ABSTRACT
Appropriate clearance of dead cells generated by apoptosis is critical to the development of multicellular organisms and tissue homeostasis. In mammals, the removal of apoptotic cell is mediated by polarized monocyte/macrophage populations of the innate immune system. The innate immune system is essential for anti-viral and anti-microbial defense. However, our current understanding of the relationship between apoptotic cell clearance and the innate immune response has remained rather limited. Here, we study how apoptotic cell clearance programs contribute to the innate immune response in C. elegans. We find apoptotic cell clearance mutant worms are more resistant to pathogenic bacteria of Pseudomonas aeruginosa PA14 and Salmonella typhimurium SL1344 due to significant upregulation of innate immune-dependent pathogen response genes. In addition, genetic epistasis analysis indicates that defects in apoptotic cell clearance can activate the innate immune response through PMK-1 p38 MAPK and MPK-1/ERK MAPK pathways in C. elegans. Taken together, our results provide evidence that insufficient clearance of apoptotic cell can protect Caenorhabditis elegans from bacterial infection through innate immune response activation.
KEYWORDS: C. elegans, apoptosis, apoptotic cell clearance, innate immune, lysosomes
Introduction
Apoptosis is the major process of programmed cell death and is evolutionarily conservative, which eliminates abnormal or damaged cells during development [1,2]. The appropriate clearance of apoptotic cell is indispensable for apoptosis and is crucial for the development of multicellular organisms and homeostatic [3]. If apoptotic cell cannot be removed in time and effectively, the harmful contents released by the apoptotic cell will lead to inflammation or autoimmune diseases, such as systemic lupus erythematosus and rheumatoid arthritis [4].
Caenorhabditis elegans has been used as an excellent model organism for studying the mechanism of apoptotic cell (corpse) clearance. C. elegans lacks any “professional” phagocytes, corpses are quickly engulfed and degraded by neighboring cells [5]. When cells undergo apoptosis, Caspase-mediated activation of CED-8 promotes phosphatidylserine (PS) externalization in apoptotic cell, which is an “eat-me” signal exposed on the surface of apoptotic cell and recognized by phagocytes [6]. The engulfment of apoptotic cell is mainly accomplished by two parallel redundant genetic pathways that are highly evolutionarily conserved [4,7]. In the ced-1/6/7 pathway, the phagocytic receptor CED-1 recognizes PS [8]. Then CED-6 interacts with CED-1 to transduce engulfment signals to downstream effectors. CED-7, an ABC transporter homolog, can expose PS to the surface of apoptotic cell [5,9]. In the ced-2/5/12 pathway, CED-2, an adaptor protein containing SH2 and SH3 domains, activates the CED-5/CED-12 complex [5,9]. The CED-5/CED-12 complex acts as a bipartite nucleotide exchange factor to activate Rac GTPase CED-10, which in turn causes rearrangement of the actin cytoskeleton for cell corpse engulfment [5,9]. The ced-1/6/7 and ced-2/5/12 pathways lead to the internalization of apoptotic cell and the formation of phagosomes [7]. Then, the phagosomes are fused with lysosomes to form phagolysosomes. In this process, VPS-18 is necessary for the endosome and lysosome biogenesis and fusion of phagosome and lysosome [10]. Finally, the corpses are digested by lysosomal acid hydrolases in the phagolysosome, such as NUC-1, a C. elegans DNase II homolog [11].
Besides apoptotic cell clearance, studies have demonstrated a certain evolutionary conservation between the mammalian innate immune and that of invertebrates [12,13]. The invertebrate C. elegans relies entirely on its innate immune to defend against pathogens, has been widely used to study the interaction between host and pathogen. For example, the human opportunistic pathogens can establish persistent infection in the intestine of C. elegans [14–16]. Researches on nematodes infected with various pathogens help people understand the mechanism of innate immunity [16]. The innate immunity in C. elegans is regulated by several major pathways to defense against pathogens including PMK-1 p38 MAPK pathway and MPK-1/ERK MAPK pathway [16,17]. In the PMK-1 pathway, NSY-1 phosphorylates SEK-1, SEK-1 phosphorylates PMK-1, and finally activates PMK-1. In the MPK-1 pathway, LIN-45 phosphorylates MEK-2, MEK-2 phosphorylates MPK-1, and finally activates MPK-1. Studies have shown that activation of the MPK-1/ERK MAPK pathway in C. elegans can facilitate the germ cells apoptosis, depending at least in part on the phagocytic mechanism of apoptotic cell [17]. However, little is known about how apoptotic cell clearance affects innate immunity in C. elegans.
In mammals, apoptotic cell clearance is mainly mediated by professional phagocytes of innate immune system [18]. It has been demonstrated that apoptotic cell can be engulfed by macrophages in DNase II-knockout mice, but the DNA of the dead cells is not properly degraded in the lysosomes, which activate the innate immune response leading to severe anemia and chronic arthritis [19]. Similarly, antibacterial peptide genes are expressed when fragmented DNA is not cleared from apoptotic germ cells by loss of DNase II in Drosophila and C. elegans [20,21]. However, evidence from other studies suggests that DNase II deficiency impairs innate immune function in Drosophila [22]. Studies have also shown that regulation of unfolded protein response by CED-1 is essential for innate immunity in C. elegans [23]. Thus, there are still a lot of work required to determine the relationship between apoptotic cell clearance and innate immunity.
In the present study, we systematically investigate the relationship between apoptotic cell clearance and innate immunity in C. elegans. We found that defective apoptotic cell clearance can trigger the innate immune response of C. elegans through PMK-1 p38 MAPK and MPK-1/ERK MAPK pathways, which protect worms against pathogenic bacteria infection. Our results provide an important basis for further elucidating the underlying mechanism of how clearance of apoptotic cell regulates innate immune responses.
Materials and methods
C. elegans and bacterial strains
C. elegans maintenance was performed using standard protocols [24]. The strains used in this study were mostly obtained from the Caenorhabditis Genetics Center, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440).
RNA interference (RNAi)
RNA interference was performed by feeding C. elegans with RNAi bacteria that express double-stranded RNA (dsRNA) targeting the gene of interest. 129.36 RNAi, ced-1 RNAi, ced-2 RNAi, nsy-1 RNAi, pmk-1 RNAi, mpk-1 RNAi were obtained from the RNAi library. The tir-1 RNAi and sek-1 RNAi were provided by our laboratory.
Lifespan assays
Lifespan assays by P. aeruginosa PA14 infection were conducted at 25°C [25]. Lifespan assays by S. typhimurium SL1344 infection were conducted at 20°C [15]. A total of 100 worms were quantified in each lifespan assay, and all experiments were repeated at least three times. Detailed procedures are provided in Supplementary Materials and methods.
Real-time quantitative RT-PCR (qRT-PCR) assays
Total RNA was extracted using the Trizol method. cDNA was synthesized using Transcriptor First Strand cDNA Synthesis Kit (Roche). Quantitative real-time PCR was performed using a StepOne Plus Real-Time PCR system and SYBR qPCR Master Mix (Vazyme). RNA fold change was calculated by comparing mRNA levels of the gene of interest with mRNA levels of the reference gene tbg-1. The primers used in this study are listed in Supplementary Table S6.
Fluorescence microscopy
For imaging fluorescence, worms were mounted onto 2% agar pads, paralyzed with levamisole. The slides were viewed using fluorescence microscope (Zeiss M2) and processed with ImageJ (http://rsb.info.nih.gov/ij/).
Immunoblot analyses
Protein quality was determined by Immunoblot analysis. Detailed procedures are provided in Supplementary Materials and methods.
Statistical analysis
Statistical analysis was performed with the two-tailed student’s t-test with GraphPad Prism 8 software. Values of P < 0.05 were considered statistically significant.
Results
Defective clearance of apoptotic cell in C. elegans enhanced resistance to infection by P. aeruginosa PA14 and S. typhimurium SL1344
To examine the interaction between apoptotic cell clearance and innate immune response in C. elegans, we used apoptotic cell clearance defective mutant nematodes and analyzed their resistance to pathogenic bacteria. As expected, when infected by P. aeruginosa PA14 and S. typhimurium SL1344, the lifespans of pmk-1(km25) mutants were significantly reduced compared to the wild-type N2 (Figure 1(a) and S1A; Table S1 and S2) [26,27]. Next, we conducted lifespan analysis on strong loss-of-function mutants of ced-1/6/7 and ced-2/5/12 pathway components and found that lifespans of these engulfment mutants were significantly extended than N2 after P. aeruginosa PA14 and S. typhimurium SL1344 infection (Figure 1(b and c); Figure S1B and S1C; Table S1 and S2). To test whether resistance to pathogenic bacteria infections was limited to engulfment mutants, we performed lifespan analysis on ced-8(n1891), vps-18(tm1125), and nuc-1(e1994), and observed that these mutants lifespans were also significantly extended (Figure 1(d) and S1D; Table S1 and S2). Moreover, the lifespan of the ced-1(e1735); ced-2(n1994) double mutant feeding on SL1344 but not on PA14 was significantly extended when compared to the single mutant (Figure S2B). Furthermore, using a GFP expressing PA14(PA14-GFP), we examined bacterial accumulation in the intestine of worms, and found no significant difference between ced-1(e1735), ced-2(n1994), ced-8(n1891), and wild-type N2 (Figure S3A-C). To corroborated the fluorescence data, we performed colony-forming units (CFUs) experiment and found there was more bacterial accumulation in the intestine of ced-2(n1994) and ced-8(n1891) but not in ced-1(e1735), which indicate apoptotic cell clearance does not block the accumulation of P. aeruginosa PA14 in the intestine. These results demonstrate that apoptotic cell clearance defective mutants are resistant to infection by pathogenic bacteria.
Figure 1.
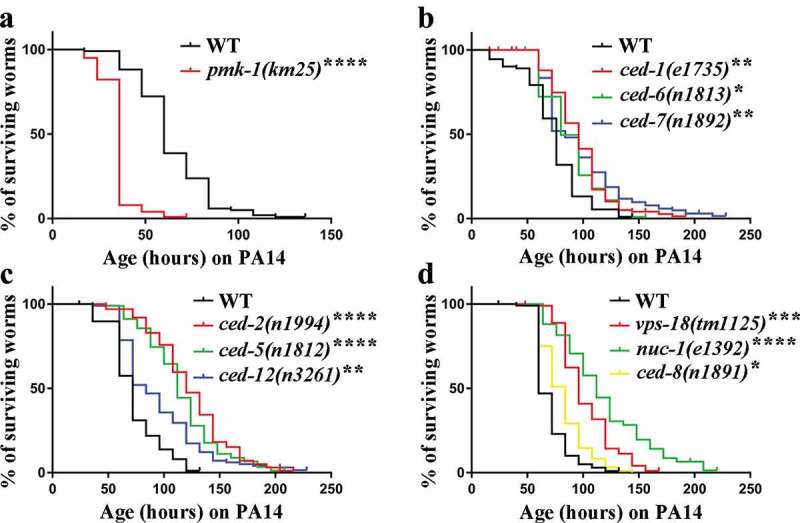
Defective clearance of apoptotic cell extends C. elegans lifespan on P. aeruginosa PA14. (a-d) Lifespan analyses in the indicated strains. Two-tailed student’s t-test method was performed to compare all the other datasets with wild type (WT). See Supplementary Table S1 for detailed statistical analysis of lifespan data. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Error bars represent SEM
Defects in apoptotic cell clearance upregulate defense gene expression in C. elegans
C. elegans does have an innate immune system and responds to pathogenic bacterial by expression of defense genes [28]. To test whether defects in apoptotic cell clearance activate the innate immune response in C. elegans, we analyzed the mRNA level of pathogen response genes by Real-time quantitative RT-PCR (qRT-PCR). In C. elegans, clec-7, clec-60, and clec-82 encode C-type lectin proteins, lys-5 encodes lysozyme, nlp-29 and F53A9.8 encode antimicrobial peptides [29,30]. Intriguingly, we observed that not all but most of the pathogen response genes listed above were upregulated in apoptotic cell clearance mutants without pathogenic bacterial infections (Figure 2). The expression of NLP-29 is regulated by a conserved innate immune signaling cascade specifically in the epidermis during infection [30]. To confirm the above findings, we knocked down ced-1 or ced-2 by RNAi in transgenic worms carrying a pnlp-29::gfp and a pcol-12::dsRed transcriptional reporters constructs, and investigated the expression level of nlp-29. Transgenic worms appear predominantly red in the absence of infection due to the constitutively expressed Pcol-12::dsRED under the control of hypodermis-specific promoter [31]. We observed the reporter green fluorescent protein (GFP) fluorescence in transgenic worms was notably increased by RNAi treatment of ced-1 or ced-2 without bacterial pathogens infections, which colocalized with Pcol-12::dsRED (Figure S4 A and B). The increased level of Pnlp-29::GFP in transgenic worms was confirmed by Western blotting (Figure S4 C and D). Taken together, these results show that the defective clearance of apoptotic cell results in upregulation of pathogen defense genes in the absence of pathogenic bacterial infections in C. elegans.
Figure 2.
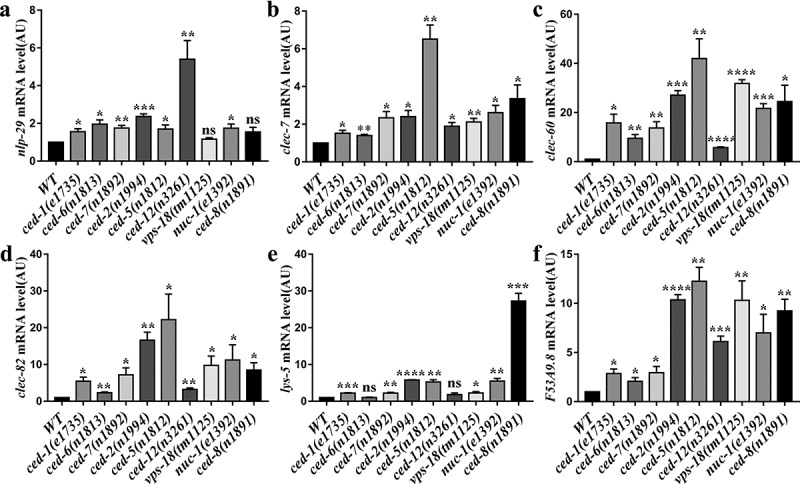
Increased expression of pathogen response genes in apoptotic cell clearance mutants without bacteria pathogens infection. (a-f) Bars represent mRNA levels for nlp-29 (a), clec-7 (b), clec-60 (c), clec-82 (d), lys-5 (e) and F53A9.8 (f) pathogen response genes determined by qRT-PCR in WT and apoptotic cell clearance defective mutant cultured on live OP50. Values in arbitrary units (AU) are the average of at least 3 biological replicates. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; not significant (ns), P > 0.05. Two-tailed Student’s t-test. Error bars represent SEM
Apoptotic cell clearance deficiency activates PMK-1 p38 MAPK and MPK-1/ERK MAPK pathways in C. elegans
Our results in line with previous studies that loss- and reduction-of-function mutations of p38 MAPK PMK-1 pathway components lead to a reduction in the levels of activated PMK-1 protein (phosphorylated PMK-1) [26,32] (Figure 3). To explore whether apoptotic cell clearance participates in the innate immune response through the PMK-1 p38 MAPK and MPK-1/ERK MAPK pathways, we measured the expression level of PMK-1 and MPK-1 in apoptotic cell clearance mutant worms. We found there is no significant difference between PMK-1 and MPK-1 in these mutants by using actin as loading control (Figure S5). We then measured activated PMK-1 and MPK-1 levels by immunoblotting in C. elegans. Our results showed that both activated PMK-1 and activated MPK-1 in apoptotic cell clearance defective mutants were significantly increased compared to wild-type controls (Figure 3 and Figure S6). To further confirm PMK-1 and MPK-1 pathways were activated in apoptotic cell clearance mutants, we detected the expression level of irg-5 and sysm-1, which are used as canonical PMK-1 effector reporters [33,34], and detected the expression level of mpk-1 dependent genes lin-39 and egl-5 [35,36]. We found irg-5, sysm-1, lin-39, and egl-5 were upregulated in most of apoptotic cell clearance mutant worms (Figure S7). To test whether generic stress is involved in apoptotic cell clearance mutants against pathogens, we detected the expression levels of prdx-3 and lgg-1, the endogenous targets for DAF-16 in ced-1 and ced-2 mutants [37], and found no significant difference when compared to wild type (Figure S8). These data prove that defects in apoptotic cell clearance can activate PMK-1 p38 MAPK and MPK-1/ERK MAPK pathways in C. elegans, suggesting that defective apoptotic cell clearance may be involved in the regulation of innate immune response through the PMK-1 p38 MAPK and MPK-1/ERK MAPK pathways.
Figure 3.
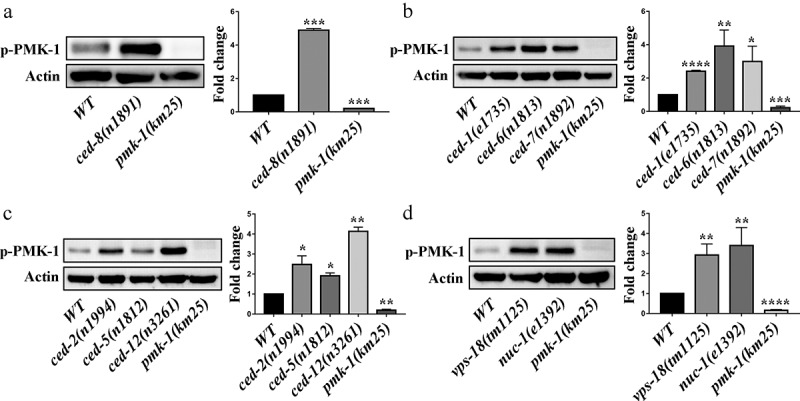
Defective clearance of apoptotic cell increases the level of activated PMK-1 in C. elegans. (a-e) Immunoblot analysis of lysates from worms using antibodies that recognize PMK-1 (p-PMK-1) and Actin (loading control). Worms at the L4 stage were cultured on plates containing live OP50 for approximately 48 hours and lysates were prepared. The blot is typical of three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Two-tailed Student’s t-test. Error bars represent SEM
PMK-1 p38 MAPK and MPK-1/ERK MAPK pathways are required for the resistance of apoptotic cell clearance mutants against pathogens
To better define how apoptotic cell clearance participates in the innate immune response through the MPK-1/ERK MAPK pathway, we measured the lifespan of ced-1(e1735) and ced-2(n1994) mutants infected by P. aeruginosa PA14. The ced-1 (e1735) and ced-2(n1994) mutants showed the significantly reduced lifespan by pmk-1 RNAi and mpk-1 RNAi treatment, respectively (Figure 4; Table S3 and Table S4). These imply that pmk-1 and mpk-1 are required for ced-1 (e1735) and ced-2(n1994) mutants lifespan extension after P. aeruginosa PA14 infection. Moreover, we found pmk-1 RNAi or mpk-1 RNAi treatment partially suppressed the extended lifespan phenotype of ced-1(e1735) mutants as well as ced-2(n1994) mutants when compared to wild-type control (Figure 4; Table S3 and Table S4), which indicate there is functional redundancy among p38 MAPK and MPK-1 MAPK for apoptotic cell clearance mutants against pathogens. To test whether there is functional redundancy among p38 MAPK and MPK-1 MAPK for apoptotic cell clearance mutants against pathogens, we performed the mpk-1(RNAi);pmk-1(RNAi) double knockdown in ced-1(e1735) and ced-2(n1994) backgrounds. We found mpk-1;pmk-1 double RNAi treatment still partially suppressed the extended lifespan phenotype of ced-1(e1735) mutants as well as ced-2(n1994) mutants when compared to wild-type control (Figure 4; Table S5), which indicates other innate immune pathways are involved in protecting apoptotic cells clearance mutants against pathogen. However, we can’t exclude that the redundancy between these two MAPK pathways, as double feeding is less reliable than single feeding. In some cases, only one gene may be significantly inhibited, or both genes may be only slightly knocked down. While our genetic epistasis analysis suggested that ced-1 and ced-8 act upstream or parallel to nsy-1, while ced-2 acts upstream or parallel to pmk-1 to regulate the PMK-1 p38 MAPK pathway (Figure S9). Therefore, we propose that apoptotic cell clearance defects actively regulate the innate immune response through the PMK-1 p38 MAPK and MPK-1/ERK MAPK pathways in C. elegans.
Figure 4.
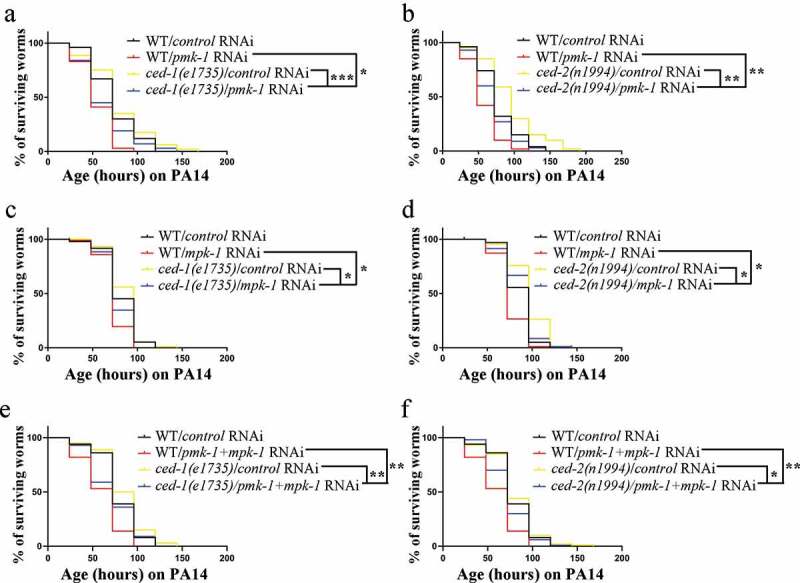
Pmk-1 and mpk-1 are required for lifespan extension of ced-1(e1735) and ced-2(n1994) mutants after exposure to P.aeruginosa PA14. (a-f) Lifespan analyses in the indicated strains. ced-1(e1735) (a), (c), (e)and ced-2(n1994) (b), (d), (f) were fed the indicated RNAi bacteria and transferred at the L4 stage to plates containing P. aeruginosa PA14 bacteria. Two-tailed student’s t-test method was performed to compare all the other datasets with wild type N2 (WT). All lifespan assays were carried out at 25°C and were repeated at least three times. *, P < 0.05; **, P < 0.01; ns, P > 0.05.
Discussion
Apoptotic cell clearance in C. elegans can be divided into three main steps: recognition of corpse, corpse internalization/engulfment, and corpse degradation [5]. Here, we show that defects in these three key steps of apoptotic cell clearance can protect worms against infection by pathogenic bacterial P. aeruginosa PA14 and S. typhimurium SL1344. We observed an accumulation of GFP-expressing PA14 in the intestine of ced-1(e1735) and ced-2(n1994) mutants. These suggest the protective effect of engulfment genes loss of function is not caused by inadequate phagocytosis of bacterial pathogens. In addition, we identify that defective clearance of apoptotic cell activates the innate immune response through conserved PMK-1 p38 MAPK and MPK-1/ERK MAPK pathways in C. elegans.
We found defects in apoptotic cell clearance upregulated innate immune response gene expression in the absence of bacterial pathogens in C. elegans. Despite lacking both Toll and Imd pathways as well as the immunological memory of vertebrate adaptive immunity, C. elegans is considered to have an innate immune defense system and respond to pathogen infection by expression of antimicrobial peptides [31]. In mammals, the swift and efficient clearance of apoptotic cell by professional phagocytes is crucial to avoid the loss of plasma membrane integrity and release of cellular contents to prevent unwanted immune responses to self-antigens that are derived from these dying cells [18]. On the other hand, macrophages that ingested apoptotic cell secrete anti-inflammatory cytokines and actively suppress the secretion of pro-inflammatory cytokines [38]. Apoptotic cell digestion in lysosomes has long been the least considered stage, possibly because it was believed to function as garbage disposal. However, recent studies have shown that macrophages lacking lysosome DNase II activated innate immunity via a STING-dependent pathway leading to severe anemia and chronic polyarthritis [39]. Innate immunity activation has also been detected in DNase II-deficient flies and worms which constitutively expressed antibacterial peptides genes [20,21]. Our results are consistent with the above-described finding that pathogen response genes were upregulated in nuc-1(n1994) mutants without pathogenic bacterial infections. NUC-1 is implicated in removing apoptotic DNA and in digesting ingested bacterial DNA in the intestine in C. elegans [11]. Upregulation of pathogen response in nuc-1(n1994) mutants was likely not due to failure to digest ingested bacterial DNA, First, similar results have been found in engulfment mutant, apoptotic cell PS externalization mutant, and apoptotic cell degradation mutant, indicating the general effects in upregulating innate immune response gene expression by defects in apoptotic cell clearance in C. elegans. Second, study has shown that loss of DNase II function in other tissues such as the gonad is associated with increased expression of antimicrobial genes [20]. In C. elegans, cell corpses are rapidly removed by neighboring cells such as hypodermal cells, muscle cells, intestinal cells, and sheath cells [7]. It will be necessary to study when and which cells express the antibacterial peptide genes in apoptotic cell clearance-deficient worms.
We found defective clearance of apoptotic cell activated PMK-1 p38 MAPK and MPK-1/ERK MAPK pathways but not the DAF-16 pathway in C. elegans. Furthermore, we demonstrated that PMK-1 p38 MAPK and MPK-1/ERK MAPK pathways were required for apoptotic cell clearance mutants against pathogens. In C. elegans, the expression level of nlp-29 is regulated in the epidermis by the innate immune signaling cascade, consisting of TIR-1/SARM, NSY-1/MAP3K, SEK-1/MAP2K, and PMK-1/p38 MAPK [31]. We observed the upregulation of nlp-29 expression in apoptotic cell clearance mutant worms. In line with these results, our immunoblotting experiments showed that activated PMK-1 and MPK-1 in apoptotic cell clearance-deficient nematodes were significantly increased compared to wild-type controls. Meanwhile, our lifespan analysis showed that ced-1(e1735) and ced-2(n1994) mutants exhibited enhanced susceptibility to P. aeruginosa PA14 infection after knockdown of pmk-1 or mpk-1 when compared to control RNAi. These indicate pmk-1 and mpk-1 are essential for apoptotic cell clearance mutants against pathogens. Further studies will be needed to clarify whether other innate immune pathways are involved in protecting apoptotic cell clearance mutants against pathogens. Moreover, by genetic epistasis analysis, we found that ced-1 and ced-8 act upstream or parallel to nsy-1, while ced-2 acts upstream or parallel to pmk-1 to regulate the p38 MAPK pathway. As the ced-1(e1735);ced-2(n1994) double mutants were more resistant to bacterial pathogens SL1344 but not PA14 compared to the single mutants. We proposed that ced-1 and ced-2 may function redundantly in the regulation of p38 MAPK pathway against SL1344. Further research should be undertaken to investigate how apoptotic cell clearance regulates innate immune responses in C. elegans. However, the findings of the current study are different from the previous research that unfolded protein response genes regulated by CED-1 and loss of function of ced-1 lead to compromised innate immune response in C. elegans and are rapidly killed by live bacteria [23,40]. There are possible explanations for this might be that they performed C. elegans killing assay by S. typhimurium at 25°C [23] instead of at 20°C and use different S. typhimurium strain [40]. At a higher temperature, S. typhimurium may grow faster to colonize and proliferate in the worm intestine, which likely masks the protective effect of ced-1 loss of function. Another possible explanation for this is that high temperatures are known to induce unfolded protein response. Unfolded protein response may become more important for innate immune response in C. elegans at a higher temperature. During apoptotic cell clearance, CED-1 is internalized from and recycled back to the cell membrane. Previous study demonstrated that loss of retromer function results in lysosomal accumulation of CED-1 [41]. It is also interesting to test whether retromer would affect innate immunity in C. elegans. Future studies on determining the mechanisms of current differences are therefore recommended.
In summary, our results provide evidence that defective apoptotic cell clearance can actively regulate the innate immune response through PMK-1 p38 MAP\K and MPK-1/ERK MAPK pathways in C. elegans, establish the link between the apoptotic cell clearance and the innate immune response. Inefficient clearance of dead cells activates the innate immune system that has been documented in mammalian [19]. Thus, our research provides important clues for further dissecting the underlying mechanism of how apoptotic cell clearance regulates innate immune responses.
Supplementary Material
Acknowledgments
We thank Drs. Chonglin Yang, Huimin Zhang as well as the C. elegans Genetic Center for C. elegans strains and Dr. Cheng-Gang Zou for P. aeruginosa PA14 and S. typhimurium SL1344 strains.
Funding Statement
This work was partially supported by the National Natural Science Foundation of China (Grant No.31671439 to H. Xiao), Natural Science Foundation of Shaanxi Province, China (Grant No. 2020JM-271 to H. Xiao), the program of Innovative Research Team for the Central Universities (Grant No. GK202001004 to H. Xiao), the Fundamental Research Key Project Funds for the Central Universities (Grant No. GK202007009 to H. Xiao), the Innovation Fund for undergraduate students (Grant No. 2019CBLZ004 to L.Yuan).
Author contributions
H. Xiao and Q. Zheng conceived the study. J. Wan and L. Yuan did most of the experiments. H. Jing contributed to materials. H. Xiao and J. Wan wrote the manuscript with feedback from all authors.
Disclosure statement
The authors declare no competing financial interests.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Jacobson MD, Weil M, Raff MC.. Programmed cell death in animal development. Cell. 1997;88:347–354. [DOI] [PubMed] [Google Scholar]
- [2].Vaux DL, Korsmeyer SJ. Cell death in development. Cell. 1999;96:245–254. [DOI] [PubMed] [Google Scholar]
- [3].Fadeel B, Xue D, Kagan V. Programmed cell clearance: molecular regulation of the elimination of apoptotic cell corpses and its role in the resolution of inflammation. Biochem Biophys Res Commun. 2010;396:7–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Nagata S, Hanayama R, Kawane K. Autoimmunity and the clearance of dead cells. Cell. 2010;140:619–630. [DOI] [PubMed] [Google Scholar]
- [5].Wang X, Yang C. Programmed cell death and clearance of cell corpses in Caenorhabditis elegans. Cell Mol Life Sci. 2016;73:2221–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Suzuki J, Denning DP, Imanishi E, et al. Xk-related protein 8 and CED-8 promote phosphatidylserine exposure in apoptotic cell. Science. 2013;341:403–406. [DOI] [PubMed] [Google Scholar]
- [7].Reddien PW, Horvitz HR. The engulfment process of programmed cell death in caenorhabditis elegans. Annu Rev Cell Dev Biol. 2004;20:193–221. [DOI] [PubMed] [Google Scholar]
- [8].Zhou Z, Hartwieg E, Horvitz HR. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell. 2001;104:43–56. [DOI] [PubMed] [Google Scholar]
- [9].Reddien PW, Horvitz HR. CED-2/CrkII and CED-10/Rac control phagocytosis and cell migration in Caenorhabditis elegans. Nat Cell Biol. 2000;2:131–136. [DOI] [PubMed] [Google Scholar]
- [10].Xiao H, Chen D, Fang Z, et al. Lysosome biogenesis mediated by vps-18 affects apoptotic cell degradation in Caenorhabditis elegans. Mol Biol Cell. 2009;20:21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wu YC, Stanfield GM, Horvitz HR. NUC-1, a caenorhabditis elegans DNase II homolog, functions in an intermediate step of DNA degradation during apoptosis. Genes Dev. 2000;14:536–548. [PMC free article] [PubMed] [Google Scholar]
- [12].Kim DH, Ausubel FM. Evolutionary perspectives on innate immunity from the study of Caenorhabditis elegans. Curr Opin Immunol. 2005;17:4–10. [DOI] [PubMed] [Google Scholar]
- [13].Hoffmann JA, Kafatos FC, Janeway CA, et al. Phylogenetic perspectives in innate immunity. Science. 1999;284:1313–1318. [DOI] [PubMed] [Google Scholar]
- [14].Mahajan-Miklos S, Tan MW, Rahme LG, et al. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell. 1999;96:47–56. [DOI] [PubMed] [Google Scholar]
- [15].Aballay A, Yorgey P, Ausubel FM. Salmonella typhimurium proliferates and establishes a persistent infection in the intestine of Caenorhabditis elegans. Curr Biol. 2000;10:1539–1542. [DOI] [PubMed] [Google Scholar]
- [16].Ermolaeva MA, Schumacher B. Insights from the worm: the C. elegans model for innate immunity. Semin Immunol. 2014;26:303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Doll MA, Soltanmohammadi N, Schumacher B. ALG-2/AGO-dependent mir-35 family regulates DNA damage-induced apoptosis through MPK-1/ERK MAPK signaling downstream of the core apoptotic machinery in Caenorhabditis elegans. Genetics. 2019;213:173–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ravichandran KS, Lorenz U. Engulfment of apoptotic cell: signals for a good meal. Nat Rev Immunol. 2007;7:964–974. [DOI] [PubMed] [Google Scholar]
- [19].Kawane K, Ohtani M, Miwa K, et al. Chronic polyarthritis caused by mammalian DNA that escapes from degradation in macrophages. Nature. 2006;443:998–1002. [DOI] [PubMed] [Google Scholar]
- [20].Yu H, Lai HJ, Lin TW, et al. Loss of DNase II function in the gonad is associated with a higher expression of antimicrobial genes in Caenorhabditis elegans. Biochem J. 2015;470:145–154. [DOI] [PubMed] [Google Scholar]
- [21].Mukae N, Yokoyama H, Yokokura T, et al. Activation of the innate immunity in Drosophila by endogenous chromosomal DNA that escaped apoptotic degradation. Genes Dev. 2002;16:2662–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Seong CS, Varela-Ramirez A, Aguilera RJ. DNase II deficiency impairs innate immune function in Drosophila. Cell Immunol. 2006;240:5–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Haskins KA, Russell JF, Gaddis N, et al. Unfolded protein response genes regulated by CED-1 are required for Caenorhabditis elegans innate immunity. Dev Cell. 2008;15:87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Tan MW, Mahajan-Miklos S, Ausubel FM. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc Natl Acad Sci U S A. 1999;96:715–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Cheesman HK, Feinbaum RL, Thekkiniath J, et al. Aberrant activation of p38 MAP kinase-dependent innate immune responses is toxic to Caenorhabditis elegans. G3 (Bethesda). 2016;6:541–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Troemel ER, Chu SW, Reinke V, et al. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2006;2:e183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Irazoqui JE, Troemel ER, Feinbaum RL, et al. Distinct pathogenesis and host responses during infection of C. elegans by P. aeruginosa and S. aureus. PLoS Pathog. 2010;6:e1000982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kumar S, Egan BM, Kocsisova Z, et al. Lifespan extension in C. elegans caused by bacterial colonization of the intestine and subsequent activation of an innate immune response. Dev Cell. 2019;49:100–117 e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Couillault C, Pujol N, Reboul J, et al. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat Immunol. 2004;5:488–494. [DOI] [PubMed] [Google Scholar]
- [31].Ewbank, J. J. Signaling in the immune response, WormBook, ed. C . elegans. Research Community, Worm Book, January 23, 2006. doi: 10.1895/wormbook.1.83.1. [DOI] [Google Scholar]
- [32].Kim DH, Feinbaum R, Alloing G, et al. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science. 2002;297:623–626. [DOI] [PubMed] [Google Scholar]
- [33].McEwan DL, Kirienko NV, Ausubel FM. Host translational inhibition by Pseudomonas aeruginosa exotoxin a triggers an immune response in Caenorhabditis elegans. Cell Host Microbe. 2012;11:364–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Shapira M, Hamlin BJ, Rong J, et al. A conserved role for a GATA transcription factor in regulating epithelial innate immune responses. Proc Natl Acad Sci U S A. 2006;103:14086–14091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lackner MR, Kornfeld K, Miller LM, et al. A MAP kinase homolog, mpk-1, is involved in ras-mediated induction of vulval cell fates in Caenorhabditis elegans. Genes Dev. 1994;8:160–173. [DOI] [PubMed] [Google Scholar]
- [36].Nicholas HR, Hodgkin J. The C. elegans Hox gene egl-5 is required for correct development of the hermaphrodite hindgut and for the response to rectal infection by Microbacterium nematophilum. Dev Biol. 2009;329:16–24. [DOI] [PubMed] [Google Scholar]
- [37].Sun X, Chen WD, Wang YD. DAF-16/FOXO transcription factor in aging and longevity. Front Pharmacol. 2017;8:548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kinchen JM, Ravichandran KS. Phagosome maturation: going through the acid test. Nat Rev Mol Cell Biol. 2008;9:781–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Yoshida H, Okabe Y, Kawane K, et al. Lethal anemia caused by interferon-beta produced in mouse embryos carrying undigested DNA. Nat Immunol. 2005;6:49–56. [DOI] [PubMed] [Google Scholar]
- [40].Sahu SN, Anriany Y, Grim CJ, et al. Identification of virulence properties in Salmonella Typhimurium DT104 using Caenorhabditis elegans. PLoS One. 2013;8:e76673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Chen D, Xiao H, Zhang K, et al. Retromer is required for apoptotic cell clearance by phagocytic receptor recycling. Science. 2010;327:1261–1264. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


