric measurements were conducted in a fasting state and all participants had to void their bladder before the start. Bodyweight was measured using a calibrated digital scale (Lindells, Malmo, Sweden) to the nearest 0.1 kg with the participants wearing underwear, and height was measured without shoes to the nearest 0.5 cm using a wall-mounted stadiometer (Hultafors). BMI was calculated as weight in kilograms divided by height in meters squared (kg/m2), and waist circumference was measured twice with a non-elastic tape measure on the skin with a precision of 0.5 cm, from which an average was calculated. As described in detail previously,18 lipid and glucose markers were analyzed from fasting blood samples, and also resting energy expenditure (REE) by the ventilated hood and breath hydrogen were measured among others.
Gut microbiota
DNA was obtained from subjects’ feces at baseline and after the intervention. Initially, gut microbiota was assessed by 16 S rRNA gene amplicon sequencing as previously described.18 The taxonomy assignment was performed using the RDP Classifier v2.12.45 The baseline abundance (on rarefied data) at the genus level was obtained, and the Prevotella/Bacteroides (P/B) ratio was determined as a predictive trait for downstream analyses. To stratify subjects by the P/B ratio, we calculated log10-transformed P/B ratios and used the median value (−0.81) to divide subjects into either low (n = 15) or high (n = 14) P/B groups.
A more detailed analysis aiming at species identification was completed by using the metagenomic information derived for samples included in the study previously.46 Approximately 0.5 Tb raw data, delivered in respective paired-end fastq files, was used to identify operational taxonomic units (mOTUs) for taxonomy profiling of >7700 microbial species.47 The mOTUsv2.0 profiler was used with the -g 1-c -l 100-y insert.scaled_count parameters to set a balance between high sensitivity and high precision configurations. From the full set of mOTUs detected, we selected the log-transformed baseline relative abundances of mOTUs with reliable classification as Bacteroides and Prevotella species (n = 10).
We additionally assessed the abundance of Prevotella copri clades by mapping the available metagenomic data of this subject cohort46 against the gene marker database generated by the Segata’s group previously.19 For reading mapping we used the Usearch v8.0.1623 algorithm with the following parameters: -usearch_local, -id 0.9, -strand both, -top_hit_only. Then, the relative abundance among P. copri clades was calculated.
Finally, the P/B ratios generated from the amplicon sequencing (genus level), the abundance of Bacteroides and Prevotella species obtained from shotgun sequencing as well as the distribution of P. copri clades were used as traits to perform predictions and associations with the weight gain/loss phenotypes (see ‘Statistical analyses’ section).
Quantitative PCR
Absolute quantification of Bacteroides cellulosilyticus was carried out on the fecal DNA of participants. The reference sequence of the rpoB gene (NZ_CP012801.1) from B. cellulosilyticus was submitted to the Primer-Blast web server (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) to retrieve specific primer pairs to amplify selectively this species-specific marker (included in the mOTUsv2 profiler). The comparison against the non-redundant NCBI database and the target organism [B. cellulosilyticus, taxid: 246,787] were fixed as checking parameters for primer prediction. As a result, we used the forward ATTTGTGGACGCTACTGTTATTCGT and reverse ACGACGCCACTTCGGAATACG primers to specifically detect and quantify the presence of B. cellulosilyticus. The single-stranded DNA (ssDNA), fully covering the region to be amplified (109 nt) was obtained from Isogen Life Science B.V (Utrecht, The Netherlands) where it was synthesized, PAGE-purified, quantified, and used for molecule titration during qPCR. The qPCR reactions were set in 96-well plates using the SYBR Green I Master Mix (Roche Lifesciences), 0.5 μM of forward primer, 0.25 μM of reverse primer, and 5 μL of the 1:10 diluted in nuclease-free water fecal DNA originally obtained for both amplicon and shotgun sequencing (final concentration in the qPCR reaction between 5 and 50 ng DNA). All samples were set in duplicate in the plate and amplified at once with standards in a LightCycler 480 II instrument (Roche Lifesciences) with the following cycling profile: initial incubation at 95°C for 5 min and 40 cycles of 10 s at 95°C, 20 s at 65°C, and 15 s at 72°C. Finally, the melting curve was set from 65°C to 97°C with a ramp rate of 0.11°/s. The absolute quantification was assessed with Ct values obtained for every sample and from titration curve (with duplicate measures) using the LightCycler® 480 Software v1.5 (Roche Lifesciences). The number of rpoB gene molecules was normalized against the total DNA concentration (ng/µL) present in the diluted DNA sample measured through high sensitive fluorometric methods such as Qubit 3.0 and the Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA).
Bacteroides species genome functional assessment
Detection of distinctive genetic traits present in B. cellulosilyticus genomes, was based on surveying CAZy genes48 dedicated to carbohydrate metabolism in more than 100 Bacteroides genomes (Table S3). This information was used to explain why weight-loss could be influenced by the proportions of these species in the baseline gut microbiota of the subjects subjected to the AXOS intervention. Gene composition was evaluated in PULs obtained from available-annotated genomes (N = 109) (April 2020) of most representative Bacteroides species in humans retrieved from the PUL database (http://www.cazy.org/PULDB/)49. B. caccae (3 genomes), B. cellulosilyticus (3 genomes), B. dorei (10 genomes), B. fragilis (24 genomes), B. ovatus (16 genomes), B. thetaiotaomicron (14 genomes), B. uniformis (6 genomes), B. vulgatus (12 genomes), and B. xylanisolvens (21 genomes) were used in this functional assessment. The composition of predicted PULs present in each genome was parsed to extract CAZy families of glycoside hydrolases (GH), glycosyl-transferases (GT), polysaccharide lyases (PL), carbohydrate esterases (CE), carbohydrate-binding modules (CBM), and any auxiliary genes present in respective PULs. Inventory of PUL-associated CAZy genes was performed for all the genomes and comparison among them was performed in a species-specific manner. Advanced functional analysis was performed by annotating the coding genes present in the Bacteroides genomes against the Pfam database through the WebMGA server.50
Statistical analyses
All statistical analyses were carried out with the use of R statistical software, version 3.6. The treatment effects on body weight between the enterotype groups were analyzed using linear-mixed models, which included a three-way interaction between treatment * time * P/B group. The difference between the intervention (AXOS) and control (PUFA) group was analyzed by pairwise comparison of the estimated mean differences between P/B groups. Linear-mixed models included age, sex, and baseline BMI as fixed effects, and subject as a random effect. Model-checking was validated by residuals and quantile-quantile probability plots. The results are reported as estimated mean change from baseline within P/B groups and differences in change between P/B groups and interventions with a 95% confidence interval (CI). The significance level was set at P < .05.
Bacteroidetes species associations with metabolic changes, and co-abundant species
Non-parametric and rank-based correlations between baseline log10-transformed Bacteroides and Prevotella species (and P. copri clades) by metagenomics and weight changes, metabolic parameters (cholesterol, HOMA-IR, REE, and breath hydrogen), and fecal SCFA (acetate, butyrate, and propionate) changes, from weeks 0 to 4 were analyzed calculating Kendall’s tau coefficient. To determine bacteria that were co-abundant with B. cellulosilyticus at baseline, a co-abundance network analysis among the 30 most abundant species (all phyla) was performed using Kendall’s tau, as described previously.20 The False-Discovery Rate (FDR) was used to adjust for multiple comparisons in the correlation tests.
To confirm the ability of B. cellulosilyticus to predict body weight change, we performed the randomForest::randomForest R v3.6 function with the baseline relative abundance of the 10 most abundant Bacteroides and Prevotella species, and P/B ratio, as variables to evaluate their importance (IncNodePurity value) for change in body weight after consuming AXOS for 4 weeks.
For qPCR analyses, a differential abundance of B. cellulosilyticus species at baseline was assessed by the Student’s t test with Welch’s correction on log10-transformed data of molecules per ng DNA derived from qPCR51.
Functional enrichment of CAZy and Pfam functions in B. cellulosilyticus genomes was evaluated using hypergeometric Fisher’s exact test with correction for multiple testing using the False-Discovery Rate (FDR) method. Genes and functions associated with B. cellulosilyticus were selected upon FDR ≤ 0.05.
Supplementary Material
Funding Statement
The European Commission 7th Framework Programme through the MyNewGut project (Grant agreement No. 613979) supported this study by sharing genetic and clinical data used in here. The Miguel Servet CP19/00132 grant from the Spanish Institute of Health Carlos III (ISCIII) to ABP is fully acknowledged.
Abbreviations
- AXOS
arabinoxylan oligosaccharides
- BMI
body mass index
- CAZy
carbohydrate-active enzymes
- CBM
carbohydrate-binding module
- CE
carbohydrate esterase
- FDR
false discovery rate
- GL
glycoside hydrolase
- GT
glycosyl transferase
- INP
IncNodePurity or Mean Decrease Gini
- mOTU
metagenomic operational taxonomic unit
- P/B
Prevotella-to-Bacteroides
- Pfam
protein families database
- PL
polysaccharide lyase
- PUFA
polyunsaturated fatty acids
- PUL
polysaccharide utilization loci
- SCFA
short-chain fatty acid
Disclosure of Potential Conflicts of Interest
No, potential conflicts of interest were disclosed.
Authors’ contribution
LC, MFH, and ABP designed the study. CVS and FUW assisted in the data analysis. LK, YS, AA, and ABP produced and shared clinical and genomic data were used in this study. ABP performed genome-wide analysis and qPCR approach. LC, MFH, and ABP drafted the manuscript, and all the authors reviewed and approved the final version submitted to this journal.
Disclosure statement
MFH, LC, and AA are co-inventors on a pending provisional patent application for the use of biomarkers to predict responses to weight-loss diets. AA is a consultant or member of the advisory boards of Groupe Ethique et Sante, France; Weight Watchers, United States; BioCare, Copenhagen; Novo Nordisk, Denmark; and Saniona, Denmark. MFH and AA are co-authors of the book Spis dig slank efter dit bloodsucker (Eat healthily according to your blood sugar), published by Politikens Forlag, Denmark, and of other books about personalized nutrition for weight loss. The remaining authors reported no conflict of interest.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
Data availability
The raw sequencing data used in this study is publicly available in the MG-RAST server upon accession number mgp84629 (for 16 S rRNA gene amplicon sequencing) 51, and in the European Nucleotide Archive (ENA), upon accession number PRJEB25727 (for shotgun metagenome sequencing).
References
- 1.Blüher M. Obesity: global epidemiology and pathogenesis. Nat Rev Endocrinol. 2019;15(5):288–298. doi: 10.1038/s41574-019-0176-8. [DOI] [PubMed] [Google Scholar]
- 2.Gardner CD, Trepanowski JF, Gobbo LCD, Hauser ME, Rigdon J, Ioannidis JPA, Desai M, King AC. Effect of low-fat VS low-carbohydrate diet on 12-month weight loss in overweight adults and the association with genotype pattern or insulin secretion the DIETFITS randomized clinical trial. J Am Med Assoc. 2018;319(7):667–679. doi: 10.1001/jama.2018.0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hjorth MF, Zohar Y, Hill JO, Astrup A. Personalized dietary management of overweight and obesity based on measures of insulin and glucose. Annu Rev Nutr. 2018;38(1):245–272. doi: 10.1146/annurev-nutr-082117-051606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeevi D, Korem T, Zmora N, Israeli D, Rothschild D, Weinberger A, Ben-Yacov O, Lador D, Avnit-Sagi T, Lotan-Pompan M, et al. Personalized nutrition by prediction of glycemic responses. Cell. 2015;163(5):1079–1095. doi: 10.1016/j.cell.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 5.Costea PI, Hildebrand F, Manimozhiyan A, Bäckhed F, Blaser MJ, Bushman FD, De Vos WM, Ehrlich SD, Fraser CM, Hattori M, et al. Enterotypes in the landscape of gut microbial community composition. Nat Microbiol. 2017;3(1):8–16. doi: 10.1038/s41564-017-0072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto J-M, et al. Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen Y, Sue A, Bewtra M, Knights D, Walters WA, Knight R, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science (80-). 2011;334(6052):105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kovatcheva-Datchary P, Nilsson A, Akrami R, Lee YS, De Vadder F, Arora T, Hallen A, Martens E, Björck I, Bäckhed F. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of prevotella. Cell Metab. 2015;22(6):971–982. doi: 10.1016/j.cmet.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 9.Hjorth MF, Christensen L, Kjølbæk L, Larsen LH, Roager HM, Kiilerich P, Kristiansen K, Astrup A. Pretreatment Prevotella-to-Bacteroides ratio and markers of glucose metabolism as prognostic markers for dietary weight loss maintenance. Eur J Clin Nutr. 2019. doi: 10.1038/s41430-019-0466-1. [DOI] [PubMed] [Google Scholar]
- 10.Christensen L, Vuholm S, Roager HM, Nielsen DS, Krych L, Kristensen M, Astrup A, Hjorth MF. Prevotella abundance predicts weight loss success in healthy, overweight adults consuming a whole-grain diet ad libitum: a post hoc analysis of a 6-wk randomized controlled trial. J Nutr. 2019;149(12):2174–2181. doi: 10.1093/jn/nxz198. [DOI] [PubMed] [Google Scholar]
- 11.Hjorth MF, Blædel T, Bendtsen LQ, Lorenzen JK, Holm JB, Kiilerich P, Roager HM, Kristiansen K, Larsen LH, Astrup A. Prevotella-to-Bacteroides ratio predicts body weight and fat loss success on 24-week diets varying in macronutrient composition and dietary fiber: results from a post-hoc analysis. Int J Obes. 2019;43:149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hjorth MF, Roager HM, Larsen TM, Poulsen SK, Licht TR, Bahl MI, Zohar Y, Astrup A. Pre-treatment microbial Prevotella-to-Bacteroides ratio, determines body fat loss success during a 6-month randomized controlled diet intervention. Int J Obes. 2017;42(3):580–583. doi: 10.1038/ijo.2017.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poulsen SK, Due A, Jordy AB, Kiens B, Stark KD, Stender S, Holst C, Astrup A, Larsen TM. Health effect of the new nordic diet in adults with increased waist circumference: A 6-mo randomized controlled trial. Am J Clin Nutr. 2014;99(1):35–45. doi: 10.3945/ajcn.113.069393. [DOI] [PubMed] [Google Scholar]
- 14.McNulty NP, Wu M, Erickson AR, Pan C, Erickson BK, Martens EC, Pudlo NA, Muegge BD, Henrissat B, Hettich RL, et al. Effects of diet on resource utilization by a model human gut microbiota containing bacteroides cellulosilyticus WH2, a symbiont with an extensive glycobiome. PLoS Biol. 2013;11(8): e1001637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benítez-Páez A, Gómez Del Pulgar EM, Sanz Y. The glycolytic versatility of Bacteroides uniformis CECT 7771 and Its genome response to oligo and polysaccharides. Front Cell Infect Microbiol. 2017;7:1–15. doi: 10.3389/fcimb.2017.00383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roager HM, Licht TR, Poulsen SK, Larsen TM, Bahl MI. Microbial enterotypes, inferred by the Prevotella-to-Bacteroides ratio, remained stable during a 6-month randomized controlled diet intervention with the new nordic diet. Appl Environ Microbiol. 2014;80(3):1142–1149. doi: 10.1128/AEM.03549-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patnode ML, Beller ZW, Han ND, Cheng J, Peters SL, Terrapon N, Henrissat B, Le Gall S, Saulnier L, Hayashi DK, et al. Interspecies competition impacts targeted manipulation of human gut bacteria by fiber-derived glycans. Cell. 2019;179(1):59–73. doi: 10.1016/j.cell.2019.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kjølbæk L, Benítez-Páez A, Gómez Del Pulgar EM, Brahe LK, Liebisch G, Matysik S, Rampelli S, Vermeiren J, Brigidi P, LH L, et al. Arabinoxylan oligosaccharides and polyunsaturated fatty acid effects on gut microbiota and metabolic markers in overweight individuals with signs of metabolic syndrome: A randomized cross-over trial. Clin Nutr. 2020;39(1):67–79. doi: 10.1016/j.clnu.2019.01.012. [DOI] [PubMed] [Google Scholar]
- 19.Tett A, Huang KD, Asnicar F, Fehlner-Peach H, Pasolli E, Karcher N, Armanini F, Manghi P, Bonham K, Zolfo M, et al. The prevotella copri complex comprises four distinct clades underrepresented in westernized populations. Cell Host Microbe. 2019;26(5):666–679. doi: 10.1016/j.chom.2019.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rampelli S, Guenther K, Turroni S, Wolters M, Veidebaum T, Kourides Y, Molnár D, Ler L, Benitez-Paez A, Sanz Y, et al. Pre-obese children’s dysbiotic gut microbiome and unhealthy diets may predict the development of obesity. Commun Biol. 2018;1(1):222. doi: 10.1038/s42003-018-0221-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen NK, Deehan EC, Zhang Z, Jin M, Baskota N, Perez-Muñoz ME, Cole J, Tuncil YE, Seethaler B, Wang T, et al. Gut microbiota modulation with long-chain corn bran arabinoxylan in adults with overweight and obesity is linked to an individualized temporal increase in fecal propionate. Microbiome. 2020;8(1):1–21. doi: 10.1186/s40168-020-00887-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christensen L, Roager HM, Astrup A, Hjorth MF. Microbial enterotypes in personalized nutrition and obesity management. Am J Clin Nutr. 2018;108(4):1–7. doi: 10.1093/ajcn/nqy175. [DOI] [PubMed] [Google Scholar]
- 23.Rivière A, Selak M, Lantin D, Leroy F, De Vuyst L. Bifidobacteria and butyrate-producing colon bacteria: importance and strategies for their stimulation in the human gut. Front Microbiol. 2016;7:979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Centanni M, Hutchison JC, Carnachan SM, Daines AM, Kelly WJ, Tannock GW, Sims IM. Differential growth of bowel commensal Bacteroides species on plant xylans of differing structural complexity. Carbohydr Polym. 2017;157:1374–1382. doi: 10.1016/j.carbpol.2016.11.017. [DOI] [PubMed] [Google Scholar]
- 25.Chung WSF, Walker AW, Vermeiren J, Sheridan PO, Bosscher D, Garcia-Campayo V, Parkhill J, Flint HJ, Duncan SH. Impact of carbohydrate substrate complexity on the diversity of the human colonic microbiota. FEMS Microbiol Ecol. 2018;95:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burger-van Paassen N, Vincent A, Puiman PJ, van der Sluis M, Bouma J, Boehm G, van Goudoever JB, Van Seuningen I, Renes IB. The regulation of intestinal mucin MUC2 expression by short-chain fatty acids: implications for epithelial protection. Biochem J. 2009;420(2):211–219. doi: 10.1042/BJ20082222. [DOI] [PubMed] [Google Scholar]
- 27.Hatayama H, Iwashita J, Kuwajima A, Abe T. The short chain fatty acid, butyrate, stimulates MUC2 mucin production in the human colon cancer cell line, LS174T. Biochem Biophys Res Commun. 2007;356(3):599–603. doi: 10.1016/j.bbrc.2007.03.025. [DOI] [PubMed] [Google Scholar]
- 28.Jung TH, Park JH, Jeon WM, Han KS. Butyrate modulates bacterial adherence on LS174T human colorectal cells by stimulating mucin secretion and MAPK signaling pathway. Nutr Res Pract. 2015;9(4):343–349. doi: 10.4162/nrp.2015.9.4.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vital M, Howe AC, Tiedje JM. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. MBio. 2014;5(2):1–11. doi: 10.1128/mBio.00889-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Louis P, Young P, Holtrop G, Flint HJ. Diversity of human colonic butyrate-producing bacteria revealed by analysis of the butyryl-CoA: acetateCoA-transferase gene. Environ Microbiol. 2010;12(2):304–314. doi: 10.1111/j.1462-2920.2009.02066.x. [DOI] [PubMed] [Google Scholar]
- 31.Yamada T, Hino S, Iijima H, Genda T, Aoki R, Nagata R, Han KH, Hirota M, Kinashi Y, Oguchi H, et al. Mucin O-glycans facilitate symbiosynthesis to maintain gut immune homeostasis. EBioMedicine. 2019;48:513–525. doi: 10.1016/j.ebiom.2019.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32..Birchenough G, Schroeder BO, Bäckhed F, Hansson GC. Dietary destabilisation of the balance between the microbiota and the colonic mucus barrier. Gut Microbes. 2019;10(2):246–250. Internet]; :. Available from. doi: 10.1080/19490976.2018.1513765. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Christiansen CB, Gabe MBN, Svendsen B, Dragsted LO, Rosenkilde MM, Holst JJ. The impact of short-chain fatty acids on glp-1 and pyy secretion from the isolated perfused rat colon. Am J Physiol - Gastrointest Liver Physiol. 2018;315(1):G53–65. doi: 10.1152/ajpgi.00346.2017. [DOI] [PubMed] [Google Scholar]
- 34.Psichas A, Sleeth ML, Murphy KG, Brooks L, Bewick GA, Hanyaloglu AC, Ghatei MA, Bloom SR, Frost G. The short chain fatty acid propionate stimulates GLP-1 and PYY secretion via free fatty acid receptor 2 in rodents. Int J Obes. 2015;39(3):424–429. doi: 10.1038/ijo.2014.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Larraufie P, Martin-Gallausiaux C, Lapaque N, Dore J, Gribble FM, Reimann F, Blottiere HM. SCFAs strongly stimulate PYY production in human enteroendocrine cells. Sci Rep. 2018;8(1):1–9. doi: 10.1038/s41598-017-18259-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kulkarni SS, Johnston JJ, Zhu Y, Hying ZT, McBride MJ. The carboxy-terminal region of flavobacterium johnsoniae sprb facilitates its secretion by the type IX secretion system and propulsion by the gliding motility machinery. J Bacteriol. 2019;201:e00218-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fehlner-Peach H, Magnabosco C, Raghavan V, Scher JU, Tett A, Cox LM, Gottsegen C, Watters A, Wiltshire-Gordon JD, Segata N, et al. Distinct polysaccharide utilization profiles of human intestinal prevotella copri isolates. Cell Host Microbe. 2019;26(5):680–690. doi: 10.1016/j.chom.2019.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Paepe K, Verspreet J, Courtin CM, Van de Wiele T. Microbial succession during wheat bran fermentation and colonisation by human faecal microbiota as a result of niche diversification. Isme J. 2019;14:584–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pedersen HK, Gudmundsdottir V, Nielsen HB, Hyotylainen T, Nielsen T, Jensen BAH, Forslund K, Hildebrand F, Prifti E, Falony G, et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016;535(7612):376–381. doi: 10.1038/nature18646. [DOI] [PubMed] [Google Scholar]
- 40.Munoz J, James K, Bottacini F, Van Sinderen D. Biochemical analysis of cross-feeding behaviour between two common gut commensals when cultivated on plant-derived arabinogalactan. Microb Biotechnol. 2020;13(6):1733–1747. doi: 10.1111/1751-7915.13577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Franke T, Deppenmeier U. Physiology and central carbon metabolism of the gut bacterium Prevotella copri. Mol Microbiol. 2018;109(4):528–540. doi: 10.1111/mmi.14058. [DOI] [PubMed] [Google Scholar]
- 42.Rios-Covian D, Salazar N, Gueimonde M, de Los Reyes-gavilan CG. Shaping the metabolism of intestinal Bacteroides population through diet to improve human health. Front Microbiol. 2017;8:1–6. doi: 10.3389/fmicb.2017.00376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patrascu O, Béguet-crespel F, Marinelli L, Chatelier EL, Abraham A, Leclerc M, Klopp C, Terrapon N, Henrissat B, Blottière HM, et al. OPEN A fibrolytic potential in the human ileum mucosal microbiota revealed by functional metagenomic. Nat Publ Gr. 2017;7:40248. DOI: 10.1038/srep40248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun W, Chung F, Walker AW, Bosscher D, Garcia-campayo V, Wagner J, Parkhill J, Duncan SH, Flint HJ. Relative abundance of the Prevotella genus within the human gut microbiota of elderly volunteers determines the inter-individual responses to dietary supplementation with wheat bran arabinoxylan-oligosaccharides. BMC Microbiology. 2020;20:283.2020;1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73(16):5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Benítez-Páez A, Kjølbæk L, Gómez Del Pulgar EM, Brahe LK, Astrup A, Matysik S, Schött H-F, Krautbauer S, Liebisch G, Boberska J, et al. A multi-omics approach to unraveling the microbiome-mediated effects of arabinoxylan oligosaccharides in overweight humans. mSystems. 2019;4(4):1–16. doi: 10.1128/mSystems.00209-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Milanese A, Mende DR, Paoli L, Salazar G, Ruscheweyh HJ, Cuenca M, Hingamp P, Alves R, Costea PI, Coelho LP, et al. Microbial abundance, activity and population genomic profiling with mOTUs2. Nat Commun. 2019;10:1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42(D1):490–495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Terrapon N, Lombard V, Drula É, Lapébie P, Al-Masaudi S, Gilbert HJ, Henrissat B. PULDB: the expanded database of Polysaccharide Utilization Loci. Nucleic Acids Res. 2018;46(D1):D677–83. doi: 10.1093/nar/gkx1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu S, Zhu Z, Fu L, Niu B, WebMGA: LW. A customizable web server for fast metagenomic sequence analysis. BMC Genomics. 2011;12:444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meyer F, Paarmann D, D’Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, et al. The metagenomics RAST server – a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinform. 2008;9(1):1–8. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing data used in this study is publicly available in the MG-RAST server upon accession number mgp84629 (for 16 S rRNA gene amplicon sequencing) 51, and in the European Nucleotide Archive (ENA), upon accession number PRJEB25727 (for shotgun metagenome sequencing).


