ABSTRACT
The natural killer group 2 member D (NKG2D) receptor is a C-type lectin-like activating receptor mainly expressed by cytotoxic immune cells including NK, CD8+ T, γδ T and NKT cells and in some pathological conditions by a subset of CD4+ T cells. It binds a variety of ligands (NKG2DL) whose expressions is finely regulated by stress-related conditions. The NKG2DL/NKG2D axis plays a central and complex role in the regulation of immune responses against diverse cellular threats such as oncogene-mediated transformations or infections. We generated a panel of seven highly specific anti-human NKG2D single-domain antibodies targeting various epitopes. These single-domain antibodies were integrated into bivalent and bispecific antibodies using a versatile plug-and-play Fab-like format. Depending on the context, these Fab-like antibodies exhibited activating or inhibitory effects on the immune response mediated by the NKG2DL/NKG2D axis. In solution, the bivalent anti-NKG2D antibodies that compete with NKG2DL potently blocked the activation of NK cells seeded on immobilized MICA, thus constituting antagonizing candidates. Bispecific anti-NKG2DxHER2 antibodies that concomitantly engage HER2 on tumor cells and NKG2D on NK cells elicited cytotoxicity of unstimulated NK in a tumor-specific manner, regardless of their apparent affinities and epitopes. Importantly, the bispecific antibodies that do not compete with ligands binding retained their full cytotoxic activity in the presence of ligands, a valuable property to circumvent immunosuppressive effects induced by soluble ligands in the microenvironment.
KEYWORDS: NKG2D/NKG2DL axis, NK cells, HER2, single-domain antibody, cell engagers
Introduction
In recent years, the complex two-edged role of the immune system, controlling or shaping/promoting tumor development, has become evident. Indeed, the tumor microenvironment including the infiltrated immune cells plays an important role in the tumor aggressiveness and the response to treatments.1 Tumor escape partly results from the modeling of its microenvironment and the creation of an immunosuppressive environment leading to ineffective antitumor immune responses.2,3 Strategies interfering with this tumor-induced immune tolerance, although challenging, hold much promise.4,5 Among them, targeting immune cells via immune checkpoint inhibitors have recently revolutionized the therapeutic approaches for several cancers with a poor prognosis.4 Several antibodies blocking different inhibitory receptors (PD-1/PD-L1 axis and CTLA-4)6,7 expressed by dysfunctional T-cells have been approved worldwide. However, a majority of patients do not respond to such treatments, stressing the need to explore new tracks and/or new immune checkpoints. Targeting of the innate immune effector cells, including NK cells, macrophages, and dendritic cells, is becoming increasingly promising and many immunomodulatory antibodies are being developed.8,9
NK cells are critical actors for immunosurveillance through their capacity to eliminate transformed cells (i.e. tumor or infected cells) without antigen priming or prior sensitization. Most importantly they secrete inflammatory mediators (cytokines (IFN-γ, TNF-α) and/or chemokines) that participate to the recruitment and priming of other types of immune cells.10,11 NK cell effector function is finely tuned by a balance of inhibitory and activating receptors.12 In humans, inhibitory receptors include the immunoglobulin–like receptors (KIRs) with a long cytoplasmic tail13 and the lectin-like CD94/NKG2A heterodimer14 against which antagonist antibodies are currently being developed in various cancer indications.15,16 As a counterpart, NK cells constitutively express activating receptors including FcγRIIIa (CD16A), well characterized as the effector of antibody-dependent cell-mediated cytotoxicity (ADCC), natural cytotoxicity receptors (NCRs) such as NKp30, NKp46, or KIRs with a short cytoplasmic tail and NKG2D.17,18
Natural killer group 2, member D (NKG2D) receptor is a type II transmembrane protein with a C-type lectin-like extracellular domain, expressed as a disulfide-linked homodimer on cell surface. Beside NK cells, NKG2D is expressed by several subsets of T cells such as γδ T cells, CD8+ T cells, and invariant NKT cells representing a bridge between innate and adaptive immunity.19,20 NKG2D functions as an hexameric complex made of an NKG2D homodimer in association with two DAP10 homodimers19 and has the unique particularity of binding a diversity of highly polymorphic ligands due to a conformational plasticity.21 NKG2D ligands (MICA, MICB, and UL16-binding proteins (ULBPs)) are cell-surface proteins, structurally related to major histocompatibility complex (MHC) class I proteins that are expressed in response to cellular stress, infection, or disease including cancer.22 Their expression, which is restricted or absent in normal tissues, directly correlates with cell sensitivity to NK cell-mediated lysis.23 Engagement of NKG2D by its ligands triggers cytotoxicity and cytokine secretion (GM-CSF, TNF-α, IFN-γ, MIP-1b) in cytokine-activated human NK cells, while NKG2D-mediated activation of resting NK cells requires co-ligation of other activating receptors such as 2B4 or NKp46.24,25 In human CD8+ T cells and γδ T cells, NKG2D ligation provides a co-activation signal that contributes to cytotoxicity and cytokine production.26
Numerous studies unraveled the role of the NKG2D/NKG2DL axis in the immune surveillance of damaged, infected, or transformed cells.18,19 However, NKG2D/NKG2DL functionality can be compromised by different strategies developed by tumor cells such as down-regulation or shedding of NKG2DL.27,28 It was also recently reported that cancer cells can appropriate NKG2D for their own benefit, thereby promoting tumor progression.29,30 Furthermore, depending on the environmental context, immune actors such as NK, macrophages, dendritic cells, and T cells can express NKG2DL, which may contribute to the down-modulation of immune response and/or fratricide.31,32 Altogether, these data underlie the importance and the complexity of the NKG2D/NKG2DL axis in pathophysiology, especially in anti-tumor responses. Several strategies are currently being developed to restore or stimulate the NKG2D/NKG2DL axis functionality including protein fusions involving recombinant NKG2DL or NKG2D and antibody fragments.33 For instance, NKG2DL fused to antigen-binding fragment of anti-tumor antibodies targeting a tumor-associated antigen has been used to artificially enhance the NKG2DL expression at the surface of cancer cells, leading to NK-mediated cell lysis.34,35 However, the development and use of these bispecific formats have several limitations, including lack of epitope diversity on NKG2D, and improper folding or instability.
In the last few years, single-domain antibodies derived from camelid heavy chain-only antibodies have proven to be of interest as building blocks to generate innovative therapeutics against various human diseases.36 Interestingly, ISVDs (immunoglobulin single variable domains) are able to bind epitopes not accessible to conventional antibodies, a natural property that makes them ideally suited to generate blocking or agonist molecules. Moreover, their small size (15 kDa) allows the generation of multivalent, multispecific, or multiparatopic constructs that can display distinctive advantages over conventional monoclonal antibodies.37–39
Based on the above-mentioned unique characteristics of ISVDs, we generated and characterized a set of NKG2D-specific ISVDs with either blocking or non-blocking properties and incorporated them successfully into bivalent or bispecific antibodies in association with an anti-tumor ISVD. We further investigated the potential of these novel NKG2D-specific antibodies to induce activation and trigger cytotoxicity of NK cells in a tumor-specific manner.
Materials and methods
Cell lines and human donor-derived cells
Cell lines were obtained from the American Type Culture Collection (Manassas, VA), submitted to no more than 20 passages and cultured in a humidified environment at 37°C and 5% CO2. Human breast cancer cell line BT-474 (ATCC® HTB-20™) was cultured in RPMI-GlutaMAX™ (Gibco) supplemented with 10% (v/v) of fetal bovine serum (FBS – Eurobio). NK92 cells (ATCC® CRL-2407™) were cultured in RPMI-GlutaMAX™ supplemented with 10% FBS (v/v) and 200 U/ml IL-2 (Proleukin – Novartis, gift of Cancer Research Center of Marseille). HEK293-T (HEK 293 T/17 ATCC® CRL-11268™) cells were cultured in DMEM-GlutaMAX™ (Gibco) supplemented with 10% FBS. FreeStyle™ HEK293-F cells (Invitrogen) were cultured in Freestyle™ 293 expression medium (Gibco). Human peripheral blood mononuclear cells (PBMC) were isolated from fresh peripheral blood of healthy donors (Etablissement Français du Sang (EFS), Marseille, France) by Ficoll LSM 1077 (Sigma) gradient centrifugation. Human NK cells were isolated by negative selection (human NK cell isolation kit, Miltenyi Biotec) according to the manufacturer’s instructions. Purity and activation status of NK cells were determined by staining with anti-CD3 PE, anti-CD56 APC, and anti-CD107a FITC antibodies (Miltenyi Biotec). Freshly purified NK cells were grown at 106 cells/mL in RPMI-GlutaMAX™ supplemented with 10% (v/v) FBS with or without 1000 UI/mL of IL-2. Expanded human γδT cells isolated from healthy PBMC were obtained from C. Fauriat (CRCM, Marseille, France). Human NK cells numerically expanded in the presence of artificial antigen-presenting cells and cytokines were described elsewhere.40
The following antibodies were used for human immune cells phenotyping: Vioblue-conjugated anti-CD3 or CD4, APC-conjugated anti-TCRγδ or CD56 or CD8, and FITC-conjugated anti-CD3 mAb (Miltenyi Biotec).
Cell transfection for NKG2D expression
FreeStyle 293-F cells in suspension were co-transfected with high-quality plasmid preparations encoding human NKG2D and DAP10 using 293fectinTM (Gibco) according to the manufacturer’s instructions. Adherent HEK293-T cells were co-transfected at 70–80% confluence using Lipofectamine™ 2000 Transfection Reagent (Invitrogen) diluted in Opti-MEM (Gibco) according to the manufacturer’s instructions. For both HEK293-derived cell lines, NKG2D expression was evaluated 24 hours post-transfection by flow cytometry using PE-conjugated anti-human NKG2D mAb (Miltenyi Biotec).
Llama immunization and library construction
Immunization and library construction were conducted by the VIB Nanobody Service Facility (Brussels, Belgium) as follows. A llama (Lama glama) was immunized through five subcutaneous injections of about 200 µg of human His-tagged NKG2D recombinant protein (Sino Biological) mixed with Gerbu LQ#3000 adjuvant at weekly intervals. On day 40, anticoagulated blood was collected and peripheral blood lymphocytes (PBLs) were prepared. The ISVD library was constructed as previously described.41 Briefly, total RNA was extracted from PBLs and used as template for first-strand cDNA synthesis with oligo(dT) primers. ISVD encoding sequences were amplified by PCR from cDNA, digested with PstI and NotI, and cloned between the PstI and NotI sites of the phagemid vector pHEN4 upstream the decapeptide HA-tag. An ISVD library of about 108 independent transformants was obtained.
Phage display panning
A ready-to-use phage-ISVD preparation was obtained as previously described.42 Briefly, the bacteria library was grown in 2YTAG medium (2xYT medium, Ampicillin 100 μg/mL, Glucose 2%) until the OD600nm reached 0.5 and infected with M13K07 helper phage (Invitrogen). After centrifugation, bacteria were resuspended in 2YTAK (2xYT medium, Ampicillin 100 μg/mL, Kanamycin 50 µg/mL) medium and grown overnight. Phage particles were precipitated from culture supernatant by addition of PEG8000 20% (w/v) and 2.5 mM NaCl, centrifuged and resuspended in PBS. Phages were submitted to one more wash and precipitation steps and finally resuspended in cold PBS/glycerol 15% (v/v).
Panning on recombinant protein
M-450 Epoxybeads (Dynabeads – Invitrogen) were coated with His-tagged NKG2D recombinant protein (Sino Biological) following manufacturer’s recommendations. The phage-ISVD library (1011 phages/selection round) and the beads (coated and naked) were saturated in PBS/milk 2% (w/v) for 1 h at room temperature. The phage-ISVD library was first depleted twice by 30 min incubation on naked beads to eliminate nonspecific clones. Unbound phage-ISVDs were recovered and incubated with NKG2D-coupled beads during 2 h, in PBS/milk 2%, at room temperature. After 10 washes with PBS/Tween 0.1% (v/v) and 2 washes with PBS, bound phage-ISVDs were resuspended in PBS (output selection), added to exponentially growing TG1 bacteria and either amplified overnight in 2YTAG medium for a new round of panning or plated on 2YTAG plates.
Panning on cells
Panning was performed at 4°C on either NK92 cells or NKG2D/DAP10-transfected HEK293-F cells. The phage-ISVD library was saturated in PBS/BSA 2% and incubated with 2 × 107 cells for 2 h at 4°C. After two washes with PBS, the cell pellet was resuspended in PBS and loaded on an FBS/Percoll gradient as previously described.43 After centrifugation cell layer was collected and washed twice with PBS. Recovered cells (with bound phages) were added into a second FBS/Percoll gradient and washed before mechanical lysis using beads (Dynabeads – Invitrogen). Recovered phage-ISVDs were used to infect exponentially growing E. coli TG1 bacteria and either amplified overnight in 2YTAG medium for a new round of panning or plated on 2YTAG plates.
Phage-ISVD and ISVD production in 96-well plates
Individual TG1 colonies from the selection outputs were randomly picked using a colony picker (Molecular Device) and grown in 96-well plates as follows.
ISVD production
Each colony was grown overnight in 2YTAG at 37°C. Overnight culture was then used to inoculate 2YTA medium in new 96-well plates. After growing for 2 h at 37°C, the production of ISVD was induced by the addition of 1 mM IPTG (isopropyl β-D-1-thiogalactopyranoside) followed by overnight incubation at 30°C. Supernatants containing ISVDs were harvested and used for screening.
Phage-ISVD production
Each colony was grown in 2YTA at 37°C until the OD600nm reached 0.5. Cells were then infected with M13K07 helper phage and grown overnight in 2YTAK at 30°C. Supernatants containing phage-ISVDs were harvested and used for screening.
Bispecific and bivalent Fab-like constructions, production and purification
After amplification by PCR, cDNA of the anti-NKG2D ISVD, anti-FMDV (Foot-and-Mouth Disease Virus) ISVD44 or anti-HER2 ISVD45 were cloned into a proprietary mammalian expression vector upstream and in frame with either the human CL domain or the human IgG1 CH1 domain fused to HA and 6-His tags. Plasmids were purified using NucleoBond Macherey-Nalgel kits and Sanger sequenced. Bispecific (bsFab) or bivalent Fab-like (bvFab) antibodies were produced by cotransfecting FreeStyle 293-F cells with a mix of 2 plasmids encoding two distinct (bsFab) or two identical (bvFab) ISVDs fused to each of the Fab constant domains. Supernatants were harvested 7 days later, purified on Nickel affinity columns and analyzed on CALIPER GXII (Perkin Elmer).
ELISA binding and competition assays
ELISA assays were performed on Nunc® MaxiSorp™ 96-well plates (Sigma) pre-coated overnight with 1 µg/ml of human His-tagged NKG2D recombinant protein in PBS at 4°C and further saturated with PBS/milk 2% for 1 h at room temperature. For binding assays, bacteria supernatants containing ISVDs or purified Fab-like antibodies were incubated for 1 h at room temperature. For competition assays, serial dilutions (8 pM-500 nM) of bvFab were incubated for 1 h at room temperature then phage-ISVDs at their EC90 were added and the incubation was extended by 45 min at room temperature. Alternatively, bvFabs or anti-hNKG2D mAb (149810- R&D Systems) or human His tagged-MICA protein (Sino Biological) were incubated for 1 h at room temperature before adding NKG2D ligands (MICA, MICB, ULBP1 and ULBP2 fused to Fc from R&D Systems) at a concentration corresponding to their EC90 for 45 min at room temperature. After several washes in PBS/Tween 0.1%, the following HRP-conjugated antibodies were added: anti-HA tag mAb (Sigma) for detection of bound Fab-like formats and ISVDs, anti-M13 mAb (Santa Cruz Biotechnology) to detect bound phage-ISVDs and anti-human IgG (Fc-specific) mAb (Sigma) to detect bound NKG2DL-Fc fusions. Detection of peroxidase activity was performed using TMB (3,3ʹ,5,5ʹ-Tétraméthylbenzidine – KPL) substrate and OD450nm was measured on a SpectraMax microplate reader after addition of sulfuric acid stop solution.
Flow cytometry binding and competition assays
All flow cytometry assays were performed on a MACSQuant cytometer (Miltenyi Biotec) using V-bottom 96-well microtiter plates. Cells were gated on viable cells (Propidium Iodure staining) and on single-cell populations and 104 events were collected for each sample. Data were analyzed with the MACSQuant software and the results were expressed as median of fluorescence intensity.
Binding assays
BT-474 or expanded NK cells or IL2-pre stimulated cells (1.5x105 cells/well) were incubated with purified bvFabs or bsFabs or supernatants containing ISVDs for 1 h at 4°C. Bound antibodies were detected by staining with either a PE-conjugated anti-HA mAb (Miltenyi Biotec) or a mouse anti-HA mAb (Sigma) followed by Alexa647-conjugated goat anti-mouse mAb (Life Technologies). Three washes in PBS/BSA 2% were performed between each incubation step.
Competitive binding assays
hNKG2D/hDAP10 transiently transfected FreeStyle 293-T cells were incubated with serial dilutions of bvFabs and phage-ISVDs at their EC90 for 90 min at 4°C. Phage-ISVDs were detected by Alexa-647 conjugated anti-M13 mAb (LifeTechnologies). Alternatively, cells were first incubated with serial dilutions of bvFabs for 1 h at 4°C and then with human NKG2D ligands fused to Fc at their EC90 for 45 min at 4°C. The ELISA competition assay was first used to confirm that all NKG2D ligands competed with each other for NKG2D binding as expected from structural and binding studies46 (Supplementary Figure 1). Bound ligands were detected by an anti-human IgG (Fc-specific) mAb followed by Alexa647-conjugated goat-anti mouse mAb.
Expression of NKG2D ligands on cancer cell lines
BT-474 cells were incubated 45 min at 4°C with the following mAbs against hNKG2D ligands: MAB1300 (MICA), MAB1599 (MICB), MAB1380 (ULBP1) and MAB1298 (ULBP2,5,6) (all from R&D Systems). After several washes, bound antibodies were detected with Alexa647-conjugated goat anti-mouse mAb for 45 min at 4°C. Cell viability was evaluated by propidium iodide staining. The fluorescence of cells was acquired on a MACSQuant analyzer.
Functional assays
Freshly isolated human NK cells (resting cells) were pre-stimulated for 12 h with 1000UI/mL of IL-2 before the assay. Addition of 10 ng/ml phorbol myristate acetate (PMA – Sigma) and 1 µg/ml ionomycin (Sigma) was used as positive control for NK cell activation. All functional assays were performed in Nunc Maxisorp 96-well microplates pre-coated overnight at 4°C with different combinations of antibodies (ratio 1/1, total mAb concentration: 10 µg/mL) depending on the desired readouts. Washes were performed with PBS/Tween 0.1% before the saturation step in PBS/milk 2% for 1 h at room temperature and the addition of pre-stimulated NK cells for 2 h at 37°C, 5% CO2. NK cells were harvested, stained with FITC-conjugated anti-CD107a or FITC-conjugated isotype control antibodies (Miltenyi Biotec) and analyzed by flow cytometry to assess their viability (Propidium iodide staining (Miltenyi Biotec)) and their degranulation status. The fluorescence was acquired on a MACSQuant analyzer and expressed as the percentage of viable CD107a-positive NK cells. Supernatants were harvested, stored at −20°C and analyzed for secretion of TNF-α using the Human TNF-α ELISA kit (Invitrogen – eBioscience) according to the manufacturer’s instructions.
For ISVD screening, 96-well plates were precoated with anti-HA tag mAb (Sigma) in combination with either mouse IgG1 (Biolegend) or anti-human 2B4 mAb (C1.7 – Biolegend) at ratio 1/1. Supernatants containing NKG2D ISVDs harvested from bacterial productions were added for 1 h at room temperature. Controls with supernatants from irrelevant and anti-human CD16 ISVD production were added.
For Fab-like format analysis, the constructs were directly coated on microplates in combination with IgG1 or anti-2B4 mAb. Wells precoated with a mix of anti-hNKG2D mAb/anti-2B4 mAb, anti-hNKG2D mAb/mIgG1 or anti-hCD16 (3G8 – Biolegend) mAb/mIgG1 were added as controls.
Blockade of NK cell activation by soluble bvFab-like antibodies
To test the blocking properties of bvFabs, 96-well microplates were precoated with HIS-tagged MICA protein in combination with anti-2B4 mAb. IL2-stimulated human NK cells (1.5x105 cells/well) were added in the presence or absence of 150 nM NKG2D bvFab, FMDV bvFab or anti-NKG2D mAb. After 2 h incubation at 37°C, NK cell activation was assessed by detection of CD107a expression by flow cytometry as described above. The percentage of CD107a-positive NK cells obtained in the condition [HIS-tagged MICA/anti-2B4 mAb combination] in the absence of soluble antibodies was set at 100%, to facilitate the comparison between experiments. The combinations [His-tagged MICA/mIgG1] and [anti-2B4/mIgG1 antibodies] were introduced as controls.
Flow cytometry-based cytotoxicity assay
Target cells (BT-474) were stained with carboxyfluorescein diacetate succinimidyl ester (CFSE – Life Technologies) according to manufacturer’s instructions. Briefly CFSE (1 µM) was added to the target cell suspension (106 cells/ml) in PBS at 37°C for 20 min in 5% CO2 in the dark. Cells were then washed and analyzed by flow cytometry for viability (>95%) and staining. Unstimulated NK cells were mixed with CFSE-labeled tumor cells (3.4x105/well) at the indicated effector/target (E:T) ratios in RPMI-GlutaMAX™ medium supplemented with 10% FBS in the presence of soluble bsFabs and/or His-tagged MICA and incubated for 2 h at 37°C (5% CO2) or 24 h for TNF-α secretion analysis. Cells were washed with PBS/BSA 2%, stained with TO-PRO®-3 (LifeTechnologies) and analyzed by flow cytometry on a MACSQuant cytometer. Maximal signal corresponding to total lysis of target cells was obtained by addition of Triton™ X-100 (1% v/v) (Sigma). Gating on TO-PRO®-3-positive population within the CFSE-positive population indicated dead target cells in percentage (%). Supernatants were stored at −20°C and analyzed for secretion of TNF-α using the Human TNF-α ELISA kit and human granzyme B using the Human Granzyme B Platinum kit (Invitrogen™ – eBioscience™).
Statistical analysis
Data were analyzed with GraphPad Prism software (V5.01) and are presented as mean ± SD or SEM. Statistical analysis data were performed by one-way ANOVA with repeated measures followed by Bonferroni-Holm adjustment or by two-way ANOVA on log- or rank-transformed data with repeated measures followed by two-tailed Dunnett’s test. P-values below 0.05 were considered statistically significant.
Results
Selection of anti-human NKG2D single-domain antibodies by phage display
NKG2D-specific ISVDs were selected from the library by a first round of panning on recombinant human NKG2D immobilized on beads followed by a second round of panning using three NKG2D sources as bait: (i) human recombinant NKG2D protein-coated beads, (ii) NK92 cells, or (iii) HEK293-F cells transiently transfected with hNKG2D/DAP10 (Supplementary Table 1). After each round of selection, 95 clones were randomly picked and screened for NKG2D binding by ELISA on recombinant human NKG2D and by flow cytometry on hNKG2D/DAP10-transfected HEK293-F cells. A total of 287 positive clones on recombinant NKG2D and/or membrane-bound NKG2D were identified. From sequence analysis and production yields, 23 clones were selected and further characterized (AR1 to AR23). Based on a sequence identity rate higher than 95%, 4 different clusters were identified (C1-C4) across 12 clones and the remaining 11 clones were present as singletons (supplementary Table 2). Cluster C1 dominated the panning on recombinant NKG2D whereas clones belonging to clusters C2 and C3 were mainly amplified from selections on NKG2D-expressing cells. Clones of cluster C4 were less frequent and appeared essentially after the first round of selection. All clones are VHHs except AR23 that holds hallmarks of VH.
Functional screening of NKG2D ISVDs
We developed a reliable and rapid functional test allowing medium-throughput screening (96-well plates) of NKG2D ISVDs for their ability to activate IL-2 prestimulated human NK cells. The test relies on the engagement of NK cell surface receptors via plate-bound antibodies and on the synergy between NKG2D and 2B4 (CD244), another co-activation NK cell receptor.24 NK cell activation was assessed by flow cytometry through the detection of CD107a (LAMP-1), a marker of NK cell degranulation (Figure 1).
Figure 1.
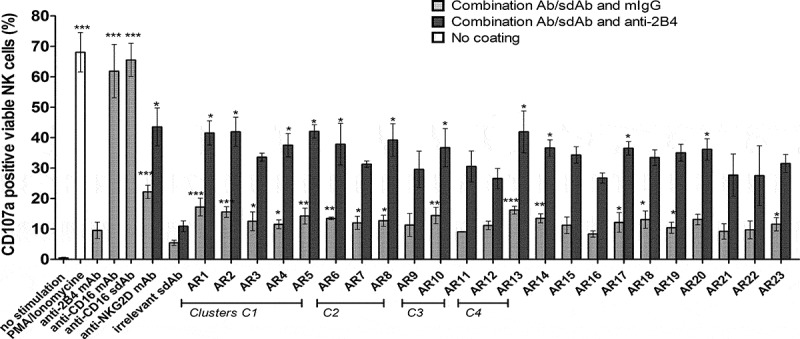
NK cell activation by immobilized anti-NKG2D ISVDs. Bacterial supernatants containing soluble ISVDs were added onto 96-well microplates pre-coated with a 1:1 mix of either anti-HA tag mAb and mIgG1 (gray bars) or anti-HA tag mAb and anti-2B4 mAb (dark gray bars). IL-2 pre-stimulated NK cells were incubated onto plates for 2 h, harvested and the percentage of CD107a-positive cells was quantified by flow cytometry. PMA/Ionomycin was used as positive control (white bar). Control conditions with anti-CD16 ISVD, anti-CD16, anti-2B4, anti-NKG2D mAbs were included. Each bar represents the mean value ± SEM of 3 independent assays (different donors). One-way analysis of variance with repeated measures and Bonferroni-Holm adjustment were used to compare each compound with irrelevant ISVD. * P < .05, ** p < .001, ***P < .0001
To evaluate the activating potential of individual NKG2D-specific ISVDs, HA-tagged ISVDs were captured from bacterial culture supernatants via plate-bound anti-HA mAb with either control IgG or anti-2B4 mAb. The presence of ISVDs in the bacterial medium was ascertained by dot-blot (data not shown). PMA/ionomycin, plate-bound anti-CD16 mAb or anti-CD16 ISVD induced a strong response with more than 60% of NK cells expressing CD107a, whereas immobilized irrelevant ISVD was unable to activate NK cells. Engagement of NKG2D by an anti-NKG2D mAb induced a small but significant increase of CD107a surface expression whereas 2B4 receptor engagement alone did not. By contrast, the co-engagement of both receptors, NKG2D and 2B4, triggered a strong activation of NK cells with more than 40% of cells expressing CD107a at their surface. Fifteen out of the 23 NKG2D binders were able to induce a low but significant increase of CD107a expression when compared to an irrelevant ISVD. Co-activation with the anti-2B4 mAb dramatically increased the capacity of all of them to elicit NK cell degranulation compared to an irrelevant ISVD (Figure 1).
Generation of bivalent and bispecific Fab-like antibodies
Bivalent or bispecific antibodies were generated using the compact and linker-free Fab-like format depicted in Figure 2.47 This format takes advantage of the natural heterodimerization of human IgG CH1 and CL domains. To generate bsFabs, the NKG2D ISVDs were associated with the previously described anti-HER2 ISVD C7b45 or with the irrelevant control FMDV ISVD M3, 44 whereas bvFabs were obtained using the same NKG2D ISVD in fusion with both CH1 and CL. Based on production yields and sequence diversity, constructs derived from seven ISVDs (AR3, AR6, AR11, AR18, AR19, AR20 and AR23) were selected for further characterization. A total of 21 Fab-like antibodies were produced as soluble molecules and purified successfully.
Figure 2.
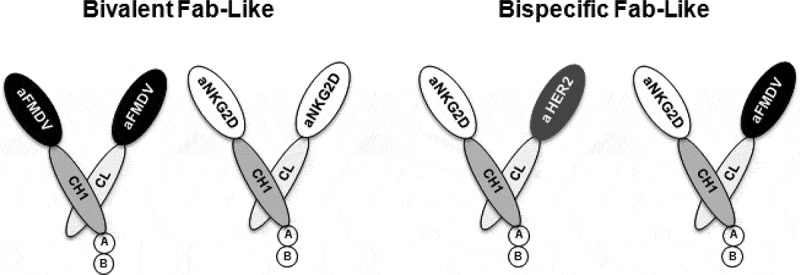
Schematic representation of bivalent and bispecific Fab-like antibodies. The CH1 constant domain of human IgG1 (dark gray) was fused to anti-NKG2D ISVD (white) whereas the CL constant domain (light gray) was fused to the same anti-NKG2D ISVD to generate bivalent Fab-like antibodies or to an anti-HER2 (C7b) or anti-FMDV ISVDs (gray and black respectively) to build bispecific Fab-like antibodies. (a): HA tag, (b): hexahistidine tag
BvFabs bind different epitopes on NKG2D
To analyze the epitope diversity of the NKG2D ISVDs, pairwise competition experiments between bvFabs (competitor) and phage particles displaying anti-NKG2D ISVDs (phage-ISVDs, tracer) were performed by flow cytometry using NKG2D/DAP10-transfected HEK293-T cells. In these experiments, each ISVD was thus assessed both as tracer and competitor in combination with all other ISVDs. Competition of ISVD with themselves for binding to plate-bound recombinant NKG2D had been previously assessed by ELISA (Supplementary Figure 2). Representative competition profiles obtained with bvFabsAR20 (Figure 3a), AR23 (Figure 3b), and AR6 (Figure 3c) are shown. Whereas bvFabAR6 competed with all phage-ISVDs, bvFabAR20 competed with the phage-ISVDs AR3, 11, 18 but not AR6, 19 and 23. BvFab AR23 competed only with the phage-ISVDs AR6 and AR19. Finally, all bvFabs but AR6 were split into two distinct epitope bins; bin A gathers AR3, AR11, AR18, and AR20 whereas AR19 and AR23 belong to bin B. Epitope binning by ELISA led to the same conclusions (data not shown).
Figure 3.
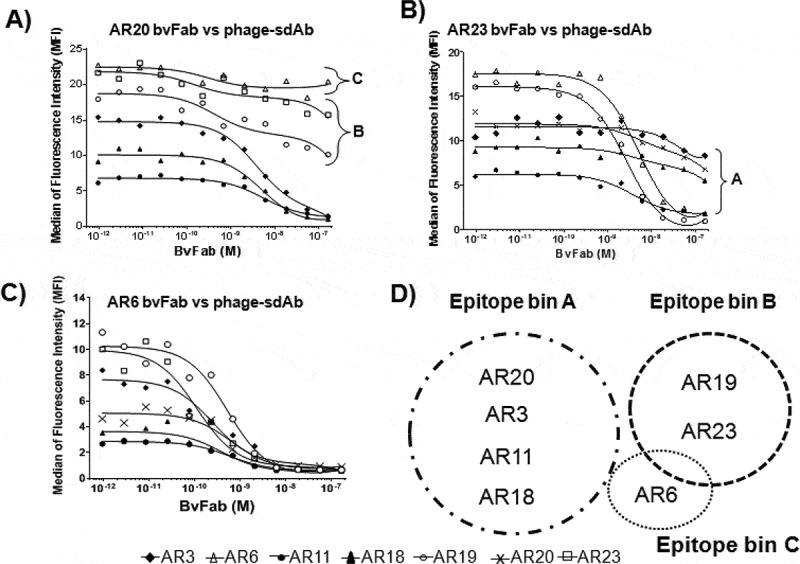
Epitope binning experiments. NKG2D/DAP10-transfected FreeStyle 293-T cells were incubated at 4°C for 90 min with ISVDs-on-phage added at their predetermined EC90 concentration and increasing concentrations of NKG2D bvFab. Phage binding was detected by flow cytometry using an Alexa-647 conjugated anti-M13 mAb. Representative profiles obtained with bvFab AR20 (a), bvFab AR23 (b) and bvFab AR6 (c) are shown. (d) Schematic representation of NKG2D ISVD epitope bins
Bispecific and bivalent Fab-like antibodies bind specifically NKG2D
The apparent affinity of bsFabs and bvFabs was evaluated by ELISA on recombinant NKG2D or by flow cytometry on expanded human NK cells (Table 1).
Table 1.
Binding properties of bivalent or bispecific Fab-like antibodies
| VHH αNKG2D | KDapp (nM) on rNKG2D |
KDapp (nM) on NK cells |
KDapp (nM) on BT-474 cells |
||
|---|---|---|---|---|---|
| bvFab | bsFab | bvFab | bsFab | bsFab | |
| AR3 | 0.2 ± 0.06 | 25 ± 9 | 0.40 ± 0.07 | 25 ± 8 | 102 ± 17 |
| AR6 | 0.3 ± 0.12 | 1.3 ± 0.5 | 0.40 ± 0.07 | 13 ± 5 | 101 ± 13 |
| AR11 | 0.4 ± 0.17 | 5 ± 1 | 6 ± 3 | ND>100 | 104 ± 14 |
| AR18 | 12 ± 4 | ND | 20 ± 4 | ND | 69 ± 8 |
| AR19 | 0.50 ± 0.10 | 12 ± 6 | 1.65 ± 0.48 | 30 ± 6 | 89 ± 12 |
| AR20 | 0.3 ± 0.05 | 7 ± 1 | 3.2 ± 0.95 | ND | 94 ± 19 |
| AR23 | 0.4 ± 0.13 | 20 ± 1 | 0.30 ± 0.09 | 20 ± 6 | 92 ± 23 |
Apparent affinities of bvFabs and bsFabs were determined by ELISA on recombinant NKG2D and by flow cytometry on expanded human NK and BT-474 HER2-positive cells. Values of KDapp (nM) are means ± SEM of independent experiments. ND = Values not determined due to the absence of a plateau.
All Fab-like antibodies bound recombinant NKG2D with apparent KD (KDapp) values ranging from 1.3 to 25 nM in bispecific format and 0.2 to 12 nM in bivalent format. AR6 and AR18 display the highest and the lowest apparent affinities, respectively. More variability was observed when determining their apparent affinity in a native context by flow cytometry. Most bsFabs bound NK cells, reaching more than 95% of positively stained expanded NK cells at the maximal-tested concentration. BsFabs AR3, AR19, and AR23 displayed KDapp values similar to those determined by ELISA whereas bsFab AR6 showed a 10-fold lower apparent affinity as compared to its affinity for recombinant NKG2D. Binding of bsFabs AR18, AR11, and AR20 did not reach saturation in the range of tested concentrations.
As observed by ELISA, the bivalent formats showed a 20- to 60-fold affinity gain compared to their monovalent cognate formats. BvFabs AR3, AR6, AR18, AR19, and AR23 exhibited similar apparent affinity on both recombinant and membrane-bound NKG2D, whereas bvFabs AR11 and AR20 displayed a 10-fold lower apparent affinity for membrane-bound NKG2D.
We further tested the binding of one representative bvFab from epitope bins A (AR3) and B (AR19) on PBMC from healthy donors. As expected from NKG2D expression profile on human PBMC, both bvFabs stained NK, CD8+ T and γδ T cells but not CD4+ T lymphocytes (Figure 4).
Figure 4.
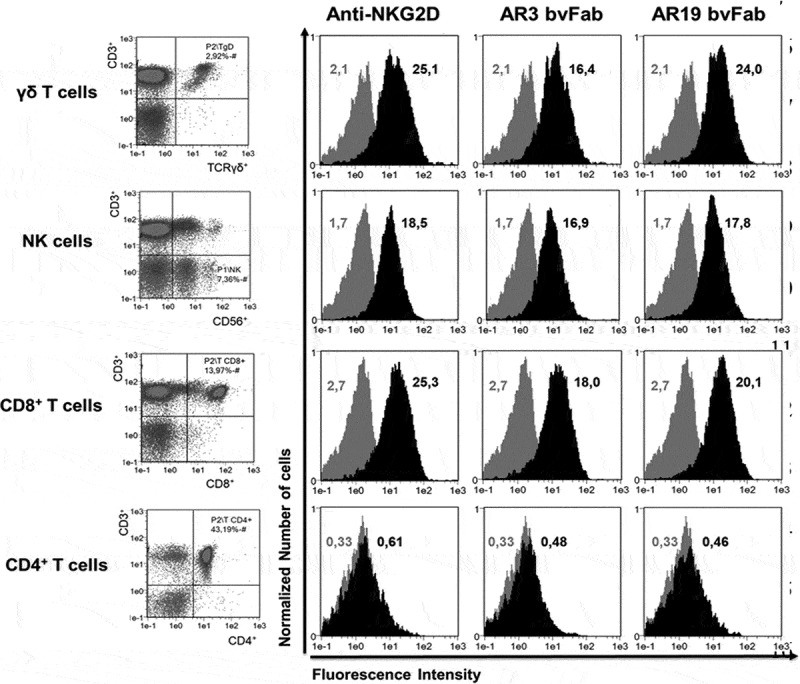
Binding of anti-NKG2D bvFabs on different immune cell populations. Cell subsets (CD3−CD56+ NK cells, CD3+CD8+ and CD3+ CD4+ T cells, CD3+ TCRγδ+ T cells) of human PBMC were identified by a combination of surface markers (left panel). Binding of anti-NKG2D bvFabs on the different cell subsets was detected by flow cytometry using PE-conjugated anti-HA mAb. PE-conjugated anti-human NKG2D mAb was used as positive control. Representative histograms (black) of AR3 and AR19 bvFabs binding are shown and compared to PE-isotype binding (gray). Numbers displayed in each histogram are the median of fluorescence intensity of 2 independent experiments
Finally, no binding of bvFab was observed on either murine NKG2D or human NKG2A recombinant proteins, demonstrating a high level of specificity (data not shown). Binding properties of all NKG2DxHER2 bsFabs were also investigated by flow cytometry on the HER2-positive cell line BT-474 (Table 1). All of them bound similarly the tumor cell line and displayed an apparent affinity around 93 ± 15 nM, in agreement with previously reported data.48
Blocking properties of soluble anti-NKG2D-bvFabs
To further explore the epitope diversity, we evaluated the ability of the different bvFabs to interfere with the interaction between NKG2D and some of its ligands, namely MICA, MICB, ULBP1, and ULBP2.
The binding of MICA-, MICB-, ULBP1- and ULBP2-Fc fusions was analyzed on recombinant NKG2D and on NKG2D/DAP10-transfected HEK293-T cells in the presence of increasing concentrations of bvFabs. Bivalent Fabs belonging to bins B and C, as well as the control anti-NKG2D mAb, hampered the binding of all ligands (Table 2, supplementary figure 3). In contrast, bvFabs from bin A did not block the binding of MICA, MICB, and ULBP1 but partially interfered with the binding of ULBP2 on recombinant NKG2D. Surprisingly, some bvFabs from bin A behaved differently depending on the source of NKG2D. For example, AR11 bvFab interfered with the interaction of MICA and MICB on membrane-bound NKG2D but not on recombinant NKG2D and AR20 bvFab partially inhibited the binding of all ligands at the highest tested concentration. Of note, all bvFabs prevented (AR6, AR19, AR23) or at least interfered (AR3, AR11, AR18, and AR20) with the binding of ULBP2.
Table 2.
Blocking properties of bvFab on NKG2D ligands, MICA, MICB, ULBP1, and ULBP2. Dark: competition, dark gray: partial competition, light gray: partial competition at high concentration, white: no competition
 |
To investigate whether these competition properties translate functionally and impact MICA-mediated NK cell activation, IL2-stimulated NK cells were incubated with or without soluble anti-NKG2D bvFabs on plates coated with MICA and anti-2B4 mAb (Figure 5).
Figure 5.
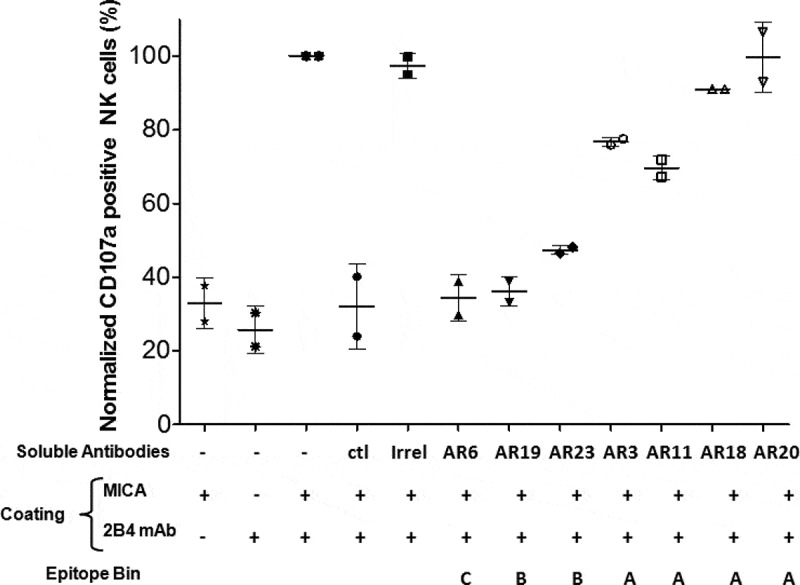
Blockade of MICA-mediated NK cell activation by soluble NKG2D bvFab. Human recombinant MICA protein and/or anti-2B4 mAb (ratio 1/1) were immobilized on microplates. IL2-stimulated human NK cells and soluble antibodies (150 nM) were then added. After 2 h of incubation, NK cell activation was assessed by quantification of cell surface CD107a by flow cytometry. The percentage of CD107a-positive NK cells obtained in the presence of MICA and anti-2B4 mAb without any soluble anti-NKG2D mAb was normalized to 100%. Ctl = soluble anti-NKG2D mAb. Irrel = FMDV bvFab. Each bar represents the mean value ± SD of 2 independent experiments
In agreement with their ability to compete with MICA for NKG2D binding (Table 2), soluble bvFabs from bins B and C drastically decreased the percentage of CD107a-positive NK cells. Of note, although belonging to bin A, bvFabs AR3, and AR11 partially inhibited MICA-induced activation of NK cells.
Combining immobilized anti-NKG2D Fab-like and anti-2B4 mAb induces degranulation and cytokine release by human NK cells
The capacity of immobilized NKG2D Fab-like antibodies to induce NK cell degranulation and TNF-α secretion was explored. IL2-prestimulated human NK cells were incubated with plate-bound Fab-like antibodies in combination with either mIgG or anti-2B4 mAb. Cell surface expression of CD107a and production of TNF-α were evaluated (Figure 6).
Figure 6.
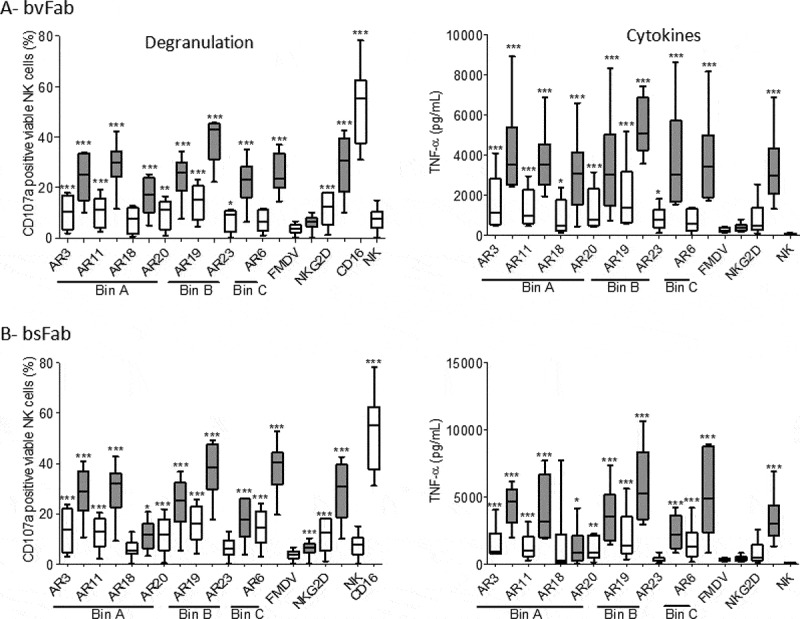
NK cell activation by NKG2D bvFabs or bsFabs. IL2-prestimulated human NK cells were added to 96-well microplates pre-coated with a mix of (a) bvFab or (b) bsFab either with mIgG1 (white bars) or anti-2B4 mAb (grey bars). Monoclonal anti-NKG2D and anti-CD16 antibodies, and FMDV bvFab were used as positive and negative controls, respectively. After 2 h of incubation, NK cell activation was evaluated by quantification of cell surface CD107a by flow cytometry (left panel) and of secreted TNF-α levels by ELISA (right panel). Assays were performed 6 times (n = 6 donors). Data are represented as box-and-whisker plots with min and max. Two-way ANOVA with repeated measures on rank-transformed data with two-tailed Dunnett’s test was used to compare CD107a values for each compound versus FMDV bvFab. Two-way ANOVA with repeated measures on log-transformed data with two-tailed Dunnett’s test was used to compare TNF-α levels for each compound with FMDV bvFab. * P < .05, **p < .01, ***P < .0001
In the absence of anti-2B4 mAb, most anti-NKG2D Fab-like antibodies induced a modest but significant NK cell degranulation and TNF-α secretion compared to an irrelevant Fab-like. The co-engagement of 2B4 strongly stimulated these processes as demonstrated by the higher percentage of CD107a-positive NK cells and higher TNF-α release induced by all bivalent (Figure 6(a)) and bispecific (Figure 6(b)) constructs except for AR18 bsFab, when compared to the irrelevant Fab.
Soluble bsFabs drive NK cell-mediated killing of HER2-positive tumor cells
Redirected cell lysis experiments using unstimulated human NK cells and HER2-positive BT-474 tumor cells were performed to evaluate the capacity of bsFabs to mediate NKG2D-driven cell cytotoxicity. As shown in Figure 7a, the HER2xCD16 bsFab48 used as positive control induced a strong killing of BT-474 tumor cells (80% with an EC50 around 10 pM) at an E/T ratio of 6. All anti-NKG2D bsFabs elicited significant NK cytotoxicity in a dose-dependent manner as compared to their cognate FMDV bsFabs tested at the highest concentration (Figure 7(b,c)). However, their potency varied considerably. No EC50 could be determined within the tested concentration range for bsFabs AR18, AR20, AR19, and AR23 while the EC50 of bsFabs AR3, AR11, and AR6 varied from 1 to 16 nM (Table 3).
Figure 7.
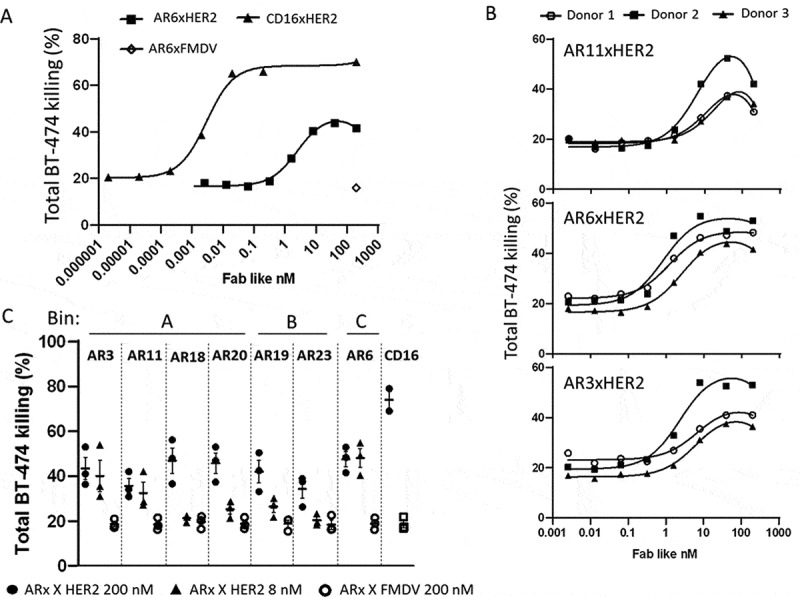
Redirected NK cell lysis against BT-474 tumor cells by NKG2DxHER2 bsFabs. CFSE-stained BT-474 cells (3.4x105 cells/well) were incubated for 2 h with unstimulated human NK cells (E/T:6/1) in the presence or absence of serial dilutions of NKG2DxHER2, NKG2DxFMDV or HER2xCD16 bsFabs. TO-PRO®-3 was used as dead cell indicator. Percentage of total BT-474 cell lysis was determined by flow cytometry as the percentage of double positive CFSE/TO-PRO-3 cells. (a) Representative curves for CD16XHER2 and AR6xHER2 bsFabs (1 donor). (b) Tumor cell lysis curves for the 3 most potent HER2xNKG2D bsFabs. (c) Killing of BT-474 tumor cells mediated by 200 nM and 8 nM HER2xNKG2D bsFabs (resp. black circle and triangle) and 200 nM of their cognate FMDVxNKG2D (open circle) bsFabs. (b) and c) represent data of 3 independent experiments (3 donors)
Table 3.
Potency of NKG2D-bsFabs against BT-474 tumor cells. Values of EC50 (nM) are means ± SEM of three independent experiments
| bsFab | EC50 (nM) |
|---|---|
| CD16xHER2 | 0.0067 ± 0.0019 |
| AR3xHER2 | 5.2 ± 1.4 |
| AR6xHER2 | 1.6 ± 0.5 |
| AR11xHER2 | 15.4 ± 5.2 |
Importantly, the cognate irrelevant NKG2DxFMDV bsFab did not enhance tumor cell death above the baseline killing induced by NK cells alone (~20%) at the highest tested concentration (200 nM).
Finally, in contrast to HER2xCD16 bsFab, none of the anti-NKG2D bsFabs triggered measurable secretion of TNF-α under the tested conditions even when the cytotoxicity reaction was extended for 24 hours (data not shown).
Taken together, these results demonstrate that within 2 hours, all HER2xNKG2D bsFabs were able to promote specific killing of HER2-overexpressing tumor cells by NK cells but did not lead to the secretion of pro-inflammatory cytokines.
BsFab-mediated NK cell cytotoxicity in the presence of soluble human MICA
Because shedding of NKG2D ligands is a common mechanism used by tumor cells to escape recognition and killing by NK cells, we investigated the ability of the 7 bsFabs to mediate NK cytotoxicity against BT-474 tumor cells in the presence of soluble human MICA (Figure 8).
Figure 8.
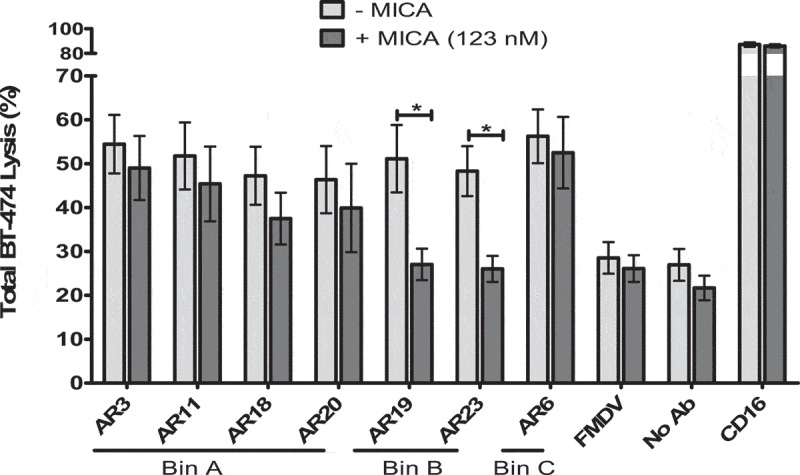
Effect of soluble NKG2D ligand on redirected NK cell lysis mediated by bsFabs. CFSE-stained BT-474 cells were incubated for 2 h with unstimulated human NK cells (E/T:6/1) and 40 nM bsFabs in the presence (dark gray bar) or absence (light gray bar) of 123 nM human His-tagged MICA. TO-PRO®-3 was used as dead cell indicator. BT-474 cell killing was quantified by flow cytometry as the percentage of double positive CFSE/TO-PRO-3 cells. Each bar represents the mean value ± SEM of 4 independent experiments. The p values were calculated with one-tailed Student’s t-test for bsFab in the presence or absence of soluble MICA. * P < .05
In these experiments, the concentration of soluble MICA was set to 123 nM, far above the soluble MICA concentration found in serum samples of patients with tumors (0.4–0.8 nM49) but approaching the estimated KD (~0.5 µM).50 The presence of soluble MICA abolished the functionality of AR19 and AR23 bsFabs (epitope bin B) shown to compete with MICA. Intriguingly, the NK cell cytotoxicity mediated by the AR6 bsFab was not altered although this bsFab was shown to partially compete with MICA in our experiments. As expected, the presence of soluble MICA did not interfere with the cytotoxic activity of HER2xCD16 bsFab.
Discussion
The activating NKG2D receptor is increasingly recognized as a potent receptor for the immune surveillance of diverse cellular stresses such as viral infection or cell transformation and emerges as a promising target for immunotherapy. NKG2D engagement triggers NK, CD8+ and/or γδ T cell cytotoxicity and cytokine secretion leading to the killing of damaged or dysfunctional cells. However, such cells have developed strategies to escape NKG2D-mediated immunosurveillance.51 In the oncology field, various therapeutic strategies are currently explored for restoring the functionality of the NKG2D/NKG2DL axis such as drug-controlled induction of NKG2DL expression on cancer cells, blockade of soluble NKG2DL activity by suppression of its shedding or its depletion with specific antibodies, tumor-specific reactivation of NKG2D by bispecific proteins made of the extracellular domain of NKG2D or NKG2DL linked to cytokines or antibodies, or infusion of donor-derived immune cells expressing NKG2D (NK cells or NKG2D-CARs T cells)33,52
Here, we report the design of novel immunotherapeutics targeting NKG2D with different functional and binding properties. These immunotherapeutics are based on single-domain antibodies derived from camelid heavy chain-only antibodies and modulate NK cell functions. To our knowledge, this is the first study using single-domain antibodies to target NKG2D. So far, most NKG2D engagers have been made of extracellular domains of the NKG2D ligands fused to tumor-targeting moieties. One recent study reports the promising in vitro and in vivo results of a single-chain variable fragments (scFvs)-based construct cotargeting NKG2D and CS1, a tumor-associated antigen in multiple myeloma.53 This scFv construct is prone to aggregation and instability, and derives from an antagonist NKG2D mAb, thus competing with NKG2D ligands. Here, we offer a panel of anti-NKG2D ISVD, a highly stable format, which contrary to scFv fragments, does not rely on a peptidic linker for VH/VL pairing. This panel offers various epitopes and affinities as building blocks easy to incorporate into a plug-and-play bispecific format to specifically switch on the cytolytic activity of NK cells against tumor cells.
Alternative panning strategies combined with NK cell activation assays were applied to ensure the identification of NKG2D binders with the broadest epitope coverage and NK cell activation capacities. We showed that 15 ISVDs among the 23 identified were able to significantly activate a low proportion of IL2-prestimulated human NK cells. Reminiscent of the observations made by different laboratories, 24,25,54 we showed that, in addition to cytokine-mediated activation of NK cells, the synergy between activating signals induced by the co-engagement of NKG2D and 2B4 triggered a 2–3 fold increase in the number of CD107a-positive NK cells. The choice of 2B4 was guided by the expression of this receptor on some NKG2D-expressing T cells subsets of CD8+ T and γδ T cells.55,56 Altogether this early functional screening established the capacity of plate-bound anti-NKG2D ISVDs to activate cytokine-stimulated NK cells.
Seven ISVDs, representative of three clusters (AR3, 6, and 11) or unique (AR18, 19, 20, and 23), were selected for the construction of bivalent monospecific and monovalent bispecific antibodies47,48 for in-depth characterization. All of them proved to be highly specific for the human receptor with no cross-reactive binding to mouse NKG2D, despite high sequence similarity. Apparent affinities of bispecific antibodies for NK cells varied from 13 nM to over 100 nM whereas bvFabs displayed higher apparent affinity ranging from 0.3 to 20 nM, a phenomenon explained by avidity.
The seven ISVDs were grouped into three epitope bins based upon their ability to block one another’s binding in a pairwise fashion. Whereas bins A and B appear clearly different, bin C is less distinct as AR6 binding hindered the binding of class B ISVDs and also altered the binding of class A ISVDs in an asymmetric fashion. Abdiche et al57 reported a phenomenon of antibody displacement due to kinetic perturbation of binding when two antibodies target very close or minimally overlapping epitopes. Based on this report, one could speculate that epitopes from classes B and C, if not the same, are overlapping and that epitope A and C are closely adjacent or minimally overlapping. However, it does not explain the asymmetric competition between AR6 and class A clones. That AR6 engraftment in bvFab but not on phage prevents binding of the class A ISVDs may be due to allosteric effect, or more likely, to steric hindrance or even favorable binding parameters such as disparate affinity/avidity or kinetics of association/dissociation.
To go further, we demonstrated that bvFabs from classes B and C totally blocked the binding of all tested soluble ligands (MICA, MICB, ULBP1, and ULBP2) on NKG2D-transfected cells. By contrast, class A ISVD could be refined into three subclasses according to their variable capacity to alter ligand binding. Our results also suggest that conformational or post-translational differences between plate-bound and membrane-bound NKG2D can affect bvFab binding to NKG2D and hence their capacity to compete with the ligands. Altogether, these results demonstrate that the seven selected ISVDs bind selectively to NKG2D at distinct epitopes and with a range of affinity.
Evaluation of the activating potential of immobilized NKG2D Fab-like molecules demonstrated that all of them significantly elicited degranulation and TNF-α secretion by IL-2-prestimulated NK cells when combined with an anti-2B4 co-activating antibody, highlighting the synergy between NKG2D and 2B4 in NK cell activation. Of note, in our experimental setting, antibody immobilization on a surface was an absolute prerequisite since no cytokine secretion was observed in the presence of soluble Fab-like molecules (data not shown), thus uncontrolled killing triggered by Fab-like molecules in the absence of target cells should not be a concern in vivo.
All bsFabs further proved to be able to induce significant killing of the HER2-positive BT-474 cell line by resting NK cells. No clear benefit related to the targeted epitope or the apparent affinity emerged. Even the AR18 bsFab turned out to be active despite its modest apparent affinity for NK cells. Importantly, the cognate irrelevant NKG2DxFMDV bsFabs were unable to elicit target cell lysis. bsFabs/bvFabs can be considered as mimics of natural NKG2D ligands, that trigger NKG2D activation when bound to membrane, but not when soluble. Further investigations would be required to explore the impact of Fab-like molecules binding on the clustering of NKG2D at the membrane compared to NKG2DL, the intracellular signaling mediated by their ligation to the receptor and the modulation of NKG2D expression. Numerous data suggest that the internalization of NKG2D/DAP10 triggered by ligand binding is required for signal propagation and downstream NK cell cytotoxic activity.28
It should also be emphasized that the cytolytic activity of resting NK cells was triggered by soluble HER2xNKG2D bvFab without exogenous 2B4 co-activation, indicating that the bridging of NK and tumor cells by the bsFabs allows the formation of an active immune synapse, a feature that might be linked to the compact Fab-like format geometry. Indeed, it has been reported that the overall binding geometry of bispecific T-cell engagers (antibody format, size and angle, antigen size and distance of epitope to membrane), by influencing the tightness of the immune synapse, contributes in part to their in vitro potency.58,59 By contrast, the drivers of NK cell engager efficacy have been so far much less studied. Generating affinity variants of the NKG2D ISVDs while keeping the same binding geometry and intercellular distance might help deciphering the relative contribution of affinity and NKG2D epitope on the in vitro potency of anti-NKG2D bsFabs. In the same vein, targeting two different epitopes with a biparatopic antibody could be studied to strengthen NKG2D clustering and ultimately increase both potency and efficacy of ISVD-based therapeutics.
The shedding of NKG2D ligands by proteolytic cleavage or the release of NKG2DL-bearing exosomes are strategies used by tumor cells to protect themselves against immune cell recognition and killing. Although conflicting results exist, it has been described that both soluble and exosomal NKG2D ligands down-regulate NKG2D on NK and T cells, leading to impaired NKG2D-mediated cytotoxicity, the exosomal ligands having a strongest effect.28,60 Our results demonstrate that bsFabs with no or low capacity to compete with NKG2D ligands may circumvent the immunosuppressive effect of soluble NKG2DL since they retained their cytotoxic activity in the presence of soluble MICA. Of note, the killing mediated by AR6 bsFab, the bsFab with highest affinity in the ligand competitor bin, was not affected by the presence of soluble MICA. This suggests that increasing the affinity of NKG2D ISVD beyond some threshold could be an efficient alternative to counteract a locally high NKG2DL concentration and associated immunosuppressive effect.
The capacity of some clones to compete with ligand binding translated functionally as they were able to block MICA/2B4-induced NK cell activation when tested as soluble reagents. Interestingly, our results also show that such clones can behave as activating molecules when bound to a surface (plastic or cell), suggesting that the format – mono or bispecific – can drive the mechanism of action of NKG2D-targeting antibodies. The dual activity of ligand-blocking anti-NKG2D antibodies has been previously reported by Kwong et al61 and Steigerwald et al62 who concluded that the ambivalent characteristics may translate into a wide therapeutic benefit.
More recently, the expression of NKG2D has been observed at the surface of ligand-bearing tumor cells and correlated with tumor progression.30,63 Even if the level of NKG2D expression is low, this phenomenon seems to be widespread in many cancers. It has been proposed that by interacting in cis with its ligands, NKG2D could behave as a tumor growth factor receptor, playing a role in cancer cell plasticity and in metastasis disease.63 In this context, antagonist NKG2D bvFabs may be attractive not only as therapeutics in inflammatory diseases but also as tools to investigate this new role of NKG2D in cancer.
In agreement with the intrinsic nature of the NKG2D receptor, all bsFabs were less effective and potent than the reference anti-HER2xCD16 bsFab in triggering tumor cell lysis by NK cells. However, numerous studies have demonstrated that the NK cell infiltrate in human tumor samples comprises poorly cytotoxic cells with low levels of CD16. Therefore, looking for other activating receptors than CD16 is an attractive strategy for potentiating the NK anti-tumor response and NKp46 has been recently proposed in that respect.64 We show here that NKG2D, whose expression is not limited to NK cells in the battery of tumor-infiltrated effector cells (CD8-positive T cells and γδ T cells), holds also great promise for cancer immunotherapy.
Supplementary Material
Acknowledgments
We thank F. Duffieux (Sanofi) for purification of Fab-like formats, Valerie Martin (Sanofi) for support to statistical analysis.
Funding Statement
This work was financially supported by SANOFI (Collaboration agreement SANOFI/INSERM Transfert), INSERM and CNRS. AR was supported by a CIFRE fellowship (N°2015/0738) funded in part by ANRT (National Association for Research and Technology) on the behalf of the French Ministery of Education and Research and in part by SANOFI.
Disclosure of potential conflicts of interest
AR, EV, KD and LV are employees of Sanofi, a company which develops therapeutic antibodies. The authors have no additional conflict of interest.
Authors’ contributions
Conception and design of the project: AR, BK, EV; Analysis and interpretation of the data: AR, LV, KD, BK, EV; Generation and acquisition of data: AR with inputs of LV, ET and KD; Study methodology: AR, LV, KD, BK, EV. Statistical analysis: EV and BK with support of Valérie Martin (Sanofi). Writing, reviewing and editing were performed by EV and BK with inputs from AR, PC, LV, DB, and KD.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Hegmans JP, Aerts JG.. Immunomodulation in cancer. Curr Opin Pharmacol. 2014;17:17–14. doi: 10.1016/j.coph.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 2.Fridman WH, Zitvogel L, Sautès–Fridman C, Kroemer G.The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol. 2017;14(12):717–734. doi: 10.1038/nrclinonc.2017.101. [DOI] [PubMed] [Google Scholar]
- 3.Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541:321–330. [DOI] [PubMed] [Google Scholar]
- 4.Mahoney KM, Rennert PD, Freeman GJ.. Combination cancer immunotherapy and new immunomodulatory targets. Nat Rev Drug Discov. 2015;14:561–584. [DOI] [PubMed] [Google Scholar]
- 5.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12(4):252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong Y, Sun Q, Zhang X. PD-1 and its ligands are important immune checkpoints in cancer. Oncotarget. 2017;8(2):2171–2186. doi: 10.18632/oncotarget.13895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grosso JF, Jure-Kunkel MN. CTLA-4blockade in tumor models: an overview of preclinical and translational research. Cancer Immun. 2013;13:5. [PMC free article] [PubMed] [Google Scholar]
- 8.Davis ZB, Vallera DA, Miller JS, Felices M. Natural killer cells unleashed: checkpoint receptor blockade and BiKE/TriKE utilization in NK-mediated anti-tumor immunotherapy. Semin immunol. 2017; 31:64–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devillier R, Chretien AS, Pagliardini T, Salem N, Blaise D, Olive D. Mechanisms of NK cell dysfunction in the tumor microenvironment and current clinical approaches to harness NK cell potential for immunotherapy. J Leukoc Biol. 2020. doi: 10.1002/JLB.5MR0920-198RR. [DOI] [PubMed] [Google Scholar]
- 10.Hamerman JA, Ogasawara K, Lanier LL. NK cells in innate immunity. Curr Opin Immunol. 2005;17(1):29–35. doi: 10.1016/j.coi.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 11.Souza-Fonseca-Guimaraes F, Cursons J, Huntington ND. The emergence of natural killer cells as a major target in cancer immunotherapy. Trends Immunol. 2019;40(2):142–158. doi: 10.1016/j.it.2018.12.003. [DOI] [PubMed] [Google Scholar]
- 12.Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol. 2008;9(5):503–510. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- 13.Pende D, Falco M, Vitale M, Cantoni C, Vitale C, Munari E, Bertaina A, Moretta F, Del Zotto G, Pietra G, et al. Killer Ig-Like Receptors (KIRs): their role in NK cell modulation and developments leading to their clinical exploitation. Front Immunol. 2019;10:1179. doi: 10.3389/fimmu.2019.01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McWilliams EM, Mele JM, Cheney C, Timmerman EA, Fiazuddin F, Strattan EJ, Mo X, Byrd JC, Muthusamy N, Awan FT. Therapeutic CD94/NKG2A blockade improves natural killer cell dysfunction in chronic lymphocytic leukemia. Oncoimmunology. 2016;5(10):e1226720. doi: 10.1080/2162402X.2016.1226720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andre P, Denis C, Soulas C, Bourbon-Caillet C, Lopez J, Arnoux T, Blery M, Bonnafous C, Gauthier L, Morel A, et al. Anti-NKG2A mAb is a checkpoint inhibitor that promotes anti-tumor immunity by unleashing both T and NK cells. Cell. 2018;175(7):1731–1743 e1713. doi: 10.1016/j.cell.2018.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kohrt HE, Thielens A, Marabelle A, Sagiv-Barfi I, Sola C, Chanuc F, Fuseri N, Bonnafous C, Czerwinski D, Rajapaksa A, et al. Anti-KIR antibody enhancement of anti-lymphoma activity of natural killer cells as monotherapy and in combination with anti-CD20 antibodies. Blood. 2014;123(5):678–686. doi: 10.1182/blood-2013-08-519199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y, Lu D, Churov A, Fu R. Research Progress on NK Cell Receptors and Their Signaling Pathways. Mediators Inflamm. 2020;2020:6437057. doi: 10.1155/2020/6437057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lazarova M, Steinle A. The NKG2D axis: an emerging target in cancer immunotherapy. Expert Opin Ther Targets. 2019;23(4):281–294. doi: 10.1080/14728222.2019.1580693. [DOI] [PubMed] [Google Scholar]
- 19.Lanier LL. NKG2D receptor and its ligands in host defense. Cancer Immunol Res. 2015;3(6):575–582. doi: 10.1158/2326-6066.CIR-15-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jamieson AM, Diefenbach A, McMahon CW, Xiong N, JR C, Raulet DH. The role of the NKG2D immunoreceptor in immune cell activation and natural killing. Immunity. 2002;17(1):19–29. doi: 10.1016/S1074-7613(02)00333-3. [DOI] [PubMed] [Google Scholar]
- 21.Zingoni A, Molfetta R, Fionda C, Soriani A, Paolini R, Cippitelli M, Cerboni C, Santoni A. NKG2D and its ligands: “One for All, All for One”. Front Immunol. 2018;9:476. doi: 10.3389/fimmu.2018.00476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carapito R, Bahram S. Genetics, genomics, and evolutionary biology of NKG2D ligands. Immunol Rev. 2015;267:88–116. [DOI] [PubMed] [Google Scholar]
- 23.Pende D, Rivera P, Marcenaro S, Chang CC, Biassoni R, Conte R, Kubin M, Cosman D, Ferrone S, Moretta L, et al. Major histocompatibility complex class I-related chain A and UL16-binding protein expression on tumor cell lines of different histotypes: analysis of tumor susceptibility to NKG2D-dependent natural killer cell cytotoxicity. Cancer Res. 2002;62:6178–6186. [PubMed] [Google Scholar]
- 24.Bryceson YT, March ME, HG L, Long EO. Synergy among receptors on resting NK cells for the activation of natural cytotoxicity and cytokine secretion. Blood. 2006;107(1):159–166. doi: 10.1182/blood-2005-04-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fauriat C, Long EO, HG L, Bryceson YT. Regulation of human NK-cell cytokine and chemokine production by target cell recognition. Blood. 2010;115(11):2167–2176. doi: 10.1182/blood-2009-08-238469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wensvee FM, Jelencic C, Polic B. NKG2D:A master regulator of immune cell responsiveness. Front Immunol. 2018;9:441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferrari de Andrade L, Tay RE, Pan D, Luoma AM, Ito Y, Badrinath S, Tsoucas D, Franz B, May KF Jr., Harvey CJ, et al. Antibody-mediated inhibition of MICA and MICB shedding promotes NK cell-driven tumor immunity. Science. 2018;359(6383):1537–1542. doi: 10.1126/science.aao0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Molfetta R, Quatrini L, Santoni A, Paolini R. Regulation of NKG2D-dependent NK cell functions: the Yin and the Yang of receptor endocytosis. Int J Mol Sci. 2017;18(8):1677. doi: 10.3390/ijms18081677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benitez AC, Dai Z, Mann HH, Reeves RS, Margineantu DH, Gooley TA, Groh V, Spies T. Expression, signaling proficiency, and stimulatory function of the NKG2D lymphocyte receptor in human cancer cells. Proc Natl Acad Sci U S A. 2011;108(10):4081–4086. doi: 10.1073/pnas.1018603108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cai X, Caballero-Benitez A, Gewe MM, Jenkins IC, Drescher CW, Strong RK, Spies T, Groh V. Control of tumor initiation by NKG2D naturally expressed on ovarian cancer cells. Neoplasia. 2017;19(6):471–482. doi: 10.1016/j.neo.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stojanovic A, Correia MP, Cerwenka A. The NKG2D/NKG2DL axis in the crosstalk between lymphoid and myeloid cells in health and disease. Front Immunol. 2018;9:827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trembath AP, Markiewicz MA. More than decoration: roles for natural killer group 2 member D ligand expression by immune cells. Front Immunol. 2018;9:231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ding H, Yang X, Wei Y. Fusion proteins of NKG2D/NKG2DL in cancer immunotherapy. Int J Mol Sci. 2018;19:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rothe A, Jachimowicz RD, Borchmann S, Madlener M, Keßler J, Reiners KS, Sauer M, Hansen HP, Ullrich RT, Chatterjee S, et al. The bispecific immunoligand ULBP2-aCEA redirects natural killer cells to tumor cells and reveals potent anti-tumor activity against colon carcinoma. Int J Cancer. 2014;134(12):2829–2840. doi: 10.1002/ijc.28609. [DOI] [PubMed] [Google Scholar]
- 35.Kellner C, Hallack D, Glorius P, Staudinger M, Mohseni Nodehi S, de Weers M, van de Winkel JG, Parren PW, Stauch M, Valerius T, et al. Fusion proteins between ligands for NKG2D and CD20-directed single-chain variable fragments sensitize lymphoma cells for natural killer cell-mediated lysis and enhance antibody-dependent cellular cytotoxicity. Leukemia. 2012;26(4):830–834. doi: 10.1038/leu.2011.288. [DOI] [PubMed] [Google Scholar]
- 36.Bannas P, Hambach J, Koch-Nolte F. Nanobodies and nanobody-based human heavy chain antibodies as antitumor therapeutics. Front Immunol. 2017;8:1603. doi: 10.3389/fimmu.2017.01603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arbabi-Ghahroudi M. Camelid single-domain antibodies: historical perspective and future outlook. Front Immunol. 2017;8:1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Del Bano J, Chames P, Baty D, Kerfelec B. Taking up cancer immunotherapy challenges: bispecific antibodies, the path forward? Antibodies. 2016;5(1):1. doi: 10.3390/antib5010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steeland S, Vandenbroucke RE, Libert C. Nanobodies as therapeutics: big opportunities for small antibodies. Drug Discov Today. 2016;21:1076–1113. [DOI] [PubMed] [Google Scholar]
- 40.Vidard L, Dureuil C, Baudhuin J, Vescovi L, Durand L, Sierra V, Parmantier E. CD137 (4-1BB) engagement fine-tunes synergistic IL-15- and IL-21-driven NK cell proliferation. J Immunol. 2019;203(3):676–685. doi: 10.4049/jimmunol.1801137. [DOI] [PubMed] [Google Scholar]
- 41.Arbabi Ghahroudi M, Desmyter A, Wyns L, Hamers R, Muyldermans S. Selection and identification of single domain antibody fragments from camel heavy-chain antibodies. FEBS Lett. 1997;414(3):521–526. doi: 10.1016/S0014-5793(97)01062-4. [DOI] [PubMed] [Google Scholar]
- 42.Even-Desrumeaux K, Nevoltris D, Lavaut MN, Alim K, Borg JP, Audebert S, Kerfelec B, Baty D, Chames P. Masked selection: a straightforward and flexible approach for the selection of binders against specific epitopes and differentially expressed proteins by phage display. Mol Cell Proteomics. 2014;13(2):653–665. doi: 10.1074/mcp.O112.025486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williams BR, Sharon J. Polyclonal anti-colorectal cancer Fab phage display library selected in one round using density gradient centrifugation to separate antigen-bound and free phage. Immunol Lett. 2002;81(2):141–148. doi: 10.1016/S0165-2478(02)00004-4. [DOI] [PubMed] [Google Scholar]
- 44.Harmsen MM, van Solt CB, Fijten HP, van Keulen L, Rosalia RA, Weerdmeester K, Cornelissen AH, De Bruin MG, Eblé PL, Dekker A. Passive immunization of guinea pigs with llama single-domain antibody fragments against foot-and-mouth disease. Vet Microbiol. 2007;120(3–4):193–206. doi: 10.1016/j.vetmic.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 45.Even-Desrumeaux K, Fourquet P, Secq V, Baty D, Chames P. Single-domain antibodies: a versatile and rich source of binders for breast cancer diagnostic approaches. Mol Biosyst. 2012;8(9):2385–2394. doi: 10.1039/c2mb25063b. [DOI] [PubMed] [Google Scholar]
- 46.McFarland BJ, Strong RK. Thermodynamic analysis of degenerate recognition by the NKG2D immunoreceptor: not induced fit but rigid adaptation. Immunity. 2003;19(6):803–812. doi: 10.1016/S1074-7613(03)00320-0. [DOI] [PubMed] [Google Scholar]
- 47.Rozan C, Cornillon A, Petiard C, Chartier M, Behar G, Boix C, Kerfelec B, Robert B, Pelegrin A, Chames P, et al. Single-domain antibody-based and linker-free bispecific antibodies targeting FcgammaRIII induce potent antitumor activity without recruiting regulatory T cells. Mol Cancer Ther. 2013;12(8):1481–1491. doi: 10.1158/1535-7163.MCT-12-1012. [DOI] [PubMed] [Google Scholar]
- 48.Turini M, Chames P, Bruhns P, Baty D, Kerfelec B. A FcgammaRIII-engaging bispecific antibody expands the range of HER2-expressing breast tumors eligible to antibody therapy. Oncotarget. 2014;5(14):5304–5319. doi: 10.18632/oncotarget.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Groh V, Wu J, Yee C, Spies T. Tumour-derived soluble MIC ligands impair expression of NKG2D and T-cell activation. Nature. 2002;419(6908):734–738. doi: 10.1038/nature01112. [DOI] [PubMed] [Google Scholar]
- 50.Spear P, Wu MR, Sentman ML, Sentman CL. NKG2D ligands as therapeutic targets. Cancer Immun. 2013;13:8. [PMC free article] [PubMed] [Google Scholar]
- 51.Schmiedel D, Mandelboim O. NKG2D ligands-critical targets for cancer immune escape and therapy. Front Immunol. 2018;9:2040. doi: 10.3389/fimmu.2018.02040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lazarova M, Wels WS, Steinle A. Arming cytotoxic lymphocytes for cancer immunotherapy by means of the NKG2D/NKG2D-ligand system. Expert Opin Biol Ther. 2020;1–11. doi: 10.1080/14712598.2020.1803273. [DOI] [PubMed] [Google Scholar]
- 53.Chan WK, Kang S, Youssef Y, Glankler EN, Barrett ER, Carter AM, Ahmed EH, Prasad A, Chen L, Zhang J, et al. A CS1-NKG2D bispecific antibody collectively activates cytolytic immune cells against multiple myeloma. Cancer Immunol Res. 2018;6(7):776–787. doi: 10.1158/2326-6066.CIR-17-0649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kwon HJ, Kim HS. Signaling for synergistic activation of natural killer cells. Immune Netw. 2012;12(6):240–246. doi: 10.4110/in.2012.12.6.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McNerney ME, Lee KM, Kumar V. 2B4 (CD244) is a non-MHC binding receptor with multiple functions on natural killer cells and CD8+ T cells. Mol Immunol. 2005;42(4):489–494. doi: 10.1016/j.molimm.2004.07.032. [DOI] [PubMed] [Google Scholar]
- 56.Simões AE, Di Lorenzo B, Silva-Santos B. Molecular determinants of target cell recognition by human γδ T cells. Front Immunol. 2018;9:929. doi: 10.3389/fimmu.2018.00929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abdiche YN, Yeung AY, Ni I, Stone D, Miles A, Morishige W, Rossi A, Strop P. Antibodies targeting closely adjacent or minimally overlapping epitopes can displace one another. PLoS One. 2017;12(1):e0169535. doi: 10.1371/journal.pone.0169535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bluemel C, Hausmann S, Fluhr P, Sriskandarajah M, Stallcup WB, Baeuerle PA, Kufer P. Epitope distance to the target cell membrane and antigen size determine the potency of T cell-mediated lysis by BiTE antibodies specific for a large melanoma surface antigen. Cancer Immunol Immunother. 2010;59(8):1197–1209. doi: 10.1007/s00262-010-0844-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li J, Stagg NJ, Johnston J, Harris MJ, Menzies SA, DiCara D, Clark V, Hristopoulos M, Cook R, Slaga D, et al. Membrane-proximal epitope facilitates efficient T Cell synapse formation by Anti-FcRH5/CD3 and is a requirement for myeloma cell killing. Cancer Cell. 2017;31(3):383–395. doi: 10.1016/j.ccell.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chitadze G, Bhat J, Lettau M, Janssen O, Kabelitz D. Generation of soluble NKG2D ligands: proteolytic cleavage, exosome secretion and functional implications. Scand J Immunol. 2013;78(2):120–129. doi: 10.1111/sji.12072. [DOI] [PubMed] [Google Scholar]
- 61.Kwong KY, Baskar S, Zhang H, Mackall CL, Rader C. Generation, affinity maturation, and characterization of a human anti-human NKG2D monoclonal antibody with dual antagonistic and agonistic activity. J Mol Biol. 2008;384(5):1143–1156. doi: 10.1016/j.jmb.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Steigerwald J, Raum T, Pflanz S, Cierpka R, Mangold S, Rau D, Hoffmann P, Kvesic M, Zube C, Linnerbauer S, et al. Human IgG1 antibodies antagonizing activating receptor NKG2D on natural killer cells. MAbs. 2009;1(2):115–127. doi: 10.4161/mabs.1.2.7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cai X, Dai Z, Reeves RS, Caballero-Benitez A, Duran KL, Delrow JJ, Porter PL, Spies T, Groh V. Autonomous stimulation of cancer cell plasticity by the human NKG2D lymphocyte receptor coexpressed with its ligands on cancer cells. PLoS One. 2014;9(10):e108942. doi: 10.1371/journal.pone.0108942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gauthier L, Morel A, Anceriz N, Rossi B, Blanchard-Alvarez A, Grondin G, Trichard S, Cesari C, Sapet M, Bosco F, et al. Multifunctional natural killer cell engagers targeting NKp46 trigger protective tumor immunity. Cell. 2019;177(7):1701–1713 e1716. doi: 10.1016/j.cell.2019.04.041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


