ABSTRACT
Introduction
Antimicrobial drugs are known to have effects on the human gut microbiota. We studied the long-term temporal relationship between several antimicrobial drug groups and the composition of the human gut microbiota determined in feces samples.
Methods
Feces samples were obtained from a community-dwelling cohort of middle-aged and elderly individuals (Rotterdam Study). Bacterial DNA was isolated and sequenced using V3/V4 16 S ribosomal RNA sequencing (Illumina MiSeq). The time between the last prescription of several antimicrobial drug groups and the day of sampling was categorized into 0–12, 12–24, 24–48 and >48 months. The effects of the antimicrobial drug groups on the Shannon alpha-diversity (diversity), the Bray–Curtis beta-diversity (community structure), the Firmicutes/Bacteroidetes (F/B) ratio and individual genera were determined.
Results
We studied the gut microbiota of 1413 individuals (57.5% female, median age 62.6 years). The alpha-diversity was significantly lower up to 4 years after prescriptions of macrolides and lincosamides. It was also lower in the first year after the use of beta-lactams. The community structure (beta-diversity) of the microbiota was significantly different up to 4 years for macrolides and lincosamides, the first year for beta-lactams and at least the first year for quinolones. For the F/B ratio, drugs with a high anaerobic activity shifted the ratio toward Firmicutes in the first year whereas other antimicrobial drugs shifted the ratio toward Bacteroidetes.
Conclusion
Use of antimicrobial drugs is associated with a shift in the composition of the gut microbiota.These effects differ in strength and duration, depending on the antimicrobial drug group used. These findings should be considered when prescribing antimicrobial drugs.
KEYWORDS: Gut microbiota, antimicrobial use, macrolides and lincosamides, quinolones, beta-lactams, sulfonamides and trimethoprim, nitrofuran derivatives, tetracyclines
Introduction
The gut microbiota plays a role in a variety of processes, such as protection against overgrowth of pathogenic micro-organisms, in the development of the host immune response, in neurologic signaling and in the synthesis and metabolism of several compounds, such as short-chain fatty acids (SCFAs).1,2 In particular, the SCFA butyrate is said to have an important function in the maintenance of a healthy colonic epithelium.3
The composition of the gut microbiota may differ with age,4 gender and BMI5 and can change under the influence of diet,6,7 physical activity,8 diabetes9 and use of drugs, such as proton pump inhibitors,10 corticosteroids11 and statins.12 Furthermore, it is known that it can be influenced by the use of antimicrobial drugs (post-antibiotic dysbiosis). The use of antimicrobial drugs has been reported to increase the vulnerability to overgrowth of potentially pathogenic bacteria, such as Clostridium difficile, with the risk of pseudomembranous colitis. Moreover, it has been described to cause a loss of diversity of the gut microbiota, cause a decrease of important taxa, alter gene expression, select for intrinsically resistant bacteria, and select for new mutations.13–15 Additionally, dysbiosis has also been designated as a factor that promotes horizontal gene transfer, thereby increasing the probability of spreading antibiotic resistance genes.16
In adults, some information is available about the effects of specific antimicrobial drug groups on the composition of the gut microbiota. Short-term exposure to clindamycin was shown to cause a shift of the gut microbiota, for example a decline in the diversity of Bacteroidetes.17 Furthermore, Lachnospiraceae abundancy in the gut was decreased up to 6 months after the use of amoxicillin or azithromycin.18 Also, one study showed the effects of using beta-lactam antibiotics in the 12 months before sampling in a population-based cohort.19 However, most studies have investigated these effects in small populations, studying rather short-term effects and using a variety of methods to investigate the microbiota. Therefore, the objective of this study was to describe and compare the effects and the duration of the effects of different antimicrobial drug groups on the composition of the human microbiota in feces samples from a large population of community-dwelling middle-aged and elderly individuals using different outcomes that characterize the microbiota.
Results
We obtained a dataset with microbiota data from 1427 participants, of whom 14 (1.0%) were excluded, because no pharmacy data were available. From the remaining 1413 participants, 812 (57.5%) were female and 601 (42.5%) were male with a median age of 62.6 years (IQR 58.6–66.1), a median BMI of 26.8 (IQR 24.5–29.7) and the feces sample had been in the mail for a median time of 1 day (IQR 1–2 days). Furthermore, 323 individuals used proton pump inhibitors and 252 used a statin. There was no use of tacrolimus and the use of antineoplastic agents (3 participants, 0.2%) was very low; therefore, these drugs were not included in the models. A total of 1281 (90.6%) participants had received at least 1 prescription of an antimicrobial drug during follow-up (at least 17 years). Most participants (73.7%) had 1 or more prescriptions of beta-lactam antibiotics, other frequently used antibacterial drugs were tetracyclines (57.7%) and macrolides and lincosamides (44.0%). The number of prescriptions for amphenicols (J01B), other beta-lactam antibacterials (J01D), aminoglycoside antibacterials (J01G), glycopeptide antibacterials (J01XA), polymyxins (J01XB), steroid antibacterials (J01XC), imidazole derivatives (J01XD) and other antibacterials (J01XX) was very low and these groups could not be analyzed separately (Table 1). The correlations between the different antimicrobial drug groups were low (Table S1).
Table 1.
Use of antimicrobial drugs in the study population.
| Antimicrobial drug group | 0–12 months* | 12–24 months | 24–48 months | >48 months | None |
|---|---|---|---|---|---|
| Antibacterial for systemic use (J01) | 355 | 217 | 236 | 473 | 132 |
| Tetracyclines (J01A) | 100 | 103 | 152 | 461 | 597 |
| Amphenicols (J01B) | 0 | 0 | 0 | 0 | 1413 |
| Beta-lactam antibacterials (J01C) | 179 | 131 | 163 | 569 | 371 |
| Other beta-lactam antibacterials (J01D) | 1 | 2 | 1 | 11 | 1398 |
| Sulfonamides and trimethoprim (J01E) | 20 | 23 | 55 | 267 | 1048 |
| Macrolides, lincosamides and streptogramins (J01F) | 62 | 49 | 106 | 405 | 791 |
| Aminoglycoside antibacterials (J01 G) | 1 | 0 | 0 | 1 | 1411 |
| Quinolone antibacterials (J01 M) | 35 | 38 | 48 | 154 | 1138 |
| Glycopeptide antibacterials (J01XA) | 0 | 0 | 0 | 1 | 1412 |
| Polymyxins (J01XB) | 0 | 0 | 0 | 0 | 1413 |
| Steroid antibacterials (J01XC) | 0 | 0 | 0 | 0 | 1413 |
| Imidazole derivatives (J01XD) | 0 | 0 | 0 | 2 | 1411 |
| Nitrofuran derivatives (J01XE) | 76 | 38 | 67 | 121 | 1111 |
| Other antibacterials (J01XX) | 13 | 3 | 5 | 3 | 1389 |
Use of antibacterial drugs per group and overall for each time period of prescription to fecal sampling (0–12, 12–24, 24–48, >48 months, or no use). The use of amphenicols (J01B), other beta-lactam antibacterials (J01D), aminoglycoside antibacterials (J01G), glycopeptide antibacterials (J01XA), polymyxins (J01XB), steroid antibacterials (J01XC), imidazole derivatives (J01XD) and other antibacterials (J01XX) is too low to analyze further.
*Time period between prescription of antimicrobial drug and fecal sampling.
Shannon alpha-diversity
The median overall diversity was 4.10 (IQR 3.73–4.37). The strongest and most prolonged effect on diversity was seen in the group of the macrolides and lincosamides (J01F). Transforming the beta’s back to physiological values (for an average person) resulted in a significantly lower diversity of 0.48 for 0–12 months after use of macrolides and lincosamides (J01F); a lower diversity of 0.28 (which was not significant when adjusting for all other antimicrobial drug use) for 12–24 months after use, a significantly lower diversity of 0.35 for 24–48 months after use and a significantly lower diversity of 0.17 for 48 months or longer after use. We also showed a significantly lower diversity of 0.24 after the use of beta-lactam antibacterials (J01C) within 1 year before feces sampling. No change in diversity was seen after the use of tetracyclines (J01A), sulfonamides and trimethoprim (J01E), quinolones (J01M) and nitrofurans (J01XE). (untransformed beta’s of model 2 in Figure 1)
Figure 1.
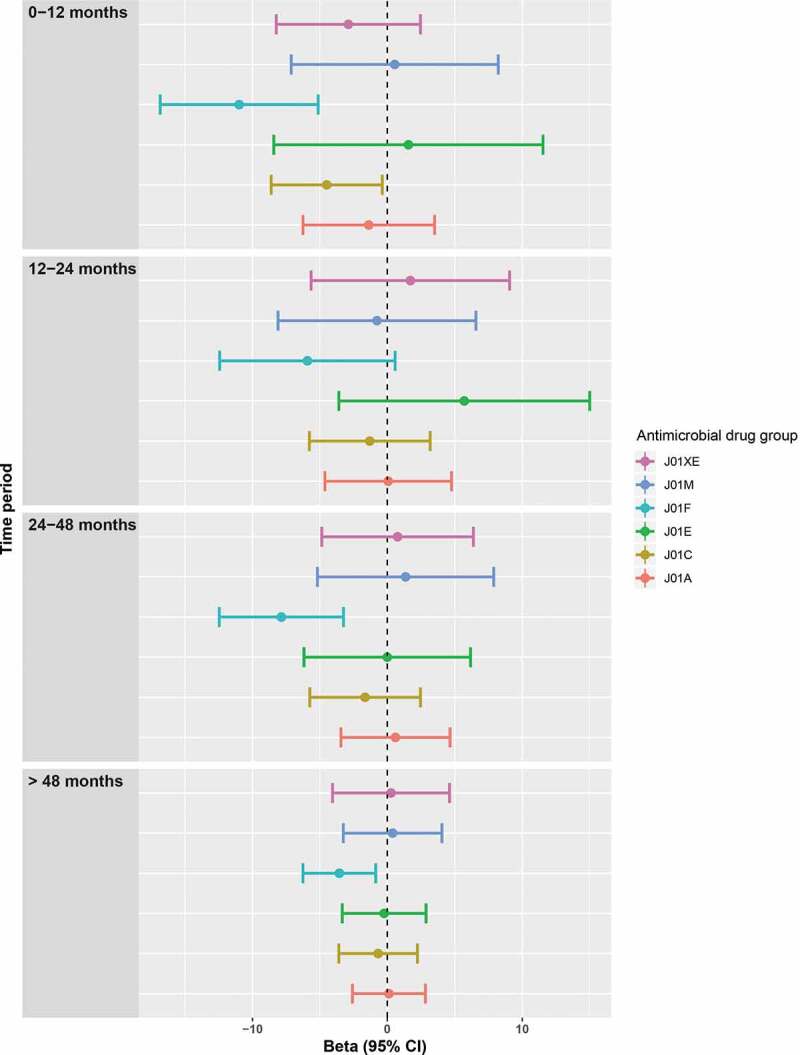
Diversity after antimicrobial drug use. Plots of the beta’s with 95% confidence intervals of the linear regression with as dependent variable the transformed (cube) Shannon alpha-diversity and as independent variables the different antimicrobial drug groups. All antimicrobial drug groups were analyzed with dummy variables with categories of use of 0–12, 12–24, 24–48 and >48 months compared to no use before sampling. The analyses were adjusted for age, sex, BMI, diabetes, time in mail, batch number, use of statins, PPIs, SSRIs, antipsychotics and systemic corticosteroids and (categorized) use of all other antimicrobial drugs.
We also classified the antimicrobial drugs in antimicrobial drugs with anaerobic activity (consisting of combinations of penicillins, including beta-lactamase inhibitors (J01CR), lincosamides (J01 FF) and imidazole derivatives (metronidazole) (J01XD): anaerobic+) and a group without this activity (all other antimicrobial drugs: anaerobic-). The use of anaerobic+ antimicrobial drugs was associated with a stronger and more prolonged effect on diversity than the use of antimicrobials without this activity. For an average person, diversity after the use of anaerobic+ antimicrobial drugs was 0.51 lower for the 0–12 months period and 0.36 lower for the 12–24 months period. Diversity was only 0.23 lower (for an average person) 0–12 months after the use of anaerobic-antimicrobial drugs. (untransformed beta’s in Figure 2) We also performed two sensitivity analyses with additional adjustment for diet and smoking, which slightly shifted the use of beta-lactams in the first year before sampling, resulting in a not significant difference. (Fig. S1 and S2)
Figure 2.
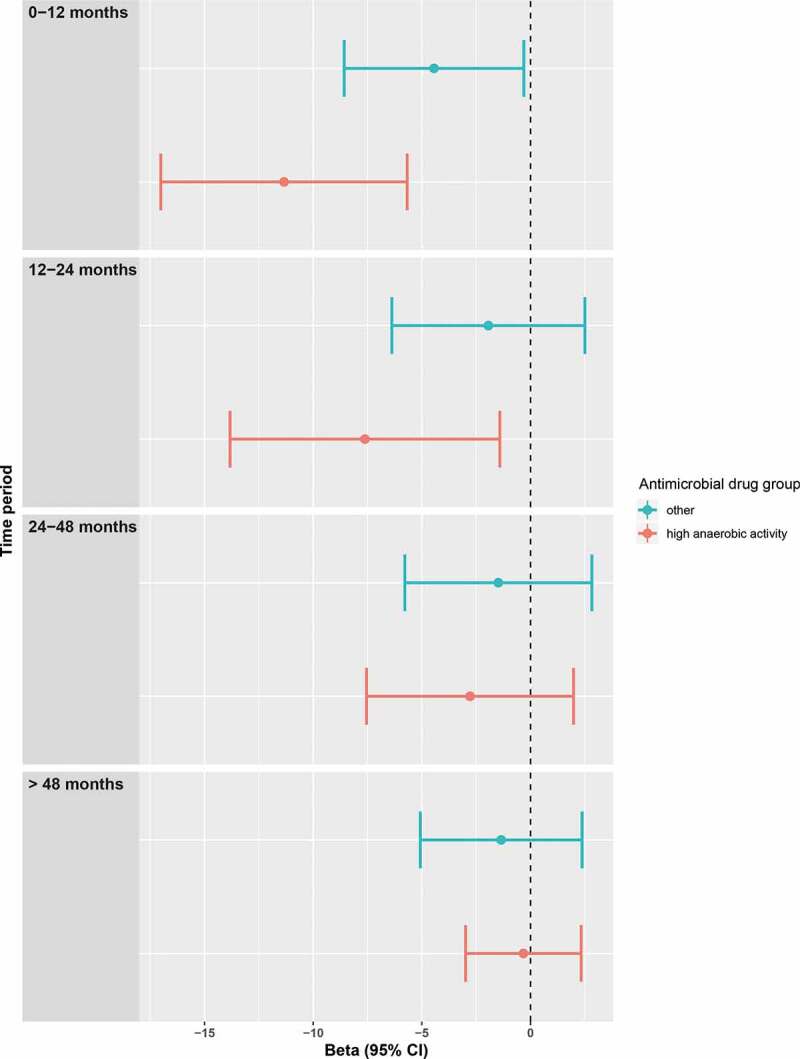
Diversity after using antimicrobial drugs with high anaerobic activity vs other antimicrobial drugs. Plots of the beta’s with 95% confidence intervals of the linear regression with as dependent variable the transformed (cube) Shannon alpha-diversity and as independent variables the combined antimicrobial drugs that have a strong anaerobic activity (anaerobic + drugs) and the combined remaining antimicrobial drugs (anaerobic- drugs). Both were analyzed with dummy variables with categories of 0–12, 12–24, 24–48 and >48 months compared to no use. The analyses were adjusted for age, sex, BMI, diabetes, time in mail, batch number, use of statins, PPIs, SSRIs, antipsychotics and systemic corticosteroids and (categorized) use of all other antimicrobial drugs.
Firmicutes/Bacteroidetes ratio
The median F/B ratio was 0.085 (IQR 0.037–0.21). We could not show any significant differences for any of the different antimicrobial drug groups on the F/B ratio, both in model 1 and in model 2, in which we additionally adjusted for all antimicrobial drug use. (untransformed beta’s for model 2 in Figure S3). However, the F/B ratio significantly shifted toward Firmicutes in the 0–12 months before sampling after the use of anaerobic+ antimicrobial drugs. Furthermore, a significant shift could be demonstrated 12–24 months, 24–48 months and 48 months after the use of anaerobic- antimicrobial drugs toward Bacteroidetes of respectively 0.18, 0.20 and 0.16. (untransformed beta’s in Figure 3).
Figure 3.
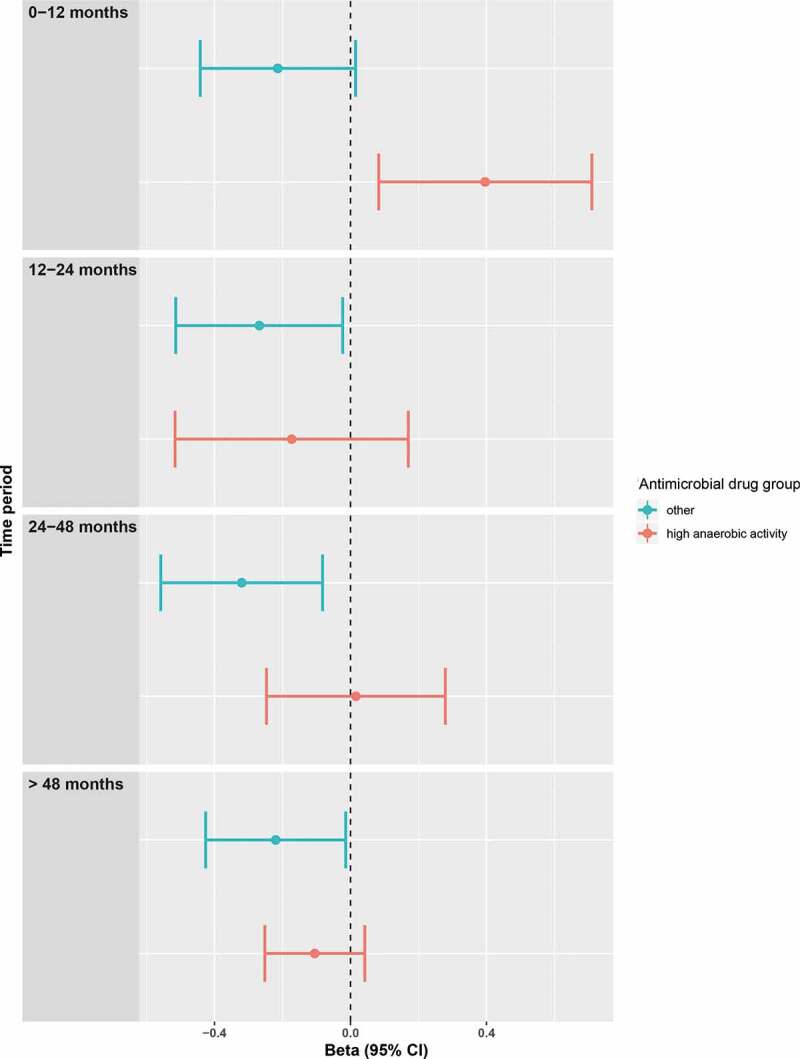
Firmicutes/Bacteroidetes ratio after using antimicrobial drugs with high anaerobic activity vs other antimicrobial drugs. Forest plots of the relative risks with 95% confidence intervals of the linear regression with as dependent variable the transformed (logarithmic) Firmicutes/Bacteroidetes ratio and as independent variables the combined antimicrobial drugs that have a strong anaerobic activity and the combined remaining antimicrobial drugs (“other”). Both were analyzed with dummy variables with categories of 0–12, 12–24, 24–48 and >48 months compared to no use. The analyses were adjusted for age, sex, BMI, diabetes, time in mail, batch number, use of statins, PPIs, SSRIs, antipsychotics and systemic corticosteroids and (categorized) use of all other antimicrobial drugs. A positive beta indicates a shift toward Firmicutes, whereas a negative beta indicates a shift toward Bacteroidetes.
Community structure
Concerning the community structure (beta-diversity), we again found significant differences for macrolides and lincosamides (J01F) in all time categories. We also found a difference for beta-lactams (J01C) in 0–12 months before sampling. Finally, we found significant differences for quinolones both for 0–12 and 24–48 months before sampling, but not for 12–24 months (Figure 4). Most and strongest differences in genera were seen after the use of macrolides and lincosamides (Supplementary Table 2).
Figure 4.
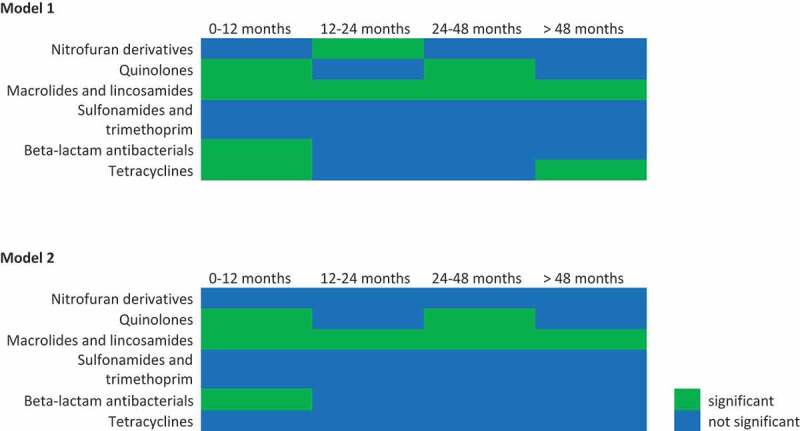
Effects of antimicrobial drug use on community structure. Significance table for all antimicrobial drug groups for all time categories studied in model 1, thus adjusted for age, sex, BMI, diabetes, time in mail, batch number, use of statins, PPIs, SSRIs, antipsychotics and systemic corticosteroids. (Top) Significance table for all antimicrobial drug groups for all time categories studied in model 2, thus adjusted for age, sex, BMI, diabetes, time in mail, batch number, use of statins, PPIs, SSRIs, antipsychotics and systemic corticosteroids and (categorized) use of all other antimicrobial drugs. (Bottom) In these significance tables green indicates significant (p < 0.05), blue indicates not significant.
Discussion
In this study, we showed an association between the use of different oral antimicrobial drugs and the diversity of the gut microbiota in feces samples of a middle-aged and elderly community-dwelling population. The strongest and most prolonged effects on the microbiota diversity were shown for macrolides and lincosamides, and these effects lasted up to several years after use. Also, diversity was lower and community structure was different in the first year after beta-lactam use. Furthermore, the use of antimicrobial drugs with a high anaerobic activity was associated with a shift toward Firmicutes. This, in contrary to the use of antimicrobial drugs without this activity, which resulted in s shift toward Bacteroidetes.
Increasing evidence shows that changes in the microbiota by antimicrobial drugs are associated with a variety of diseases. A study in Finnish school children reported an association between frequent macrolide use in early life (<2 years) and the development of asthma.20 Furthermore, the use of several antimicrobial drug groups was shown to be associated with several cancers in a large-nested case–control study, possibly acting via the gut microbiota.21 Also, but still unproven, it has been suggested to prescribe probiotics simultaneously with antimicrobial drugs. A large systemic review showed that there is evidence that probiotics are effective in preventing Clostridium difficile-associated diarrhea in not-immunocompromised individuals with a high baseline risk.22 Furthermore, in mice concurrent probiotics treatment during or after antibiotic therapy caused suppression of Enterobacteriaceae outgrowth, while promoting blooming of Firmicutes.23 Therefore, the influence on the gut microbiota should be taken into consideration when prescribing antibiotic drugs.
Many studies investigated the microbiota with different outcome parameters, included few participants or reported only short-term effects.17–19 A strength of our study is that we studied the gut microbiota in a large population-based cohort with detailed information on antimicrobial drug prescriptions and over more than a 4-year time period using different outcome parameters. Furthermore, we adjusted our models for several potential confounders, such as sex, age, BMI, diabetes and use of co-medication (statins, systemic corticosteroids, proton pump inhibitors, SSRIs and antipsychotics). Diversity is the most frequently reported outcome in microbiota studies and loss of diversity appears as the most consistent finding of intestinal dysbiosis.24 We also report on the Firmicutes/Bacteroidetes ratio, and the community structure, using the beta-diversity. The different outcome parameters enable a broader interpretation of the effect of antibiotic use on gut microbiota. Unfortunately, we could not compare the microbiota after with the microbiota before antimicrobial drug use, but the time frame, the cohort size and the fact that the participants were prescribed antibiotics by their physician for an infection and not specifically for this study made such a study design not feasible. Also, because of the length of the study time, other factors, such as intestinal surgery, could also have influenced the microbiota. However, because of the size of the cohort, we assume that the effect of these factors is small. Another limitation is that the feces was sent to the study center by the participants via mail, which might have influenced changes of the microbiota composition by environmental factors, such as temperature. However, the effects of our collection method have been studied and has resulted in the exclusion of samples that were in the mail longer than three days. Furthermore, the time in the mail was included as a covariate in the analyses. Also, the microbiota within our cohort had similar profiles as those in two other large population-based cohorts.25 Another limitation may be that our results were obtained from feces samples and may not reflect the microbiota more proximal in the digestive tract.26 Furthermore, we only used the last prescription before sampling, not taking into account the antimicrobial drug prescriptions used previously. However, we showed that correlations between the use of antimicrobial drugs of different classes were low and additionally, we adjusted for all other antimicrobial drug use.
All our results pointed to macrolides and lincosamides as the antimicrobial drugs with the highest ability to cause changes in the composition of the gut microbiota. Of these two types of antibiotics, lincosamides such as clindamycin probably have the strongest effect, since we also showed that antibiotics with a high anaerobic activity (which included clindamycin) had strong associations with the diversity of the microbiota. Another study has also shown long-lasting effects of clindamycin on the gut microbiota but only up to 2 years.17 Furthermore, the macrolide azithromycin was shown to have effects up to 6 months,18 and a shift of the gut microbiota at phylum level was found in Finnish children of 2–7 years old after macrolide use in the 2 years before sampling.20 Our data indicate that a shift in the composition of the gut microbiota persists for a longer time period.
Beta-lactam antibiotics caused a lower diversity and differences in community structure in the first year after use. Beta-lactams have been associated with effects on the composition of the gut microbiota in several small studies.17 Use of tetracyclines has been associated with a relative increase in the abundance of Bacteroidetes.27 Doxycycline use was shown to be associated with a lower diversity and a relative increase of Bacteroidetes in mice.28 Nitrofurantoin was shown to have only minor effects in a study in patients with urinary tract infections.29 These studies, however, investigated the effects after a maximum of a few months. Since we did not find any effects in our longer time periods, this might suggest that the effects of these antimicrobial drugs on the gut microbiota are restored after a few months. We also did not find any effects for sulfonamides and trimethoprim, but the use of these drugs (singly or in combination products) was very low.
Although we could not find an effect on the F/B ratio for separate antimicrobial drug classes, we found that the use of antimicrobial drugs with a high anaerobic activity was associated with a shift toward Firmicutes in the first year. In contrast, the use of antimicrobial drugs without this anaerobic activity was associated with a shift toward Bacteroidetes up to several years. Others also described effects on this ratio, but only directly after treatment, showing relatively more Bacteroidetes after the use of antimicrobial drugs.30,31
In conclusion, we showed that antimicrobial drugs, especially macrolides and lincosamides, are associated with a long-lasting shift in the gut microbiota. Further research is needed to explore the interaction and effect of specific antibiotics on the gut microbiota, considering the consequences of the use of antimicrobial drugs on the gut microbiota.
Patients and methods
Source population
The feces samples that were used in this study were obtained from study participants of the third cohort (RSIII) of The Rotterdam Study (RS), a prospective population-based study. This cohort includes 3122 individuals, who were recruited in the period March 2012 to June 2014 and who were 45 years and older, living in the Ommoord district in Rotterdam. All participants are invited every 3–4 years for follow-up interviews and examinations. More detailed information on the Rotterdam Study can be found elsewhere.32
Gut microbiota composition
Stool samples were collected at home by the participants using a Commode Specimen Collection System (Covidien, Mansfield, MA). An aliquot of approximately 1 g was transferred to a 25 × 76 mm feces collection tube (Minigrip Nederland, Lelystad, The Netherlands) and sent through regular mail to the Erasmus MC. A short questionnaire addressing amongst others date and time of defecation was filled out by the participants (response percentage 69%). After receipt, the samples were stored at −20°C. Approximately, 300 mg of feces was homogenized in stool stabilizing buffer. Automated DNA-isolation (Arrow DNA; DiaSorin S.p.A., Saluggia, Italy) was performed using the Arrow DNA kit according to the manufacturer’s instructions and included bead-beating in Lysing Matrix B tubes containing 0.1 mm silica beads (MP Biomedicals, LLC, Bio Connect Life Sciences, Huissen, The Netherlands). The hypervariable regions V3 and V4 of the (bacterial) 16 S rRNA gene were amplified and sequenced using the Illumina MiSeq 2 × 300 base pairs protocol (FADROSH, PMID: 24558975). Phylogenetic multi-sample profiling was performed using an in-house developed pipeline based on the QIIME 1.9.0 (Caporaso PMID: 20383131) and USEARCH version 8.1 (Edgar PMID: 23955772) software packages. After subsampling at 10,000 reads per sample, taxonomy was assigned using the naïve Bayesian RDP classifier (vs 2.12)33 and the SILVA database (v128; Quast PMID: 23193283). The OTU table was cleaned by filtering out low abundance OTUs (<0.005% of total reads per OTUs and OTUs present in <1% of the samples). Samples with unknown information of time in the mail, samples arriving 3 days after collection and samples from participants who used antibiotics during or just before sampling were removed.
Medication use
The date of the last prescription of an antimicrobial drug before feces sampling was obtained from a collaborative database of all community pharmacies in the Ommoord area that goes back to 1 January 1995. The antimicrobial drug prescriptions were grouped on Anatomical Therapeutic Chemical (ATC) code, which included: tetracyclines (J01A), amphenicols (J01B), beta-lactam antibacterials (J01C), other beta-lactam antibacterials (J01D) (which includes all generations of cephalosporins and carbapenems), sulfonamides and trimethoprim (J01E), macrolides and lincosamides (J01F) (J01F also includes streptogramins, but these were not prescribed in the study period), aminoglycoside antibacterials (J01G), quinolone antibacterials (J01M), glycopeptide antibacterials (J01XA), polymyxins (J01XB), steroid antibacterials (J01XC), imidazole derivatives (J01XD), nitrofuran derivatives (J01XE) and other antibacterials (J01XX). For each antimicrobial drug group, the time interval between the date of the last prescription and feces sampling was calculated and categorized into the use of 0–12, 12–24, 24–48 and >48 months before sampling or no use of the antimicrobial drug group. Additionally, the antimicrobial drugs were classified in a group antimicrobial drugs with a high activity against anaerobic species (anaerobic+), consisting of combinations of penicillins, including beta-lactamase inhibitors (J01CR), lincosamides (J01FF) and imidazole derivatives (metronidazole) (J01XD) and a group without this activity (anaerobic-) (all other antimicrobial drugs).
Confounders
The following potential confounders (at the time of feces sampling) were taken into account in the analyses: age, sex, BMI, diabetes (use of anti-diabetic drugs (A10)), use of co-medication, time in the mail of the feces sample and batch number representing two batches of DNA isolation: the first 102 DNA isolation runs with a relatively high yield were labeled 0 and the last 32 runs with a relatively low yield were labeled 1. Patients who were prescribed a drug within 90 days before feces sampling were considered as current user of that drug. Drugs that possibly influence the composition of the microbiota according to literature: proton pump inhibitors (A02BC),10 statins (C10AA),12 systemic corticosteroids (H02),11 antipsychotics (N05A),34 selective serotonin reuptake inhibitors (SSRIs) (N06AB),35 antineoplastic agents (L01),36 and tacrolimus (L04AD).37 Since proton pump inhibitors may have been sold over the counter, participants were also asked if they used proton pump inhibitors.
Other potential confounders for studying the association with the gut microbiota are diet and smoking. Adjustment for diet was performed by adjusting for the dietary guidelines score (DGS), which is a score that varies from 0 to 14 and represents the adherence to the Dutch dietary guidelines which include 14 items: vegetables (≥200 g/day), fruit (≥200 g/day), whole-grains (≥90 g/day), legumes (≥135 g/week), nuts (≥15 g/day), dairy (≥350 g/day), fish (≥100 g/week), tea (≥450 mL/day), ratio whole-grains:total grains (≥50%), ratio unsaturated fats and oils:total fats (≥50%), red and processed meat (<300 g/week), sugar-containing beverages (≤150 mL/day), alcohol (≤10 g/day) and salt (≤6 g/day).38 Adjustment for smoking was performed by adjusting for the smoking status (never, ever, current).
Statistical analyses
We performed several analyses in order to study the association between antimicrobial drug use and the composition of the gut microbiota, using several measures described below. For the diversity, Firmicutes/Bacteroidetes ratio (F/B ratio) and community structure analysis, we performed two models. In model 1, we adjusted for the above-mentioned confounders (sex, age, BMI, diabetes, use of co-medication, time in the mail and batch number). In model 2, we additionally adjusted for the categorized use of other antimicrobial drug groups (thus, for example: the association between tetracyclines and the gut microbiota was adjusted for the mentioned confounders and for categorized use of beta-lactam antibacterials, categorized use of sulfonamides and trimethoprim, categorized use of macrolides and lincosamides, categorized use of quinolones, and categorized use of nitrofuran derivatives). For diet and smoking, the data were not available for all participants: 269 (19.0%) were missing for diet and 108 (7.6%) for smoking. Therefore, adjustment for these confounders was only performed in two sensitivity analyses.
Diversity analysis
The Shannon index was used to calculate the alpha-diversity (measure of diversity of species within a sample). In order to obtain a normal distribution (according to the Kolgomorov–Smirnov test), it was transformed by calculating the cube. For each antimicrobial drug group, a linear regression was performed with the transformed Shannon alpha-diversity as the dependent variable, and four dummies for antimicrobial drug use of the specific group and the confounding variables as independent variables. The outcome was back transformed for male gender, batch 0, with median age, median BMI, for those who had no diabetes, who used no co-medication, of whom the sample was a median time in the mail and who used no other antimicrobial drugs than the one of interest (= average person). P-values <0.05 were considered to be significant.
The Firmicutes/Bacteroidetes ratio
The F/B ratio was calculated and logarithmically transformed to obtain a normal distribution. Different linear regressions were performed and transformed back to the average person as described above but now with as a dependent variable the transformed F/B ratio. P-values <0.05 were considered to be statistically significant.
Community structure analysis
MiRKAT is a recently developed package, available in the statistical program R, that tests for associations between microbiota composition and an outcome, using a semi-parametric kernel machine regression.39 MiRKAT (using 100,000 permutations) was used to investigate differences in the composition of the fecal microbiota using the Bray–Curtis beta-diversity distance (measure of dissimilarity of species composition between sample pairs). P-values <0.05 were considered to be statistically significant.
Single genera analyses
The genera that were significantly different in individuals who had used antimicrobial drugs were determined using the MaAsLin (Multivariate Association with Linear Models) function.40 The categorized variable for the use of each antimicrobial drug group was linearly included in the model (0 for no prescription at all, 4 for latest prescription in 0–12 months before sampling). For the analysis, the default settings were used: a false discovery rate of 25% and q-values <0.05 were considered to be statistically significant.
Supplementary Material
Acknowledgments
The authors are grateful to the study participants, the staff from the Rotterdam Study and the participating general practitioners and pharmacists.
Funding Statement
The Rotterdam Study is supported by the Erasmus MC and Erasmus University Rotterdam; the Netherlands Organisation for Scientific Research (NWO); the Netherlands Organisation for Health Research and Development (ZonMw); the Research Institute for Diseases in the Elderly (RIDE); the Netherlands Genomics Initiative (NGI); the Ministry of Education, Culture and Science, the Ministry of Health, Welfare and Sports; the European Commission (DG XII) and the Municipality of Rotterdam. None of the funders had any role in design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript.
Authors contributions
The microbiota dataset was made by DR under the supervision of AU and RK. The study design was made by MM with the aid of BS and AV and the critical input of all co-authors. The statistical analyses were performed by MM with the aid of DR, RK, JK, BS and AV. The manuscript was written by MM with the critical input of all co-authors.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Ethics approval and consent to participate
The Rotterdam Study has been approved by the Medical Ethics Committee of the Erasmus University Medical Centre and by the Ministry of Health, Welfare and Sport of the Netherlands, on the basis of the Wet Bevolkingsonderzoek ERGO (Population Studies Act: Rotterdam Study). All participants provided written informed consent to participate in the study and to obtain additional information from their medical files.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
Data availability
The datasets generated and/or analyzed during the current study are not publicly available due to privacy agreements of the participants of the study but are available from the corresponding author on reasonable request.
References
- 1.Lynch SV, Pedersen O.. The human intestinal microbiome in health and disease. N Engl J Med. 2016;375:2369–12. [DOI] [PubMed] [Google Scholar]
- 2.den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res. 2013;54:2325–2340. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morrison DJ, Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes. 2016;7:189–200. doi: 10.1080/19490976.2015.1134082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saraswati S, Sitaraman R, Ge M-Y, Ibrahim M, Li B, Zhao W-J, Chen G-Y, Zhu B, Xie G-L. Aging and the human gut microbiota-from correlation to causality. Front Microbiol. 2014;5:764. doi: 10.3389/fmicb.2014.00547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haro C, Rangel-Zuniga OA, Alcala-Diaz JF, Gomez-Delgado F, Perez-Martinez P, Delgado-Lista J, Quintana-Navarro GM, Landa BB, Navas-Cortés JA, Tena-Sempere M, et al. Intestinal microbiota is influenced by gender and body mass index. PLoS One. 2016;11(5):e0154090. doi: 10.1371/journal.pone.0154090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scott KP, Gratz SW, Sheridan PO, Flint HJ, Duncan SH. The influence of diet on the gut microbiota. Pharmacol Res. 2013;69(1):52–60. doi: 10.1016/j.phrs.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 8.Bressa C, Bailen-Andrino M, Perez-Santiago J, Gonzalez-Soltero R, Perez M, Montalvo-Lominchar MG, Maté-Muñoz JL, Domínguez R, Moreno D, Larrosa M, et al. Differences in gut microbiota profile between women with active lifestyle and sedentary women. PLoS One. 2017;12(2):e0171352. doi: 10.1371/journal.pone.0171352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sircana A, Framarin L, Leone N, Berrutti M, Castellino F, Parente R, De Michieli F, Paschetta E, Musso G. Altered gut microbiota in type 2 diabetes: just a coincidence? Curr Diab Rep. 2018;18(10):98. doi: 10.1007/s11892-018-1057-6. [DOI] [PubMed] [Google Scholar]
- 10.Minalyan A, Gabrielyan L, Scott D, Jacobs J, Pisegna JR. The gastric and intestinal microbiome: role of proton pump inhibitors. Curr Gastroenterol Rep. 2017;19:42. doi: 10.1007/s11894-017-0577-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tetel MJ, de Vries GJ, Melcangi RC, Panzica G, O’Mahony SM. Steroids, stress, and the gut microbiome-brain axis. J Neuroendocrinol. 2017;30(2):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caparros-Martin JA, Lareu RR, Ramsay JP, Peplies J, Reen FJ, Headlam HA, Ward NC, Croft KD, Newsholme P, Hughes JD, et al. Statin therapy causes gut dysbiosis in mice through a PXR-dependent mechanism. Microbiome. 2017;5:95. doi: 10.1186/s40168-017-0312-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lange K, Buerger M, Stallmach A, Bruns T. Effects of antibiotics on gut microbiota. Dig Dis. 2016;34:260–268. doi: 10.1159/000443360. [DOI] [PubMed] [Google Scholar]
- 14.Modi SR, Collins JJ, Relman DA. Antibiotics and the gut microbiota. J Clin Invest. 2014;124:4212–4218. doi: 10.1172/JCI72333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Francino MP. Antibiotics and the human gut microbiome: dysbioses and accumulation of resistances. Front Microbiol. 2015;6:1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stecher B, Maier L, Hardt WD. ‘Blooming’ in the gut: how dysbiosis might contribute to pathogen evolution. Nat Rev Microbiol. 2013;11:277–284. doi: 10.1038/nrmicro2989. [DOI] [PubMed] [Google Scholar]
- 17.Jernberg C, Lofmark S, Edlund C, Jansson JK. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. Isme J. 2007;1:56–66. doi: 10.1038/ismej.2007.3. [DOI] [PubMed] [Google Scholar]
- 18.Abeles SR, Jones MB, Santiago-Rodriguez TM, Ly M, Klitgord N, Yooseph S, Nelson KE, Pride DT. Microbial diversity in individuals and their household contacts following typical antibiotic courses. Microbiome. 2016;4:39. doi: 10.1186/s40168-016-0187-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, Kurilshikov A, Bonder MJ, Valles-Colomer M, Vandeputte D, et al. Population-level analysis of gut microbiome variation. Science. 2016;352(6285):560–564. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- 20.Korpela K, Salonen A, Virta LJ, Kekkonen RA, Forslund K, Bork P, de Vos WM. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat Commun. 2016;7(1):10410. doi: 10.1038/ncomms10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boursi B, Mamtani R, Haynes K, Yang YX. Recurrent antibiotic exposure may promote cancer formation–another step in understanding the role of the human microbiota? Eur J Cancer. 2015;51:2655–2664. doi: 10.1016/j.ejca.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldenberg JZ, Yap C, Lytvyn L, Lo CK, Beardsley J, Mertz D, Johnston BC. Probiotics for the prevention of Clostridium difficile-associated diarrhea in adults and children. Cochrane Database Syst Rev. 2017;12:CD006095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grazul H, Kanda LL, Gondek D. Impact of probiotic supplements on microbiome diversity following antibiotic treatment of mice. Gut Microbes. 2016;7:101–114. doi: 10.1080/19490976.2016.1138197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mosca A, Leclerc M, Hugot JP. Gut microbiota diversity and human diseases: should we reintroduce key predators in our ecosystem? Front Microbiol. 2016;7:455. doi: 10.3389/fmicb.2016.00455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Radjabzadeh D, Boer CG, Beth SA, van der Wal P, Kiefte-De Jong JC, Jansen MAE, Konstantinov SR, Peppelenbosch MP, Hays JP, Jaddoe VWV, et al. Diversity, compositional and functional differences between gut microbiota of children and adults. Sci Rep. 2020;10(1):1040. doi: 10.1038/s41598-020-57734-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stearns JC, Lynch MD, Senadheera DB, Tenenbaum HC, Goldberg MB, Cvitkovitch DG, Croitoru K, Moreno-Hagelsieb G, Neufeld JD. Bacterial biogeography of the human digestive tract. Sci Rep. 2011;1(1):170. doi: 10.1038/srep00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jung JY, Ahn Y, Khare S, Gokulan K, Pineiro SA, Cerniglia CE. An in vitro study to assess the impact of tetracycline on the human intestinal microbiome. Anaerobe. 2018;49:85–94. doi: 10.1016/j.anaerobe.2017.12.011. [DOI] [PubMed] [Google Scholar]
- 28.Boynton FDD, Ericsson AC, Uchihashi M, Dunbar ML, Wilkinson JE. Doxycycline induces dysbiosis in female C57BL/6NCrl mice. BMC Res Notes. 2017;10:644. doi: 10.1186/s13104-017-2960-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vervoort J, Xavier BB, Stewardson A, Coenen S, Godycki-Cwirko M, Adriaenssens N, Kowalczyk A, Lammens C, Harbarth S, Goossens H, et al. Metagenomic analysis of the impact of nitrofurantoin treatment on the human faecal microbiota. J Antimicrob Chemother. 2015;70:1989–1992. doi: 10.1093/jac/dkv062. [DOI] [PubMed] [Google Scholar]
- 30.Claesson MJ, Cusack S, O’Sullivan O, Greene-Diniz R, de Weerd H, Flannery E, Marchesi JR, Falush D, Dinan T, Fitzgerald G, et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4586–4591. doi: 10.1073/pnas.1000097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Panda S, El Khader I, Casellas F, Lopez Vivancos J, Garcia Cors M, Santiago A, Cuenca S, Guarner F, Manichanh C, et al. Short-term effect of antibiotics on human gut microbiota. PLoS One. 2014;9(4):e95476. doi: 10.1371/journal.pone.0095476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ikram MA, Brusselle GGO, Murad SD, van Duijn CM, Franco OH, Goedegebure A, Klaver CW, Nijsten TEC, Peeters RP, Stricker, BH, et al. The rotterdam study: 2018 update on objectives, design and main results. Eur J Epidemiol. 2017;32:807–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flowers SA, Evans SJ, Ward KM, McInnis MG, Ellingrod VL. Interaction between atypical antipsychotics and the gut microbiome in a bipolar disease cohort. Pharmacotherapy. 2017;37:261–267. doi: 10.1002/phar.1890. [DOI] [PubMed] [Google Scholar]
- 35.Munoz-Bellido JL, Munoz-Criado S, Garcia-Rodriguez JA. Antimicrobial activity of psychotropic drugs: selective serotonin reuptake inhibitors. Int J Antimicrob Agents. 2000;14:177–180. doi: 10.1016/S0924-8579(99)00154-5. [DOI] [PubMed] [Google Scholar]
- 36.Montassier E, Gastinne T, Vangay P, Al-Ghalith GA, Bruley Des Varannes S, Massart S, Moreau P, Potel G, de La Cochetière MF, Batard E, et al. Chemotherapy-driven dysbiosis in the intestinal microbiome. Aliment Pharmacol Ther. 2015;42(5):515–528. doi: 10.1111/apt.13302. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Z, Liu L, Tang H, Jiao W, Zeng S, Xu Y, Zhang Q, Sun Z, Mukherjee A, Zhang X, et al. Immunosuppressive effect of the gut microbiome altered by high-dose tacrolimus in mice. Am J Transplant. 2018;18(7):1646-1656. [DOI] [PubMed] [Google Scholar]
- 38.Voortman T, Kiefte-de Jong JC, Ikram MA, Stricker BH, van Rooij FJA, Lahousse L, Tiemeier H, Brusselle GG, Franco OH, Schoufour JD, et al. Adherence to the 2015 Dutch dietary guidelines and risk of non-communicable diseases and mortality in the Rotterdam Study. Eur J Epidemiol. 2017;32(11):993–1005. doi: 10.1007/s10654-017-0295-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao N, Chen J, Carroll IM, Ringel-Kulka T, Epstein MP, Zhou H, Zhou J, Ringel Y, Li H, Wu M, et al. Testing in microbiome-profiling studies with MiRKAT, the microbiome regression-based kernel association test. Am J Hum Genet. 2015;96(5):797–807. doi: 10.1016/j.ajhg.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13(9):R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are not publicly available due to privacy agreements of the participants of the study but are available from the corresponding author on reasonable request.


