ABSTRACT
Colorectal cancer (CRC) is a heterogeneous disease with different gene expression patterns. There are two major colorectal carcinogenesis pathways: conventional adenoma-carcinoma pathway and alternative serrated neoplasia pathway. Apart from the conventional pathway that is typically initiated by characteristic APC mutation and chromosomal instability, the serrated neoplasia pathway is mainly characterized by mutations of BRAF or KRAS, microsatellite instability (MSI), and CpG island methylator phenotype (CIMP). Despite the malignant potential of serrated lesions, they can be easily overlooked during endoscopy screening and even in pathological assessment due to its anatomical location, morphology, and histological features. It has been shown that environmental factors especially the gut microbial composition play a key role in CRC pathogenesis. Thus, the preferential localization of serrated lesions in specific intestine areas suggest that niche-specific microbiota composition might intertwined with host genetic perturbations during the development of serrated lesions. Although serrated lesions and conventional adenomas are biologically different, most studies have focused on conventional adenomas, while the pathophysiology and role of microorganisms in the development of serrated lesions remain elusive. In this review, we discuss on the role of gut microbiota in the serrated neoplasia pathway of colorectal carcinogenesis and its specific clinical and molecular features, and summarize the potential mechanisms involved.
KEYWORDS: Serrated pathway, microbiota, colorectal cancer, neoplasia
Introduction
Colorectal cancer (CRC) is the third most common cancer and the second leading death of cancer worldwide.1 In 2018, CRC was the most commonly diagnosed gastrointestinal cancer, constituting 10.2% and 9.2% cancer cases and deaths respectively worldwide.2 In the United States, CRC is estimated to make up 8.2% and 8.8% of total cancer incidence and mortality in 2020, respectively.3,4 Malignant changes in the intestinal tract are often developed from a focal dysplastic polypoid precursor, the adenoma, which accumulates further genetic mutations and progresses following the adenoma-carcinoma sequence.5 Similar to conventional adenomas, serrated lesions in the colorectum have a potential to transform into malignant CRC,6 especially large serrated lesions that are located in the proximal colon.7
The development of CRC follows several distinct mechanistic pathways, including the adenoma–carcinoma pathway and serrated neoplasia pathway.8 While the conventional adenoma-carcinoma pathway is more common, a small subset of CRC occurs through the serrated pathway. In the past, these serrated lesions were considered as relatively benign lesions;9 however, emerging evidences suggested that certain sessile lesions are non-adenomatous precursors of malignant cancers.10,11 In the fifth edition of the World Health Organization classification of digestive tumors, sessile serrated polyp/adenoma was renamed as sessile-serrated lesion (SSL). In the British pathological classification system, serrated lesions can be classified into several lesion types, including hyperplastic polyp (HP), SSL, SSL with dysplasia, traditional-serrated adenoma (TSA) and mixed polyp.10 SSLs and TSAs have been recognized as important precancerous lesions of CRC.
Because of their indistinctive morphological and histological features, serrated lesions can be easily overlooked during colonoscopy and even in pathological assessment. SSLs are typically flat or sessile under endoscopic visualization, and are occasionally covered by a mucus cap.10 Many CRCs derived from SSLs are located in the right side of the colon, with molecular features of BRAF mutation, high microsatellite instability (MSI), and CpG island methylator phenotype (CIMP). These cancers are thought to account for a large proportion of interval cancers and may represent the main cause of cancer screening failure. Thus, it is important to study the serrated pathway to develop better management strategies for these cancers.
Various genetic and environmental factors contribute to colorectal carcinogenesis. Previous twin studies showed that the heritability of CRC is only around 12–35%,12 suggesting that environmental factors may play a greater role in sporadic CRC.8 Certain environmental factors are associated with serrated colorectal neoplasia. Systematic reviews found that smoking, alcohol, and body mass index were more strongly associated with serrated polyps than conventional adenomas.13,14 A strong association between red meat consumption and risk of SSLs was also shown in a colonoscopy-based case–control study.15 These epidemiological findings could enhance our mechanistic understanding and help identify mitigating strategies for serrated neoplasia.
Furthermore, the microbiota has recently received increasing attention as a non-genetic factor in colorectal neoplasia. Tens of trillion microorganisms colonize the human gastrointestinal tract,16 to interact with our epithelial cell as part of the host–microbe interaction.17,18 Research in recent years showed that several bacteria is associated with CRC, including Fusobacterium nucleatum, Bacteroides fragilis, and other CRC-enriched bacteria,19 through different pro-inflammatory and pro-carcinogenic mechanisms.20 Despite this, the role of gut microbiota in the serrated neoplasia remains largely unknown.
In this article, we review the role of microbiota and molecular pathways pertinent to the formation of serrated neoplasm.
The serrated neoplasia pathway
Our knowledge on the molecular pathways of colorectal adenomas and other precancerous lesions has increased substantially over the past few years. With the advent of molecular testing for MSI, RAS (KRAS, NRAS) and BRAF mutations, accurate and tailored treatment for advanced CRC is possible.21 These tumor genetic insights have shed light on their precursor lesions as well. There are two main pathways of carcinogenesis: the conventional adenoma-carcinoma pathway (also known as chromosomal instability pathway) and the alternative serrated neoplasia pathway.22 Conventional adenomas are typically initiated by APC mutations, followed by RAS activation or loss of function mutations in TP53.22 In contrast, the serrated neoplasia pathway is mainly characterized by mutations of BRAF or KRAS, chromosomal stability, and CIMP.22 Most CRC develop through the conventional adenoma-carcinoma pathway, while approximately 10–20% of CRC cases occur through the alternative serrated neoplasia pathway.22 Autopsy studies showed that the prevalence of serrated lesion varies, but in general about 25% of adults have one or more serrated lesions.23 Recently, a systematic review identified 74 relevant colonoscopy studies and found that SSL prevalence greatly varied by geographical regions, ranging from 2.6% in Asia to 10.5% in Australia.24
In 2007, Makinen evaluated three molecular alterations to help further subtype serrated lesions.25 By combining the RAS mutations, the degree of MSI, and the level of CIMP, two separate serrated pathways26 could be classified:11,27 (1) Sessile serrated pathway with BRAF mutation, MSI-H/L and CIMP-H, typical lesions being SSLs, and (2) Traditional serrated pathway with KRAS mutation, low-level MSI (MSI-L) or microsatellite stability (MSS), and CIMP-L, typical lesions being TSAs (Figure 1).
Figure 1.
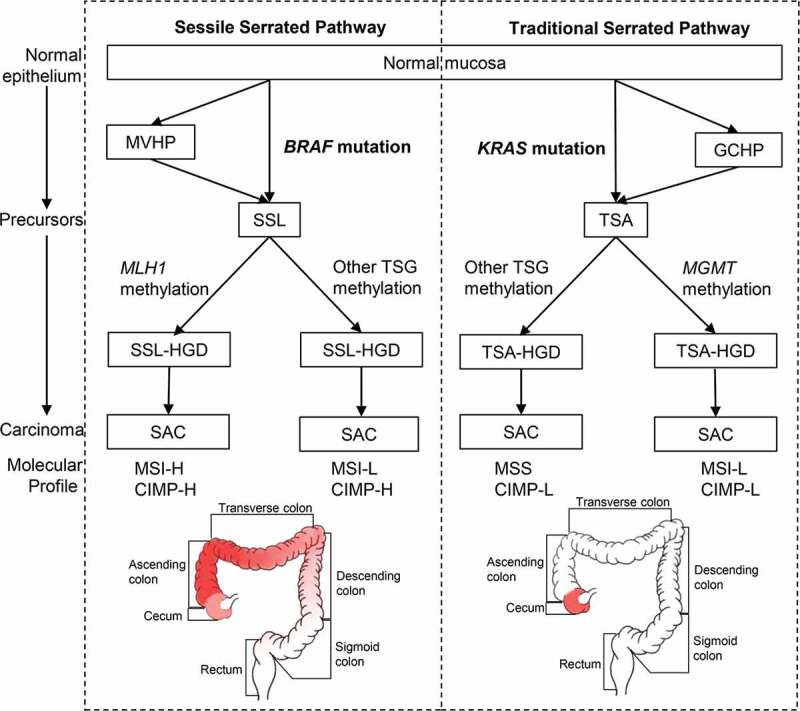
The sessile (left) and traditional (right) serrated pathways. Frequently affected areas for colorectal tumors in each pathway are highlighted in red and the color depth represents the frequency of CIMP-H, MSI-H and BRAF/KRAS mutations in CRC. Abbreviations: MVHP, microvascular hyperplastic polyp; GCHP, goblet cell-rich hyperplastic polyps; SSL, sessile serrated lesion; TSA, traditional-serrated adenoma; MLH1, MutL homolog 1; MGMT, O-6-methylguanine-DNA methyltransferase; TSG, tumor suppressor genes; SSL-HGD, sessile serrated lesion with high-grade dysplasia; TSA-HGD, traditional-serrated adenoma with high-grade dysplasia; SAC, serrated adenocarcinoma; MSI-H, high-level microsatellite instability; MSI-L, low-level microsatellite instability; MSS, microsatellite stability; CIMP-H, high-level CpG island methylator phenotype; CIMP-L, low-level CpG island methylator phenotype
Further studies have investigated the anatomical locations of these colorectal lesions. Although Bufill et al. divided the colorectal tumor location at splenic flexure into proximal and distal colons in 1990,28 the frequencies of the molecular signatures, including CIMP-H, high-level MSI (MSI-H), and BRAF mutations do not change abruptly at the splenic flexure.29 Instead, these frequencies increased gradually from the rectum to ascending colon, followed by a relatively decrease in the cecum,29 challenging the common conception of discrete molecular features of proximal (right-sided) versus distal (left-sided) CRC30,31 (Figure 1). Nevertheless, cecal cancers harbor a high frequency of KRAS mutations.29
Consensus molecular subtypes (CMSs)
CRC is a heterogeneous disease with distinctive gene expression patterns.32–38 In the genomic analysis of 276 samples in the Cancer Genome Atlas Project, three-quarters among the hypermutated tumors had high MSI, usually with hypermethylation and MLH1 silencing, were located in the right colon and were frequently associated with CIMP.38 Schlicker et al. first reported an epithelial-mesenchymal-transition (EMT) expression signature defined subgroup in 2012.34 Subsequent molecular classifications of CRCs based on its stemness, Wnt pathway expression,35 and clinicopathological features36 have been proposed. Marisa et al. identified six molecular subtypes associated with distinct clinicopathological characteristics, molecular alterations, specific enrichments of supervised gene expression signatures (stem cell phenotype-like, normal-like, serrated colon cancer phenotype-like), and deregulated signaling pathways.37 Budinska et al. distinguished five different gene expression CRC subtypes, which are surface crypt-like, lower crypt-like, CIMP-H-like, mesenchymal, and mixed.32 A molecular classification associated with prognosis and chemotherapy response was developed by Roepman et al. in 2014, which consist of three major intrinsic subtypes (A-, B- and C-type) based on three tumor biological hallmarks: EMT, mismatch repair genes deficiency, and cellular proliferation.33 To better consolidate the biological findings and enhance international communications, the consensus molecular subtypes (CMS) was proposed in 2015 to unify six independent transcriptome-based CRC subtyping strategies as abovementioned.32–37,39 The four subtypes with distinguishing features include: CMS1 (MSI immune) tumors that are immunogenic, microsatellite unstable, and hyper-mutated; CMS2 (canonical) tumors that show WNT and MYC signaling activation; CMS3 (metabolic) tumors that have metabolic dysregulation; and CMS4 (mesenchymal) tumors that have stromal infiltration, TGF-β activation, angiogenesis39 (Figure 2). Samples with mixed features are transition phenotypes or may represent intra-tumoral heterogeneity.
Figure 2.

Consensus molecular subtypes (CMS) in CRC and their precursor lesions. Abbreviations: MSI, microsatellite instability; CIMP, CpG island methylator phenotype; SCNA, somatic copy number alterations
This molecular scheme raised an immediate question to how the pathological precursor types are related to the cancer subtypes. To address this question, Fessler et al. investigated the role of premalignant lesions using organoid culture and found that SSLs overexpressed TGF-β signaling, a key molecular characteristic of CMS4 subtype of CRC.40 Besides, Chang et al. analyzed the transcriptomes of 311 sporadic and 78 hereditary adenomatous and serrated lesions by a random forest classifier, and found that adenomatous polyps showed a highly similar transcriptomic profile to the CMS2 subtype, whereas the transcriptomic profiles of HP and SSL resemble that of the CMS1 subtype. Together with their right-sided anatomic location and BRAF mutations,41 this suggests a strong relationship between serrated lesions and the CMS1 subtype of CRC. Nevertheless, significant KRAS mutations were not observed probably because of the small number of precursor lesions resembling CMS3 in their study. The relationships between premalignant lesions (SSLs versus tubular adenomas42) and CMS3 tumors42,43 remain uncertain. Furthermore, a recent systematic review suggested tubulovillous adenomas with serrated features to be precursors of KRAS mutant tumors.44 Tsai et al. evaluated the pathological and molecular features of 60 TSAs with cytologic dysplasia and/or invasive carcinoma, and shown that tubulovillous adenoma with serrated features had higher frequencies of KRAS mutations than TSAs with serrated dysplasia.44,45 Potential precursor lesions assigned to the CMSs based on the above research results are shown in Figure 2.
Gut microbiota in serrated lesions
Recent literature has provided evidence that microorganisms can promote colorectal carcinogenesis.20 Nevertheless, these studies have focused on CRC and premalignant polyps derived from the conventional pathway,20 and the role of microorganisms in the serrated neoplasia is less clear. Peters et al. compared the stool microbiota between conventional adenoma and serrated lesions of 540 colonoscopy-screened adults by 16S rDNA gene sequencing and observed a significant depletion of Erysipelotrichi in 33 SSL cases.46 The increase of this bacterial class is associated with impenetrable mucus layer in mice47 and may play a protective role in SSL development. However, in a study from Iran, researchers analyzed the changes of fecal microbiota in patients with different precursor lesions including serrated lesions (21 HP and 16 SSL cases) and failed to observe significant differences in the microbiota.48 Similarly, a Korean study did not identify significant microbiota changes in rectal mucosae from healthy controls and patients with conventional adenoma, SSL, and CRC, respectively.49 However, both studies were limited by their small sample size. Thus, further studies with more samples could provide insight into the metagenomic landscape of SSLs.
There is a close association between F. nucleatum and CRC progression,50 and high level of F. nucleatum was associated with poor survival in metastatic CRC.51 Yu et al. examined the invasive F. nucleatum using 16S rRNA fluorescence in situ hybridization (FISH) and observed significantly more invasive F. nucleatum in proximal HPs and SSLs than that of conventional adenomas.52 On the contrary, Ito et al. detected F. nucleatum by quantitative PCR in HPs, SSLs, TSAs, and non-serrated adenomas, and found that this bacterium was not significantly associated with lesion histology, but rather was associated with right-sided premalignant lesions with BRAF mutation, CIMP-high, and MSI.53 Because of these features pointing to serrated neoplasia,11,27 the existence of colorectal F. nucleatum may influence CRC progression through serrated pathway. Another similar study by Park et al. compared the gut microbiota between tubular adenoma (TA) and SSLs and found that the relative abundance of Fusobacteria did not differ significantly between these patients.54 These two similar results suggested that Fusobacteria may contribute to carcinogenesis regardless of the molecular pathway.53,54 However, the small sample sizes and lack of multi-omics platforms have again limited these studies.
Furthermore, a study has associated CRC microbiota with tumor CMS type and identified some bacterial species specific to CMS155 characterized by MSI and immune activation.39 Given the connection between CMS1 and serrated neoplasia,41 these species might contribute to the serrated pathway of CRC development. In this study,55 16S rRNA analysis showed that the relative abundances of Fusobacteria and Bacteroidetes increased and the levels of Firmicutes and Proteobacteria decreased in CMS1. Species-level analysis showed that Fusobacterium hwasookii and Porphyromonas gingivalis are the most highly enriched species associated with CMS1, as well as oral pathogens such as F. nucleatum, Parvimonas micra, and Peptostreptococcus stomatis.
Lastly, there was a case report that human intestinal spirochetosis may be responsible for colonic adenomas or HPs.4 In a retrospective case–control study, the rate of human intestinal spirochetosis infection was significantly higher in SSL at 52.6% (10/19) compared to controls at 8.1% (14/172), which suggested a possible association between human intestinal spirochetosis and SSL.56 Nevertheless, this finding is yet to be validated in larger studies preferably from more diverse populations.
Gut microbiota and specific molecular features
Many studies explored the microbial community of CRC samples in different cohorts, and established the associations of F. nucleatum with important clinical and molecular features.53,57–63 For instance, F. nucleatum was shown to be significantly associated with MLH1 methylation,53,57,59,60,63 high-level MSI,53,57,59–63 high-level CIMP53,59,60,63 and BRAF mutation57,59,61,62 (Table 1). However, controversial data have been reported on whether KRAS mutations associated with F. nucleatum abundance.53,58–65 In a Brazilian study analyzing 43 fresh CRC tissues by qPCR and direct sequencing, Proenca et al. found that KRAS mutations occurred more frequently in F. nucleatum-infected CRC.64 Yamaoka et al. measured F. nucleatum copy numbers by droplet digital PCR and found a significant correlation between F. nucleatum abundance and KRAS mutations.65 Higher abundance of intra-tumoral F. nucleatum was also reported in CRC with proximal tumor location,57,59,60 higher clinical stage (T3/T4),57,59,60 poorer tumor differentiation,57,59,60 and worse survival.57,59,66 In addition, CIMP high cases were characterized by a high rate of mutations in MSI, BRAF67 and chromatin regulator genes, especially CHD7 and CHD8,68 and rarely KRAS and TP53 mutations.67 F. nucleatum abundance was found to be associated with CHF7/8 mutation and TP53 wild-type status.63 KRAS mutation was also detected, but there was no statistical difference between the mutation state and F. nucleatum abundance.53,58–63
Table 1.
Serrated pathways associated with molecular features in Fusobacterium nucleatum (Fn) high expression CRC tissues. + indicates Fn-high CRC tissues exhibiting more frequent molecular features than Fn-low/negative ones (P < .05); whereas – indicates no significant difference of serrated pathway associated molecular features between Fn-high and Fn-low/negative tissues. Abbreviations: FFPE, formalin-fixed paraffin-embedded; Fn-high, high amount of Fusobacterium nucleatum DNA in tissues; Fn-low, low amount of Fusobacterium nucleatum DNA in tissues; MSI-H, high-level microsatellite instability; CIMP-H, high-level CpG island methylator phenotype
| Authors | Year | Cohort | Specimen Type | Detection Method | Molecular Features in Fn-high Tissues |
||||
|---|---|---|---|---|---|---|---|---|---|
| MLH1 Methylated | MSI-H | CIMP-H | BRAF Mutation | ||||||
| Tahara et al.[63] | 2014 | United States | Fresh-frozen tissue | qPCR | + | + | + | - | |
| Ito et al.[53] | 2015 | Japanese | FFPE tissue | qPCR | + | + | + | - | |
| Mima et al.[60] | 2015 | United States | FFPE tissue | qPCR | + | + | + | - | |
| Mima et al.[59] | 2016 | United States | FFPE tissue | qPCR | + | + | + | + | |
| Nosho et al.[61] | 2016 | Japanese | FFPE tissue | qPCR | / | + | / | + | |
| Park et al.[62] | 2017 | Korean | FFPE tissue | qPCR | - | + | - | + | |
| de Carvalho et al.[57] | 2019 | Brazilian | Fresh-frozen tissue | 16S rDNA sequencing, qPCR | + | + | / | + | |
Besides F. nucleatum, correlations between other microbial species with the status of MLH1, BRAF, KRAS were also reported. Immunohistochemical analysis indicated that KRAS and BRAF expressions were obvious in tumor with high abundance of F. nucleatum and Bacteroides fragilis, while tumors with MLH1 mutation showed lower abundance of these species.66 Moreover, a high abundance of F. nucleatum and B. fragilis were independent indicators of poor survival.66 A positive correlation between Ruminococcus gnavus and KRAS mutation in aberrant crypt foci samples was also described, although this finding was only reported in one study with a limited sample size.69 As described previously, serrated neoplasia is characterized by high MLH1 deficiency, KRAS and BRAF mutation,6,11,25,27 yet the association with F. nucleatum, B. fragilis, or R. gnavus remains unclear and needs to be explored in future studies.
Potential mechanisms of microbial dysbiosis in serrated neoplasm formation
The fact that serrated lesions are preferentially localized in specific colonic locations43 suggested that non-genetic factors, such as niche-specific microbiota, may interplay with genetic perturbations to affect their development. To verify this hypothesis, Lira et al. have modeled a series of transgenic mice.70–73 Based on the immunohistochemical and immunoblot analyses, they found that the EGFR signaling pathway is activated in human-serrated lesions.70 Activation of EGFR signaling by transgenic expression of the EGFR ligand heparin-binding epidermal growth factor-like growth factor (HBEGF) in mice intestine promotes the development of cecal-serrated lesions.70 It showed that host-specific microbiota was associated with serrated polys, and microbiota alteration induced by antibiotics or by embryo transfer rederivation suppressed the formation of serrated lesions in the cecum of HBEGF transgenic mice.72 The development of serrated lesions was associated with epithelial barrier breakdown, bacterial invasion, and overexpression of several inflammatory factors.72,73 The release of IL1B from inflammatory macrophages stimulate subsets of cecal platelet-derived growth factor receptor alpha+ (PDGRFA+) fibroblasts during an early stage of serrated lesion development, resulting in upregulation of Matrix Metallopeptidase 3 (MMP3), which can promote inflammation and accelerate serrated lesion development by facilitating HBEGF/EGFR signaling.73 Using 16S rDNA sequencing, the authors showed that the bacterial phylum of Verrucomicrobia was enriched, whereas Deferribacteres was decreased in the mouse cecal mucosa of serrated lesions compared to rederived HBUS mice.72
As discussed previously, F. nucleatum is an important bacterium in CRC and shows association with serrated neoplasia. F. nucleatum attaches and invades human epithelial cells via adhesion (FadA).74 Another virulence factor from F. nucleatum, an autotransporter protein (Fap2), has been shown to promote CRC progression by suppressing immune cell activity.75 Kostic et al. reported that F. nucleatum selectively recruits myeloid-derived immune cells (MDSCs) in CRC.76 F. nucleatum increases the production of reactive oxygen species (ROS), 76,77 possibly by MDSCs recruit. Tumor-associated MDSCs promote carcinogenesis through oxidative metabolism, including the production of ROS in human CRC.78 ROS induction is correlated with DNA methylation.79 Interestingly, methylation could also occur in promoter regions of MLH1 gene and lead to MSI,61,80 which are the characters of sessile-serrated pathway.
Another mechanism for serrated neoplaia progression related to F. nucleatum is a tumor immunosuppressive microenvironment. F. nucleatum is associated with a lower density of CD3 + T cells in a US cohort,60 and F. nucleatum high MSI-H CRC was significantly associated with a high density of CD68+ tumor-infiltrating macrophages, a special subtype of MDSC.62 A study by Hamada et al. found that the presence of F. nucleatum in CRC tissues was associated with MSI, lower-level tumor-infiltrating lymphocytes (TIL), and poor clinical outcomes.81 Therefore, F. nucleatum may promote immune evasion by suppressing anti-tumor immune responses in MSI-H CRC. Moreover, the F. nucleatum derived FadA can interact with E-cadherin to promote CRC cells proliferation.74 This may be relevant to serrated lesions, as altered expression and localization of E-cadherins and its associated β-catenin have been described in hyperplastic polyps and serrated adenomas.82 The change in E-cadherin expression may be related to epithelial remodeling and stratification implicated in serrated adenoma formation.
Finally, F. nucleatum can also impact serrated carcinogenesis by generating a pro-inflammatory microenvironment. Lipopolysaccharide (LPS) is a virulence factor present on F. nucleatum, which is recognized by Toll-like receptors to activate the TLR4/MYD88 pathway, leading to nuclear factor-κB (NF-κB) activation64 and release of inflammatory cytokines such as TNF-α, IL-6, IL-8, IL-18.64,66,74,76,83,84 IL8 was upregulated in MSI-H CRC.64 Inflammation reduces the enzymatic activity of mismatch repair (MMR) proteins and causes MLH1 silencing, leading to MSI.85 The potential F. nucleatum associated mechanisms involved in the pathogenesis of serrated neoplasm is presented in Figure 3.
Figure 3.

Potential mechanisms of gut microbiota dysbiosis on serrated neoplasm formation. F. nucleatum presents the virulence factors of FadA,74 Fap275 and LPS,64 mediating its invasion and the promotion of serrated tumors. F. nucleatum can increase cell proliferation by binding of FadA74 to E-cadherin to activate the Wnt/β-catenin pathway.74 The TLR4/MYD88 pathway is stimulated in response to LPS on F. nucleatum,64 activating NF-κB64 and resulting in a pro-inflammatory microenvironment.64,66,74,76,83,84 F. nucleatum modifies the tumor microenvironment by attracting MDSC76 and suppressing anti-tumoral immune responses.60,81 MDSCs can produce ROS,76–78 inducing MLH1 methylation79 and leading to MSI.61,80 Other microorganisms, like spirochetes,4,56 may also participate in the serrated pathway of cancer formation. EGFR signaling activation was observed in human-serrated polyps70 and the role of gut microbiota was confirmed in transgenic HBUS mice.72,73 Subsets of cecal PDGFRA+ fibroblasts are activated by IL1B released from inflammatory macrophages during an early stage of serrated lesions development.73 Proinflammatory genes and MMP3 are upregulated in activated fibroblasts, which can promote inflammation and SP development by facilitating HBEGF/EGFR signaling.73 Abbreviations: Fap2, F. nucleatum autotransporter protein 2; LPS, lipopolysaccharide; FadA, F. nucleatum adhesin; NF-κB, nuclear factor-κB; MDSC, myeloid-derived immune cell; ROS, reactive oxygen species; MLH1, mutL homolog 1; MSI, microsatellite instability; EGFR, epidermal growth factor receptor; HBEGF, heparin-binding epidermal growth factor-like growth factor; PDGRFA+, platelet-derived growth factor receptor alphaþ positive; MMP3, matrix metallopeptidase 3.
Conclusion and future perspectives
This review summarized the potential association between the gut microbiota and the serrated pathways and proposed putative mechanisms of how gut microorganisms might participate in colorectal carcinogenesis. Although serrated lesions-derived CRC is not the most common type of CRC, its invasiveness and relatively favorable response to target therapy and immunotherapy render it a distinct patient group to be further studied. Most interval cancers in CRCs are proximal tumors with molecular features of MLH1 methylation, MSI-H, CIMP-H and BRAF mutation, and these patients are often diagnosed at advanced stages, with poor prognosis and low survival rates. Early detection of these serrated lesions as premalignant precursors is essential for clinicians. Besides histological and molecular features, the gut microbiota emerges as a critical environmental factor that should be studied to improve the tumor biology, diagnosis, and treatment response of this cancer subtype. Further studies would be necessary to determine the exact role of the gut microbiota in the serrated neoplasia pathway with specific murine models, such as the BRAFV637E mutant mice,86,87 and to identify specific biomarkers for screening, diagnosis, prognosis, and prediction of serrated cancers.
Funding Statement
This project was supported by Science and Technology Program Grant Shenzhen (JCYJ20170413161534162), HMRF Hong Kong (17160862), RGC-CRF Hong Kong (C4039-19G), RGC-GRF Hong Kong (14163817), and National Natural Science Foundation of China Grant (31871340, 81922082).
Abbreviations
CIMP, CpG island methylator phenotype;
CIMP-H, high-level CpG island methylator phenotype;
CIMP-L, low-level CpG island methylator phenotype;
CMSs, consensus molecular subtypes;
CRC, colorectal cancer;
EGFR, epidermal growth factor receptor;
EMT, epithelial-mesenchymal-transition;
FadA, Fusobacterium nucleatum adhesin;
Fap2, Fusobacterium nucleatum autotransporter protein 2;
FFPE, formalin-fixed paraffin-embedded;
FISH, fluorescence in situ hybridization;
Fn-high, high amount of Fusobacterium nucleatum DNA in tissues;
Fn-low, low amount of Fusobacterium nucleatum DNA in tissues;
GCHP, goblet cell-rich hyperplastic polyps;
HBEGF, heparin-binding epidermal growth factor-like growth factor;
HP, hyperplastic polyp;
LPS, lipopolysaccharide;
MDSCs, myeloid-derived immune cells;
MGMT, O-6-methylguanine-DNA methyltransferase;
MLH1, MutL homolog 1;
MMP3, matrix metallopeptidase 3;
MMR, mismatch repair;
MSI, microsatellite instability;
MSI-H, high-level MSI;
MSI-L, low-level MSI;
MSS, microsatellite stability;
MVHP, microvascular hyperplastic polyp;
NF-κB, nuclear factor-κB;
PDGRFA+, platelet-derived growth factor receptor alphaþ positive;
ROS, reactive oxygen species;
SAC, serrated adenocarcinoma;
SSL, sessile serrated lesion;
SSL-HGD, sessile serrated lesion with high-grade dysplasia;
TIL, tumor-infiltrating lymphocytes;
TSA, traditional-serrated adenoma;
TSA-HGD, traditional-serrated adenoma with high-grade dysplasia;
TSG, tumor suppressor genes.
Authors’ contributions
XK, JY and SHW meditated the project, wrote the primary and following drafts. The co-authors revised the drafts and critically edited the manuscript. All authors participated in the drafting and agreed with this final manuscript for submission.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A.. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–12. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Nguyen LH, Goel A, Chung DC. Pathways of colorectal carcinogenesis. Gastroenterology. 2020;158:291–302. doi: 10.1053/j.gastro.2019.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 4.Calderaro A, Gorrini C, Montecchini S, Villanacci V, Bassotti G, Dettori G, Chezzi C. Intestinal spirochaetosis associated with hyperplastic and adenomatous colonic polyps. Pathol Res Pract. 2012;208:177–180. doi: 10.1016/j.prp.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 5.Day DW. The adenoma-carcinoma sequence. Scand J Gastroenterol Suppl. 1984;104:99–107. [PubMed] [Google Scholar]
- 6.Jass JR. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology. 2007;50:113–130. doi: 10.1111/j.1365-2559.2006.02549.x. [DOI] [PubMed] [Google Scholar]
- 7.Ng SC, Ching JY, Chan VC, Wong MC, Tang R, Wong S, Luk AK, Lam TY, Gao Q, Chan AW, et al. Association between serrated polyps and the risk of synchronous advanced colorectal neoplasia in average-risk individuals. Aliment Pharmacol Ther. 2015;41:108–115. doi: 10.1111/apt.13003. [DOI] [PubMed] [Google Scholar]
- 8.Keum N, Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol. 2019;16:713–732. doi: 10.1038/s41575-019-0189-8. [DOI] [PubMed] [Google Scholar]
- 9.Lane N. The precursor tissue of ordinary large bowel cancer. Cancer Res. 1976;36:2669–2672. [PubMed] [Google Scholar]
- 10.East JE, Atkin WS, Bateman AC, Clark SK, Dolwani S, Ket SN, Leedham SJ, Phull PS, Rutter MD, Shepherd NA, et al. British society of gastroenterology position statement on serrated polyps in the colon and rectum. Gut. 2017;66(7):1181–1196. doi: 10.1136/gutjnl-2017-314005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bettington M, Walker N, Clouston A, Brown I, Leggett B, Whitehall V. The serrated pathway to colorectal carcinoma: current concepts and challenges. Histopathology. 2013;62:367–386. doi: 10.1111/his.12055. [DOI] [PubMed] [Google Scholar]
- 12.Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K. Environmental and heritable factors in the causation of cancer–analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 13.Bailie L, Loughrey MB, Coleman HG. lifestyle risk factors for serrated colorectal polyps: a systematic review and meta-analysis. Gastroenterology. 2017;152:92–104. doi: 10.1053/j.gastro.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 14.He X, Wu K, Ogino S, Giovannucci EL, Chan AT, Song M. Association between risk factors for colorectal cancer and risk of serrated polyps and conventional adenomas. Gastroenterology. 2018;155:355–373 e318. doi: 10.1053/j.gastro.2018.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davenport JR, Su T, Zhao Z, Coleman HG, Smalley WE, Ness RM, Zheng W, Shrubsole MJ. Modifiable lifestyle factors associated with risk of sessile serrated polyps, conventional adenomas and hyperplastic polyps. Gut. 2018;67:456–465. doi: 10.1136/gutjnl-2016-312893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sender R, Fuchs S, Are MR. We really vastly outnumbered? revisiting the ratio of bacterial to host cells in humans. Cell. 2016;164:337–340. doi: 10.1016/j.cell.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 17.Hold GL, Allen-Vercoe E. Gut microbial biofilm composition and organisation holds the key to CRC. Nat Rev Gastroenterol Hepatol. 2019;16:329–330. doi: 10.1038/s41575-019-0148-4. [DOI] [PubMed] [Google Scholar]
- 18.Vonaesch P, Anderson M, Sansonetti PJ. Pathogens, microbiome and the host: emergence of the ecological Koch’s postulates. FEMS Microbiol Rev. 2018;42:273–292. doi: 10.1093/femsre/fuy003. [DOI] [PubMed] [Google Scholar]
- 19.Dai Z, Coker OO, Nakatsu G, Wu WKK, Zhao L, Chen Z, Chan FKL, Kristiansen K, Sung JJY, Wong SH, et al. Multi-cohort analysis of colorectal cancer metagenome identified altered bacteria across populations and universal bacterial markers. Microbiome. 2018;6:70. doi: 10.1186/s40168-018-0451-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong SH, Yu J. Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nat Rev Gastroenterol Hepatol. 2019;16:690–704. doi: 10.1038/s41575-019-0209-8. [DOI] [PubMed] [Google Scholar]
- 21.Nagtegaal ID, Odze RD, Klimstra D, Paradis V, Rugge M, Schirmacher P, Washington KM, Carneiro F, Cree IA. The 2019 WHO classification of tumours of the digestive system. Histopathology. 2020;76:182–188. doi: 10.1111/his.13975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. The Lancet. 2019;394:1467–1480. doi: 10.1016/s0140-6736(19)32319-0. [DOI] [PubMed] [Google Scholar]
- 23.Rex DK, Ahnen DJ, Baron JA, Batts KP, Burke CA, Burt RW, Goldblum JR, Guillem JG, Kahi CJ, Kalady MF, et al. Serrated lesions of the colorectum: review and recommendations from an expert panel. Am J Gastroenterol. 2012;107:1315–1329; quiz 1314, 1330. doi: 10.1038/ajg.2012.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meester RGS, van Herk M, Lansdorp-Vogelaar I, Ladabaum U. Prevalence and clinical features of sessile serrated polyps: a systematic review. Gastroenterology. 2020;159(1):105–118.e25. doi: 10.1053/j.gastro.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makinen MJ. Colorectal serrated adenocarcinoma. Histopathology. 2007;50:131–150. doi: 10.1111/j.1365-2559.2006.02548.x. [DOI] [PubMed] [Google Scholar]
- 26.Park SJ, Rashid A, Lee J-H, Kim SG, Hamilton SR, Wu -T-T. Frequent CpG island methylation in serrated adenomas of the colorectum. Am J Pathol. 2003;162:815–822. doi: 10.1016/S0002-9440(10)63878-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noffsinger AE. Serrated polyps and colorectal cancer: new pathway to malignancy. Annu Rev Pathol. 2009;4:343–364. doi: 10.1146/annurev.pathol.4.110807.092317. [DOI] [PubMed] [Google Scholar]
- 28.Bufill JA. Colorectal cancer: evidence for distinct genetic categories based on proximal or distal tumor location. Ann Intern Med. 1990;113:779–788. doi: 10.7326/0003-4819-113-10-779. [DOI] [PubMed] [Google Scholar]
- 29.Yamauchi M, Morikawa T, Kuchiba A, Imamura Y, Qian ZR, Nishihara R, Liao X, Waldron L, Hoshida Y, Huttenhower C, et al. Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut. 2012;61:847–854. doi: 10.1136/gutjnl-2011-300865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wynter, CV, Walsh MD, Higuchi T, Leggett BA, Young J, Jass JR. Methylation patterns define two types of hyperplastic polyp associated with colorectal cancer. Gut. 2004;53:573–580. doi: 10.1136/gut.2003.030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gervaz P, Bucher P, Morel P. Two colons-two cancers: paradigm shift and clinical implications. J Surg Oncol. 2004;88:261–266. doi: 10.1002/jso.20156. [DOI] [PubMed] [Google Scholar]
- 32.Budinska E, Popovici V, Tejpar S, D’Ario G, Lapique N, Sikora KO, Di Narzo AF, Yan P, Hodgson JG, Weinrich S, et al. Gene expression patterns unveil a new level of molecular heterogeneity in colorectal cancer. J Pathol. 2013;231:63–76. doi: 10.1002/path.4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roepman P, Schlicker A, Tabernero J, Majewski I, Tian S, Moreno V, Snel MH, Chresta CM, Rosenberg R, Nitsche U, et al. Colorectal cancer intrinsic subtypes predict chemotherapy benefit, deficient mismatch repair and epithelial-to-mesenchymal transition. Int J Cancer. 2014;134:552–562. doi: 10.1002/ijc.28387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schlicker A, Beran G, Chresta CM, McWalter G, Pritchard A, Weston S, Runswick S, Davenport S, Heathcote K, Castro DA, et al. Subtypes of primary colorectal tumors correlate with response to targeted treatment in colorectal cell lines. BMC Med Genomics. 2012;5:66. doi: 10.1186/1755-8794-5-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sadanandam A, Lyssiotis CA, Homicsko K, Collisson EA, Gibb WJ, Wullschleger S, Ostos LCG, Lannon WA, Grotzinger C, Del Rio M, et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med. 2013;19:619–625. doi: 10.1038/nm.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Sousa EMF, Wang X, Jansen M, Fessler E, Trinh A, de Rooij LP, de Jong JH, de Boer OJ, van Leersum R, Bijlsma MF, et al. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat Med. 2013;19:614–618. doi: 10.1038/nm.3174. [DOI] [PubMed] [Google Scholar]
- 37.Marisa L, de Reyniès A, Duval A, Selves J, Gaub MP, Vescovo L, Etienne-Grimaldi M-C, Schiappa R, Guenot D, Ayadi M, et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med. 2013;10(5):e1001453. doi: 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, Marisa L, Roepman P, Nyamundanda G, Angelino P, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fessler E, Drost J, van Hooff SR, Linnekamp JF, Wang X, Jansen M, De Sousa EMF, Prasetyanti PR, I Jspeert JE, Franitza M, et al. TGFbeta signaling directs serrated adenomas to the mesenchymal colorectal cancer subtype. EMBO Mol Med. 2016;8:745–760. doi: 10.15252/emmm.201606184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang K, Willis JA, Reumers J, Taggart MW, San Lucas FA, Thirumurthi S, Kanth P, Delker DA, Hagedorn CH, Lynch PM, et al. Colorectal premalignancy is associated with consensus molecular subtypes 1 and 2. Ann Oncol. 2018;29:2061–2067. doi: 10.1093/annonc/mdy337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fessler E, Medema JP. Colorectal Cancer Subtypes: developmental Origin and Microenvironmental Regulation. Trends Cancer. 2016;2:505–518. doi: 10.1016/j.trecan.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 43.Lee MS, Menter DG, Kopetz S. Right versus left colon cancer biology: integrating the consensus molecular subtypes. J Natl Compr Canc Netw. 2017;15:411–419. doi: 10.6004/jnccn.2017.0038. [DOI] [PubMed] [Google Scholar]
- 44.Thanki K, Nicholls ME, Gajjar A, Senagore AJ, Qiu S, Szabo C, Hellmich MR, Chao C. Consensus molecular subtypes of colorectal cancer and their clinical implications. Int Biol Biomed J. 2017;3:105–111. [PMC free article] [PubMed] [Google Scholar]
- 45.Tsai JH, Liau JY, Lin YL, Lin LI, Cheng YC, Cheng ML, Jeng YM. Traditional serrated adenoma has two pathways of neoplastic progression that are distinct from the sessile serrated pathway of colorectal carcinogenesis. Mod Pathol. 2014;27:1375–1385. doi: 10.1038/modpathol.2014.35. [DOI] [PubMed] [Google Scholar]
- 46.Peters BA, Dominianni C, Shapiro JA, Church TR, Wu J, Miller G, Yuen E, Freiman H, Lustbader I, Salik J, et al. The gut microbiota in conventional and serrated precursors of colorectal cancer. Microbiome. 2016;4:69. doi: 10.1186/s40168-016-0218-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jakobsson HE, Rodriguez-Pineiro AM, Schutte A, Ermund A, Boysen P, Bemark M, Sommer F, Backhed F, Hansson GC, Johansson ME. The composition of the gut microbiota shapes the colon mucus barrier. EMBO Rep. 2015;16:164–177. doi: 10.15252/embr.201439263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rezasoltani S, Asadzadeh Aghdaei H, Dabiri H, Akhavan Sepahi A, Modarressi MH, Nazemalhosseini Mojarad E. The association between fecal microbiota and different types of colorectal polyp as precursors of colorectal cancer. Microb Pathog. 2018;124:244–249. doi: 10.1016/j.micpath.2018.08.035. [DOI] [PubMed] [Google Scholar]
- 49.Yoon H, Kim N, Park JH, Kim YS, Lee J, Kim HW, Choi YJ, Shin CM, Park YS, Lee DH, et al. Comparisons of gut microbiota among healthy control, patients with conventional adenoma, sessile serrated adenoma, and colorectal cancer. J Cancer Prev. 2017;22:108–114. doi: 10.15430/JCP.2017.22.2.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, Neuberg D, Huang K, Guevara F, Nelson T, et al. Analysis of fusobacterium persistence and antibiotic response in colorectal cancer. Science. 2017;358:1443–1448. doi: 10.1126/science.aal5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee DW, Han SW, Kang JK, Bae JM, Kim HP, Won JK, Jeong SY, Park KJ, Kang GH, Kim TY. Association between fusobacterium nucleatum, pathway mutation, and patient prognosis in colorectal cancer. Ann Surg Oncol. 2018;25:3389–3395. doi: 10.1245/s10434-018-6681-5. [DOI] [PubMed] [Google Scholar]
- 52.Yu J, Chen Y, Fu X, Zhou X, Peng Y, Shi L, Chen T, Wu Y. Invasive fusobacterium nucleatum may play a role in the carcinogenesis of proximal colon cancer through the serrated neoplasia pathway. Int J Cancer. 2016;139:1318–1326. doi: 10.1002/ijc.30168. [DOI] [PubMed] [Google Scholar]
- 53.Ito M, Kanno S, Nosho K, Sukawa Y, Mitsuhashi K, Kurihara H, Igarashi H, Takahashi T, Tachibana M, Takahashi H, et al. Association of fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. Int J Cancer. 2015;137:1258–1268. doi: 10.1002/ijc.29488. [DOI] [PubMed] [Google Scholar]
- 54.Park CH, Han DS, Oh YH, Lee AR, Lee YR, Eun CS. Role of Fusobacteria in the serrated pathway of colorectal carcinogenesis. Sci Rep. 2016;6:25271. doi: 10.1038/srep25271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Purcell RV, Visnovska M, Biggs PJ, Schmeier S, Frizelle FA. Distinct gut microbiome patterns associate with consensus molecular subtypes of colorectal cancer. Sci Rep. 2017;7:11590. doi: 10.1038/s41598-017-11237-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Omori S, Mabe K, Hatanaka K, Ono M, Matsumoto M, Takahashi M, Yoshida T, Ono S, Shimizu Y, Sugai N, et al. Human intestinal spirochetosis is significantly associated with sessile serrated adenomas/polyps. Pathol Res Pract. 2014;210:440–443. doi: 10.1016/j.prp.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 57.Carvalho AC, de Mattos Pereira L, Datorre JG, Dos Santos W, Berardinelli GN, Matsushita MM, Oliveira MA, Duraes RO, Guimaraes DP, Reis RM. Microbiota profile and impact of fusobacterium nucleatum in colorectal cancer patients of barretos cancer hospital. Front Oncol. 2019;9:813. doi: 10.3389/fonc.2019.00813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gao R, Kong C, Huang L, Li H, Qu X, Liu Z, Lan P, Wang J, Qin H. Mucosa-associated microbiota signature in colorectal cancer. Eur J Clin Microbiol Infect Dis. 2017;36:2073–2083. doi: 10.1007/s10096-017-3026-4. [DOI] [PubMed] [Google Scholar]
- 59.Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, Yang J, Dou R, Masugi Y, Song M, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65:1973–1980. doi: 10.1136/gutjnl-2015-310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mima K, Sukawa Y, Nishihara R, Qian ZR, Yamauchi M, Inamura K, Kim SA, Masuda A, Nowak JA, Nosho K, et al. Fusobacterium nucleatum and T cells in colorectal carcinoma. JAMA Oncology. 2015;1:653–661. doi: 10.1001/jamaoncol.2015.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nosho K, Sukawa Y, Adachi Y, Ito M, Mitsuhashi K, Kurihara H, Kanno S, Yamamoto I, Ishigami K, Igarashi H, et al. Association of fusobacterium nucleatum with immunity and molecular alterations in colorectal cancer. World J Gastroenterol. 2016;22:557–566. doi: 10.3748/wjg.v22.i2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park HE, Kim JH, Cho NY, Lee HS, Kang GH. Intratumoral Fusobacterium nucleatum abundance correlates with macrophage infiltration and CDKN2A methylation in microsatellite-unstable colorectal carcinoma. Virchows Arch. 2017;471:329–336. doi: 10.1007/s00428-017-2171-6. [DOI] [PubMed] [Google Scholar]
- 63.Tahara T, Yamamoto E, Suzuki H, Maruyama R, Chung W, Garriga J, Jelinek J, Yamano HO, Sugai T, An B, et al. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res. 2014;74:1311–1318. doi: 10.1158/0008-5472.can-13-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.He X, Wu K, Ogino S, Giovannucci EL, Chan AT, Song M. Relationship between fusobacterium nucleatum, inflammatory mediators and microRNAs in colorectal carcinogenesis. World J Gastroenterol. 2018;24:5351–5365. doi: 10.3748/wjg.v24.i47.5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamaoka Y, Suehiro Y, Hashimoto S, Hoshida T, Fujimoto M, Watanabe M, Imanaga D, Sakai K, Matsumoto T, Nishioka M, et al. Fusobacterium nucleatum as a prognostic marker of colorectal cancer in a Japanese population. J Gastroenterol. 2018;53:517–524. doi: 10.1007/s00535-017-1382-6. [DOI] [PubMed] [Google Scholar]
- 66.Wei Z, Cao S, Liu S, Yao Z, Sun T, Li Y, Li J, Zhang D, Zhou Y. Could gut microbiota serve as prognostic biomarker associated with colorectal cancer patients’ survival? A pilot study on relevant mechanism. Oncotarget. 2016;7:46158–46172. doi: 10.18632/oncotarget.10064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shen L, Toyota M, Kondo Y, Lin E, Zhang L, Guo Y, Hernandez NS, Chen X, Ahmed S, Konishi K, et al. Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer. Proc Natl Acad Sci U S A. 2007;104:18654–18659. doi: 10.1073/pnas.0704652104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tahara T, Yamamoto E, Madireddi P, Suzuki H, Maruyama R, Chung W, Garriga J, Jelinek J, Yamano HO, Sugai T, et al. Colorectal carcinomas with CpG island methylator phenotype 1 frequently contain mutations in chromatin regulators. Gastroenterology. 2014;146:530–538 e535. doi: 10.1053/j.gastro.2013.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hong BY, Ideta, T, Lemos BS, Igarashi Y, Tan Y, DiSiena M, Mo A, Birk JW, Forouhar F, Devers TJ, et al. Characterization of mucosal dysbiosis of early colonic neoplasia. NPJ Precis Oncol. 2019;3:29. doi: 10.1038/s41698-019-0101-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bongers G, Muniz LR, Pacer ME, Iuga AC, Thirunarayanan N, Slinger E, Smit MJ, Reddy EP, Mayer L, Furtado GC, et al. A role for the epidermal growth factor receptor signaling in development of intestinal serrated polyps in mice and humans. Gastroenterology. 2012;143:730–740. doi: 10.1053/j.gastro.2012.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bongers G, Maussang D, Muniz LR, Noriega VM, Fraile-Ramos A, Barker N, Marchesi F, Thirunarayanan N, Vischer HF, Qin L, et al. The cytomegalovirus-encoded chemokine receptor US28 promotes intestinal neoplasia in transgenic mice. J Clin Invest. 2010;120:3969–3978. doi: 10.1172/JCI42563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bongers G, Pacer ME, Geraldino TH, Chen L, He Z, Hashimoto D, Furtado GC, Ochando J, Kelley KA, Clemente JC, et al. Interplay of host microbiota, genetic perturbations, and inflammation promotes local development of intestinal neoplasms in mice. J Exp Med. 2014;211:457–472. doi: 10.1084/jem.20131587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.He Z, Chen L, Chen G, Smaldini P, Bongers G, Catalan-Dibene J, Furtado GC, Lira SA. Interleukin 1 beta and matrix metallopeptidase 3 contribute to development of epidermal growth factor receptor-dependent serrated polyps in mouse cecum. Gastroenterology. 2019;157:1572–1583 e1578. doi: 10.1053/j.gastro.2019.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, Enk J, Bar-On Y, Stanietsky-Kaynan N, Coppenhagen-Glazer S, et al. Binding of the Fap2 protein of fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42:344–355. doi: 10.1016/j.immuni.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang LS, Kuo CT, Huang YW, Stoner GD, Lechner JF. Gene-diet interactions on colorectal cancer risk. Curr Nutr Rep. 2012;1:132–141. doi: 10.1007/s13668-012-0023-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.OuYang LY, Wu XJ, Ye SB, Zhang RX, Li ZL, Liao W, Pan ZZ, Zheng LM, Zhang XS, Wang Z, et al. Tumor-induced myeloid-derived suppressor cells promote tumor progression through oxidative metabolism in human colorectal cancer. J Transl Med. 2015;13:47. doi: 10.1186/s12967-015-0410-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lim SO, Gu JM, Kim MS, Kim HS, Park YN, Park CK, Cho JW, Park YM, Jung G. Epigenetic changes induced by reactive oxygen species in hepatocellular carcinoma: methylation of the E-cadherin promoter. Gastroenterology. 2008;135:2128–2140, 2140 e2121-2128. doi: 10.1053/j.gastro.2008.07.027. [DOI] [PubMed] [Google Scholar]
- 80.Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, Jessup JM, Kolodner R. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57:808–811. [PubMed] [Google Scholar]
- 81.Hamada T, Zhang X, Mima K, Bullman S, Sukawa Y, Nowak JA, Kosumi K, Masugi Y, Twombly TS, Cao Y, et al. Fusobacterium nucleatum in colorectal cancer relates to immune response differentially by tumor microsatellite instability status. Cancer Immunol Res. 2018;6:1327–1336. doi: 10.1158/2326-6066.CIR-18-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hardy RG, Tselepis C, Hoyland J, Wallis Y, Pretlow TP, Talbot I, Sanders DS, Matthews G, Morton D, Jankowski JA. Aberrant P-cadherin expression is an early event in hyperplastic and dysplastic transformation in the colon. Gut. 2002;50:513–519. doi: 10.1136/gut.50.4.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Quah SY, Bergenholtz G, Tan KS. Fusobacterium nucleatum induces cytokine production through Toll-like-receptor-independent mechanism. Int Endod J. 2014;47:550–559. doi: 10.1111/iej.12185. [DOI] [PubMed] [Google Scholar]
- 84.Dharmani P, Strauss J, Ambrose C, Allen-Vercoe E, Chadee K. Fusobacterium nucleatum infection of colonic cells stimulates MUC2 mucin and tumor necrosis factor alpha. Infect Immun. 2011;79:2597–2607. doi: 10.1128/IAI.05118-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schetter AJ, Heegaard NH, Harris CC. Inflammation and cancer: interweaving microRNA, free radical, cytokine and p53 pathways. Carcinogenesis. 2010;31:37–49. doi: 10.1093/carcin/bgp272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rad R, Cadinanos J, Rad L, Varela I, Strong A, Kriegl L, Constantino-Casas F, Eser S, Hieber M, Seidler B, et al. A genetic progression model of Braf(V600E)-induced intestinal tumorigenesis reveals targets for therapeutic intervention. Cancer Cell. 2013;24:15–29. doi: 10.1016/j.ccr.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lannagan TRM, Lee YK, Wang T, Roper J, Bettington ML, Fennell L, Vrbanac L, Jonavicius L, Somashekar R, Gieniec K, et al. Genetic editing of colonic organoids provides a molecularly distinct and orthotopic preclinical model of serrated carcinogenesis. Gut. 2019;68:684–692. doi: 10.1136/gutjnl-2017-315920. [DOI] [PMC free article] [PubMed] [Google Scholar]


