Abstract
There are several hurdles to overcome before implementing pharmacogenomics (PGx) in precision medicine. One of the hurdles is unawareness of PGx by clinicians due to insufficient pharmacogenomic information on drug labels. Therefore, it might be important to implement PGx that reflects pharmacogenomic information on drug labels, standard of prescription for clinicians. This study aimed to evaluate the level at which PGx was being used in clinical practice by comparing the Clinical Pharmacogenetics Implementation Consortium and Dutch Pharmacogenetics Working Group guidelines and drug labels of the US Food and Drug Administration (FDA) and the Korea Ministry of Food and Drug Safety (MFDS). Two PGx guidelines and drugs labels were scrutinized, and the concordance of the pharmacogenomic information between guidelines and drug labels was confirmed. The concordance of the label between FDA and MFDS was analyzed. In FDA labels, the number of concordant drug with guidelines was 24, while 13 drugs were concordant with MFDS labels. The number of drugs categorized as contraindication, change dose, and biomarker testing required was 7, 12 and 12 for the FDA and 8, 5 and 4 for the MFDS, respectively. The pharmacogenomic information of 9 drugs approved by both FDA and MFDS was identical. In conclusion, pharmacogenomic information on clinical implementation guidelines was limited on both FDA and MFDS labels because of various reasons including the characteristics of the guidelines and the drug labels. Therefore, more effort from pharmaceutical companies, academia and regulatory affairs needs to be made to implement pharmacogenomic information on drug labels.
Keywords: Pharmacogenomics, Guideline, Drug Labeling, United States Food and Drug Administration, Korea Ministry of Food and Drug Safety
INTRODUCTION
In 2015, President Barack Obama launched the precision medicine initiative, and the United States Food and Drug Administration (FDA) has gathered large-scaled biologic databases such as human genome sequence, metabolomics, and clinical studies for drugs. Like Obama's precision medicine initiative, modern medicine is heading towards precision medicine, a model that pursues personalization of healthcare that covers medical decisions, treatments, practices or products based on individual genotypic, phenotypic or psychosocial characteristics [1].
The concept of personalization of healthcare has existed before. For example, therapeutic drug monitoring has been practiced for a long time in clinical fields to adjust the drug dose according to the individual patients' status [2]. By identifying the pharmacokinetics and pharmacodynamics of a drug and the characteristics of the individual variances of a drug, clinicians or pharmacists can adjust the dosage of drugs or change to another drug for an individual patient. Nowadays, precision medicine is also implemented based on the concept of pharmacogenomics (PGx), and crowdsourcing platforms have made it possible for clinicians to access healthcare sources to improve precision medicine [3].
Many organizations associated with PGx such as the Clinical Pharmacogenetics Implementation Consortium (CPIC), Dutch Pharmacogenetics Working Group (DPWG), Pharmacogenomics Research Network and Ubiquitous Pharmacogenomics have been contributing to the implementation of PGx in clinical practice by establishing pharmacogenomic information and developing implementation tools [4,5]. CPIC and DPWG have released public guidelines to implement PGx in clinical practice. These PGx guidelines help clinicians and pharmacists understand how to interpret and apply the results of a genetic test for optimizing pharmacotherapy. Moreover, various tools have been designed like a personalized pocket card that integrates genetic test results into electronic medical records and a clinical decision support system. The card contains personal pharmacogenomic information and a quick response code connected to the individual's personalized dosing recommendations, and it could help clinicians to make a decision for personalized pharmacotherapy [6].
Not only academia but also regulatory affairs have made an effort to encourage PGx implementation. In Unite States, cloud-based next-generation DNA sequencing (NGS) data platform called precision FDA was established. The platform enables researchers to upload and compare data against reference genomes, bioinformatics pipelines, and genomic data [7]. In addition, the FDA released a “Guidance for Industry: Clinical Pharmacogenomics: Premarket Evaluation in Early-Phase Clinical Studies and Recommendations for Labeling” to recommend a prospective collection of samples for PGx research during early phase clinical studies [8]. In the Republic of Korea, the cost of some NGS-based oncology panel tests has been covered by the Korea National Health Insurance Service.
However, there are several huddles for implementing PGx in precision medicine. Cost of genetic tests and reimbursements are common problems facing implementation of PGx in clinical practice around the world [9]. Moreover, lack of education and unawareness of PGx by physicians and pharmacists who actually work with drugs are other obstacles [10,11]. Insufficient pharmacogenomic information on drug labels could be the main reason for the unawareness of PGx by physicians and pharmacists. Therefore, it might be important to implement PGx that reflects evidence-based pharmacogenomic information on drug labels which should be the standard of prescription labels for clinicians.
This study aimed to evaluate the level at which PGx was being used in clinical practice by comparing the PGx guidelines and drug labels of the FDA and Korea Ministry of Food and Drug Safety (MFDS). We reviewed two major public PGx implementation guidelines and identified whether the pharmacogenomic information was reflected on the drug labels. Furthermore, we discussed scientific and regulatory reasons underlying the discordance between the guidelines and drug labels.
METHODS
Data collection
Pharmacogenomic information of drugs listed on the CPIC (https://cpicpgx.org/guidelines/) and DPWG (https://www.knmp.nl/patientenzorg/medicatiebewaking/farmacogenetica/pharmacogenetics-1/pharmacogenetics) guidelines were reviewed and collected based on the following criteria: generic name of drug, drug related gene, genotype, and phenotype in accordance with genetic information. Based on all the drugs listed in the CPIC and DPWG guidelines, the drug labels of original or reference drugs were reviewed through the FDA (https://www.accessdata.fda.gov/scripts/cder/daf/) and MFDS (https://nedrug.mfds.go.kr/searchDrug) databases. The following information was collected: contraindication, change of dose (e.g., dose reduction), biomarker testing requirement, and consistent recommendation with the CPIC or DPWG guidelines. All information and data were collected as of September 2020.
Definition
“Concordance” indicated that the pharmacogenomic information between the PGx guidelines and drug labels was identical. When pharmacogenomic information between the guidelines and drug labels was not identical or there was no pharmacogenomic information on the drug labels, it was classified as “discordance.” Pharmacogenomic information belonging “concordance” was categorized as “contraindication,” “change dose” and “biomarker testing required” depending on the PGx related indication on the drug labels. Indication of both ‘changing dosage’ or ‘not changing dosage’ depending on genotypes was included in “change dose.”
Data analysis
All drugs listed in either the CPIC or DPWG guidelines were included in the analysis set, except drug categories classified as “This is not a gene-drug interaction” on the DPWG guideline. The drugs included in the analysis set were classified based on the anatomical therapeutic chemical (ATC) code by using the WHO-ATC/DDD Index (https://www.whocc.no/atc_ddd_index/) and PGx related genes.
To match the pharmacogenomic information in the two guidelines to the drug labels, the PGx related section on the drugs labels of the FDA and MFDS was scrutinized, and the concordance of the pharmacogenomic information between the guidelines and the drug labels was confirmed. The ratio of concordance and discordance, indication category of concordance, and ATC code of concordant drugs were analyzed by FDA and MFDS. Drugs that were not approved by the FDA and MFDS were excluded from the analysis, respectively. Among the pharmacogenomic information of drugs that matched the guidelines to the drug labels, the concordance of the label between the FDA and MFDS was analyzed.
RESULTS
Classification of drugs in the CPIC and DPWG guidelines
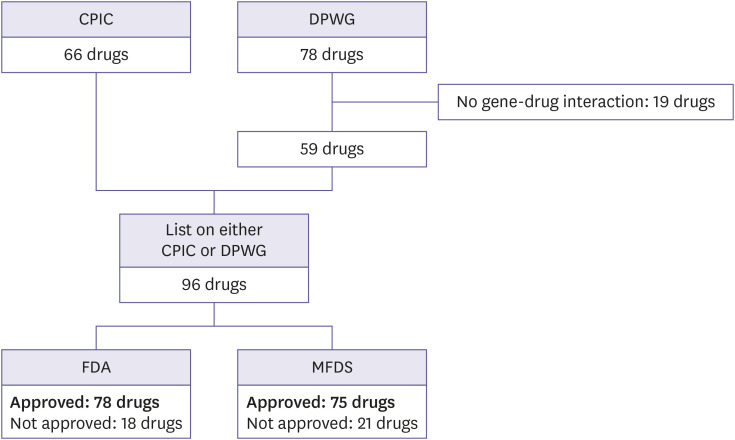
A total of 66 and 78 drugs were screened from the CPIC and DPWG guidelines, respectively. Among the 78 drugs listed on the DPWG, 19 drugs were excluded because they were classified as “this is not a gene-drug interaction.” The total number of drugs listed on either the CPIC or DPWG were 96 (Table 1 and Fig. 1).
Table 1. Summary of concordant drugs listed in the PGx guidelines and on the drug labels.
| Drug | Related Genes | Indication in the guidelines | Label indication | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CPIC | DPWG | FDA (concordance) | MFDS (concordance) | ||||||||
| Contraindication | Change dose | Biomarker testing required | Discordance | Contraindication | Change dose | Biomarker testing required | Discordance | ||||
| Abacavir | HLA-B | O | O | O | X | O | X | O | X | O | X |
| Aceclofenac | CYP2C9 | O | X | NA | X | X | X | O | |||
| Acenocoumarol* | CYP2C9 | X | O | NA | NA | ||||||
| VKORC1 | |||||||||||
| Allopurinol | HLA-B | O | X | X | X | X | O | O | X | O | X |
| Amitriptyline* | CYP2C9 | O | O | X | X | X | O | X | X | X | O |
| CYP2C19 | |||||||||||
| Aripiprazole | CYP2D6 | X | O | X | O | X | X | X | X | X | O |
| Aspirin | CYP2C9 | O | X | X | X | X | O | X | X | X | O |
| Atazanavir | UGT1A1 | O | X | X | X | X | O | X | X | X | O |
| Atomoxetine | CYP2D6 | O | O | X | O | X | X | X | X | X | O |
| Atorvastatin | SLCO1B1 | X | O | X | X | X | O | X | X | X | O |
| Azathioprine | TPMT | O | O | X | O | O | X | X | X | X | O |
| Brexpiprazole | CYP2D6 | X | O | X | O | X | X | NA | |||
| Carvedilol* | CYP2D6 | X | O | X | X | X | O | X | X | X | O |
| Capecitabine | DPYD | O | O | X | X | X | O | O | X | O | X |
| Carbamazepine | HLA-B | O | X | O | X | O | X | O | X | O | X |
| HLA-A | |||||||||||
| Celecoxib | CYP2C9 | O | X | X | O | X | X | X | O | X | X |
| Citalopram | CYP2C19 | O | O | X | O | X | X | X | O | X | X |
| Clomipramine* | CYP2D6 | O | O | X | X | X | O | X | X | X | O |
| CYP2C19 | |||||||||||
| Clopidogrel | CYP2C19 | O | O | X | X | O | X | X | X | X | O |
| Codeine | CYP2D6 | O | O | O | X | X | X | O | X | X | X |
| Desflurane | RYR1 | O | X | X | X | X | O | X | X | X | O |
| Desipramine | CYP2C19 | O | X | X | X | X | O | NA | |||
| CYP2D6 | |||||||||||
| Dexlansoprazole | CYP2C19 | O | X | X | X | X | O | X | X | X | O |
| Diclofenac | CYP2C9 | O | X | X | X | X | O | X | X | X | O |
| Doxepine | CYP2D6 | O | O | X | X | X | O | X | X | X | O |
| Efavirenz | CYP2B6 | O | O | X | X | X | O | X | X | X | O |
| Eliglustat | CYP2D6 | X | O | X | O | O | X | NA | |||
| Enflurane | RYR1 | O | X | X | X | X | O | NA | |||
| Escitalopram | CYP2C19 | O | O | X | X | X | O | X | O | X | X |
| Esomeprazole* | CYP2C19 | X | O | X | X | X | O | X | X | X | O |
| Flecainide | CYP2D6 | X | O | X | X | X | O | X | X | X | O |
| Flucloxacillin | HLA-B | X | O | NA | NA | ||||||
| Flucytosine | DPYD | X | O | X | X | X | O | NA | |||
| Fluorouracil | DPYD | O | O | O | X | X | X | O | X | X | X |
| Fluoxetine* | CYP2D6 | X | O | X | X | X | O | X | X | X | O |
| Flurbiprofen | CYP2C9 | O | X | NA | X | X | X | O | |||
| Fluvoxamine | CYP2D6 | O | X | X | X | X | O | X | X | X | O |
| Gefitinib* | CYP2D6 | X | O | X | O | X | X | X | O | X | X |
| Glibenclamide* | CYP2C9 | X | O | NA | X | X | X | O | |||
| Gliclazide* | CYP2C9 | X | O | NA | X | X | X | O | |||
| Glimepiride* | CYP2C9 | X | O | X | X | X | O | X | X | X | O |
| Haloperidol | CYP2D6 | X | O | X | X | X | O | X | X | X | O |
| Halothane | RYR1 | O | X | NA | NA | ||||||
| Ibuprofen | CYP2C9 | O | X | X | X | X | O | X | X | X | O |
| Imipramine | CYP2C19 | O | O | X | X | X | O | X | X | X | O |
| CYP2D6 | |||||||||||
| Indomethacin | CYP2C9 | O | X | X | X | X | O | X | X | X | O |
| Irinotecan* | UGT1A1 | X | O | X | O | O | X | X | X | X | O |
| Isoflurane | RYR1 | O | X | X | X | X | O | X | X | X | O |
| Ivacaftor | CFTR | O | X | O | X | O | X | NA | |||
| Lansoprazole* | CYP2C19 | O | O | X | X | X | O | X | X | X | O |
| Lornoxicam | CYP2C9 | O | X | NA | X | X | X | O | |||
| Lumiracoxib | CYP2C9 | O | X | NA | NA | ||||||
| Meloxicam | CYP2C9 | O | X | X | X | X | O | X | X | X | O |
| Methoxyflurane | RYR1 | O | X | NA | NA | ||||||
| Metamizole | CYP2C9 | O | X | NA | NA | ||||||
| Mercaptopurine | TPMT | O | O | X | X | O | X | O | X | X | X |
| Metoprolol | CYP2D6 | X | O | X | X | X | O | X | X | X | O |
| Moclobemide* | CYP2C19 | X | O | NA | X | X | X | O | |||
| Nabumetone | CYP2C9 | O | X | X | X | X | O | X | X | X | O |
| Naproxen | CYP2C9 | O | X | X | X | X | O | X | X | X | O |
| Nortriptyline | CYP2C19 | O | O | X | X | X | O | X | X | X | O |
| CYP2D6 | |||||||||||
| Omeprazole* | CYP2C19 | O | O | X | X | X | O | X | X | X | O |
| Ondansetron | CYP2D6 | O | X | X | X | X | O | X | X | X | O |
| Oxcarbazepine | HLA-B | O | X | X | X | O | X | X | X | X | O |
| HLA-A | |||||||||||
| Oxycodone* | CYP2D6 | X | O | X | X | X | O | X | X | X | O |
| Pantoprazole* | CYP2C19 | O | O | X | X | X | O | X | X | X | O |
| Paroxetine* | CYP2D6 | O | O | X | X | X | O | X | X | X | O |
| Peginterferon alfa-2a | IFNL3 | O | X | X | X | X | O | X | X | X | O |
| Peginterferon alfa-2b | IFNL3 | O | X | X | X | X | O | NA | |||
| Phenprocoumon* | VKORC1 | X | O | NA | NA | ||||||
| Phenytoin | CYP2C9 | O | O | X | X | X | O | X | X | X | O |
| Pimozide | CYP2D6 | X | O | X | X | O | X | X | X | X | O |
| Piroxicam | CYP2C9 | O | X | X | X | X | O | X | X | X | O |
| Propafenone | CYP2D6 | X | O | X | X | X | O | X | X | X | O |
| Rabeprazole* | CYP2C19 | X | O | X | X | X | O | X | X | X | O |
| Rasburicase | G6PD | O | X | O | X | O | X | O | X | X | X |
| Ribavirin | IFNL3 | O | X | X | X | X | O | X | X | X | O |
| Risperidone* | CYP2D6 | X | O | X | X | X | O | X | X | X | O |
| Sertraline | CYP2C19 | O | O | X | X | X | O | X | X | X | O |
| Sevoflurane | RYR1 | O | X | X | X | X | O | X | X | X | O |
| Simvastatin | SLCO1B1 | O | O | X | X | X | O | X | X | X | O |
| Siponimod | CYP2C9 | X | O | X | O | X | X | NA | |||
| Succinylcholine | RYR1 | O | X | X | X | X | O | X | X | X | O |
| Tacrolimus | CYP3A5 | O | O | X | X | X | O | X | X | X | O |
| Tamoxifen | CYP2D6 | O | O | X | X | X | O | X | X | X | O |
| Tegafur | DPYD | O | O | NA | X | X | X | O | |||
| Tenoxicam | CYP2C9 | O | X | NA | NA | ||||||
| Thioguanine | TPMT | O | O | X | O | O | X | NA | |||
| Tolbutamide* | CYP2C9 | X | O | NA | NA | ||||||
| Tramadol | CYP2D6 | X | O | O | X | X | X | X | X | X | O |
| Trimipramine | CYP2D6 | O | X | NA | NA | ||||||
| CYP2C19 | |||||||||||
| Tropisetron | CYP2D6 | O | X | NA | NA | ||||||
| Venlafaxine | CYP2D6 | X | O | X | X | X | O | X | X | X | O |
| Voriconazole | CYP2C19 | O | O | X | X | X | O | X | X | X | O |
| Warfarin | CYP2C9 | O | O | X | O | X | X | X | O | X | X |
| VKORC1 | |||||||||||
| Zuclopenthixol | CYP2D6 | X | O | NA | NA | ||||||
| Total No. | 66 | 59 | 7 (9.0%) | 12 (15.4%) | 12 (15.4%) | 54 (69.2%) | 8 (10.7%) | 5 (6.7%) | 4 (5.3%) | 62 (82.7%) | |
Data are presented as the total number and ratio (%) of drugs and total number of drugs with pharmacogenetic information in the CPIC and/or DPWG guideline except for not approved drugs (total FDA = 78, MFDS = 75).
CPIC, Clinical Pharmacogenetics Implementation Consortium; DPWG, Dutch Pharmacogenetics Working Group; FDA, US Food and Drug Administration; MFDS, Korea Ministry of Food and Drug Safety.
*Drug represents that no action is required for this gene-drug interaction. NA indicates that the drug is not approved in United States and/or Republic of Korea.
Figure 1. Selection of drugs for comparison between the pharmacogenomics guidelines and drug labels of the regulatory affairs.
CPIC, Clinical Pharmacogenetics Implementation Consortium; DPWG, Dutch Pharmacogenetics Working Group; FDA, US Food and Drug Administration; MFDS, Korea Ministry of Food and Drug Safety.
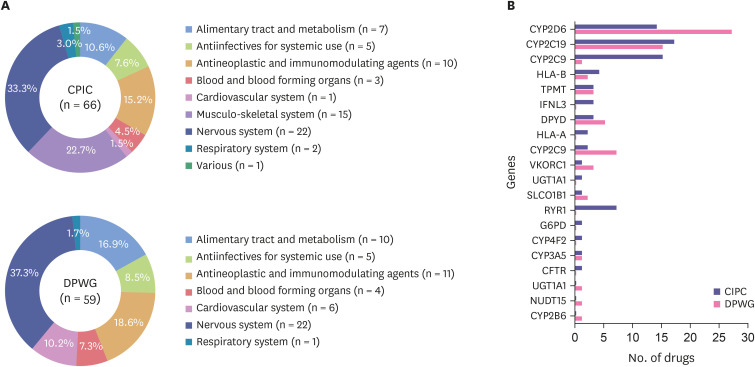
The most common type of drug classified by ATC code was nervous system in both the CPIC (33.3%) and DPWG (37.3%) guidelines (Fig. 2A). In the case of PGx related genes, CYP2D6 and CYP2C19 were dominant genes in both guidelines (Fig. 2B).
Figure 2. Classification of drugs listed in the CPIC and DPWG guidelines by (A) the anatomical therapeutic chemical code and (B) pharmacogenomics related genes.
Drug categories classified as “This is not a gene-drug interaction” on the DPWG guideline.
CPIC, Clinical Pharmacogenetics Implementation Consortium; DPWG, Dutch Pharmacogenetics Working Group.
Concordance of the pharmacogenomic information between the guidelines and drug labels
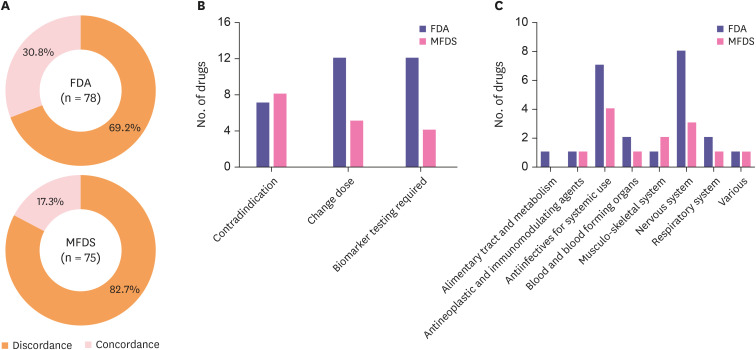
Among the 96 drugs listed on the PGx guidelines, 78 and 75 drugs were approved by the FDA and MFDS, respectively. In the FDA labels, the number of concordant drugs was 24 (30.8%), while 13 drugs (17.3%) were concordant to the MFDS labels (Fig. 3A). In detail, the number of drugs categorized as contraindication, change dose and biomarker testing required was 7, 12 and 12 in the FDA and 8, 5 and 4 in the MFDS, respectively. Nervous system and antineoplastic and immunomodulation agents were dominant drugs among concordant drugs.
Figure 3. Concordance between the pharmacogenomic information in the guidelines and on the drug labels (A) pie chart showing the ratio of the concordance and discordance of the FDA and MFDS (B) bar chart representing the number of concordant drugs categorized by indication (C) bar chart representing the number of concordant drugs categorized by the anatomical therapeutic chemical code.
FDA, US Food and Drug Administration; MFDS, Korea Ministry of Food and Drug Safety.
Concordance of pharmacogenomic information between the FDA and MFDS labels
The drugs that had the same pharmacogenomic information between the FDA and MFDS were as follow: abacavir, carbamazepine, celecoxib, citalopram, codeine, fluorouracil, geftinib, rasburicase and warfarin (Table 2). Abacavir, carbamazepine, codeine, fluorouracil and rasburicase were prohibited to people with certain genotypes. In the case of carbamazepine, biomarker test for HLA-B*1502 should be conducted prior to treatment. The dosage of celecoxib, citalopram and warfarin should be changed depending on the genotype, while the dosage of geftinib did not need to be adjusted regardless of the genotype.
Table 2. List of concordant drugs for which the pharmacogenetic information on the FDA and MFDS labels was identical.
| Drug | Gene | CPIC | DPWG | FDA/MFDS | Indication |
|---|---|---|---|---|---|
| Abacavir | HLA-B | Abacavir is not recommended for carrier of HLA-B*57:01 because of significantly increased risk of abacavir hypersensitivity | Abacavir is contra-indicated for HLA-B*5701-positive patients | Patients who carry the HLA-B*5701 allele are at a higher risk of experiencing a hypersensitivity reaction to abacavir | Contraindication |
| Carbamazepine | HLA-B | If patient is carbamazepine-naive and alternative agents are available, do not use carbamazepine | NA | Patients with ancestry in genetically at-risk populations should be screened for the presence of HLA-B*1502 prior to initiating treatment with carbamazepine. Patients testing positive for the allele should not be treated with carbamazepine unless the benefit clearly outweighs the risk | Contraindication, Biomarker testing required |
| Celecoxib | CYP2C9 | Initiate therapy with 25–50% of the lowest recommended starting dose | NA | In patients who are known or suspected to be poor CYP2C9 metabolizers (i.e., CYP2C9*3/*3), based on genotype or previous history/experience with other CYP2C9 substrates administer celecoxib starting with half the lowest recommended dose | Change dose |
| Citalopram | CYP2C19 | Consider a 50% reduction of the recommended starting dose. Utilize therapeutic drug monitoring to guide dose adjustments | Do not exceed the following daily doses (50% of the standard maximum dose) | Dosage adjustment is recommended in CYP2C19 poor metabolizers | Change dose |
| Codeine | CYP2D6 | Avoid codeine use due to potential for toxicity | Codeine is contra-indicated | Ultra-rapid metabolism of codeine and other risk factors for life-threatening respiratory depression in children | Contraindication |
| Fluorouracil | DYPD | Avoid use of 5-fluorouracil or 5-fluorouracil prodrug-based regimens | Start with 50% of the standard dose or avoid fluorouracil | Increased risk of serious or fatal adverse reactions in patients with low or absent dipyrimidine dehydrogenase activity | Contraindication |
| Geftinib | CYP2D6 | NA | NO action is needed for this gene-drug interaction | No dose adjustment is recommended in patients with a known CYP2D6 poor metabolizer genotype, but these patients should be closely monitored for adverse reactions | Change dose |
| Rasburicase | G6PD | Rasburicase is contraindicated | NA | Do not administer rasburicase to patients with glucose-6phosphate dehydrogenase (G6PD) deficiency | Contraindication |
| Warfarin | CYP2C9 | Dosing Recommendations with Consideration of Genotype | Dosing Recommendations with Consideration of Genotype | Dosing Recommendations with Consideration of Genotype | Change dose |
NA indicates that the drug is not approved in United States and/or Republic of Korea.
FDA, US Food and Drug Administration; MFDS, Korea Ministry of Food and Drug Safety; CPIC, Clinical Pharmacogenetics Implementation Consortium; DPWG, Dutch Pharmacogenetics Working Group.
DISCUSSION
The comparison of pharmacogenomic information between the guidelines and drug labels was conducted to identify whether the pharmacogenomic information in guidelines was well reflected on the drug labels which was easily accessible information for physicians and pharmacists. As a result of the study, a total of 96 drugs listed in either the CPIC or DPWG guidelines were screened, and 78 and 75 drug labels of the FDA and MFDS, respectively, were scrutinized. Less than half of the pharmacogenomic information from the approved drugs was consistent with the PGx guidelines.
Inconsistency between the CPIC and DPWG guidelines could be one of the factors why PGx related recommendations by guidelines were not reflected on the drug labels. Bank et al. reported that CPIC and DPWG showed a high level of consistency; however, some differences between them existed such as methodology and terminology [12]. Methodology for the scaling level of evidence, utilizing the source of information and processing establishment of dose recommendation between the two PGx guidelines was inconsistent. Like the example that “increased function” referred to “greater than normal function” in the CPIC guideline had a similar meaning of “gain-of-function” in the DPWG guideline; thus, there was subtle gap for the terminology between the two guidelines. These methodological and terminological differences possibly hindered the integration of pharmacogenomic information, leading to inconsistency between the guidelines and drug labels.
Above all, the main reason of discordance of the pharmacogenomic information between the guidelines and labels is different characteristics as informative tools. PGx guidelines are generally established and updated by various experts in the field based on the latest knowledge. The underlying assumption for the guidelines is that genotyping will be common, and establishing guidelines enables clinicians to translate genotyping results into actionable clinical decisions [5]. Drug labels are released after permission from the reviewers of regulatory affairs under a comprehensive and meticulous regulatory principle. The drug label can be revised if strong evidence and the potential impact of the gene-drug response are supported [13]. Therefore, PGx guidelines tend to reflect up-to-date knowledge, whereas drug labels are relatively slowly updated.
Among approved drugs from both the FDA and MFDS, the pharmacogenomic information of only 9 drugs was identical. One of the reasons for the inconsistent information between the FDA and MFDS might be the ethnic characteristics of US and South Korean citizens in that South Korea is close to a mono-ethnicity, while the United States is a poly-ethnicity with a small percentage of Asians. Ethnicity is one of the factors that affect the pharmacokinetics, pharmacodynamics and PGx of a drug, resulting in variability of the drug response, and a difference in the frequency of genetic polymorphisms of the drug-metabolizing enzymes between Caucasian and East Asians were reported [11,14]. In addition, a difference in the system of establishing or revising drug labeling between the two agencies possibly triggered a gap between the FDA and MFDS drug labels.
One of the limitations of this study was that the labels of the European Medicines Agency and Pharmaceuticals and Medical Devices Agency were not considered. Also, the other limitation is that some labeling information between original and generic drugs containing same active pharmaceutical ingredient is inconsistent [15]. To minimize bias, the original and reference drug were selected in the process of collecting the pharmacogenomic information of the drug labels in this study.
In conclusion, pharmacogenomic information on clinical implementation guidelines was limited in both FDA and MFDS labels because of various reasons including the characteristics of the guidelines and drug labels. Therefore, more effort from pharmaceutical companies, academia and regulatory affairs needs to be made to implement pharmacogenomic information onto drug labels.
Footnotes
Reviewer: This article was reviewed by peer experts who are not TCP editors.
Conflict of Interest: - Authors: Nothing to declare
- Reviewers: Nothing to declare
- Editors: Nothing to declare
- Conceptualization: Yoon DY, Jang IJ, Lee SH.
- Data curation: Yoon DY, Lee SH.
- Formal analysis: Yoon DY, Lee SH.
- Investigation: Yoon DY, Lee SH.
- Methodology: Yoon DY, Jang IJ, Lee SH.
- Project administration: Yoon DY, Lee SH.
- Supervision: Lee S, Lee SH.
- Validation: Yoon DY.
- Visualization: Yoon DY, Lee S.
- Writing - original draft: Yoon DY.
- Writing - review & editing: Lee S, Ban MS, Jang IJ, Lee SH.
References
- 1.Jameson JL, Longo DL. Precision medicine--personalized, problematic, and promising. N Engl J Med. 2015;372:2229–2234. doi: 10.1056/NEJMsb1503104. [DOI] [PubMed] [Google Scholar]
- 2.Cremers S, Guha N, Shine B. Therapeutic drug monitoring in the era of precision medicine: opportunities! Br J Clin Pharmacol. 2016;82:900–902. doi: 10.1111/bcp.13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weinshilboum RM, Wang L. Pharmacogenomics: precision medicine and drug response. Mayo Clin Proc. 2017;92:1711–1722. doi: 10.1016/j.mayocp.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Wouden CH, Cambon-Thomsen A, Cecchin E, Cheung KC, Dávila-Fajardo CL, Deneer VH, et al. Implementing pharmacogenomics in Europe: design and implementation strategy of the ubiquitous pharmacogenomics consortium. Clin Pharmacol Ther. 2017;101:341–358. doi: 10.1002/cpt.602. [DOI] [PubMed] [Google Scholar]
- 5.Caudle KE, Klein TE, Hoffman JM, Muller DJ, Whirl-Carrillo M, Gong L, et al. Incorporation of pharmacogenomics into routine clinical practice: the Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline development process. Curr Drug Metab. 2014;15:209–217. doi: 10.2174/1389200215666140130124910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blagec K, Romagnoli KM, Boyce RD, Samwald M. Examining perceptions of the usefulness and usability of a mobile-based system for pharmacogenomics clinical decision support: a mixed methods study. PeerJ. 2016;4:e1671. doi: 10.7717/peerj.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altman RB, Prabhu S, Sidow A, Zook JM, Goldfeder R, Litwack D, et al. A research roadmap for next-generation sequencing informatics. Sci Transl Med. 2016;8:335ps10. doi: 10.1126/scitranslmed.aaf7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.US Food and Drug Administration. Guidance for industry: clinical pharmacogenomics: premarket evaluation in early-phase clinical studies and recommendations for labeling. 2013. Jan, [Accessed August 8, 2020]. https://www.fda.gov/media/84923/download.
- 9.Abou Diwan E, Zeitoun RI, Abou Haidar L, Cascorbi I, Khoueiry Zgheib N. Implementation and obstacles of pharmacogenetics in clinical practice: An international survey. Br J Clin Pharmacol. 2019;85:2076–2088. doi: 10.1111/bcp.13999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klein ME, Parvez MM, Shin JG. Clinical implementation of pharmacogenomics for personalized precision medicine: barriers and solutions. J Pharm Sci. 2017;106:2368–2379. doi: 10.1016/j.xphs.2017.04.051. [DOI] [PubMed] [Google Scholar]
- 11.Yasuda SU, Zhang L, Huang SM. The role of ethnicity in variability in response to drugs: focus on clinical pharmacology studies. Clin Pharmacol Ther. 2008;84:417–423. doi: 10.1038/clpt.2008.141. [DOI] [PubMed] [Google Scholar]
- 12.Bank PC, Caudle KE, Swen JJ, Gammal RS, Whirl-Carrillo M, Klein TE, et al. Comparison of the guidelines of the Clinical Pharmacogenetics Implementation Consortium and the Dutch Pharmacogenetics Working Group. Clin Pharmacol Ther. 2018;103:599–618. doi: 10.1002/cpt.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drozda K, Pacanowski MA, Grimstein C, Zineh I. Pharmacogenetic labeling of FDA-approved drugs: a regulatory retrospective. JACC Basic Transl Sci. 2018;3:545–549. doi: 10.1016/j.jacbts.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim K, Johnson JA, Derendorf H. Differences in drug pharmacokinetics between East Asians and Caucasians and the role of genetic polymorphisms. J Clin Pharmacol. 2004;44:1083–1105. doi: 10.1177/0091270004268128. [DOI] [PubMed] [Google Scholar]
- 15.Pfistermeister B, Saß A, Criegee-Rieck M, Bürkle T, Fromm MF, Maas R. Inconsistencies and misleading information in officially approved prescribing information from three major drug markets. Clin Pharmacol Ther. 2014;96:616–624. doi: 10.1038/clpt.2014.156. [DOI] [PubMed] [Google Scholar]