ABSTRACT
The early life immune system is characterized by unique developmental milestones. Functionally diverse immune cells arise from distinct waves of hematopoietic stem cells, a phenomenon referred to as ‘layered’ immunity. This stratified development of immune cells extends to lineages of both innate and adaptive cells. The defined time window for the development of these immune cells lends itself to the influence of specific exposures typical of the early life period. The perinatal immune system develops in a relatively sterile fetal environment but emerges into one filled with a multitude of antigenic encounters. A major burden of this comes in the form of the microbiota that is being newly established at mucosal surfaces of the newborn. Accumulating evidence suggests that early life microbial exposures, including those arising in utero, can imprint long-lasting changes in the offspring’s immune system and determine disease risk throughout life. In this review, I highlight unique features of early life immunity and explore the role of intestinal bacteria in educating the developing immune system.
KEYWORDS: Perinatal immune system, window of opportunity, layered immunity, microbiota, immune education
At the time of writing this review, the world was coming to grips with the scale of the COVID-19 pandemic caused by the novel coronavirus, SARS-CoV-2. The virus had infected millions of people worldwide and claimed hundreds of thousands of lives. Epidemiological data indicated, however, that the disease affected far fewer children than adults. Children and infants that did test positive for SARS-CoV-2 were largely asymptomatic or experienced a mild form of the disease. Children are usually more susceptible to respiratory viral infections and not less prone to developing severe acute respiratory distress syndrome (ARDS), a grim presentation of COVID-19. As demonstrated in the H1N1 flu pandemic of 2009, being under the age of 1 year was a significant risk factor for ARDS and the more severe form of the disease.1 Numerous hypotheses have been proposed for the mild COVID-19 disease in children including unique characteristics of the early life immune system, circulating maternal antibodies, and protective memory T and B cells from prior coronavirus infections that may limit damage from SARS-CoV-2 infection.2 Newer reports also indicate that some children infected with SARS-CoV-2 may progress to a multisystem inflammatory syndrome (MISC) resembling Kawasaki disease.3 The pathophysiology is yet unknown and this appears to be a rare complication affecting some children over others. The bottom line is that many infectious diseases affect children differently than adults. Understanding these differences, including how the immune system develops and the environmental and host factors that dictate plasticity in early life immune responses, will yield important insight into disease pathogenesis and inform development of new therapeutics.
The early life represents a period of unique immune development during which the foundation for lifelong immunity is laid. Epidemiological data link pre- and post-natal encounters with susceptibility to chronic inflammatory and metabolic diseases in later life.4–6 Distinct immune cell subsets arise in early life that are wired to function in an environment with unique exposures. One exposure of critical importance is the burgeoning intestinal microbiota, the collection of diverse bacteria, archaea, viruses, and fungi, that regulate aspects of immune system development and function. Here, I review the premise for an early life time window when microbial encounters influence the development of select immune phenotypes.
Early life window of opportunity
The Developmental Origins of Health and Disease (DOHaD) hypothesis proposes that exposure to certain environmental stimuli during critical periods of prenatal development have significant consequences on an individual’s short- and long-term health.7 The developing fetus may respond to hostile uterine environments, resulting, for example, from poor maternal nutrition or infection, by changing endocrine and metabolic activity that slow down its growth rate. Resulting irreversible changes in the structure and function of vital organs are exacerbated by subsequent environmental exposures during infancy and childhood that condition the risk of disease in later life. A critical foundation of the DOHaD hypothesis is the concept of developmental plasticity, or the ability of an organism to develop in diverse ways, depending on the particular environment and setting.8 The immune system displays considerable developmental plasticity. Hematopoietic stem cells (HSCs) that give rise to numerous blood and immune cells are able to sense diverse stimuli to influence the type and function of cell lineages generated.9–11 Plasticity is also observed during the generation of antigen-specific immune responses, the quality of which are determined by numerous macro- and micro-environmental factors that regulate the differentiation of specific effector cells.12,13 Thus, the principles of the DOHaD hypothesis are applicable to the immune system too, and numerous studies have established links between maternal nutritional or infectious status and the offspring’s immunity and subsequent disease risk.14,15 For example, maternal malnutrition results in decreased placental transfer of maternal antibody to the fetus. This has long-lasting effects on B cell development in offspring that correlates with susceptibility to allergies and autoimmune diseases.16,17 Chronic helminthiasis during pregnancy promotes Th2 cytokine responses and regulatory T cell differentiation in offspring18,19 that correlates with diminished vaccine responses and increased susceptibility to tuberculosis.20
However, while unfavorable early life exposures certainly increase the risk of disease in offspring, nonpathogenic microbial exposures may in fact promote the generation of protective immunity. This idea was first proposed by Strachan in 1989 to explain the inverse correlation between birth order and incidence of allergic diseases in families in Britain.21 Shared living spaces and air was presumed to increase microbial spread in younger children of large families, which favored the development of protective immunity to hay fever in younger siblings. Similar observations have since been made for a variety of environmental conditions that promote microbial exposure early in life. The prevalence of atopy was found to be lower in children who started to attend day care at a younger age (6–11 months) compared to those who started at an older age.22,23 Farm living is associated with exposure to higher diversity of bacteria and fungi that appear to protect from atopy.24 Conversely, higher incidences of asthma and hypersensitivity disorders were observed in children raised in ‘cleaner’ urban dwellings compared to rural and farm environments.25–27 Thus, while the DOHaD hypothesis proposes a prenatal developmental period during which adversarial exposures increase the risk of disease, the ‘early life window of opportunity’ argument advances the notion of a defined perinatal period during which specific microbial exposures lead to a favorable imprint on immunity that reduces risk of disease later in life. As windows of opportunity go, it is predicted that this period of immune malleability would be short, would not appear again in later life and would lead to long-term beneficial health consequences. It is of interest, therefore, to understand1 when the window of opportunity arises in early life,2 the identity of immune cells generated during this period that are likely to be receptive to microbial influence,3 the nature of microbes and their components that play a role in educating the early life immune system and of course,4 the practical implications of this phenomenon to ultimately develop strategies that promote health and prevent the incidence of chronic inflammatory diseases. I discuss these in some detail below.
Perinatal microbial encounters
An appreciation of the duration of the window of opportunity that establishes trajectories of disease risk begins with an understanding of when microbial exposures occur early in life. Three windows of influential microbial encounters may be distinguished in the pre- and post-natal period: microbial exposure in utero during pregnancy, microbes acquired at the time of delivery, and microbes established postnatally by the acquisition of maternal and environmental bacteria (Figure 1). Thus, the first 1000 days of life, that is, the period from conception to 2 years of age, may provide opportunities for microbial imprint of immunity that set thresholds of reactivity throughout life.
Figure 1.
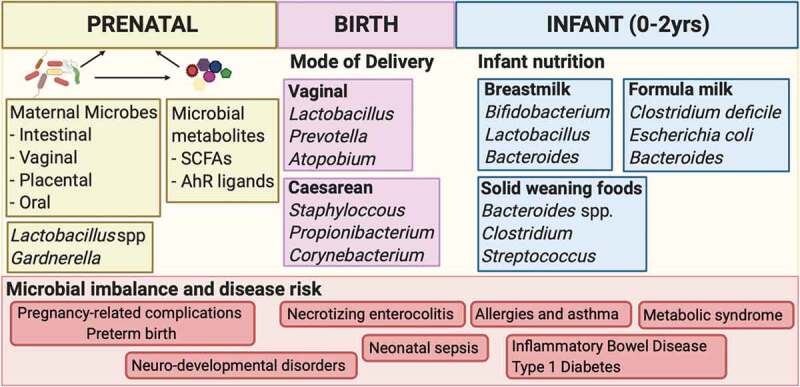
Perinatal microbial encounters. Three windows of influential microbial encounters may be distinguished in the pre- and post-natal period. Maternal microbes as well as their metabolites may influence the uterine environment in which the fetus develops. Delivery mode shapes the initial microbial inoculum of the newborn while nutrition drives microbial composition during the postnatal period. Microbial imbalance during these periods alter susceptibility to pathogens and inflammatory disease in early and later life.
Pregnancy-associated microbiota
The maternal microbiota in the gut and reproductive tract undergoes significant changes during pregnancy that is influenced by a combination of hormonal, metabolic, and immunological factors as well as by maternal diet, supplement intake, and antibiotics use.28 Alterations in the vaginal microbiota have been linked to preterm birth, a major cause of worldwide neonatal morbidity and mortality.29 Activation of maternal immunity leading to preterm birth has been well documented;30 however, an impact on the developing fetal immune system and subsequent long-term health consequences in offspring cannot be ruled out. Maternal gut microbes metabolize dietary components that is passed to the developing fetus across the placenta. The placenta is the highly adapted primary interface between the mother and the developing fetus that facilitates the exchange of nutrients, gases, xenobiotics, and waste products while protecting the fetus from rejection by the maternal immune system. It is also the focus of a controversial and debated topic about the existence of an intra-uterine microbiome in a healthy pregnancy. The long-held premise that the fetus develops in a sterile environment in utero is being challenged.31 Some studies have demonstrated the presence of low abundance commensal bacteria in the healthy placenta,32,33 while others refute this premise of a placental microbiome.34,35 Distinct bacterial profiles were also reported in gestational week 20 fetal intestines, findings that need to be corroborated in other independent cohorts.36 While exposure of the fetus to live bacteria in utero remains to be confirmed, the fetal tissues are likely exposed to numerous microbial metabolites and microbial fragments of maternal origin that are transferred across the placenta from the maternal serum. Gut microbes can induce an IgG antibody response in hosts that protect from systemic infections.37 Importantly, these commensal-specific IgG antibodies can be transferred across the placenta to the offspring where they regulate mucosal CD4+T cell responses to commensal antigens early after birth.38 Maternal IgG antibodies can also facilitate transfer of bacterial compounds themselves across the placenta.39 In a mouse model where bacteria were only present during pregnancy, maternal antibodies enhanced transfer of bacterial compounds, including ligands for the aryl hydrocarbon receptor (AhR), from the mother to the offspring where they primed the developing immune system. Maternal enteric microbes also ferment dietary fibers to the short-chain fatty acid (SCFA) acetate, that suppresses allergic airway disease, a mouse model for human asthma, by enhancing Treg cell numbers and function in adult offspring.40 In the context of a maternal infection, bacterial peptidoglycan, which is a ligand for Toll-like Receptor 2 (TLR2), traversed the placenta to influence fetal neuro-proliferation.41 Passage of other microbial TLR ligands across the placenta that influence fetal immune development remains a formal possibility.
Delivery associated microbiota
Parturition is the next major point of microbial exposure for the developed fetus. The first microbial contacts of a newborn outside of the uterus are influenced by the mode of delivery and the presence of microbial communities on the maternal skin and in the reproductive tract. While the composition of the neonatal microbiota across different body sites early after birth is not impacted by the delivery mode, gut bacterial communities display significant differences in babies born by either vaginal or Cesarean-section (CS) delivery.42–44 Vaginally delivered newborns display early enteric communities resembling the maternal vaginal and fecal microbiota, a result reflective of their prolonged transit through the birth canal. CS babies bypass contact with maternal mucosal surfaces and their initial microbial exposure consists of maternal skin bacteria and other environmental bacteria.45 The use of antibiotics perinatally by mother-infant dyads significantly impacts the neonatal microbiome and likely contributes to the decreased diversity of the enteric microbiota observed in CS infants.44,46 Studies have attempted to establish a correlation between mode of delivery to disease susceptibility in later life. CS babies may be at higher risk for developing allergies and asthma compared to vaginally delivered babies.47,48 A study of a Danish register-based cohort reported CS babies to have increased susceptibility for chronic inflammatory diseases including diabetes, arthritis, inflammatory bowel disease, and celiac disease.49 However, another study found no link between delivery mode and risk for celiac disease.50 Numerous factors accompanying birth may complicate interpretation of such studies, including the use of antibiotics, dominant mode of newborn nutrition immediately after birth and maternal stress and health.51 Further, while it is evident that the mode of delivery affects the quality of pioneer microbes introduced into newborns after birth, whether it also impacts subsequent patterns of colonization in the first 3 months of life remains unclear.45,52 Irrespective of birth mode, any observed differences in the earliest microbial community structure typically disappear after 6 months of life with the diversification of the infant diet.53,54
Postnatal microbiota
Nutrition, in the form of breastmilk, formula milk, and solid weaning foods, has a defining role in establishing colonization patterns of the infant gut microbiota after initial maternal inoculum at delivery.55,56 The TEDDY study established that breast milk consumption was the most significant factor associated with the microbiome structure early in life.57 Numerous epidemiologic studies have documented differences in the composition of the gut microbiota in breastfed and formula-fed infants.58,59 Breastmilk itself contains viable bacteria originating from the maternal gut and infant oral cavity that influences the composition of the enteric microbiome.60 Exclusive breast-feeding in the first 4 months of life was found to prevent atopic dermatitis (AD) in infants at high risk of atopy.61 However, in another study, breast-feeding for at least 3 months was not significantly protective against the development of AD compared with partial breast-feeding or use of conventional formula.62 In addition to nutrition, numerous environmental and host factors also contribute to the step-wise progression of the infant microbiome toward adult-like configurations that are typically reached by 2–3 years of age.63
Perinatal immune development
The early life mammalian immune system has evolved to develop in functional layers, likely to counter an array of exposures over ontogeny.64 Distinct hematopoietic stem cells give rise to unique lineages of lymphoid and myeloid cells at specified times during development, creating layers of immune cell populations that collectively endow the immune system with vast capabilities (Figure 2). Immune function in the perinatal period is defined by two sets of antigenic experiences. First, the immune system must tolerate continuous exposure to an array of non-inherited maternal antigens as well as a growing repertoire of self-antigens in utero to ensure fetal viability during pregnancy and to avoid autoimmunity postnatally.65 Second, it must enable the colonization of mucosal surfaces by trillions of bacteria, a process initiated immediately after birth, to ensure the establishment of a diverse, robust microbiota that is restrained and well tolerated.66 Numerous components of the innate and adaptive immune systems play specialized roles in these processes and cell subsets with suppressive activity as well as potent pro-inflammatory activity may be found at various tissue sites in the fetus and in the newborn. These mechanisms establish fetal tolerance and also prepare for the transition to the postnatal milieu to jumpstart an immune program that contends with the establishment of the microbiota and encounters with a multitude of antigens. Given these specific developmental demands on perinatal immunity, it is not surprising, therefore, that susceptibility to certain infections and severe inflammatory diseases such as necrotizing enterocolitis (NEC) and neonatal sepsis are a significant threat in the early life period.67 This susceptibility tracks with the development of the immune system, occurring within specific time windows that parallel waves of immune cell generation and tissue colonization.68 For example, while immune cells migrate to the skin and gut during gestation, lung resident Th2 effector cells and ILCs are only found after birth,69,70 contributing to the increased susceptibility to RSV infection in newborns.71,72 A discussion of the perinatal generation of immune lineages follows to provide a framework for assessing impact of microbial exposures on developmental immune imprinting.
Figure 2.
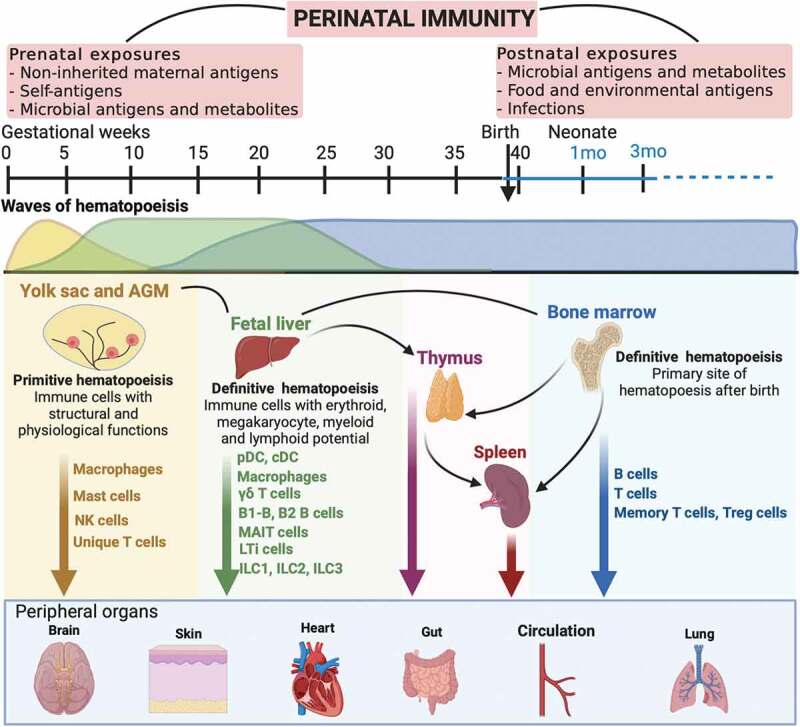
Perinatal immune development. Immune function in early life is shaped by specific prenatal and postnatal exposures. Waves of hematopoietic stem cells arise from the yolk sac, fetal liver and the bone marrow over gestation and generate distinct lineages of immune cells that serve the early life milieu.
Fetal immunity
Yolk sac and aorta-gonad mesonephros (AGM) hematopoiesis: Immune cells that arise from the first wave of hematopoiesis in the yolk sac have unique structural and physiological functions and equip the embryo with a repertoire of innate effectors consisting primarily of macrophages, mast cells, and Natural Killer (NK) cells. Long-lived macrophages generated from yolk-sac progenitors persist in tissues such as the liver, lung, brain, and epidermis,73,74 while those from HSC-derived monocytes are found in the gut, lung, and heart.75 Mast cells also display a similar ontogeny to macrophages and may arise from both yolk-sac and HSC-derived precursors that imprint them with distinct patterns of tissue residency.76,77 Pre-thymic lymphoid progenitors with long-term T cell engraftment capacity are found in the AGM region and arise before the appearance of embryonic HSCs in humans.78–80 In murine embryos, a subset of innate-like B cells called B-1 cells arise from hematogenic endothelia in both the yolk sac and the AGM region prior to HSCs, and seed the fetal liver.81
Fetal liver hematopoiesis: The next wave of hematopoiesis originates from the fetal liver where definitive HSCs with the full complement of erythroid, megakaryocyte, myeloid, and lymphoid potential may be found. Numerous innate and innate-like lymphocytes arise from common lymphoid progenitors in the fetal liver including innate lymphoid cells (ILCs), γδ T cells, and mucosal-associated invariant T cells (MAIT cells).82 While most mature ILC subsets are absent from lymphoid and non-lymphoid tissues such as the skin, kidneys, lungs, and spleen in early fetal development,82 ILC2s and ILC3s are present in the fetal gut.83,84 Antigen-presenting cells (APCs) including monocytes/macrophages, plasmacytoid dendritic cells (pDC), and conventional DC (cDC) subsets may be found in the fetal spleen, skin, thymus, and lungs by 12 weeks post-conception (PCW).82,85 HSC-derived innate B-1 cells in mice81 and adaptive B cell (B-2) precursors in humans are found in the fetal liver from 7PCW and mature B cells by 9PCW.82
Bone marrow (BM) hematopoiesis: At mid-gestation in humans and just before birth in mice, the BM becomes the primary site of hematopoiesis and the major source of B cells. B cells and follicular DCs form follicles in the fetal spleen,86,87 enabling antigen presentation to B cells. B cells are also found in the fetal intestine where they display phenotypes of follicular and transitional B cells but not plasma cells.84 While neutrophil progenitors abound in the fetal liver and BM, mature neutrophils are only found in the BM by 14 PCW where their numbers remain extremely low88 until immediately after birth.89,90
Thymus: The human thymus begins to develop by 8PCW and the first T cells appear in the periphery by 12–14 PCW.91,92 Thymus specific progenitors from the fetal liver and bone marrow seed the thymus and both γδ and αβ T cells including regulatory T (Treg) cells arise in the human fetus.93–96 In mice, however, except for the Vγ3/Vδ5 TCR expressing γδ T cell population that arises by embryonic day 15 (E15), most γδ and αβ T cell subsets arise in the thymus after birth in the first week of life.65,97 Memory T cells have also been found in the fetal spleen98,99 and in tissues including the skin100 and intestine,101 suggesting that complete T cell maturation can occur in utero. A unique feature of naïve T cells arising from fetal HSCs is their increased propensity to differentiate into Treg cells upon antigen encounter, a mechanism that likely establishes tolerance and prevents damaging alloreactivity in utero.102,103 CD1d-restricted Vα24+ invariant natural killer T (iNKT) cells are found in the early fetal thymus104 but their numbers decline with gestational age such that they are very rare in the neonatal thymus. However, iNKT cells are highly prevalent in the small intestines as well as the lungs, spleen, and mesenteric lymph nodes during the second trimester.105 MR-1 restricted MAIT cells are another class of innate-like T cells that have been found in the human fetal small intestine.106
Postnatal immunity
Early life represents a period of dramatic escalation in antigenic encounters by the relatively naïve and still developing newborn immune system. There is significant overlap of fetal and postnatally derived hematopoietic precursor cells in the period immediately after birth that influence the quality of immune cells generated and the overall functionality of the immune system. It is well established that newborns are highly susceptible to infections by pathogens and commensals, mostly due to insufficient, inappropriate, and biased immune responses. Dampened early life immune responses also necessitate pediatric vaccination strategies to include multi-dose immunization schedules with administration of doses later in infancy to increase immunogenicity and induce protective responses. However, overly robust immune responses are also observed in infants in developed countries, who have become increasingly sensitive to acquiring autoimmune diseases such as Type 1 Diabetes (T1D) as well as allergies and asthma. While the neonate has historically been considered to be immature in its immune characteristics, evidence clearly indicates that it is capable of mounting a range of pro- and anti-inflammatory responses.68,107,108
Innate immunity: Cells of the innate immune system are the first responders to the massive antigenic exposure that follows immediately after birth. The manner in which the innate immune system responds in early life is critical for the initiation and maintenance of host defense throughout life. The immune system recognizes microbial pathogens as well as commensal microbes via pattern-recognition receptors (PRRs) that are expressed on a variety of host cells including innate and adaptive immune cells.109 Appropriate regulation of these receptors is essential to prevent damaging inflammation at mucosal sites during the establishment of the commensal microbiota in early life as well as to generate productive responses against pathogenic viruses and bacteria. Failure of these mechanisms, especially in preterm infants, results in devastating disease such as neonatal sepsis and NEC as well as ineffective responses that hamper viral clearance.68 Decreased expression of TLR4 on monocytes from preterm and very-low birth weight infants and their subsequent lack of cytokine production has been suggested to contribute to susceptibility to infections with Gram-negative bacteria.110,111 The expression and cellular distribution of most other TLRs are similar between term newborn and adult monocytes.112 However, despite similar basal TLR expression, the functional consequences of TLR engagement in neonates are distinct. TLR stimulation of whole blood from preterm infants results in massive production of IL-10 whereas production of Th17 cell-promoting cytokines IL-6 and IL-23 dominates in term infants.112 pDCs are one of the major sources of Type 1 IFN following TLR triggering during viral infections and can be recruited efficiently to the respiratory tract of infants suffering from acute viral respiratory infections. However, they display dampened responses in RSV infected neonates and are defective in RIG-I dependent type 1 IFN responses.113 Interestingly, monocyte derived DCs as well as pDCs in human cord blood can potently produce IL-12p70 and Type 1 IFNs upon exposure to human CMV, HIV, and HSV.114–116 Thus, context dependent activation of TLRs on pDCs appears to impact innate immune responses to different viral triggers.
Adaptive Immunity: Neonatal adaptive immunity has been historically viewed as being deficient or deviant. Studies demonstrated that exposure to antigen before or during the neonatal period led to the inactivation on absence of T cells capable of mounting an immune response, a deficiency referred to as neonatal tolerance.117,118 Neonatal T cells were also shown to preferentially produce T helper type 2 (Th2) cytokines IL-4, IL-5, and IL-13 upon stimulation,119,120 a deviant response that increased susceptibility to infections and allergies during early stages of development.121 Accumulating evidence, however, has led to a revision of this paradigm and it is now appreciated that neonatal T cells are a distinct population of T cells with unique capacities that are well adapted to the perinatal period.122 Neonatal and adult T cells are derived from distinct progenitor cells that imprint in them specific functionalities; for example, a preference for Th2 and Treg cell differentiation in CD4+ T cells and the ability for rapid effector production in CD8+ T cells.103,123,124 Distinct gene expression profiles of neonatal T cells observed even prior to stimulation suggest that they may be more responsive upon antigen exposure.123,125–128 Neonatal T cells also respond to microbial ligands by virtue of their expression of numerous TLRs including TLR2 and TLR5.129–131 Thus, neonatal T cells appear wired to establish a state of tolerance to self and allogenic antigens at baseline homeostatic conditions, while simultaneously being poised to mount a rapid effector response upon infectious challenge or injury. B cell responses in the neonatal period are also distinct from adult. Fetal precursor cells preferentially give rise to B1-B cells that persist in the developing spleen at least 4 weeks after birth. These cells recognize self and microbial antigens and secrete low-affinity antibodies with broad specificities.132 The newborn B cell compartment primarily consists of naïve and transitional B cells and few memory cells.133,134
Suppressor cells: Specialized populations of suppressor cells are found in the early life period that control inflammation and also contribute to age-specific susceptibility to a pathogen and disease. IL-10 producing regulatory B cells are prevalent in human neonates and play a role in RSV clearance.135 Myeloid-derived suppressor cells are a heterogeneous population of granulocytic or monocytic cells that suppress innate and adaptive immune responses and play a role in establishing fetal tolerance in utero.136,137 CD71+ erythroid cells are enriched in the spleen of neonatal mice and in human cord blood and suppress both myeloid cell responses and T cell activation through their production of arginase 2.138
Microbial education of perinatal immunity
Unique forces drive early life immunity, and a microbial imprint of this system during the ‘window of opportunity’ may take the form of programming of either the development of immune cell subsets themselves or their functionality. Favorable microbial exposures could direct hematopoietic progenitors down immune developmental pathways that serve unique early life milieus. An alteration in their homeostasis and ability to precisely differentiate may result in losses in immune lineages, or conversely, may cause an increased persistence of fetal-derived immune cell subsets, both scenarios that would impact long-term immune repertoires. Encounters with microbes and microbial components in early life may also program the function of immune cells, some of which take up long-term residence in tissues, to influence host physiological processes and protect from disease throughout life. When and how microbial inputs are interpreted in these distinct early life environments may alter the nature of developmental immune layering, impact long-term immune function, and drive hypersensitivity.
Microbial inputs may originate either from endogenous compounds that are synthesized by microbes such as lipopolysaccharide (LPS) and flagellin or by dietary components that are metabolized by the microbiota, for example, SCFAs produced from dietary fiber. Broad manipulations of the microbiome with antibiotic depletion or germfree rearing have provided important insights into the contributions of the early life microbiome on immune development and later life disease susceptibility. However, microbiome interventions applied maternally can alter both the maternal and offspring microbiomes, rendering it difficult to distinguish gestational from neonatal influences on offspring immune outcomes. Identifying mechanisms by which specific microbes and microbial products affect offspring immune phenotypes may alleviate these concerns and numerous reductionist approaches in animal models have been employed to identify such causal relationships. These primarily involve either starting with an immune phenotype and narrowing down the microbiota to identify specific bacteria that influence the phenotype, or starting with bacteria-derived compounds and metabolites, and seeking to understand their effect on the immune system. A discussion of some of these studies follows.
Perinatal antibiotics and germfree animals
Epidemiologic and experimental data support a major impact of perinatal antibiotics on numerous aspects of immune system development and function that modulate susceptibility to inflammatory diseases in offspring. Antibiotic exposure in utero was positively associated with atopic dermatitis, food allergies and very-early onset IBD within the first year of life.139,140 Similarly, antepartum antibiotic exposure of pregnant C57BL/6 mice and colitis-prone IL-10-deficient mice led to altered intestinal bacterial colonization in offspring and increased susceptibility to DSS colitis in adulthood.141,142 We have shown that antibiotics exposure of pregnant C57BL/6 dams impacts the development of innate-like lymphocytes in the thymus of offspring that alters susceptibility to experimental colitis in later life.143 TF PLZF expressing thymic cells did not develop efficiently in mice that were treated with broad-spectrum antibiotics in the perinatal period, but mice treated in later life were spared. Microbe-induced altered development of thymic innate and innate-like cells in early life persisted into adulthood and contributed to increased susceptibility to DSS-induced colitis that could be reversed by the transfer of PLZF+ cells from mice that developed with normal microbiota in early life. Exposure of pregnant C57BL/6 mice to a cocktail of antibiotics decreased the numbers of circulating bone marrow neutrophils and granulocyte/macrophage-restricted progenitor cells (GMPs) in offspring that was associated with increased susceptibility to E. coli K1 and Klebsiella pneumoniae sepsis.89 The balance between Th17 and Treg cells, both important players in mucosal homeostasis,144 was also affected by antibiotics exposure of murine dams. There were reduced numbers of IL-17 producing cells in the intestinal lamina propria of offspring of antibiotic treated murine dams.89 In the non-obese diabetic (NOD) mouse model, antibiotics treatment of pregnant NOD mice also reduced the numbers of IL-17 producing CD4+ and CD8+ T cells in the mesenteric lymph nodes and Peyer’s patches with a concomitant increase in FOXP3+ Treg cells, that protected offspring from T1D.145–147 Interestingly, however, vancomycin treatment of C57BL/6 breeders reduced colonic Treg cells in offspring and increased their susceptibility to asthma.148 Impaired anti-viral responses and deficient production of IFNγ by CD8+ T cells were observed in neonatal offspring of antibiotic-treated murine dams.149 However, antibiotic treatment in the neonatal period lead to dysregulated vaccine responses with decreased IgG production and increased IFNγ production by T cells in adult mice.150
Similar to offspring of antibiotic treated murine dams, reduction in GMPs and lower plasma G-CSF levels were also observed in neonatal GF mice, suggesting that the immune impairments were due to alterations in the perinatal microbiome rather than confounding off-target effects of antibiotics.89 In GF mice, an early life defect in differentiation potential and self-maintenance capacity of bone marrow GMPs persisted into adulthood and could be restored partially by re-introduction of commensal microbes.151 GF mice also displayed increased serum IgE levels that were only normalized by conventionalization with normal microbiota before 4 weeks of age.152 Neonatal GF Swiss Webster (SW) mice but not GF C57BL/6 mice had higher numbers of iNKT cells in the colonic lamina propria, that increased their susceptibility in models of oxazolone-induced colitis and ovalbumin-induced allergic asthma.153 Elevations in colonic iNKT cells in GF mice were abrogated by re-introduction of conventional microbes at birth but not in adulthood, revealing an early life period during which the gut microbiota affects intestinal iNKT cell numbers. Early life microbes also regulate intestinal transcriptional programs, and alterations in expression of immune-related genes have been documented in GF mice that were only restored by colonization at birth but not in adulthood.154 Maternal microbes also imprint distinct patterns of gene expression in small intestinal mucosa of offspring in a mouse model of gestation-only microbial colonization.39
Endogenous microbial compounds
Endogenous microbial compounds can penetrate into the adult murine host, especially from bacteria in the lower small intestine.155 Isotope-labeling studies show the presence of microbial molecules of maternal origin in the placenta and the developing fetus as well as in breastmilk that drive the development of innate immune system and maturation of the intestinal epithelium in murine offspring.39 Microbial ligands for the aryl hydrocarbon receptor, which is expressed in the placenta, liver, and mucus membranes, promoted the development of intestinal NKp46+ ILC3s in the offspring. Components of bacterial cell wall and capsule are recognized by innate pattern recognition receptors expressed on a plethora of host cells in the intestines. Lipopolysaccharide (LPS) could restore impaired anti-viral CD8+ T cell responses in neonatal offspring of antibiotic-treated murine dams,149 suggesting that endogenous LPS from the neonatal microbiome might be required for proper CD8+ T cell function.156 LPS and microbial ligands for other PRRs have been shown to have diverse effect on central nervous system development and function.157 We showed that capsular polysaccharide A (PSA) from Bacteroides fragilis promoted the development of PLZF+ innate-like lymphocytes in the thymus of neonatal mice in a TLR2 dependent manner.143 B. fragilis sphingolipid also inhibited iNKT cell proliferation in the neonatal colon that protected from colitis in later life.158 Flagellin is a ligand for TLR5 that is expressed on intestinal epithelial cells primarily in early life.159 Neonatal flagellin and TLR5 interactions influence the composition of the murine microbiota throughout life and impact susceptibility to metabolic syndrome.160 In mice transgenic for a T cell receptor recognizing Lachnospiracea flagellin, antigen-specific RORγt+ Treg cells were generated in the early life period, specifically during the weaning process, and protected them from severe colitis later in life.161 In another TCR transgenic mouse model, recognition of a Staphylococcus epidermidis antigen to generate antigen-specific Treg cells also occurred during the second week of birth and was necessary to maintain tolerance to this antigen in adulthood.162 There is still a dearth of information on effector microbial products and further research is needed to understand the nature of specific microbial molecules that elicit immune modulatory effects in early life.
Diet and dietary components metabolized by microbiota
In animal models, maternal consumption of a high-fat diet (HFD) altered the microbiota and the nature of microbial exposures in both pregnant mothers and their offspring.163–165 Maternal obesity in murine dams fed a chronic HFD correlated with restricted expansion of hematopoietic stem and progenitor cells (HSPCs) in the fetal liver of offspring and altered lineage specification that resulted in distinct patterns of reconstitution upon adoptive transfer.166 Children with acute malnutrition also have impaired development of their gut microbiota (.167 Undernutrition in early life is associated with a number of adverse outcomes including immune dysfunction.168 IgA responses against specific bacterial taxa was altered in undernourished children that disrupted normal postnatal assembly of the microbiome and led to an enrichment of disease-promoting microbial communities including those containing members of Enterobacteriaceae and associated enteropathogenic strains.169 Further, transplantation of this bacterial consortium from undernourished children into gnotobiotic recipient altered gut barrier function and transmitted the weight loss phenotype.169 Dietary components may also be metabolized by the maternal and postnatal microbiota to influence immunity.170,171 It is estimated that there are 500–1000 microbial operating taxonomic units in the mammalian gut, giving the microbiome as a whole, an immense capacity to code for nutrient transporters and metabolizing enzymes.172 Maternal microbes can metabolize SCFAs by the fermentation of dietary fibers. SCFAs imprint the capacity to generate regulatory T cells in the fetal lung that protects adult murine offspring from house dust-mite induced allergy airway disease.40 Maternal supplementation with the SCFA acetate during pregnancy was also shown to rescue thymic and T cell developmental defects by upregulating AIRE (autoimmune regulator) expression and enhancing Treg cell generation in a mouse model of pre-eclampsia.173 Retinoic acid (RA) is a Vitamin A metabolite whose synthesis by intestinal epithelial cells is regulated by commensal bacteria.174 Maternal RA was shown to regulate CXCL13 production by murine fetal mesenchymal cells and the differentiation of lymphoid tissue inducer (LTi) cells.175 Vitamin B2/riboflavin metabolites produced by the microbiota before weaning promoted the development of MAIT cells in the murine thymus.176
Practical implications of the early life window of opportunity
It is evident that the immune system is amenable to beneficial programming by microbes in the perinatal period. Lack of appropriate microbial exposures, either due to microbial perturbation during pregnancy, antibiotics use in the perinatal period, malnutrition, or exposure to low diversity microbiota can lead to pathological imprinting that contributes to increased susceptibility to asthma, allergies, and other chronic inflammatory conditions.177,178 A practical approach to prevent such pathologies is to identify components of the microbiota that may be used clinically to ensure healthy imprinting in early life. A combination of probiotics, prebiotics, and immunomodulatory metabolites specific to the early life period may then be designed that promote favorable immune outcomes and decrease the incidence of inflammatory pathologies in later life. These may be administered to mothers during pregnancy, to mother-infant dyads postpartum, or directly to the infant.179 Another strategy is the transfer of mother’s vaginal and fecal microbiota at birth to introduce pioneer bacteria that build a robust microbial community that promotes offspring health. While antibiotic treatment during the neonatal period is causally linked to pathological imprinting, it is difficult to envision how one might avoid these treatments given their critical role in clearing infections in infants. Similarly, consumption of high-fat diets by mothers and weanlings can increase susceptibility to metabolic disorders; however, such high-calorie foods are crucial for meeting nutritional needs in developing countries. One strategy might be to prevent microbial alterations brought about by these treatments and behaviors with dietary prebiotics and probiotics.
Commonly administered probiotics belong to the genera Bifidobacterium and Lactobacillus and their use may prevent antibiotic-induced diarrhea and reduce the risk of NEC in preterm infants.180 However, no beneficial effect of probiotics in acute gastroenteritis in children was found.181 Bifidobacterium longum subsp. infantis EVC001 is a gut symbiont that can efficiently utilize human milk oligosaccharides (HMOs) into organic acids that lowers intestinal pH and can decrease markers of enteric inflammation in infants.182 In neonatal mice, colonization with Lactobacillus rhamnosus GG enhanced intestinal functional maturation and IgA production that protected from inflammation in later life.183 Probiotics may modulate the composition of the gut microbiota, enhance epithelial barrier function, and direct antagonism against pathogens. However, the precise mechanism of their action remains to be determined and is likely strain dependent. Prebiotics are substrates that are selectively utilized by host bacteria to confer a health benefit. Dietary fiber that is metabolized to SCFAs (acetate, propionate, butyrate) by gut microbes has been shown to influence microbial composition and the generation of immune cell subsets.184 Prebiotics such as short-chain galactooligosaccharides and long-chain fructooligosaccharides are routinely added to infant formula and have been shown to increase the abundance of Bifidobacterium.185 However, further studies are needed to determine their impact on immune function and the composition of the microbiota in early and later life. Crucial information is also needed on the possible long-term effects of early life pro- and pre-biotic administration on immune function and disease susceptibility. Probiotics in infant formula can deliver a large bacterial inoculum into a still-developing intestinal environment. Alterations in intestinal epithelial gene expression profiles, immune cell distribution, and function as well as the microbial landscape could potentially persist into adulthood leading to unwanted long-term health consequences.
Efficacy of vaccine responses can also be potentially programmed by intestinal microbes that may act as immunomodulators as well as natural adjuvants. Low antigen-specific plasma cell frequencies and IgG levels were observed in germfree and antibiotic-treated Tlr5−/- mice upon vaccination with a trivalent inactivated influenza vaccine, that was restored by reestablishment of the microbiota.186 Similarly, intestinal microbiota and recognition of the Nod2 sensor were required for inducing an immune response to cholera toxin in mice.187 In humans, variations in microbial communities may provide one explanation for the poor response to oral vaccines in low-income countries. While a causal link between specific community composition and vaccine responsiveness still needs to be established, some bacterial species have emerged as modulators of humoral and cellular immunity in responders. A robust IgA response to oral rotavirus vaccine in Ghanaian infants correlated with an increased relative abundance of stool Streptococcus bovis and a decreased abundance of Bacteroidetes.188 A higher polio-specific T cell response and serum IgG levels in Bangladeshi infants that received oral polio vaccine correlated with a higher abundance of Bifidobacteria and lower Pseudomonadales.189 Approaches to test the immunomodulatory and adjuvant effects of probiotics on beneficial vaccine responses need to be defined.190 As with any probiotic use in early life, choice of strain and dose and timing of administration must carefully be evaluated along with long-term health consequences.
Conclusion
In summary, the maternal and neonatal microbiomes are important regulators of health and influence the development of the early life immune system. The precise time frame for this ‘window of opportunity’ when immune cells are likely to be malleable to microbial influences is as yet undetermined. This period likely spans the time from fetal development to weaning or introduction of solid foods (Figure 3). Specific microbial encounters in the first 100 days of life were shown to influence the incidence of asthma in high-risk infants.177 In mice, a ‘weaning reaction’ encompassing the first 2–4 weeks of life was demonstrated to be critical for mitigating disease risk in adulthood.191 Distinct immune cell subsets are generated from precursor cells arising sequentially from the yolk sac, fetal liver, and bone marrow and may be subject to imprinting by specific exposures arising during these times. Animal models have been vital in providing insights into the perinatal development of the immune system. However, developmental timelines differ between species as do the quality of microbial exposures in the perinatal period. The influence of in utero microbial exposures on human fetal immune development and hematopoiesis remains largely unknown. Most studies have utilized cord blood at the time of parturition to make inferences on the earliest immune cells. However, human cord blood measurements are not representative of postnatal immunity, and display diverse immune phenotypes that appear to converge in the first few weeks of life.52 Further, while cell subsets such as B, NK, and DCs reach adult-like phenotypes during the first 3 months of life, T cell phenotypes do not converge during this period, suggesting that specific cell populations have different periods of calibration and time frames when they are amenable to microbial imprinting.52 Recent single-cell multi-omics technologies utilizing small numbers of cells from human fetal tissue have provided a new-detailed view of the complex immune development and structural and cellular communications occurring over ontogeny.82,192 More studies are needed to precisely define the temporal contributions of specific microbial species of maternal and neonatal origin on immune cells and their function.
Figure 3.
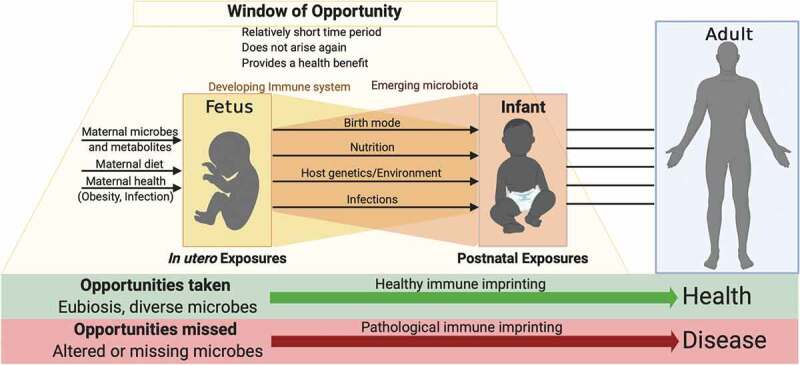
Of opportunities missed and taken. Early life microbial exposures influence developmental imprinting of the immune system that determines health and disease susceptibility in later life. Favorable imprinting of developing immunity by microbes may occur during an early life time window of opportunity. This window of opportunity, by definition, is predicted to be relatively short, to not arise again in later life and must provide a health benefit to the offspring. Numerous factors during pregnancy and in the postnatal period influence the quality of microbial exposures that have the potential to imprint immune functionality. Treatments and behaviors that disrupt these exposures may lead to pathological imprinting, altering risk for disease throughout life.
Reductionist approaches in animal models have been used to delineate causal relationships between early life microbes and disease susceptibility. However, while these studies have been and will continue to be critical for understanding basic mechanisms of early life host-microbiome interactions, how they may be translated to clinical applications and the development of microbiome based therapeutics will be key. Efforts to better model clinically relevant microbiome perturbations will be important as microbiome-based therapeutic strategies are developed. Applying pharmacologically relevant doses of antibiotics to animals, evaluating their off-target effects, and elucidating how microbiome perturbations might impact developmental processes will be important to evaluate safety, efficacy, and promise of such endeavors. Deeper mechanistic and functional insights into microbe-immune interactions at specific time periods are thus critical to inform disease mitigating strategies that modulate early life immune imprinting.
Acknowledgments
Thanks are due to Dr. Bobby Cherayil for providing critical feedback on this manuscript. NJ was supported by grants 1R21AI139735-01A1 and 1R01AI154626-01. Figures were created with BioRender.
Funding Statement
This work was supported by the National Institute of Allergy and Infectious Diseases [1R01AI154626-01]; National Institute of Allergy and Infectious Diseases [5R21AI139735-02].
Disclosure of potential conflicts of interest
The author declares no competing conflicts of interest.
References
- 1.W. H. O. C. o. C. A. o. P. I. Writing Committee , et al. Clinical aspects of pandemic 2009 influenza A (H1N1) virus infection. N Engl J Med. 2010;362:1708–1719. doi: 10.1056/NEJMra1000449. [DOI] [PubMed] [Google Scholar]
- 2.Brodin P. Why is COVID-19 so mild in children? Acta Paediatr. 2020;109:1082–1083. [DOI] [PubMed] [Google Scholar]
- 3.Rowley AH. Understanding SARS-CoV-2-related multisystem inflammatory syndrome in children. Nat Rev Immunol. 2020. doi: 10.1038/s41577-020-0367-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang KD. Perinatal programming of childhood asthma. Clin Dev Immunol. 2012;2012:438572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monasta L, Batty GD, Cattaneo A, Lutje V, Ronfani L, Van Lenthe FJ, et al. Early-life determinants of overweight and obesity: a review of systematic reviews. Obes Rev. 2010;11:695–708. [DOI] [PubMed] [Google Scholar]
- 6.Algert CS, McElduff A, Morris JM, Roberts CL. Perinatal risk factors for early onset of Type 1 diabetes in a 2000-2005 birth cohort. Diabet Med. 2009;26:1193–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gluckman PD, Hanson MA, Cooper C, Thornburg KL. Effect of in utero and early-life conditions on adult health and disease. N Engl J Med. 2008;359:61–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bateson P, Barker D, Clutton-Brock T, Deb D, D'Udine B, Foley RA, et al. Developmental plasticity and human health. Nature. 2004;430:419–421. [DOI] [PubMed] [Google Scholar]
- 9.Hirche C, Frenz T, Haas SF, Doring M, Borst K, Tegtmeyer PK, et al. Systemic Virus Infections Differentially Modulate Cell Cycle State and Functionality of Long-Term Hematopoietic Stem Cells In Vivo. Cell Rep. 2017;19:2345–2356. [DOI] [PubMed] [Google Scholar]
- 10.Matatall KA, Jeong M, Chen S, Sun D, Chen F, Mo Q, et al. Chronic Infection Depletes Hematopoietic Stem Cells through Stress-Induced Terminal Differentiation. Cell Rep. 2016;17:2584–2595. doi: 10.1016/j.celrep.2016.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haas S, Hansson J, Klimmeck D, Loeffler D, Velten L, Uckelmann H, et al. Inflammation-Induced Emergency Megakaryopoiesis Driven by Hematopoietic Stem Cell-like Megakaryocyte Progenitors. Cell Stem Cell. 2015;17:422–434. doi: 10.1016/j.stem.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 12.Lee YK, Mukasa R, Hatton RD, Weaver CT. Developmental plasticity of Th17 and Treg cells. Curr Opin Immunol. 2009;21:274–280. [DOI] [PubMed] [Google Scholar]
- 13.Zhou L, Chong MM, Littman DR. Plasticity of CD4+ T cell lineage differentiation. Immunity. 2009;30:646–655. [DOI] [PubMed] [Google Scholar]
- 14.Pullar J, Wickramasinghe K, Demaio AR, Roberts N, Perez-Blanco KM, Noonan K, et al. The impact of maternal nutrition on offspring’s risk of non-communicable diseases in adulthood: a systematic review. J Glob Health. 2019;9:020405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dauby N, Goetghebuer T, Kollmann TR, Levy J, Marchant A. Uninfected but not unaffected: chronic maternal infections during pregnancy, fetal immunity, and susceptibility to postnatal infections. Lancet Infect Dis. 2012;12:330–340. [DOI] [PubMed] [Google Scholar]
- 16.Zinkernagel RM. Maternal antibodies, childhood infections, and autoimmune diseases. N Engl J Med. 2001;345:1331–1335. [DOI] [PubMed] [Google Scholar]
- 17.Kearney JF, Patel P, Stefanov EK, King RG. Natural antibody repertoires: development and functional role in inhibiting allergic airway disease. Annu Rev Immunol. 2015;33:475–504. [DOI] [PubMed] [Google Scholar]
- 18.Figueiredo CA, Barreto ML, Rodrigues LC, Cooper PJ, Silva NB, Amorim LD, et al. Chronic intestinal helminth infections are associated with immune hyporesponsiveness and induction of a regulatory network. Infect Immun. 2010;78:3160–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Riet E, Hartgers FC, Yazdanbakhsh M. Chronic helminth infections induce immunomodulation: consequences and mechanisms. Immunobiology. 2007;212:475–490. [DOI] [PubMed] [Google Scholar]
- 20.Elias D, Britton S, Aseffa A, Engers H, Akuffo H. Poor immunogenicity of BCG in helminth infected population is associated with increased in vitro TGF-beta production. Vaccine. 2008;26:3897–3902. [DOI] [PubMed] [Google Scholar]
- 21.Strachan DP. Hay fever, hygiene, and household size. BMJ. 1989;299:1259–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kramer U, Heinrich J, Wjst M, Wichmann HE. Age of entry to day nursery and allergy in later childhood. Lancet. 1999;353:450–454. [DOI] [PubMed] [Google Scholar]
- 23.Nicolaou NC, Simpson A, Lowe LA, Murray CS, Woodcock A, Custovic A. Day-care attendance, position in sibship, and early childhood wheezing: a population-based birth cohort study. J Allergy Clin Immunol. 2008;122:500–506 e505. [DOI] [PubMed] [Google Scholar]
- 24.Ege MJ, Mayer M, Normand AC, Genuneit J, Cookson WO, Braun-Fahrlander C, et al. Exposure to environmental microorganisms and childhood asthma. N Engl J Med. 2011;364:701–709. [DOI] [PubMed] [Google Scholar]
- 25.Ege MJ, Herzum I, Buchele G, Krauss-Etschmann S, Lauener RP, Roponen M, et al. Prenatal exposure to a farm environment modifies atopic sensitization at birth. J Allergy Clin Immunol. 2008;122:407–412. [DOI] [PubMed] [Google Scholar]
- 26.Wood RA, Bloomberg GR, Kattan M, Conroy K, Sandel MT, Dresen A, et al. Relationships among environmental exposures, cord blood cytokine responses, allergy, and wheeze at 1 year of age in an inner-city birth cohort (Urban Environment and Childhood Asthma study). J Allergy Clin Immunol. 2011;127:913–919 e911-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riedler J, Braun-Fahrlander C, Eder W, Schreuer M, Waser M, Maisch S, et al. Exposure to farming in early life and development of asthma and allergy: a cross-sectional survey. Lancet. 2001;358:1129–1133. [DOI] [PubMed] [Google Scholar]
- 28.Al-Nasiry S, Ambrosino E, Schlaepfer M, Morre SA, Wieten L, Voncken JW, et al. The Interplay Between Reproductive Tract Microbiota and Immunological System in Human Reproduction. Front Immunol. 2020;11:378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fettweis JM, Serrano MG, Brooks JP, Edwards DJ, Girerd PH, Parikh HI, et al. The vaginal microbiome and preterm birth. Nat Med. 2019;25:1012–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gomez-Lopez N, StLouis D, Lehr MA, Sanchez-Rodriguez EN, Arenas-Hernandez M. Immune cells in term and preterm labor. Cell Mol Immunol. 2014;11:571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen HJ, Gur TL. Intrauterine Microbiota: missing, or the Missing Link? Trends Neurosci. 2019;42:402–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med. 2014;6:237ra265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Collado MC, Rautava S, Aakko J, Isolauri E, Salminen S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci Rep. 2016;6:23129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Goffau MC, Lager S, Sovio U, Gaccioli F, Cook E, Peacock SJ, et al. Human placenta has no microbiome but can contain potential pathogens. Nature. 2019;572:329–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lauder AP, Roche AM, Sherrill-Mix S, Bailey A, Laughlin AL, Bittinger K, et al. Comparison of placenta samples with contamination controls does not provide evidence for a distinct placenta microbiota. Microbiome. 2016;4:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rackaityte E, Halkias J, Fukui EM, Mendoza VF, Hayzelden C, Crawford ED, et al. Viable bacterial colonization is highly limited in the human intestine in utero. Nat Med. 2020;26:599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeng MY, Cisalpino D, Varadarajan S, Hellman J, Warren HS, Cascalho M, et al. Gut Microbiota-Induced Immunoglobulin G Controls Systemic Infection by Symbiotic Bacteria and Pathogens. Immunity. 2016;44:647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koch MA, Reiner GL, Lugo KA, Kreuk LS, Stanbery AG, Ansaldo E, et al. Maternal IgG and IgA Antibodies Dampen Mucosal T Helper Cell Responses in Early Life. Cell. 2016;165:827–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gomez de Aguero M, Ganal-Vonarburg SC, Fuhrer T, Rupp S, Uchimura Y, Li H, et al. The maternal microbiota drives early postnatal innate immune development. Science. 2016;351:1296–1302. [DOI] [PubMed] [Google Scholar]
- 40.Thorburn AN, McKenzie CI, Shen S, Stanley D, Macia L, Mason LJ, et al. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat Commun. 2015;6:7320. [DOI] [PubMed] [Google Scholar]
- 41.Humann J, Mann B, Gao G, Moresco P, Ramahi J, Loh LN, et al. Bacterial Peptidoglycan Traverses the Placenta to Induce Fetal Neuroproliferation and Aberrant Postnatal Behavior. Cell Host Microbe. 2016;19:388–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chu DM, Ma J, Prince AL, Antony KM, Seferovic MD, Aagaard KM. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat Med. 2017;23:314–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fouhy F, Watkins C, Hill CJ, O'Shea CA, Nagle B, Dempsey EM, et al. Perinatal factors affect the gut microbiota up to four years after birth. Nat Commun. 2019;10:1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rutayisire E, Huang K, Liu Y, Tao F. The mode of delivery affects the diversity and colonization pattern of the gut microbiota during the first year of infants’ life: a systematic review. BMC Gastroenterol. 2016;16:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107:11971–11975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi YC, Guo H, Chen J, Sun G, Ren RR, Guo MZ, et al. Initial meconium microbiome in Chinese neonates delivered naturally or by cesarean section. Sci Rep. 2018;8:3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bager P, Wohlfahrt J, Westergaard T. Caesarean delivery and risk of atopy and allergic disease: meta-analyses. Clin Exp Allergy. 2008;38:634–642. [DOI] [PubMed] [Google Scholar]
- 48.Negele K, Heinrich J, Borte M, von Berg A, Schaaf B, Lehmann I, et al. Mode of delivery and development of atopic disease during the first 2 years of life. Pediatr Allergy Immunol. 2004;15:48–54. [DOI] [PubMed] [Google Scholar]
- 49.Andersen V, Moller S, Jensen PB, Moller FT, Green A, Delivery C. Risk of Chronic Inflammatory Diseases (Inflammatory Bowel Disease, Rheumatoid Arthritis, Coeliac Disease, and Diabetes Mellitus): A Population Based Registry Study of 2,699,479 Births in Denmark During 1973-2016. Clin Epidemiol. 2020;12:287–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lionetti E, Castellaneta S, Francavilla R, Pulvirenti A, Catassi C, Weaning SWGo, et al. Mode of Delivery and Risk of Celiac Disease: risk of Celiac Disease and Age at Gluten Introduction Cohort Study. J Pediatr. 2017;184:81–86 e82. [DOI] [PubMed] [Google Scholar]
- 51.Mesa MD, Loureiro B, Iglesia I, Fernandez Gonzalez S, Llurba Olive E, Garcia Algar O, et al. The Evolving Microbiome from Pregnancy to Early Infancy: A Comprehensive Review. Nutrients. 2020;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Olin A, Henckel E, Chen Y, Lakshmikanth T, Pou C, Mikes J, et al. Stereotypic Immune System Development in Newborn Children. Cell. 2018;174:1277–1292 e1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stinson LF, Payne MS, Keelan JA, Critical A. Review of the Bacterial Baptism Hypothesis and the Impact of Cesarean Delivery on the Infant Microbiome. Front Med. 2018;5:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Backhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe. 2015;17:852. [DOI] [PubMed] [Google Scholar]
- 55.Jain N, Walker WA. Diet and host-microbial crosstalk in postnatal intestinal immune homeostasis. Nat Rev Gastroenterol Hepatol. 2015;12:14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Flint HJ, Scott KP, Louis P, Duncan SH. The role of the gut microbiota in nutrition and health. Nat Rev Gastroenterol Hepatol. 2012;9:577–589. [DOI] [PubMed] [Google Scholar]
- 57.Stewart CJ, Ajami NJ, O'Brien JL, Hutchinson DS, Smith DP, Wong MC, et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature. 2018;562:583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Azad MB, Konya T, Maughan H, Guttman DS, Field CJ, Chari RS, et al. Gut microbiota of healthy Canadian infants: profiles by mode of delivery and infant diet at 4 months. CMAJ. 2013;185:385–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gomez-Llorente C, Plaza-Diaz J, Aguilera M, Munoz-Quezada S, Bermudez-Brito M, Peso-Echarri P, et al. Three main factors define changes in fecal microbiota associated with feeding modality in infants. J Pediatr Gastroenterol Nutr. 2013;57:461–466. [DOI] [PubMed] [Google Scholar]
- 60.Moossavi S, Sepehri S, Robertson B, Bode L, Goruk S, Field CJ, et al. Composition and Variation of the Human Milk Microbiota Are Influenced by Maternal and Early-Life Factors. Cell Host Microbe. 2019;25:324–335 e324. [DOI] [PubMed] [Google Scholar]
- 61.Schoetzau A, Filipiak-Pittroff B, Franke K, Koletzko S, Von Berg A, Gruebl A, et al. Effect of exclusive breast-feeding and early solid food avoidance on the incidence of atopic dermatitis in high-risk infants at 1 year of age. Pediatr Allergy Immunol. 2002;13:234–242. [DOI] [PubMed] [Google Scholar]
- 62.Yang YW, Tsai CL, Lu CY. Exclusive breastfeeding and incident atopic dermatitis in childhood: a systematic review and meta-analysis of prospective cohort studies. Br J Dermatol. 2009;161:373–383. [DOI] [PubMed] [Google Scholar]
- 63.Tamburini S, Shen N, Wu HC, Clemente JC. The microbiome in early life: implications for health outcomes. Nat Med. 2016;22:713–722. [DOI] [PubMed] [Google Scholar]
- 64.Herzenberg LA, Herzenberg LA. Toward a layered immune system. Cell. 1989;59:953–954. [DOI] [PubMed] [Google Scholar]
- 65.Mold JE, McCune JM. Immunological tolerance during fetal development: from mouse to man. Adv Immunol. 2012;115:73–111. [DOI] [PubMed] [Google Scholar]
- 66.Maynard CL, Elson CO, Hatton RD, Weaver CT. Reciprocal interactions of the intestinal microbiota and immune system. Nature. 2012;489:231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kollmann TR, Kampmann B, Mazmanian SK, Marchant A, Levy O. Protecting the Newborn and Young Infant from Infectious Diseases: lessons from Immune Ontogeny. Immunity. 2017;46:350–363. [DOI] [PubMed] [Google Scholar]
- 68.Zhang X, Zhivaki D, Lo-Man R. Unique aspects of the perinatal immune system. Nat Rev Immunol. 2017;17:495–507. [DOI] [PubMed] [Google Scholar]
- 69.de Kleer IM, Kool M, de Bruijn MJ, Willart M, van Moorleghem J, Schuijs MJ, et al. Perinatal Activation of the Interleukin-33 Pathway Promotes Type 2 Immunity in the Developing Lung. Immunity. 2016;45:1285–1298. [DOI] [PubMed] [Google Scholar]
- 70.Saluzzo S, Gorki AD, Rana BMJ, Martins R, Scanlon S, Starkl P, et al. First-Breath-Induced Type 2 Pathways Shape the Lung Immune Environment. Cell Rep. 2017;18:1893–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Forsberg A, Bengtsson M, Eringfalt A, Ernerudh J, Mjosberg J, Jenmalm MC. GATA binding protein 3(+) group 2 innate lymphoid cells are present in cord blood and in higher proportions in male than in female neonates. J Allergy Clin Immunol. 2014;134:228–230. [DOI] [PubMed] [Google Scholar]
- 72.Cohen C, Walaza S, Moyes J, Groome M, Tempia S, Pretorius M, et al. Epidemiology of viral-associated acute lower respiratory tract infection among children <5 years of age in a high HIV prevalence setting, South Africa, 2009-2012. Pediatr Infect Dis J. 2015;34:66–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gomez Perdiguero E, Klapproth K, Schulz C, Busch K, Azzoni E, Crozet L, et al. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature. 2015;518:547–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hoeffel G, Chen J, Lavin Y, Low D, Almeida FF, See P, et al. C-Myb(+) erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity. 2015;42:665–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ginhoux F, Guilliams M. Tissue-Resident Macrophage Ontogeny and Homeostasis. Immunity. 2016;44:439–449. [DOI] [PubMed] [Google Scholar]
- 76.Gentek R, Ghigo C, Hoeffel G, Bulle MJ, Msallam R, Gautier G, et al. Hemogenic Endothelial Fate Mapping Reveals Dual Developmental Origin of Mast Cells. Immunity. 2018;48:1160–1171 e1165. [DOI] [PubMed] [Google Scholar]
- 77.Li Z, Liu S, Xu J, Zhang X, Han D, Liu J, et al. Adult Connective Tissue-Resident Mast Cells Originate from Late Erythro-Myeloid Progenitors. Immunity. 2018;49:640–653 e645. [DOI] [PubMed] [Google Scholar]
- 78.Yoshimoto M, Porayette P, Glosson NL, Conway SJ, Carlesso N, Cardoso AA,et al. Autonomous murine T-cell progenitor production in the extra-embryonic yolk sac before HSC emergence. Blood. 2012;119:5706–5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tavian M, Robin C, Coulombel L, Peault B. The human embryo, but not its yolk sac, generates lympho-myeloid stem cells: mapping multipotent hematopoietic cell fate in intraembryonic mesoderm. Immunity. 2001;15:487–495. [DOI] [PubMed] [Google Scholar]
- 80.Ivanovs A, Rybtsov S, Welch L, Anderson RA, Turner ML, Medvinsky A. Highly potent human hematopoietic stem cells first emerge in the intraembryonic aorta-gonad-mesonephros region. J Exp Med. 2011;208:2417–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yoshimoto M. The first wave of B lymphopoiesis develops independently of stem cells in the murine embryo. Ann N Y Acad Sci. 2015;1362:16–22. [DOI] [PubMed] [Google Scholar]
- 82.Popescu DM, Botting RA, Stephenson E, Green K, Webb S, Jardine L, et al. Decoding human fetal liver haematopoiesis. Nature. 2019;574:365–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bernink JH, Peters CP, Munneke M, te Velde AA, Meijer SL, Weijer K, et al. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat Immunol. 2013;14:221–229. [DOI] [PubMed] [Google Scholar]
- 84.Stras SF, Werner L, Toothaker JM, Olaloye OO, Oldham AL, McCourt CC, et al. Maturation of the Human Intestinal Immune System Occurs Early in Fetal Development. Dev Cell. 2019;51:357–373 e355. [DOI] [PubMed] [Google Scholar]
- 85.McGovern N, Shin A, Low G, Low D, Duan K, Yao LJ, et al. Human fetal dendritic cells promote prenatal T-cell immune suppression through arginase-2. Nature. 2017;546:662–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Namikawa R, Mizuno T, Matsuoka H, Fukami H, Ueda R, Itoh G, et al. Ontogenic development of T and B cells and non-lymphoid cells in the white pulp of human spleen. Immunology. 1986;57:61–69. [PMC free article] [PubMed] [Google Scholar]
- 87.Timens W, Rozeboom T, Poppema S. Fetal and neonatal development of human spleen: an immunohistological study. Immunology. 1987;60:603–609. [PMC free article] [PubMed] [Google Scholar]
- 88.Ohls RK, Li Y, Abdel-Mageed A, Buchanan G, Jr., Mandell L, Christensen RD. Neutrophil pool sizes and granulocyte colony-stimulating factor production in human mid-trimester fetuses. Pediatr Res. 1995;37:806–811. [DOI] [PubMed] [Google Scholar]
- 89.Deshmukh HS, Liu Y, Menkiti OR, Mei J, Dai N, O'Leary CE, et al. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat Med. 2014;20:524–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Manroe BL, Weinberg AG, Rosenfeld CR, Browne R. The neonatal blood count in health and disease. I. Reference values for neutrophilic cells. J Pediatr. 1979;95:89–98. [DOI] [PubMed] [Google Scholar]
- 91.Haynes BF, Heinly CS. Early human T cell development: analysis of the human thymus at the time of initial entry of hematopoietic stem cells into the fetal thymic microenvironment. J Exp Med. 1995;181:1445–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Farley AM, Morris LX, Vroegindeweij E, Depreter ML, Vaidya H, Stenhouse FH, et al. Dynamics of thymus organogenesis and colonization in early human development. Development. 2013;140:2015–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Havran WL, Allison JP. Developmentally ordered appearance of thymocytes expressing different T-cell antigen receptors. Nature. 1988;335:443–445. [DOI] [PubMed] [Google Scholar]
- 94.Ikuta K, Kina T, MacNeil I, Uchida N, Peault B, Chien YH, et al. A developmental switch in thymic lymphocyte maturation potential occurs at the level of hematopoietic stem cells. Cell. 1990;62:863–874. [DOI] [PubMed] [Google Scholar]
- 95.Cupedo T, Nagasawa M, Weijer K, Blom B, Spits H. Development and activation of regulatory T cells in the human fetus. Eur J Immunol. 2005;35:383–390. [DOI] [PubMed] [Google Scholar]
- 96.Darrasse-Jeze G, Marodon G, Salomon BL, Catala M, Klatzmann D. Ontogeny of CD4+CD25+ regulatory/suppressor T cells in human fetuses. Blood. 2005;105:4715–4721. [DOI] [PubMed] [Google Scholar]
- 97.Friedberg SH, Weissman IL. Lymphoid tissue architecture. II. Ontogeny of peripheral T and B cells in mice: evidence against Peyer’s patches as the site of generation of B cells. J Immunol. 1974;113:1477–1492. [PubMed] [Google Scholar]
- 98.Byrne JA, Stankovic AK, Cooper MD. A novel subpopulation of primed T cells in the human fetus. J Immunol. 1994;152:3098–3106. [PubMed] [Google Scholar]
- 99.Michaelsson J, Mold JE, McCune JM, Nixon DF. Regulation of T cell responses in the developing human fetus. J Immunol. 2006;176:5741–5748. [DOI] [PubMed] [Google Scholar]
- 100.Schuster C, Vaculik C, Prior M, Fiala C, Mildner M, Eppel W, et al. Phenotypic characterization of leukocytes in prenatal human dermis. J Invest Dermatol. 2012;132:2581–2592. doi: 10.1038/jid.2012.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bunders MJ, van der Loos CM, Klarenbeek PL, van Hamme JL, Boer K, Wilde JC, et al. Memory CD4(+)CCR5(+) T cells are abundantly present in the gut of newborn infants to facilitate mother-to-child transmission of HIV-1. Blood. 2012;120:4383–4390. doi: 10.1182/blood-2012-06-437566. [DOI] [PubMed] [Google Scholar]
- 102.Mold JE, Michaelsson J, Burt TD, Muench MO, Beckerman KP, Busch MP, et al. Maternal alloantigens promote the development of tolerogenic fetal regulatory T cells in utero. Science. 2008;322:1562–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mold JE, Venkatasubrahmanyam S, Burt TD, Michaelsson J, Rivera JM, Galkina SA, et al. Fetal and adult hematopoietic stem cells give rise to distinct T cell lineages in humans. Science. 2010;330:1695–1699. doi: 10.1126/science.1196509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sandberg JK, Stoddart CA, Brilot F, Jordan KA, Nixon DF. Development of innate CD4+ alpha-chain variable gene segment 24 (Valpha24) natural killer T cells in the early human fetal thymus is regulated by IL-7. Proc Natl Acad Sci U S A. 2004;101:7058–7063. doi: 10.1073/pnas.0305986101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Loh L, Ivarsson MA, Michaelsson J, Sandberg JK, Nixon DF. Invariant natural killer T cells developing in the human fetus accumulate and mature in the small intestine. Mucosal Immunol. 2014;7:1233–1243. doi: 10.1038/mi.2014.13. [DOI] [PubMed] [Google Scholar]
- 106.Leeansyah E, Loh L, Nixon DF, Sandberg JK. Acquisition of innate-like microbial reactivity in mucosal tissues during human fetal MAIT-cell development. Nat Commun. 2014;5:3143. doi: 10.1038/ncomms4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Adkins B, Leclerc C, Marshall-Clarke S. Neonatal adaptive immunity comes of age. Nat Rev Immunol. 2004;4:553–564. [DOI] [PubMed] [Google Scholar]
- 108.Georgountzou A, Papadopoulos NG. Postnatal Innate Immune Development: from Birth to Adulthood. Front Immunol. 2017;8:957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Iwasaki A, Medzhitov R. Control of adaptive immunity by the innate immune system. Nat Immunol. 2015;16:343–353. doi: 10.1038/ni.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sadeghi K, Berger A, Langgartner M, Prusa AR, Hayde M, Herkner K, et al. Immaturity of infection control in preterm and term newborns is associated with impaired toll-like receptor signaling. J Infect Dis. 2007;195:296–302. doi: 10.1086/509892. [DOI] [PubMed] [Google Scholar]
- 111.Forster-Waldl E, Sadeghi K, Tamandl D, Gerhold B, Hallwirth U, Rohrmeister K, et al. Monocyte toll-like receptor 4 expression and LPS-induced cytokine production increase during gestational aging. Pediatr Res. 2005;58:121–124. [DOI] [PubMed] [Google Scholar]
- 112.Kollmann TR, Levy O, Montgomery RR, Goriely S. Innate immune function by Toll-like receptors: distinct responses in newborns and the elderly. Immunity. 2012;37:771–783. doi: 10.1016/j.immuni.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Marr N, Wang TI, Kam SH, Hu YS, Sharma AA, Lam A, et al. Attenuation of respiratory syncytial virus-induced and RIG-I-dependent type I IFN responses in human neonates and very young children. J Immunol. 2014;192:948–957. doi: 10.4049/jimmunol.1302007. [DOI] [PubMed] [Google Scholar]
- 114.Renneson J, Dutta B, Goriely S, Danis B, Lecomte S, Laes JF, et al. IL-12 and type I IFN response of neonatal myeloid DC to human CMV infection. Eur J Immunol. 2009;39:2789–2799. [DOI] [PubMed] [Google Scholar]
- 115.Zhang X, Casartelli N, Lemoine S, Mozeleski B, Azria E, Le Ray C, Schwartz O, Launay O, Leclerc C, Lo-Man R, et al. Plasmacytoid dendritic cells engagement by influenza vaccine as a surrogate strategy for driving T-helper type 1 responses in human neonatal settings. J Infect Dis. 2014;210(3):424–434. doi: 10.1093/infdis/jiu103. [DOI] [PubMed] [Google Scholar]
- 116.Zhang X, Lepelley A, Azria E, Lebon P, Roguet G, Schwartz O, et al. Neonatal plasmacytoid dendritic cells (pDCs) display subset variation but can elicit potent anti-viral innate responses. PLoS One. 2013;8:e52003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Billingham RE, Brent L, Medawar PB. Actively acquired tolerance of foreign cells. Nature. 1953;172:603–606. [DOI] [PubMed] [Google Scholar]
- 118.Gammon G, Dunn K, Shastri N, Oki A, Wilbur S, Sercarz EE. Neonatal T-cell tolerance to minimal immunogenic peptides is caused by clonal inactivation. Nature. 1986;319:413–415. [DOI] [PubMed] [Google Scholar]
- 119.Forsthuber T, Yip HC, Lehmann PV. Induction of TH1 and TH2 immunity in neonatal mice. Science. 1996;271:1728–1730. [DOI] [PubMed] [Google Scholar]
- 120.Chen N, Field EH. Enhanced type 2 and diminished type 1 cytokines in neonatal tolerance. Transplantation. 1995;59:933–941. [DOI] [PubMed] [Google Scholar]
- 121.Prescott SL, Macaubas C, Smallacombe T, Holt BJ, Sly PD, Holt PG. Development of allergen-specific T-cell memory in atopic and normal children. Lancet. 1999;353:196–200. [DOI] [PubMed] [Google Scholar]
- 122.Rudd BD. Neonatal T Cells: A Reinterpretation. Annu Rev Immunol. 2020;38:229–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Smith NL, Patel RK, Reynaldi A, Grenier JK, Wang J, Watson NB, et al. Developmental Origin Governs CD8(+) T Cell Fate Decisions during Infection. Cell. 2018;174:117–130 e114. [DOI] [PubMed] [Google Scholar]
- 124.Wang J, Wissink EM, Watson NB, Smith NL, Grimson A, Rudd BD. Fetal and adult progenitors give rise to unique populations of CD8+ T cells. Blood. 2016;128:3073–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zens KD, Chen JK, Guyer RS, Wu FL, Cvetkovski F, Miron M, et al. Reduced generation of lung tissue-resident memory T cells during infancy. J Exp Med. 2017;214:2915–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wissink EM, Smith NL, Spektor R, Rudd BD, Grimson A. MicroRNAs and Their Targets Are Differentially Regulated in Adult and Neonatal Mouse CD8+ T Cells. Genetics. 2015;201:1017–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yu HR, Hsu TY, Huang HC, Kuo HC, Li SC, Yang KD, et al. Comparison of the Functional microRNA Expression in Immune Cell Subsets of Neonates and Adults. Front Immunol. 2016;7:615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Palin AC, Ramachandran V, Acharya S, Lewis DB. Human neonatal naive CD4+ T cells have enhanced activation-dependent signaling regulated by the microRNA miR-181a. J Immunol. 2013;190:2682–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.McCarron M, Reen DJ. Activated human neonatal CD8+ T cells are subject to immunomodulation by direct TLR2 or TLR5 stimulation. J Immunol. 2009;182:55–62. [DOI] [PubMed] [Google Scholar]
- 130.Komai-Koma M, Jones L, Ogg GS, Xu D, Liew FY. TLR2 is expressed on activated T cells as a costimulatory receptor. Proc Natl Acad Sci U S A. 2004;101:3029–3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sinnott BD, Park B, Boer MC, Lewinsohn DA, Lancioni CL, Direct TLR-2. Costimulation Unmasks the Proinflammatory Potential of Neonatal CD4+ T Cells. J Immunol. 2016;197:68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hardy RR, Hayakawa K. B cell development pathways. Annu Rev Immunol. 2001;19:595–621. [DOI] [PubMed] [Google Scholar]
- 133.Duchamp M, Sterlin D, Diabate A, Uring-Lambert B, Guerin-El Khourouj V, Le Mauff B, et al. B-cell subpopulations in children: national reference values. Immun Inflamm Dis. 2014;2:131–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Piatosa B, Wolska-Kusnierz B, Pac M, Siewiera K, Galkowska E, Bernatowska E. B cell subsets in healthy children: reference values for evaluation of B cell maturation process in peripheral blood. Cytometry B Clin Cytom. 2010;78:372–381. [DOI] [PubMed] [Google Scholar]
- 135.Zhivaki D, Lemoine S, Lim A, Morva A, Vidalain PO, Schandene L, et al. Respiratory Syncytial Virus Infects Regulatory B Cells in Human Neonates via Chemokine Receptor CX3CR1 and Promotes Lung Disease Severity. Immunity. 2017;46:301–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rieber N, Gille C, Kostlin N, Schafer I, Spring B, Ost M, et al. Neutrophilic myeloid-derived suppressor cells in cord blood modulate innate and adaptive immune responses. Clin Exp Immunol. 2013;174:45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gervassi A, Lejarcegui N, Dross S, Jacobson A, Itaya G, Kidzeru E, et al. Myeloid derived suppressor cells are present at high frequency in neonates and suppress in vitro T cell responses. PLoS One. 2014;9:e107816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Elahi S, Ertelt JM, Kinder JM, Jiang TT, Zhang X, Xin L, et al. Immunosuppressive CD71+ erythroid cells compromise neonatal host defence against infection. Nature. 2013;504:158–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Metzler S, Frei R, Schmausser-Hechfellner E, von Mutius E, Pekkanen J, Karvonen AM, et al. Association between antibiotic treatment during pregnancy and infancy and the development of allergic diseases. Pediatr Allergy Immunol. 2019;30:423–433. [DOI] [PubMed] [Google Scholar]
- 140.Ortqvist AK, Lundholm C, Halfvarson J, Ludvigsson JF, Almqvist C. Fetal and early life antibiotics exposure and very early onset inflammatory bowel disease: a population-based study. Gut. 2019;68:218–225. [DOI] [PubMed] [Google Scholar]
- 141.Munyaka PM, Eissa N, Bernstein CN, Khafipour E, Ghia JE. Antepartum Antibiotic Treatment Increases Offspring Susceptibility to Experimental Colitis: A Role of the Gut Microbiota. PLoS One. 2015;10:e0142536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Miyoshi J, Bobe AM, Miyoshi S, Huang Y, Hubert N, Delmont TO, et al. Peripartum Antibiotics Promote Gut Dysbiosis, Loss of Immune Tolerance, and Inflammatory Bowel Disease in Genetically Prone Offspring. Cell Rep. 2017;20:491–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ennamorati M, Vasudevan C, Clerkin K, Halvorsen S, Verma S, Ibrahim S, et al. Intestinal microbes influence development of thymic lymphocytes in early life. Proc Natl Acad Sci U S A. 2020;117:2570–2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pandiyan P, Bhaskaran N, Zou M, Schneider E, Jayaraman S, Huehn J. Microbiome Dependent Regulation of Tregs and Th17 Cells in Mucosa. Front Immunol. 2019;10:426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Candon S, Perez-Arroyo A, Marquet C, Valette F, Foray AP, Pelletier B, et al. Antibiotics in early life alter the gut microbiome and increase disease incidence in a spontaneous mouse model of autoimmune insulin-dependent diabetes. PLoS One. 2015;10:e0125448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hu Y, Peng J, Tai N, Hu C, Zhang X, Wong FS, et al. Maternal Antibiotic Treatment Protects Offspring from Diabetes Development in Nonobese Diabetic Mice by Generation of Tolerogenic APCs. J Immunol. 2015;195:4176–4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Tormo-Badia N, Hakansson A, Vasudevan K, Molin G, Ahrne S, Cilio CM. Antibiotic treatment of pregnant non-obese diabetic mice leads to altered gut microbiota and intestinal immunological changes in the offspring. Scand J Immunol. 2014;80:250–260. [DOI] [PubMed] [Google Scholar]
- 148.Russell SL, Gold MJ, Hartmann M, Willing BP, Thorson L, Wlodarska M, et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 2012;13:440–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gonzalez-Perez G, Hicks AL, Tekieli TM, Radens CM, Williams BL, Lamouse-Smith ES. Maternal Antibiotic Treatment Impacts Development of the Neonatal Intestinal Microbiome and Antiviral Immunity. J Immunol. 2016;196:3768–3779. [DOI] [PubMed] [Google Scholar]
- 150.Lynn MA, Tumes DJ, Choo JM, Sribnaia A, Blake SJ, Leong LEX, et al. Early-Life Antibiotic-Driven Dysbiosis Leads to Dysregulated Vaccine Immune Responses in Mice. Cell Host Microbe. 2018;23:653–660 e655. [DOI] [PubMed] [Google Scholar]
- 151.Khosravi A, Yanez A, Price JG, Chow A, Merad M, Goodridge HS, et al. Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe. 2014;15:374–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Cahenzli J, Koller Y, Wyss M, Geuking MB, McCoy KD. Intestinal microbial diversity during early-life colonization shapes long-term IgE levels. Cell Host Microbe. 2013;14:559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.El Aidy S, Hooiveld G, Tremaroli V, Backhed F, Kleerebezem M. The gut microbiota and mucosal homeostasis: colonized at birth or at adulthood, does it matter? Gut Microbes. 2013;4:118–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Uchimura Y, Fuhrer T, Li H, Lawson MA, Zimmermann M, Yilmaz B, et al. Antibodies Set Boundaries Limiting Microbial Metabolite Penetration and the Resultant Mammalian Host Response. Immunity. 2018;49:545–559 e545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Gonzalez-Perez G, Lamouse-Smith ES. Gastrointestinal Microbiome Dysbiosis in Infant Mice Alters Peripheral CD8(+) T Cell Receptor Signaling. Front Immunol. 2017;8:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Pronovost GN, Hsiao EY. Perinatal Interactions between the Microbiome, Immunity, and Neurodevelopment. Immunity. 2019;50:18–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.An D, Oh SF, Olszak T, Neves JF, Avci FY, Erturk-Hasdemir D, et al. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell. 2014;156:123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Fulde M, Sommer F, Chassaing B, van Vorst K, Dupont A, Hensel M, et al. Neonatal selection by Toll-like receptor 5 influences long-term gut microbiota composition. Nature. 2018;560:489–493. [DOI] [PubMed] [Google Scholar]
- 160.Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ohnmacht C, Park JH, Cording S, Wing JB, Atarashi K, Obata Y, et al. MUCOSAL IMMUNOLOGY. The microbiota regulates type 2 immunity through RORgammat(+) T cells. Science. 2015;349:989–993. [DOI] [PubMed] [Google Scholar]
- 162.Scharschmidt TC, Vasquez KS, Truong HA, Gearty SV, Pauli ML, Nosbaum A, et al. A Wave of Regulatory T Cells into Neonatal Skin Mediates Tolerance to Commensal Microbes. Immunity. 2015;43:1011–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chu DM, Meyer KM, Prince AL, Aagaard KM. Impact of maternal nutrition in pregnancy and lactation on offspring gut microbial composition and function. Gut Microbes. 2016;7:459–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Rando OJ, Simmons RA. I’m eating for two: parental dietary effects on offspring metabolism. Cell. 2015;161:93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Babu ST, Niu X, Raetz M, Savani RC, Hooper LV, Mirpuri J. Maternal high-fat diet results in microbiota-dependent expansion of ILC3s in mice offspring. JCI Insight. 2018;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kamimae-Lanning AN, Krasnow SM, Goloviznina NA, Zhu X, Roth-Carter QR, Levasseur PR, et al. Maternal high-fat diet and obesity compromise fetal hematopoiesis. Mol Metab. 2015;4:25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Subramanian S, Huq S, Yatsunenko T, Haque R, Mahfuz M, Alam MA, et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. 2014;510:417–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Prendergast AJ, Humphrey JH. The stunting syndrome in developing countries. Paediatr Int Child Health. 2014;34:250–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kau AL, Planer JD, Liu J, Rao S, Yatsunenko T, Trehan I, et al. Functional characterization of IgA-targeted bacterial taxa from undernourished Malawian children that produce diet-dependent enteropathy. Sci Transl Med. 2015;7:276ra224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ganal-Vonarburg SC, Hornef MW, Macpherson AJ. Microbial-host molecular exchange and its functional consequences in early mammalian life. Science. 2020;368:604–607. [DOI] [PubMed] [Google Scholar]
- 171.Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol. 2016;16:341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kim CH. Immune regulation by microbiome metabolites. Immunology. 2018;154:220–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hu M, Eviston D, Hsu P, Marino E, Chidgey A, Santner-Nanan B, et al. Decreased maternal serum acetate and impaired fetal thymic and regulatory T cell development in preeclampsia. Nat Commun. 2019;10:3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Grizotte-Lake M, Zhong G, Duncan K, Kirkwood J, Iyer N, Smolenski I, et al. Commensals Suppress Intestinal Epithelial Cell Retinoic Acid Synthesis to Regulate Interleukin-22 Activity and Prevent Microbial Dysbiosis. Immunity. 2018;49:1103–1115 e1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.van de Pavert SA, Ferreira M, Domingues RG, Ribeiro H, Molenaar R, Moreira-Santos L, et al. Maternal retinoids control type 3 innate lymphoid cells and set the offspring immunity. Nature. 2014;508:123–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Constantinides MG, Link VM, Tamoutounour S, Wong AC, Perez-Chaparro PJ, Han SJ, et al. MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science. 2019;366:445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7:307ra152. [DOI] [PubMed] [Google Scholar]
- 178.Russell SL, Gold MJ, Willing BP, Thorson L, McNagny KM, Finlay BB. Perinatal antibiotic treatment affects murine microbiota, immune responses and allergic asthma. Gut Microbes. 2013;4:158–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Davis EC, Dinsmoor AM, Wang M, Donovan SM. Microbiome Composition in Pediatric Populations from Birth to Adolescence: impact of Diet and Prebiotic and Probiotic Interventions. Dig Dis Sci. 2020;65:706–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Sansotta N, Peroni DG, Romano S, Rugiano A, Vuilleumier P, Baviera G, et al. The good bugs: the use of probiotics in pediatrics. Curr Opin Pediatr. 2019;31:661–669. [DOI] [PubMed] [Google Scholar]
- 181.Preidis GA, Weizman AV, Kashyap PC, Sadeghirad B, Morgan RL, Technical AGA. Review on the Role of Probiotics in the Management of Gastrointestinal Disorders. Gastroenterology. 2020. doi: 10.1053/j.gastro.2020.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Henrick BM, Chew S, Casaburi G, Brown HK, Frese SA, Zhou Y, et al. Colonization by B. infantis EVC001 modulates enteric inflammation in exclusively breastfed infants. Pediatr Res. 2019;86:749–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Yan F, Liu L, Cao H, Moore DJ, Washington MK, Wang B, et al. Neonatal colonization of mice with LGG promotes intestinal development and decreases susceptibility to colitis in adulthood. Mucosal Immunol. 2017;10:117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Verspreet J,Damen B, Broekaert WF, Verbeke K, Delcour JA, Courtin CM. A Critical Look at Prebiotics Within the Dietary Fiber Concept. Annu Rev Food Sci Technol. 2016;7:167–190. [DOI] [PubMed] [Google Scholar]
- 185.Davis EC, Wang M, Donovan SM. The role of early life nutrition in the establishment of gastrointestinal microbial composition and function. Gut Microbes. 2017;8:143–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Oh JZ, Ravindran R, Chassaing B, Carvalho FA, Maddur MS, Bower M, et al. TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity. 2014;41:478–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kim D, Kim YG, Seo SU, Kim DJ, Kamada N, Prescott D, et al. Nod2-mediated recognition of the microbiota is critical for mucosal adjuvant activity of cholera toxin. Nat Med. 2016;22:524–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Harris VC, Armah G, Fuentes S, Korpela KE, Parashar U, Victor JC, et al. Significant Correlation Between the Infant Gut Microbiome and Rotavirus Vaccine Response in Rural Ghana. J Infect Dis. 2017;215:34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Huda MN, Lewis Z, Kalanetra KM, Rashid M, Ahmad SM, Raqib R, et al. Stool microbiota and vaccine responses of infants. Pediatrics. 2014;134:e362–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Zimmermann P, Curtis N. The influence of probiotics on vaccine responses - A systematic review. Vaccine. 2018;36:207–213. [DOI] [PubMed] [Google Scholar]
- 191.Al Nabhani Z, Dulauroy S, Marques R, Cousu C, Al Bounny S, Dejardin F, et al. A Weaning Reaction to Microbiota Is Required for Resistance to Immunopathologies in the Adult. Immunity. 2019;50:1276–1288 e1275. [DOI] [PubMed] [Google Scholar]
- 192.Park JE, Botting RA, Dominguez Conde C, Popescu DM, Lavaert M, Kunz DJ, et al. A cell atlas of human thymic development defines T cell repertoire formation. Science. 2020;367:868. [DOI] [PMC free article] [PubMed] [Google Scholar]


