ABSTRACT
The efficacy of PD-1/PD-L1 blockades is heterogeneous in different molecular subtypes of gastric cancer (GC). In this study, we analyzed relevant clinical trials to identify the molecular subtypes associated with the efficacy of PD-1/PD-L1 blockades, and public datasets, patient samples, and GC cell lines were used for investigating potential mechanisms. We found that GC with EBV-positive, MSI-H/dMMR, TMB-H or PIK3CA mutant subtype had enhanced efficacy of PD-L1/PD-1 blockades. Also, differentially expressed genes of these molecular subtypes shared the same gene signature and functional annotations related to immunity. Meanwhile, CIBERSORT identified that the overlapping landscapes of tumor-infiltrating immune cells in the four molecular subtypes were mainly M1-like macrophages (M1). The relationships between M1 and clinical characteristics, M1, and gene signatures associated with PD-1/PD-L1 blockades also revealed that M1 was associated with improved prognosis and required for the efficacy of PD-L1/PD-1 blockades in GC. We identified that tumor-infiltrating CD68+CD163− macrophages could represent M1 calculated by CIBERSORT in clinical application, and CXCL9, 10, 11/CXCR3 axis was involved in the mechanism of CD68+CD163− macrophages in the enhanced efficacy of PD-L1/PD-1 blockades. In conclusion, CD68+CD163− macrophages are required for the efficacy of PD-L1/PD-1 blockades and expand the applicable candidates in GC patients without the molecular subtypes.
KEYWORDS: Tumor-associated macrophages, PD-L1/PD-1 blockades, gastric cancer
Background
Gastric cancer (GC) is the fifth most frequently diagnosed cancer and the third leading cause of cancer death worldwide.1 Surgery and chemotherapy are the most common treatments. For localized gastric cancer, curative resection is a standard and preferred treatment, but the recurrence rate remains high, with a five-year survival of less than 25% and median overall survival (OS) of 7 to 10 months after diagnosis.2,3 For unresectable or metastatic advanced GC, cytotoxic chemotherapy is mostly used. However, cytotoxic chemotherapy is nonspecific and has serious adverse reactions, the outcomes being usually poor.2,4 Although molecular targeted agents such as anti-HER2 monoclonal antibody and vascular endothelial growth factor receptor-2 (VEGFR-2) inhibitors have been introduced into clinical practice in recent years, the efficacy was limited.2,5
Recently, immune checkpoint inhibitors targeting PD-1/PD-L1 have become the most revolutionary treatment in solid tumors, and the PD-1/PD-L1 blockades have been recommended as first-line treatments for advanced non–small-cell lung cancer (NSCLC) and melanoma. And PD-1/PD-L1 blockades in combination with chemotherapy in patients with advanced gastric/gastroesophageal junction cancer have demonstrated encouraging efficacy.6 Meanwhile, there were also several clinical trials investigating the efficacy of PD-1/PD-L1 inhibitors for advanced GC, and a meta-analysis reported that the summary of objective response rates (ORRs) was about 12% (95% confidence interval, 6.8% to 17.1%).7 Notably, the ORRs for GC with PD-L1-positive and MSI-H/dMMR were higher than those with PD-L1-negative and Non-MSI-H/dMMR, respectively. Furthermore, the ORRs in EBV-positive, tumor mutation burden-high (TMB-H), and PIK3CA mutation of GC are higher than those in EBV-negative, TMB-L, and PIK3CA wild type, respectively.8–10 These findings suggested a high heterogeneity of GC, and the efficacy of PD-1/PD-L1 blockades might be enhanced in certain molecular subtypes of GC.
However, among multiple molecular subtypes of GC, it is not clear which one could be the best for improving the efficacy of PD-1/PD-L1 blockades, why the molecular subtypes are associated with better ORRs, and which is helpful to further improve the ORRs. In this study, we aim to analyze current clinical trials about PD-1/PD-L1 blockades used in different molecular subtypes of GC and to investigate the requirements for effective treatment of PD-1/PD-L1blockades, which will bring benefits to the improvement of ORRs of the molecular subtypes and the expansion of the applied candidates of PD-L1/PD-1 blockades in patients without the molecular subtypes.
Methods and materials
Literature search and public data analysis
We systematically searched the PubMed and Web of Science for clinical trials investigating the PD-L1/PD-1 blockades in GC before July 2020. The ORRs in the included studies were extracted to compare the efficacy of PD-L1/PD-1 blockades between different molecular subtypes in GC. Besides, we also downloaded datasets of two GC cohorts from Gene-Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) and The Cancer Genome Atlas (TCGA, https://www.cancer.gov/) respectively. All datasets were preprocessed by R (version 3.4.0) and R Bioconductor packages, and then data were subjected to cluster analysis, functional and pathway enrichment analysis, and tumor-infiltrating immune cell analysis (Online supplementary materials).
Collection of tumor samples and preoperative peripheral blood
In total, 150 GC patients were included from West China Hospital of Sichuan University between April 2017 and June 2020, whose tumor samples were collected during the open surgery and stored in liquid nitrogen. Additionally, preoperative peripheral blood samples were obtained from 40 of these 150 patients. All patients signed informed consent forms. This study was supported by the Biomedical Ethics Subcommittee of Sichuan University West China Hospital and conducted following the Declaration of Helsinki.
Immunohistochemistry (IHC) and immunofluorescence
For IHC, tumor samples were fixed with formalin and embedded with paraffin. Then, each sample was sectioned at 4 μm, and sections of tumor core were selected for staining the PD-L1. Besides, sections of tumor core were also selected for immunofluorescence analysis with the use of primary antibodies against CD68, CD163, CD8, PD-1, LAG-3, TIM-3, TIGIT, CXCL9, and CXCR3 (Online supplementary materials).
Cell culture and western blot
Two human GC cell lines, HGC-27 and MKN74, and a human monocytic cell line, THP-1, were obtained from the State Key Laboratory of Biotherapy of Sichuan University. All cells were maintained in RPMI 1640 with 10% fetal bovine serum and 1% penicillin-streptomycin. HGC-27 and MKN74 cells were treated with 100 ng/ml CXCL9 for 48 hours, and then the expressions of PD-L1 were analyzed by western blot. To induce differentiation of THP-1 into macrophage, THP-1 cells were treated with 200 nM Phorbol 12-myristate 13-acetate (PMA) for 24 hours. Then, macrophages were induced to progress toward the M1 phenotype treated with 100 ng/ml lipopolysaccharide and 20 ng/ml IFN-γ for 24 hours. Besides, macrophages were also induced to M2 phenotype treated with 20 ng/ml IL-4 for 24 hours (Online supplementary materials).
RNA-seq
Twenty GC samples, as well as M1 and M2 phenotype macrophages induced from THP-1, were subjected to RNA-seq through Illumina NovaSeq 6000 (Illumina, USA) (Online supplementary materials).
Flow cytometry
Peripheral blood mononuclear cells (PBMCs) isolated from preoperative peripheral blood of 40 GC patients were stained with primary antibodies against CD8, PD-1, LAG-3, TIM-3, TIGIT, and CXCR3. Then, cells were subjected to flow cytometry through ACEA NovoCyteTM (Agilent Biosciences, San Diego, CA, USA) (Online supplementary materials).
Assessing phosphorylation profiles of kinases and levels of cytokines
Phosphorylation profiles of kinases and cytokines of GC tissues were respectively detected by Human Phospho-Kinase Array Kit and Human XL Oncology Array Kit according to the manufacturer’s protocol.
Statistical analysis
Differences between continuous variables were analyzed through Student’s two-tailed test, Mann-Whitney U test or one-way analysis of variance, and categorical variables were analyzed using Pearson’s Chi-square test or Fisher’s exact test. Correlation coefficients were computed by Spearman and distance correlation analyses. For corresponding survival data attached to gene-expression profiles, survival curves were generated by the Kaplan–Meier method, and the log-rank (Mantel-Cox) test was used to determine the statistical significance of differences. All P values were two-tailed, and the P value of < 0.05 was considered statistically significant. The software R 3.4.0, SPSS 25.0, and GraphPad Prism 8.0 were used for data analysis and image presentation.
Results
The molecular subtypes of GC related to the efficacy of PD-L1/PD-1 blockades were mainly enriched in the same cluster
To compare the different molecular subtypes of GC and explore the requirements for the efficacy of PD-1/PD-L1 blockades, we gathered all relevant clinical trials, which involved altogether 5 molecular subtypes, including EBV-positive, MSI-H/dMMR, TMB-H, PIK3CA mutant, and PD-L1-positive (Table S1). In all the five molecular subtypes, the overall ORRs were enhanced significantly (Figure 1A), though ORRs of PD-L1-positive in one of the six trials were improved insignificantly (Figure 1B). By contrast, the outcomes of MSI-H/dMMR were meaningful in all trials (Figure 1C). To explore the common feature of four molecular subtypes (excluding PD-L1), we analyzed gene-expression profiles from GSE62254 by cluster analysis, revealing that almost all EBV-positive, 60% of TMB-H, 69% of MSI-H/dMMR, and 64% of PIK3CA mutant were enriched in the same cluster (Figure 1D, Table S2). Furthermore, the rates of the four molecular subtypes were similar to their ORRs shown in Table S1, which suggested that the cluster was related to the efficacy of PD-L1/PD-1 blockades. Also, the clinical analysis showed the analogous results: the ORR of EBV-positive was the best; the ORR of MSI-H/dMMR was better than that of PD-L1-positive, and there was no significant difference among other subtypes (Figure 1E). Meanwhile, no obvious differences in ORRs were found between different drugs or treatments, which could not interfere with the results of clinical analysis (figure 1f, G). These analyses indicated that EBV-positive, MSI-H/dMMR, TMB-H, and PIK3CA mutant possessed the common molecular phenotype related to the efficacy of PD-L1/PD-1 blockades in GC.
Figure 1.

The molecular subtypes of GC related to the efficacy of PD-L1/PD-1 blockades were mainly enriched in the same cluster. (A), The overall ORRs were enhanced significantly in the five molecular subtypes. (B), The ORR of PD-L1-positive in the first trial was improved insignificantly, but those in the other five trials were significant. (C), The ORRs of MSI-H/dMMR were improved significantly in all trials. (D), Almost all EBV-positive, 60% of TMB-H, 69% of MSI-H/dMMR, and 64% of PIK3CA mutant were enriched in Cluster A. (E), The ORR of EBV-positive was the best; the ORR of MSI-H/dMMR was better than that of PD-L1-positive. (F), No obvious differences in ORRs were found between different drugs. (G), No obvious differences in ORRs were found between different treatments
The differentially expressed genes (DEGs) of the four molecular subtypes shared the same gene signature and functional annotations
The common molecular phenotype suggested that there was an identical mechanism involving in the enhanced ORRs of the four molecular subtypes. To figure out the mechanism, we used a limma package to calculate the DEGs between EBV-positive and EBV-negative; MSI-H/dMMR and non-MSI-H/pMMR; TMB-H and TMB-L; PIK3CA mutant and PIK3CA wild type (Figure S1 A, Table S3). Then the Venn diagram of DEGs from the molecular subtypes showed 11 overlapping up-regulated genes (Figure 2A), but no overlapping down-regulated genes (Figure 2B), suggesting that the 11 genes formed a gene signature of the four molecular subtypes. To explore whether there were common signaling pathways related to the gene signature, GO and KEGG enrichment analysis of the up-regulated genes was conducted in each subtype using the R software, and significantly enriched biological processes were summarized in Table S4, Figure S1B (GO) and Figure 2C (KEGG). Further analyses indicated that four overlapping pathways were discovered via KEGG (Figure 2D), and the other one via GO (Figure S1C), all of which were the key pathways participating in immune regulation. To verify the regulating relationship of the gene signature with other DEGs in each subtype, the STRING was used to construct the regulating networks (Figure S1D), which suggested that every gene from the gene signature was in the core position in regulating networks. Furthermore, by GSEA with all transcripts ranked by the log2 (Fold Change) in each subtype, we found 15 overlapping pathways related to immunity from 21 top enriched pathways in each subtype (Figure 2E, Table S5), including the antigen processing and presentation (figure 2F), and inflammatory response (Figure 2G). These results indicated that the four molecular subtypes had the same gene signature associated with the immune relevant signature, and overlapping enriched pathways of DEGs, and all transcripts were related to the immune regulating pathways. In summary, the common mechanism was related to immunity from the results of the gene signature and functional annotations, and we speculated that the results should be associated with immune cells infiltrating in the tumor.
Figure 2.
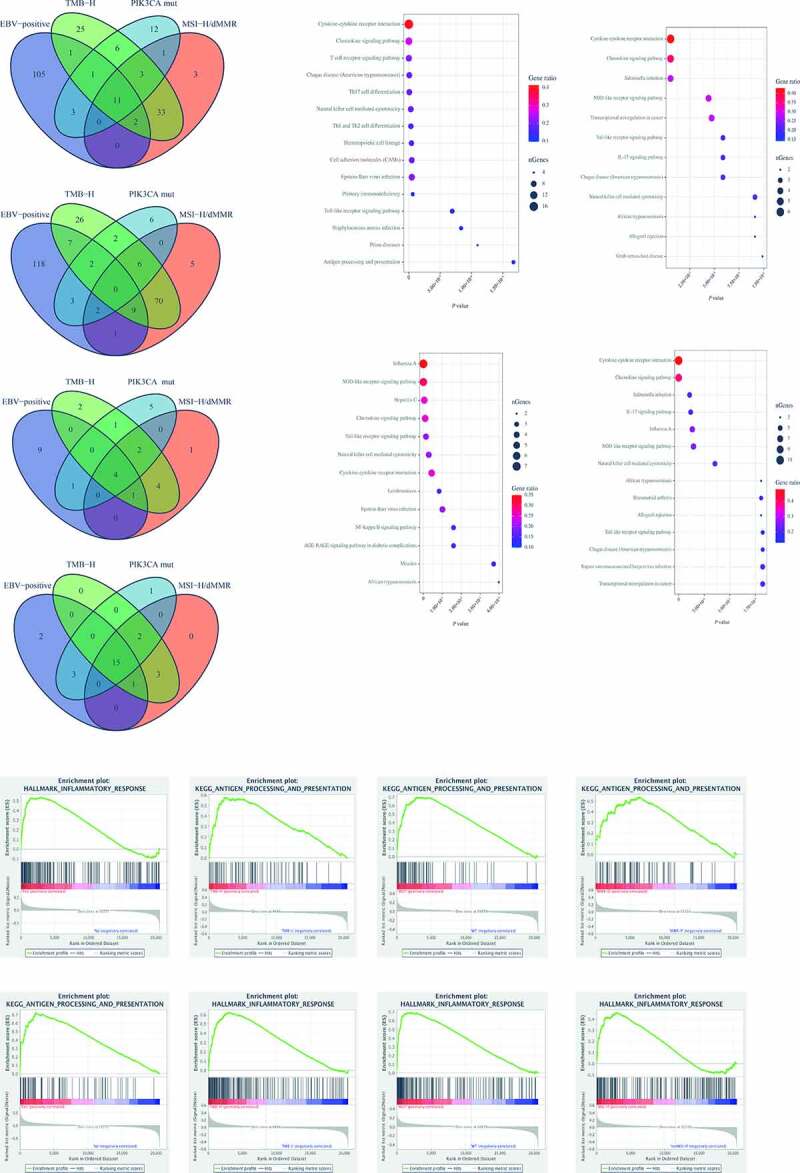
The differentially expressed genes (DEGs) of the four molecular subtypes shared the same gene signature and functional annotations. (A), The Venn diagram of DEGs from the molecular subtypes showed 11 overlapping up-regulated genes. (B), The Venn diagram of DEGs from the molecular subtypes showed no overlapping down-regulated gene. (C), KEGG enrichment analysis of biological processes related to the up-regulated genes in the different molecular subtypes. (D), The Venn diagram of biological processes related to the up-regulated genes by KEGG showed 4 overlapping pathways participating in immune regulation. (E), By GSEA with all transcripts ranked by the log2 (Fold Change) in each subtype, 15 overlapping pathways related to immunity were discovered. (F), The results of antigen processing and presentation in the different molecular subtypes, one of 15 overlapping pathways, were displayed. (G), The results of the inflammatory response in the different molecular subtypes, one of 15 overlapping pathways, were displayed
Overlapping landscapes of immune cells infiltrating in the four molecular subtypes were mainly M1-like macrophages (M1)
To verify the hypothesis, we used CIBERSORT and GSE62254 to systematically evaluate landscapes of immune cells infiltrating in GC to find out the characteristics of four molecular subtypes (Figure 3A). The heatmap from cluster analysis showed that the landscapes were different, which could be divided into four clusters and most of the four molecular subtypes gathered in the same cluster (including 95% EBV-positive, 61% of TMB-H, 69% of MSI-H/dMMR, and 65% of PIK3CA mutant), which was consistent with the result of Figure 1(C) (Figure 3A). In this cluster, the sharpest enriched cells were M1 and CD4+ T cells memory activated. Besides, Figure 3B indicated that the significant overlapping cells of the four molecular subtypes were M1. Then, we analyzed the relationships between M1 and other immune cells and found that M1 was positively correlated with CD8+ T cells and CD4+ T cells memory activated, in the GSE62254 (Figure 3C) and TCGA data (Figure S2A). However, there was a lack of information about the functional phenotype of T cells. Thus, XCELL was used to evaluate the functional phenotype of T cells infiltrating in GC (Figure 3D, S2B), showing that M1 was positively correlated with cytotoxic cells and memory CD8+ T cells, which involved in antitumor activity the same as the results in Figure 2. The findings suggested that M1 might be required for the efficacy of PD-L1/PD-1 blockades. To prove this, we explored the relationships between M1 and the gene signature identified in Figure 2(A) with GSE62254 (Figure 3E, F, S3C) and TCGA cohorts (Figure S2D, E), which displayed that levels of M1 were in positive linear correlation to the 9 genes of the gene signature. All of the results demonstrated that levels of M1 infiltrating in GC were required for the efficacy of PD-L1/PD-1 blockades.
Figure 3.
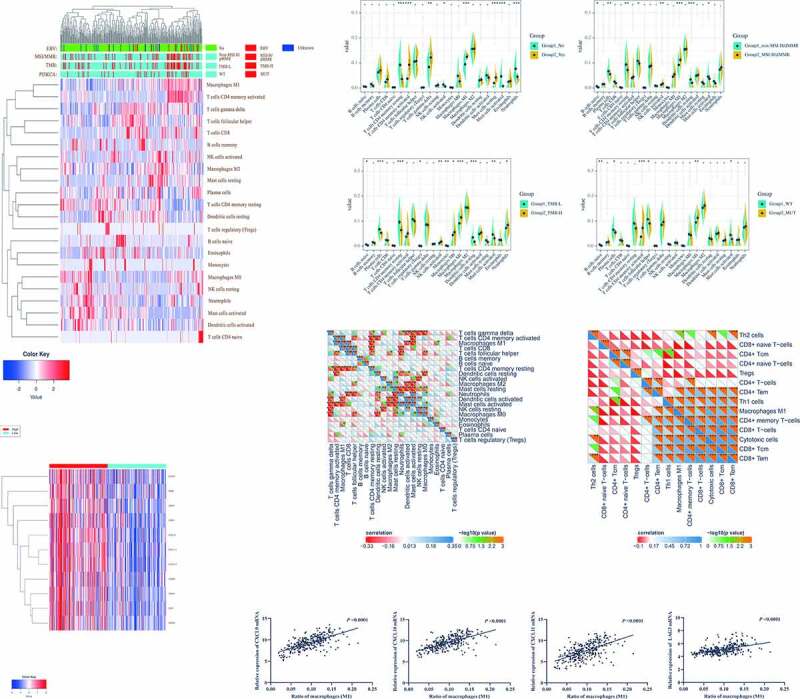
Overlapping landscapes of immune cells infiltrating in the four molecular subtypes were mainly M1-like macrophages (M1). (A), The landscapes of immune cells infiltrating in the molecular subtypes were evaluated by CIBERSORT. (B), The significant overlapping cells of the four molecular subtypes were M1. (C), M1 has positively correlated with CD8+ T cells and CD4+ T cells memory activated. (D), M1 was positively correlated with cytotoxic cells and memory CD8+ T cells. (E), There were significant differences in the expressions of the gene signature between high and low M1 groups. (F), Levels of M1 were in positive linear correlation to the 9 genes of the gene signature
The relationships between M1 and clinical characteristics, M1, and other gene signatures also revealed the important role of M1 in the four molecular subtypes
Besides, the four molecular subtypes were not only associated with the efficacy of PD-L1/PD-1 blockades but also independent favorable prognostic factors in GC, non-related to the TNM stage.11 The results from Figure 3 had suggested that M1 was required for the four molecular subtypes involved in the efficacy of PD-L1/PD-1 blockades. Thus, it raised the question of whether M1 was also related to the four molecular subtypes in prognosis. Firstly, the Kaplan-Meier analysis of M1 and CD8+ T cells in the GSE62254 and TCGA cohorts showed that patients with high M1 had significantly longer OS and Disease-free survival (DFS) than those with low M1 (Figure 4A, C, S3A). Although unable to prolong the OS of the patients (Figure 4B, S3B), high CD8+T cells could significantly improve the DFS (Figure 4D), as reported in many cancers.12,13 Then, in the univariate and multivariate Cox regression model of 22 immune cells, M1 among immune cells was an independent favorable prognostic factor in the OS and DFS (Figure 4E, F, S3C; Table S6-8), and CD8+ T cells were an independent favorable prognostic factor in DFS (figure 4F). To further explore the relationship between M1, CD8+ T cells and TNM stage, the correlation analysis was applied and showed that CD8+ T cells were not related to T, N, M or TNM stage (Figure 4H, S3E), while M1 was only significantly associated with a lower risk of tumor metastasis (Figure 4G, S3D). These results suggested that M1 was an independent favorable prognostic factor, and involved in the four molecular subtypes concerning improving prognosis.
Figure 4.

The relationships between M1 and clinical characteristics, M1, and other gene signatures also revealed the important role of M1 in the four molecular subtypes. (A), The patients with high M1 had significantly longer OS than those with low M1. (B), High CD8+T cells were unable to prolong the OS. (C), The patients with high M1 had significantly longer DFS than those with low M1. (D), High CD8+ T cells could significantly improve the DFS. E, M1 among the 22 immune cells was an independent favorable prognostic factor in the OS. (F), M1, and CD8+ T cells were the independent favorable prognostic factors in the DFS. (G), M1 was not associated with T, N, M, or TNM stage in GSE62254. (H), The CD8+ T cells were not related to T, N, M, or TNM stage in GSE62254. (I), All of the genes of the IFN-γ gene signatures and T-cell-inflamed gene expression profiles were enriched in the M1 high cluster
However, this was not enough proof that M1 was required for the efficacy of PD-L1/PD-1 blockades in GC since the conclusion was based only on the four molecular subtypes with a lack of verification. So, we reviewed all the relevant studies so far and found two gene sets IFN-γ gene signature and T-cell-inflamed gene expression profiles were also associated with the efficacy of PD-L1/PD-1 blockades in GC.4,14,15 Therefore, we used the IFN-γ gene signatures (including IDO1, CXCL10, CXCL9, HLA-DRA, STAT1, and IFNG) and T-cell-inflamed gene expression profiles (including CCL5, CD27, CD274, CD276, CD8A, CMKLR1, CXCL9, CXCR6, HLA- DQA1, HLA-DRB1, HLA-E, IDO1, LAG3, NKG7, PDCD- 1KLG2, PSMB10, STAT1, and TIGIT) to verify the conclusion. The results displayed that all of the genes were enriched in the M1 high cluster (Figure 4I, S3F), which proved that M1 was required for the efficacy of PD-L1/PD-1 blockades in GC, not limited in the four molecular subtypes.
The CD68+CD163−, a typical M1-like tumor-associated macrophages (TAMs) phenotype, could represent thoroughly M1 calculated by CIBERSORT
The bioinformatics method had provided sufficient evidence to prove the important role of M1 in the efficacy of PD-L1/PD-1 blockades. However, M1 calculated by CIBERSORT could not be used to guide the clinical treatment of PD-L1/PD-1 blockades in GC. A previous study had found that CD68+CD163− macrophages were typical phenotype of M1-like TAMs in GC.16 Thus, we adopted this definition to study the relationship between CD68+CD163− macrophages and M1 calculated by CIBERSORT. Firstly, we verified the levels of CD68+CD163− macrophages in three molecular subtypes (EBV-positive, MSI-H/dMMR, and PIK3CA mutant) because of lack of TMB information in our patients, which showed that the levels of CD68+CD163− macrophages were significantly higher in EBV-positive, MSI-H/dMMR, and PIK3CA mutant (Figure 5A, S4A), but levels of CD68+CD163+ macrophage, a typical phenotype of M2-like TAMs, were of insignificant differences in the three subtypes (Figure 5B). These results were the same as the results of CIBERSORT (Figure 3B), which suggested that the result of CD68+CD163− macrophages infiltrating in different subtypes accorded with that of M1. Then we used the ROC curve to determine the cutoff value of CD68+CD163− macrophages was 685.7/mm2 (Figure 5C) and performed survival analysis of the cutoff value by Kaplan-Meier. The cutoff value was used to distinguish the high and low groups and the patients in the high group had a longer OS than that in the low group following the outcomes of M1 in GSE62254 and TCGA (Figure 5D). Further, we performed RNA-sequencing for 20 tumor tissues, in which 10 samples were from the high CD68+CD163− macrophages group, and 10 samples were from the low CD68+CD163− macrophages group. The sequencing showed that the expressions of the 400 most representative DEGs were significantly distant between the high and low groups (Figure 5E), and the principal component analysis also revealed the same result (figure 5F). In the volcano map, the genes from the gene signature identified in Figure 2(A) had high expression in the high CD68+CD163− macrophages group (Figure 5G), and the genes were also in the core positions in regulating networks by STRING (Figure S4B). Then the up-regulated genes were conducted by KEGG analysis and mainly enriched biological processes including cytokine-cytokine receptor interaction, chemokine signaling pathway, Toll-like receptor signaling pathway, NOD-like receptor signaling pathway, and IL-17 signaling pathway (Figure 5H). To verify the results of KEGG analysis, the Protein Arrays were applied to test levels of cytokines and protein phosphorylation in the tumor from the high and low groups (Figure S4C), showing that the different levels of cytokines between high and low CD68+CD163− macrophages, especially granulocyte-macrophage colony-stimulating factor (GM-CSF) and macrophage colony-stimulating factor (M-CSF), conformed to the feature of M1-like TAMs (Figure 5I), and protein phosphorylation was the same as the KEGG analysis (Figure 5J). In summary, all the results from our RNA-sequencing revealed that features of CD68+CD163− macrophages were completely consistent with the characteristics of M1 calculated by CIBERSORT in GSE62254 and TCGA. Besides, all the genes from IFN-γ gene signatures and T-cell-inflamed gene expression profiles were enriched in the high group of CD68+CD163− macrophages (Figure 5K), which also proved that the CD68+CD163− macrophages, M1-like TAMs, could thoroughly represent M1 calculated by CIBERSORT.
Figure 5.
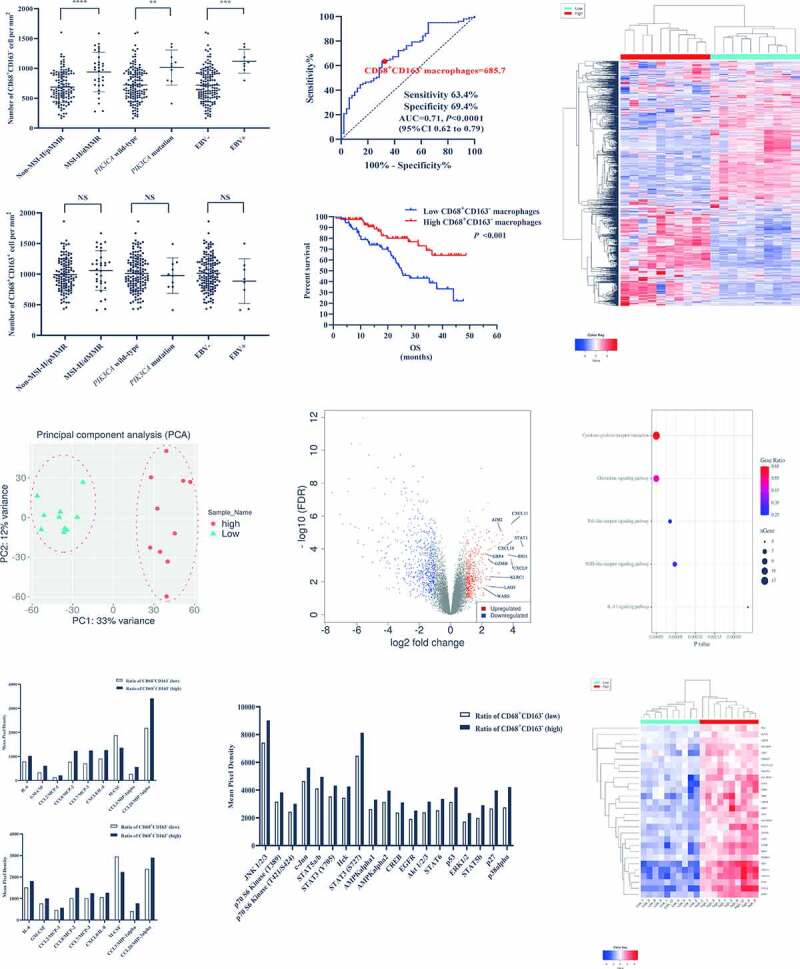
The CD68+CD163−, a typical M1-like tumor-associated macrophages (TAMs) phenotype, could represent thoroughly M1 calculated by CIBERSORT. (A), The levels of CD68+CD163− macrophages were significantly higher in EBV-positive, MSI-H/dMMR, and PIK3CA mutant. (B), The levels of CD68+CD163+ macrophages were of insignificant differences in the three subtypes. (C), The cutoff value of CD68+CD163− macrophages used by the ROC curve was 685.7/mm2. (D), The patients with the high CD68+CD163− macrophages had a longer OS than that with the low. E The expressions of the 400 most representative DEGs were significantly distant between the high and low CD68+CD163− macrophages. (F), The PCA revealed that there was a significant difference between the high and low CD68+CD163− macrophages. (G), The genes identified in figure 2A had high expressions in the high CD68+CD163− macrophages group. H, The mainly enriched biological processes conducted by up-regulated genes included cytokine-cytokine receptor interaction, chemokine signaling pathway, Toll-like receptor signaling pathway, NOD-like receptor signaling pathway, and IL-17 signaling pathway. (I), The Protein Arrays showed that the different levels of cytokines between high and low CD68+CD163− macrophages, especially GM-CSF and M-CSF. (J), The Protein Arrays showed that the protein phosphorylation was the same as the enriched biological processes. (K), All the genes from IFN-γ gene signatures and T-cell-inflamed gene expression profiles were enriched in the high CD68+CD163− macrophages
The CXCL9, 10, 11/CXCR3 axis was involved in the mechanism of M1-like TAMs in the efficacy of PD-L1/PD-1 blockades
From analyses of GSE62243, our gene arrays, and two gene sets (IFN-γ gene signatures and T-cell-inflamed gene expression profiles), the levels of CD68+CD163− macrophages infiltrating in GC were in significantly positive correlation with antigen presentation related genes (HLA-DQA1, HLA-DRB1, HLA-E, HLA-DRA, HLA-DQB1), chemokines (CXCL9, 10, 11) and T-cell-related genes (IFNG, CD8A, CD274, GZMB, CXCR3, LAG3, TIGIT, HAVCR2, TNF). In conclusion, we speculated that M1-like TAMs with antigen presentation (HLA-DQA1, HLA-DRB1, HLA-E, HLA-DRA, HLA-DQB1) could secrete more CXCL9, 10, 11 to recruit more CXCR3+CD8+ T cells infiltrating in the tumor, but CXCL9, 10, 11 and IFN-γ also raised the expression of PD-L1 (CD274) to exhaust CD8+ T cells (up-regulate the expressions of LAG3, TIGIT, and HAVCR2), as reported in previous researches.17,18 First, we did the co-culture of primary Macrophages-T cells-tumor cells from GC patients to verify M1-like TAMs could enhance the killing effect of the CD8+T cells to tumors and improve the effect of PD-1 for the CD8+T cells (Figure 6A). By the lactate dehydrogenase releasing method (LDHRM), when the effective target ratio was 10:1, the killing effect of the M1-like macrophages group was higher than that of the macrophages group, saying M1-like macrophages could enhance the killing activity of CD8+T cells to the tumor cells. However, it was not significant, when the effective target ratio was 5:1. When the PD-1 block was added, the significant increase of killing effect in PD-1 block+M1-like macrophages was about two times higher than that in PD-1 block+macrophages. The results showed that M1-like TAMs could mainly improve the effect of PD-1 block for the CD8+T cells. Then, we induced THP-1 cells to differentiate into M1 and M2 in vitro (Figure S5A). The RNA-sequencing showed that there were obvious differences in the expression of chemokines between M1 and M2 (Figure S5B). Even though expressions of CXCL9, 10, 11 were higher in M1 than those in M2, the expression of CXCL9 was the most obvious difference (Figure 6B), and the RNA-sequencing of tumors also showed that CXCL9 was the most noteworthy (Figure 6C). Based on this finding, we focused on CXCL9 in the next experiments. Furthermore, immunofluorescence was used to mark the CD68, CD163, and CXCL9, showing that most CXCL9 were labeled in the cytoplasm of CD68+CD163− macrophages (Figure 6D), which confirmed that CD68+CD163− macrophages secreted CXCL9 in the tumor. Then, immunofluorescence was used to study the relationships between CD68+CD163− macrophages and CD8+ T cells. The number of CD8+ T cells, especially the number and percentage of CXCR3+CD8+ T cells (Figure 6E, S5C, D), was positively correlated with that of CD68+CD163− macrophages (Figure 6E). Even though M1 was positively correlated with cytotoxic cells and memory CD8+ T cells by CIBERSORT and CD68+CD163− macrophages were also related to the increased CD8+ T cells, Figure 4(G), H revealed that the levels of M1 and CD8+ T were not associated with tumor progression, which might be caused by the up-regulation of PD-L1. Since results from GSE62243, our gene arrays, and two gene sets said that M1 was positively correlated with CD274, IHC (IHC) was used to test expressions of PD-L1 and demonstrated that expressions of PD-L1 in the high group of CD68+CD163− macrophages were significantly higher than those in the low group, which also explained that the ORR was enhanced in PD-L1-positive (figure 6f, S5E). Besides, in vitro, we used CXCL9 to intervene in GC cell lines and found that the expression of PD-L1 was up-regulated in GC cells (Figure 6G), which suggested that CXCL9 was secreted by CD68+CD163− macrophage up-regulated the expression of PD-L1 in GC, as reported in previous researches.17,18 Then, we studied the relationships between CD68+CD163− macrophages and states of CD8+ T cells (PD-1, LAG-3, TIGIT, and TIM-3), which indicated that the numbers of PD-1+CD8+, LAG-3+CD8+, TIM-3+CD8+, and TIGIT+CD8+ T cells increased dramatically with the rising numbers of CD68+CD163− macrophage (Figure 6H), while the percentages of them were not significantly associated with CD68+CD163− macrophage (Figure S5F, G). Furthermore, the divergences between CXCR3+CD8+ T cells and CXCR3−CD8+ T cells were intensively studied and the result showed that the percentages of PD-1+, LAG-3+, TIM-3+, and TIGIT+ on CXCR3+CD8+ T cells were higher than those on CXCR3−CD8+ T cells (Figure 6I, S6A-D). However, after we examined the CD8, CXCR3, PD-1, LAG-3, TIGIT, and TIM-3 in peripheral blood by flow cytometry (Figure 6J), it showed that the percentage of PD-1+ on CXCR3+CD8+ T cells was higher than that on CXCR3−CD8+ T cells, and the percentages of LAG-3+, TIGIT+, and TIM-3+ between two subtypes of CD8+ T cells were insignificantly different. In summary, M1-like TAMs recruit CD8+ T cells to kill GC cells by CXCL9, 10, 11/CXCR3 axis, but GC cells could utilize the PD-L1/PD-1 axis to escape the killing of CD8+ T cells. Thus PD-L1/PD-1 blockades therapy would be effective in these subtypes of GC.
Figure 6.
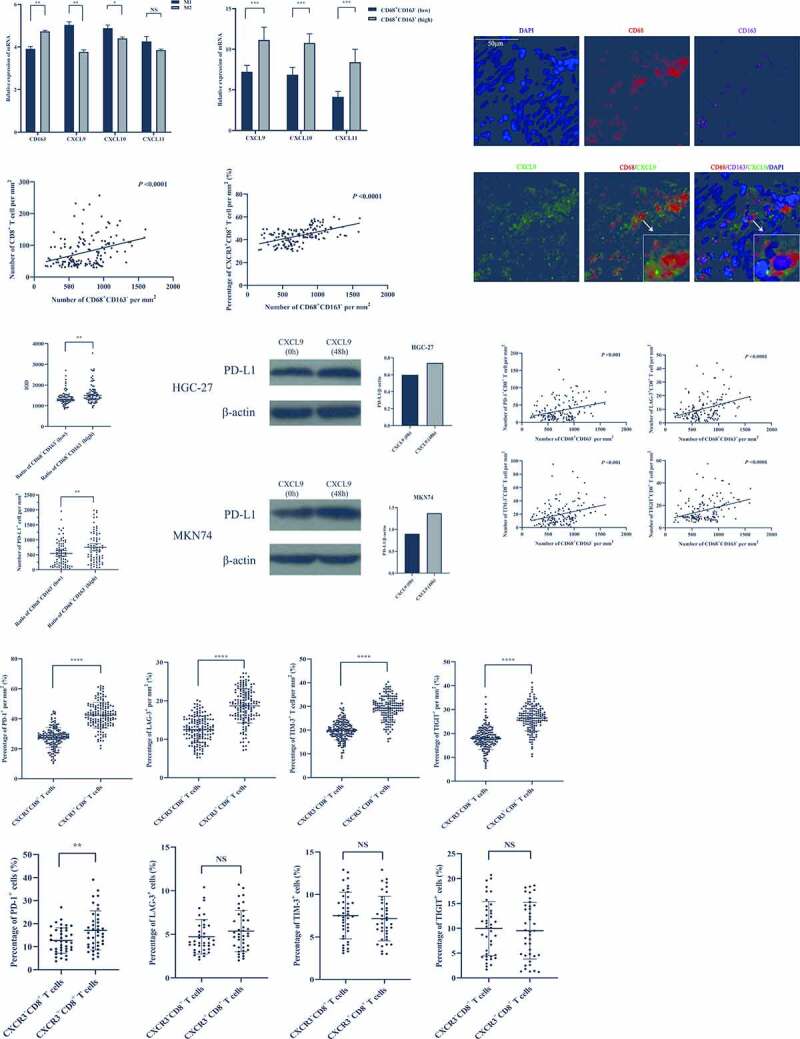
The CXCL9, 10, 11/CXCR3 axis was involved in the mechanism of M1-like TAMs for the efficacy of PD-L1/PD-1 blockades. (A), By lactate dehydrogenase releasing method (LDHRM), when the effective target ratio was 10:1, the killing effect of the M-1like macrophages group was higher than that of the macrophages group. When the PD-1 block was added, the significant increase of killing effect in PD-1 block+M1-like macrophages was about two times higher than that in PD-1 block+macrophages. (B), The expressions of CXCL9, 10, 11 mRNA were higher in M1 than those in M2. (C), The expressions of CXCL9, 10, 11 mRNA were higher in the tumor with high CD68+CD163− macrophages. (D), The most CXCL9s were labeled in the cytoplasm of CD68+CD163− macrophages. (E), The number of CD8+ T cells, especially the number and percentage of CXCR3+CD8+ T cells was positively correlated with that of CD68+CD163− macrophages. (F), The expressions of PD-L1 in the high group of CD68+CD163− macrophages were significantly higher than those in the low group. (G), The expression of PD-L1 was up-regulated in GC cells intervened by CXCL9. (H), The numbers of PD-1+CD8+, LAG-3+CD8+, TIM-3+CD8+, and TIGIT+CD8+ T cells increased dramatically with the rising numbers of CD68+CD163− macrophage. (I), The percentages of PD-1+, LAG-3+, TIM-3+, and TIGIT+ on CXCR3+CD8+ T cells were higher than those on CXCR3−CD8+ T cells in the tumor. (J), The percentage of PD-1+ on CXCR3+CD8+ T cells was higher than that on CXCR3−CD8+ T cells, and the percentages of LAG-3+, TIGIT+, and TIM-3+ between two subtypes of CD8+ T cells were insignificantly different in peripheral blood
Discussion
Through bioinformatics analysis and our experiments, it is confirmed that M1-like TAMs are required for the efficacy of PD-L1/PD-1 blockades in GC, which suggested that CD68+CD163− macrophages should be regarded as an important biomarker to predict the efficacy of PD-L1/PD-1 blockades and guide the clinical treatment of PD-L1/PD-1 blockades in GC.
In the 7 clinical trials, there were no significant differences of ORRs between different drugs and therapeutic regimens, saying that PD-L1/PD-1 blockades combined with CTLA-4 blockades cannot prolong the OS of patients with GC but may increase the side effects. By comparing the five molecular subtypes, we discover that the ORR of EBV-positive is the highest, about 100%, and that of MSI-H/dMMR, about 60%, is better than other subtypes. For the patients with EBV-positive and MSI-H/dMMR, PD-L1/PD-1 blockades treatment is the most appropriate. However, the rate of EBV-positive in GC is extremely low, only about 7%, which helps only a small number of patients. Meanwhile, the ORRs of the other four molecular subtypes range from 60% to 22%. Even though the ORRs are improved in the five molecular subtypes of GC, meaning that the five molecular subtypes are used to guide the treatment of PD-L1/PD-1 blockades, it is still not accurate enough or useful only for few appropriate candidates. Thus, CD68+CD163− macrophages can be an important biomarker to help improve the ORRs of the five molecular subtypes and expand the applicable candidates of PD-L1/PD-1 blockades in patients without the five molecular subtypes. The ORR of PD-L1-positive is the worst, only 22.6%, which is caused by different standards of PD-L1-positive. The definition in one trial without the advantage of PD-L1-positive is tumor positive score (TPS) ≥ 1, while that in other trials is combined positive score (CPS) ≥ 1.4,9,10,15,19–21 Previous studies have reported that CPS was a more useful assessment method of determining PD-L1 expression than TPS as a prognostic biomarker.22 Thus, CPS should be a unified standard in GC not only for prognosis but also for guiding treatment of PD-L1/PD-1 blockades.
Macrophages are a diverse collection of cell types with a wide range of functional roles in homeostatic and pathological conditions and its classic adaptive responses include tolerance, priming, and a wide spectrum of activation state divided into two categories, M1 and M2.23,24 M1 is pro-inflammatory and plays a major role in the host defense against infection, while M2 suppresses inflammatory responses and mainly supports tissue repair and remodeling.24 Along similar lines, infiltration of macrophages in solid tumors, generally defined as TAMs, constitute a plastic and heterogeneous cell population of the tumor microenvironment that can account for up to 50% of some solid neoplasms.23,25 In general, TAMs are double-edged swords with the capacity to exert pro- and antitumor activity, depending on the balance of cytokines, chemokines, antibodies, and myeloid checkpoints.26–28 In GC, the presence and density of TAMs are correlated with prognosis and resistance to treatment.29,30 In most researches, pro-tumor activity of TAMs, called M2-like TAMs, is usually the focus of attention, which is generally associated with a poor prognosis and promotes multiple aspects of tumorigenesis from initiation to angiogenesis and systemic dissemination.31,32 However, TAMs are a diverse population of cells in GC, some subtypes of which can also mediate antineoplastic effects, named as M1-like TAMs.33 Thus, many studies suggest that TAMs are good targets for anticancer therapy through their re-differentiation away from pro-tumoral toward antitumoral states.34 Through analyses of the five molecular subtypes and previous studies, we discover that M1-like TAMs are required for the efficacy of PD-L1/PD-1 blockades, and also associated with a favorable prognosis in GC, which suggests that it should be combined with the TNM stage to refine a risk stratification system and better stratify patients with different prognosis. Current evidence suggests that increased numbers of CD8+ T cells are a crucial determinant of favorable clinical outcomes in many cancers.35 Besides, we also find that increased numbers of CD8+ T cells can improve the DFS but not the OS in GC, suggesting that CD8+ T cells in GC can provide an indicator of tumor recurrence. Interestingly, the numbers of CD8+ T cells, especially that of CXCR3+CD8+ T cells in GC, are positively correlated with M1-like TAMs, which might be caused by chemokines released by M1-like TAMs. Extensive researches suggest that the diversity of chemokines released by M1 and M2 is one of the key factors for their different regulatory functions.36 In our vitro experiments, there are significant differences in chemokine gene expression between M1 and M2, particularly CXCL 9, 10, 11. Many studies including ours reveal that expressions of CXCL 9, 10, 11 are related to the efficacy of PD-L1/PD-1 blockades in GC.3,37 Meanwhile, all receptors of CXCL 9, 10, 11 are CXCR3 and CXCL9, 10, 11/CXCR3 axis leads migration of immune cells to their focal sites.38 The researches show that CXCL9, 10, 11/CXCR3 axis involves directing the migration of CD8+ T cells to the tumor site and inducing their potentiation and proliferation there.39 Recently Andy Luster and his group showed that anti-PD-1 efficacy is reduced in CXCR3KO mice, and suggested that the CXCR3 on CD8+ T cells enhances anti-PD-1 efficacy.40 However, we discover that the CXCL9, 10, 11/CXCR3 axis can recruit more CXCR3+ CD8+ T cells infiltrating in GC; meanwhile the dysfunction (LAG-3+, TIM-3+, and TIGIT+) of CXCR3+CD8+ T cells also increases with CXCR3+CD8+ T cells infiltrating. Nevertheless, in the peripheral blood, expressions of LAG-3+, TIM-3+, and TIGIT+, markers of dysfunction, between CXCR3+CD8+ T cells and CXCR3−CD8+ T cells are insignificantly different, and the expression of PD-1+, a marker on tumor-specific cytotoxic CD8+ T cells,41 on CXCR3+ CD8+ T cells is higher than that on CXCR3− CD8+ T cells, which suggests that CXCR3+ CD8+ T cells are a group of CD8+ T cells with tumor-killing function. In the tumor with high M1-like TAMs, even though CXCR3+ CD8+ T cells are highly infiltrated in the tumor, tumor cells can take advantage of PD-L1, up-regulated by CXCL9, 10, 11, and IFN-γ, to escape the killing of CD8+ T cells.18,30,42 Thus, in the subtypes of GC with high M1-like TAMs and pro-inflammation, it is likely that the PD-L1/PD-1 axis is the main pattern of tumor immune evasion, so the treatment of PD-L1/PD-1 blockades in this subtype of GC is effective. However, most TAMs in GC are M2-like and facilitate tumor growth by inducing immune suppression. Thus, reprogramming or repolarizing TAMs toward an anti-tumor phenotype could therefore prove a more efficacious approach to augmenting the efficacy of PD-L1/PD-1 blockades in GC. For instance, CD40 antibody is one of the most productive approaches to date, and enhance responses to PD-1 and CTLA-4 antagonists have been observed43–45
In conclusion, immune evasion is an obstacle to successful cancer therapy, which is involved in a series of complex and collaborative mechanisms, such as PD-L1/PD-1 axis, CD80, 86/CTLA-4 axis, antigen presentation deletion, metabolism, and so on.46 Only when one mechanism plays a crucial role in the immune evasion of this tumor, can the related targeted therapy achieve the clinical therapeutic effect in this tumor. Therefore, M1-like TAMs are required for the efficacy of PD-L1/PD-1 blockades in GC, since PD-L1/PD-1 blockades can break the circle: M1-like TAMs release CXCL9,10,11 to recruit more CXCR3+CD8+ T cells infiltrating in GC but also up-regulate the expression of PD-L1, and PD-L1 combines with PD-1 on CD8+ T cells to result in dysfunction of CD8+ T cells. M1-like TAMs can not only improve the ORRs of the five molecular subtypes and but also expand the applicable candidates of PD-L1/PD-1 blockades in patients without the five molecular subtypes. Meanwhile, it suggests that TAMs are good targets for anticancer therapy through their re-differentiation away from pro-tumor toward antitumor states.
Supplementary Material
Acknowledgments
The special and deepest thanks went to Miss Siming Chen for polishing the language, and the nursing and supporting staff at West China Hospital, Sichuan University, especially the Biobank of West China Hospital, Sichuan University.
Funding Statement
This work was supported by Sichuan Province Science and Technology Support Project (2012SZ0142) (2018SZ0189); Sichuan Province Science and Technology Support Project [2012SZ0142, 2018SZ0189].
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors’ contributions
Prof. Xiaoting Wu, together with Rui Zhao and Qianyi Wan, conceived the study, while all of those authors mentioned above have contributed to the research and development process of this article. Under the guidance of Prof. Xiaoting Wu and Hanshuo Yang, Rui Zhao, Qianyi Wan, Yong Wang, and Qiqi Li have conducted the experiments with the clinical samples collected by Lin Xia, Shuomeng Xiao, Yinghan Song, Xiaoding Shen, Wen Zhuang, Yong Zhou, and Yi Chen. The manuscript was completed by Rui Zhao, Qianyi Wan, and Yutao Wu. All the authors have read and approved the final manuscript.
Ethics approval
This study was supported by the Biomedical Ethics Subcommittee of West China Hospital, Sichuan University.
Patient consent for publication
Not required.
Availability of data and materials
All data generated or analyzed during this study are included in this published article
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
Abbreviations
| CPS | Combined positive score |
| DEGs | Differentially expressed genes |
| GC | Gastric cancer |
| GEO | Gene-Expression Omnibus |
| GM-CSF | granulocyte-macrophage colony-stimulating factor |
| IHC | Immunohistochemistry |
| OS | Overall survival |
| ORRs | Objective response rates |
| PMA | Phorbol 12-myristate 13-acetate |
| PBMCs | Peripheral blood mononuclear cells |
| M-CSF | Macrophage colony-stimulating factor |
| NSCLC | Non–small-cell lung cancer |
| TCGA | The Cancer Genome Atlas |
| TAMs | Tumor-associated macrophages |
| TPS | Tumor positive score |
| VEGFR-2 | Vascular endothelial growth factor receptor-2 |
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A.. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–11. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Nie S, Yang G, Lu H. Current molecular targeted agents for advanced gastric cancer. Onco Targets Ther. 2020;13:4075–4088. doi: 10.2147/OTT.S246412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kono K, Nakajima S, Mimura K. Current status of immune checkpoint inhibitors for gastric cancer. Gastric Cancer. 2020;23(4):565–578. doi: 10.1007/s10120-020-01090-4. [DOI] [PubMed] [Google Scholar]
- 4.Muro K, Chung H, Shankaran V, Geva R, Catenacci D, Gupta S, Eder JP, Golan T, Le DT, Burtness B, et al. Pembrolizumab for patients with PD-L1-positive advanced gastric cancer (KEYNOTE-012): a multicentre, open-label, phase 1b trial. Lancet Oncol. 2016;17(6):717–726. doi: 10.1016/S1470-2045(16)00175-3. [DOI] [PubMed] [Google Scholar]
- 5.Palle J, Rochand A, Pernot S, Gallois C, Taieb J, Zaanan A. Human epidermal growth factor receptor 2 (HER2) in advanced gastric cancer: current knowledge and future perspectives. Drugs. 2020;80(4):401–415. doi: 10.1007/s40265-020-01272-5. [DOI] [PubMed] [Google Scholar]
- 6.Boku N, Ryu MH, Kato K, Chung HC, Minashi K, Lee KW, Cho H, Kang WK, Komatsu Y, Tsuda M, et al. Safety and efficacy of nivolumab in combination with S-1/capecitabine plus oxaliplatin in patients with previously untreated, unresectable, advanced, or recurrent gastric/gastroesophageal junction cancer: interim results of a randomized, phase II trial (ATTRACTION-4). Ann Oncol. 2019;30(2):250–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen C, Zhang F, Zhou N, Gu Y-M, Zhang Y-T, He Y-D, Wang L, Yang L-X, Zhao Y, Li Y-M, et al. Efficacy and safety of immune checkpoint inhibitors in advanced gastric or gastroesophageal junction cancer: a systematic review and meta-analysis. Oncoimmunology. 2019;8(5):e1581547. doi: 10.1080/2162402X.2019.1581547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim ST, Cristescu R, Bass AJ, Kim K-M, Odegaard JI, Kim K, Liu XQ, Sher X, Jung H, Lee M, et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat Med. 2018;24(9):1449. [DOI] [PubMed] [Google Scholar]
- 9.Mishima S, Kawazoe A, Nakamura Y, Sasaki A, Kotani D, Kuboki Y, Bando H, Kojima T, Doi T, Ohtsu A, et al. Clinicopathological and molecular features of responders to nivolumab for patients with advanced gastric cancer. J Immunother Cancer. 2019;7(1):24. doi: 10.1186/s40425-019-0514-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang F, Wei XL, Wang FH, Xu N, Shen L, Dai GH, Yuan XL, Chen Y, Yang SJ, Shi JH, et al. Safety, efficacy and tumor mutational burden as a biomarker of overall survival benefit in chemo-refractory gastric cancer treated with toripalimab, a PD-1 antibody in phase Ib/II clinical trial NCT02915432. Ann Oncol. 2019;30(9):1479–1486. doi: 10.1093/annonc/mdz197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suh YS, Na D, Lee JS, Chae J, Kim E, Jang G, Lee J, Min J, Ock CY, Kong SH, et al. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513(7517):202–209. doi: 10.1038/nature13480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Challoner B, von Loga K, Woolston A, Griffiths B, Sivamanoharan N, Semiannikova M, Newey A, Barber LJ, Mansfield D, Hewitt LC, et al. Computational image analysis of T-cell infiltrates in resectable gastric cancer: association with survival and molecular subtypes. JNCI. 2020. doi: 10.1093/jnci/djaa051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson E, Zahurak M, Murphy A, Cornish T, Cuka N, Abdelfatah E, Yang S, Duncan M, Ahuja N, Taube JM, et al. Patterns of PD-L1 expression and CD8 T cell infiltration in gastric adenocarcinomas and associated immune stroma. Gut. 2017;66(5):794–801. doi: 10.1136/gutjnl-2015-310839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, Albright A, Cheng JD, Kang SP, Shankaran V, et al. IFN-γ–related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. 2017;127(8):2930–2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuchs C, Doi T, Jang R, Jang RW, Muro K, Satoh T, Machado M, Sun W, Jalal SI, Shah MA, et al. Safety and efficacy of pembrolizumab monotherapy in patients with previously treated advanced gastric and gastroesophageal junction cancer: phase 2 clinical KEYNOTE-059 trial. JAMA Oncol. 2018;4(5):e180013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang Y, Wang M, Sun Y, Di Costanzo N, Mitchell C, Achuthan A, Hamilton JA, Busuttil RA, Boussioutas A. Macrophage spatial heterogeneity in gastric cancer defined by multiplex immunohistochemistry. Nat Commun. 2019;10(1):3928. doi: 10.1038/s41467-019-11788-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taube J, Anders R, Young G, Xu H, Sharma R, McMiller TL, Chen S, Klein AP, Pardoll DM, Topalian SL, et al. Colocalization of inflammatory response with B7-h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci Transl Med. 2012;4(127):127ra137. doi: 10.1126/scitranslmed.3003689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang C, Li Z, Xu L, Che X, Wen T, Fan Y, Li C, Wang S, Cheng Y, Wang X, et al. CXCL9/10/11, a regulator of PD-L1 expression in gastric cancer. BMC Cancer. 2018;18(1):462. doi: 10.1186/s12885-018-4384-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doi T, Iwasa S, Muro K, Satoh T, Hironaka S, Esaki T, Nishina T, Hara H, Machida N, Komatsu Y, et al. Phase 1 trial of avelumab (anti-PD-L1) in Japanese patients with advanced solid tumors, including dose expansion in patients with gastric or gastroesophageal junction cancer: the JAVELIN Solid Tumor JPN trial. Gastric Cancer. 2019;22(4):817–827. doi: 10.1007/s10120-018-0903-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janjigian Y, Bendell J, Calvo E, Kim JW, Ascierto PA, Sharma P, Ott PA, Peltola K, Jaeger D, Evans J, et al. CheckMate-032 study: efficacy and safety of nivolumab and nivolumab plus ipilimumab in patients with metastatic esophagogastric cancer. J clin oncol. 2018;36(28):2836–2844. doi: 10.1200/JCO.2017.76.6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim S, Cristescu R, Bass A, Kim K-M, Odegaard JI, Kim K, Liu XQ, Sher X, Jung H, Lee M, et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat Med. 2018;24(9):1449–1458. doi: 10.1038/s41591-018-0101-z. [DOI] [PubMed] [Google Scholar]
- 22.Yamashita K, Iwatsuki M, Harada K, Eto K, Hiyoshi Y, Ishimoto T, Nagai Y, Iwagami S, Miyamoto Y, Yoshida N, et al. Prognostic impacts of the combined positive score and the tumor proportion score for programmed death ligand-1 expression by double immunohistochemical staining in patients with advanced gastric cancer. Gastric Cancer. 2020;23(1):95–104. doi: 10.1007/s10120-019-00999-9. [DOI] [PubMed] [Google Scholar]
- 23.DeNardo DG, Ruffell B. Macrophages as regulators of tumour immunity and immunotherapy. Nat Rev Immunol. 2019;19:369–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Locati M, Curtale G, Mantovani A. Diversity, mechanisms, and significance of macrophage plasticity. Ann Rev Pathol Mech Dis. 2020;15(1):123–147. doi: 10.1146/annurev-pathmechdis-012418-012718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vitale I, Manic G, Coussens LM, Kroemer G, Macrophages GL. Metabolism in the tumor microenvironment. Cell Metab. 2019;30(1):36–50. doi: 10.1016/j.cmet.2019.06.001. [DOI] [PubMed] [Google Scholar]
- 26.Mantovani A, Marchesi F, Malesci A, Laghi L, Allavena P. Tumour-associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol. 2017;14(7):399–416. doi: 10.1038/nrclinonc.2016.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qian B, Pollard J. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141(1):39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gui P, Ben-Neji M, Belozertseva E, Dalenc F, Franchet C, Gilhodes J, Labrousse A, Bellard E, Golzio M, Poincloux R, et al. The protease-dependent mesenchymal migration of tumor-associated macrophages as a target in cancer immunotherapy. Cancer Immunol Res. 2018;6(11):1337–1351. doi: 10.1158/2326-6066.CIR-17-0746. [DOI] [PubMed] [Google Scholar]
- 29.Ge S, Xia X, Ding C, Zhen B, Zhou Q, Feng J, Yuan J, Chen R, Li Y, Ge Z, et al. A proteomic landscape of diffuse-type gastric cancer. Nat Commun. 2018;9(1):1012. doi: 10.1038/s41467-018-03121-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harada K, Dong X, Estrella J, Correa AM, Xu Y, Hofstetter WL, Sudo K, Onodera H, Suzuki K, Suzuki A, et al. Tumor-associated macrophage infiltration is highly associated with PD-L1 expression in gastric adenocarcinoma. Gastric Cancer. 2018;21(1):31–40. doi: 10.1007/s10120-017-0760-3. [DOI] [PubMed] [Google Scholar]
- 31.Zhang H, Li R, Cao Y, Gu Y, Lin C, Liu X, Lv K, He X, Fang H, Jin K, et al. Poor clinical outcomes and immunoevasive contexture in intratumoral IL-10-producing macrophages enriched gastric cancer patients. Ann Surg. 2020. doi: 10.1097/SLA.0000000000004037. [DOI] [PubMed] [Google Scholar]
- 32.Chen Y, Zhang S, Wang Q, Tumor-recruited ZX. M2 macrophages promote gastric and breast cancer metastasis via M2 macrophage-secreted CHI3L1 protein. J Hematol Oncol. 2017;10(1):36. doi: 10.1186/s13045-017-0408-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gambardella V, Castillo J, Tarazona N, Gimeno-Valiente F, Martínez-Ciarpaglini C, Cabeza-Segura M, Roselló S, Roda D, Huerta M, Cervantes A, et al. The role of tumor-associated macrophages in gastric cancer development and their potential as a therapeutic target. Cancer Treat Rev. 2020;86:102015. doi: 10.1016/j.ctrv.2020.102015. [DOI] [PubMed] [Google Scholar]
- 34.Cassetta L, Pollard J. Targeting macrophages: therapeutic approaches in cancer. Nat Rev Drug Discov. 2018;17:887–904. [DOI] [PubMed] [Google Scholar]
- 35.van der Leun A, Thommen D, Schumacher T. CD8+ T cell states in human cancer: insights from single-cell analysis. Nat Rev Cancer. 2020;20(4):218–232. doi: 10.1038/s41568-019-0235-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pathria P, Louis T, Varner JA. Targeting tumor-associated macrophages in cancer. Trends Immunol. 2019;40(4):310–327. doi: 10.1016/j.it.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 37.Kumagai S, Togashi Y, Sakai C, Kawazoe A, Kawazu M, Ueno T, Sato E, Kuwata T, Kinoshita T, Yamamoto M, et al. An oncogenic alteration creates a microenvironment that promotes tumor progression by conferring a metabolic advantage to regulatory T cells. Immunity. 2020;53(1):187–203.e188. doi: 10.1016/j.immuni.2020.06.016. [DOI] [PubMed] [Google Scholar]
- 38.Tokunaga R, Zhang W, Naseem M, Puccini A, Berger MD, Soni S, McSkane M, Baba H, Lenz H-J. CXCL9, CXCL10, CXCL11/CXCR3 axis for immune activation – a target for novel cancer therapy. Cancer Treat Rev. 2018;63:40–47. doi: 10.1016/j.ctrv.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mikucki M, Fisher D, Matsuzaki J, Skitzki JJ, Gaulin NB, Muhitch JB, Ku AW, Frelinger JG, Odunsi K, Gajewski TF, et al. Non-redundant requirement for CXCR3 signalling during tumoricidal T-cell trafficking across tumour vascular checkpoints. Nat Commun. 2015;6(1):7458. doi: 10.1038/ncomms8458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chow M, Ozga A, Servis R, Frederick DT, Lo JA, Fisher DE, Freeman GJ, Boland GM, Luster AD. Intratumoral activity of the CXCR3 chemokine system is required for the efficacy of anti-PD-1 therapy. Immunity. 2019;50(6):1498–1512.e1495. doi: 10.1016/j.immuni.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Y, Liang X, Dong W, Fang Y, Lv J, Zhang T, Fiskesund R, Xie J, Liu J, Yin X, et al. Tumor-repopulating cells induce PD-1 expression in CD8+ T cells by transferring kynurenine and AhR activation. Cancer Cell. 2018;33(3):480–494.e487. doi: 10.1016/j.ccell.2018.02.005. [DOI] [PubMed] [Google Scholar]
- 42. Zhao R, Song Y, Wang Y, Huang Y, Li Z, Cui Y, Yi M, Xia L, Zhuang W, Wu X, et al. PD-1/PD-L1 blockade rescue exhausted CD8+ T cells in gastrointestinal stromal tumours via the PI3K/Akt/mTOR signalling pathway. Cell Prolif. 2019;52(3):e12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vonderheide R. The immune revolution: a case for priming, not checkpoint. Cancer Cell. 2018;33(4):563–569. doi: 10.1016/j.ccell.2018.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh M, Vianden C, Cantwell M, Dai Z, Xiao Z, Sharma M, Khong H, Jaiswal AR, Faak F, Hailemichael Y, et al. Intratumoral CD40 activation and checkpoint blockade induces T cell-mediated eradication of melanoma in the brain. Nat Commun. 2017;8(1):1447. doi: 10.1038/s41467-017-01572-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leblond M, Tillé L, Nassiri S, Gilfillan CB, Imbratta C, Schmittnaegel M, Ries CH, Speiser DE, Verdeil G. CD40 agonist restores the antitumor efficacy of anti-PD1 therapy in muscle-invasive bladder cancer in an IFN I/II-mediated manner. Cancer Immunol Res. 2020;8(9):1180–1192. doi: 10.1158/2326-6066.CIR-19-0826. [DOI] [PubMed] [Google Scholar]
- 46.Lawson KA, Sousa CM, Zhang X, Kim E, Akthar R, Caumanns JJ, Yao Y, Mikolajewicz N, Ross C, Brown KR, et al. Functional genomic landscape of cancer-intrinsic evasion of killing by T cells. Nature. 2020;586(7827):120–126. doi: 10.1038/s41586-020-2746-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article


