Abstract
This tutorial introduces background and methods to predict the human volume of distribution (Vd) of drugs using in vitro and animal pharmacokinetic (PK) parameters. The physiologically based PK (PBPK) method is based on the familiar equation: Vd = Vp + ∑T(VT × ktp). In this equation, Vp (plasma volume) and VT (tissue volume) are known physiological values, and ktp (tissue plasma partition coefficient) is experimentally measured. Here, the ktp may be predicted by PBPK models because it is known to be correlated with the physicochemical property of drugs and tissue composition (fraction of lipid and water). Thus, PBPK models' evolution to predict human Vd has been the efforts to find a better function giving a more accurate ktp. When animal PK parameters estimated using i.v. PK data in ≥ 3 species are available, allometric methods can also be used to predict human Vd. Unlike the PBPK method, many different models may be compared to find the best-fitting one in the allometry, a kind of empirical approach. Also, compartmental Vd parameters (e.g., Vc, Vp, and Q) can be predicted in the allometry. Although PBPK and allometric methods have long been used to predict Vd, there is no consensus on method choice. When the discrepancy between PBPK-predicted Vd and allometry-predicted Vd is huge, physiological plausibility of all input and output data (e.g., r2-value of the allometric curve) may be reviewed for careful decision making.
Keywords: Volume of Distribution, PBPK Human PK Prediction
MECHANISM OF DRUG DISTRIBUTION
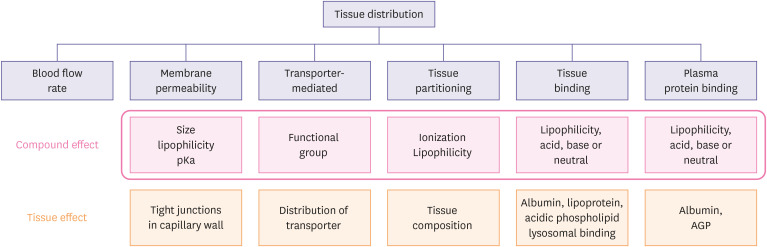
In this tutorial, we will briefly review the concept of Vd and methods to predict human Vd using in vitro and animal PK parameters. Drug distribution is defined as the reversible partitioning of drug molecules into the various tissues from plasma, and it is driven by blood flow and the tendency of the drug to cross cell membranes of the tissues [1]. Differences in the tendency are caused by drugs' chemical properties (lipophilicity, polarity, pKa, molecular weight, etc.), transporter, and tissue composition. They result in different partitioning of drugs in various tissues after the distribution phase is ended (Fig. 1).
Figure 1. Compound- and physiology-dependent factors affecting rate and extent of tissue. Redrawn from [1].
pKa, the symbol for the acid dissociation constant at logarithmic scale; AGP, alpha-1-acid glycoprotein.
Drugs are rapidly distributed to highly perfused organs with discontinuous capillaries (e.g., liver, spleen, intestine etc.) while slowly to the others. Because the capillary wall is a thin, single layer of endothelial cells, many drugs may easily pass the membrane. Those of the kidney and liver are mostly loose, allowing the extensive transfer of drugs into the interstitial space. In the phospholipid bilayers of the cell membrane, lipophilic, base compounds may better permeate through the membrane than the acidic, hydrophilic ones. The existence of efflux or influx transporters may also influence the distribution of drugs. Different tissue composition may also affect the tissue partitioning of drugs. Lipophilic drugs distribute well into fat-rich organs (e.g., adipose, liver, brain, and kidney). Drugs with extensive tissue binding also show a large Vd. Irreversible binding of drugs to some tissues (e.g., tetracycline to the bone, iodine to the thyroid) does not apply to the conventional distribution model that assumes reversible transport of drug molecules between compartments. Instead, this may be accounted for by non-specific elimination (not by the liver or kidney).
PHYSIOLOGICAL VIEW ON Vd
Unbound plasma concentration and unbound tissue concentration would be equal at equilibrium for drugs distributed only by passive diffusion without efflux or influx by transporters (equation 1).
| Ctotal, T × fut = Ctotal, p × fup | Eq. 1 |
The partition coefficient (ktp) is simply defined as in equation 2. However, the equation 2 is not used anymore in PBPK-based methods because more refined partition coefficients are needed for each of lean tissues and adipose tissues as in equations 5 and 6.
| Eq. 2 |
The fut can be obtained by in vitro and in vivo laboratory methods, but it may be predicted by the fup and the macromolecular (albumin, globulin, lipoprotein) concentration ratio (= the tissue interstitial fluid/plasma) (equation 3). The ratio (R) is known to be 0.5 in the lean tissue and 0.15 in the fat tissue [2].
| Eq. 3 |
The distribution of drugs has been described using the physiological volumes of the human body (plasma and tissue volumes) and the tendency of drugs to distribute into the tissues (ktp) as shown in equation 4.
| Vd = Vp + ∑T(VT × ktp) | Eq. 4 |
The Vp (volume of plasma) and VT (volume of tissues other than plasma) are human physiological variables. The ktp is the tissue-plasma partition coefficient of each tissue (Ctotal, plasma/Ctotal, tissue), determined by the relationship between drug and body. The ktp in animal organs can be measured experimentally after intravenous infusion, which is too costly and time-consuming to try for all drug candidates under discovery or early development steps. Thus, ktp prediction in equations 5 and 6 instead of the in vivo study is the key to PBPK modeling. Efforts to predict Vd have been focused on the improvement of methods predicting the ktp. Regression analyses in animals indicated that lipophilicity and protein binding are two main factors related to Vss [3,4].
PBPK MODELS TO PREDICT ktp (TISSUE: PLASMA PARTITION COEFFICIENT)
Among more than twenty PBPK methods published so far, we introduce the most frequently used ones in this tutorial. Poulin and Theil first reported methods to estimate ktp in the lean and adipose tissues [2] based on the assumption that the unbound drug is distributed homogeneously into lipid and water in each tissue. Tissue permeability or organ blood flow that may predict multi-compartmental distribution is rarely used in currently available PBPK models predicting Vd. Thus, Vss is the most frequently predicted parameter by PBPK models. Later, Poulin and Theil's model was corrected by Berezhkovsky et al., which is used these days (equations 5 and 6).
| Eq. 5 |
| Eq. 6 |
The Po:w is the octaol: buffer partition ratio of the non-ionized drug at pH7.4, and Dvo:w is the vegetable oil (olive oil): buffer partition ratio of both ionized and non-ionized drugs at pH7.4. The V's are fractional volumes in the human tissues: Vnl is for neutral lipid, Vph is for phospholipid, and Vw is for water. They are available from literature on human physiological values [5]. The fut and fup are the unbound fraction of tissue and plasma, respectively.
The equations assume passive diffusion of neutral compounds only without considering transporter-mediated distribution or ionization status. Neutral non-polar lipids of the lean tissue are composed of triglyceride and cholesterol. The lipophilicity of this mixture is similar to that of n-octanol. Unlike the lean tissue, triglyceride is the only component of the adipose tissue's non-polar lipid, and its lipophilicity is not represented correctly by n-octanol. Thus, the Po:w (obtained using n-octanol) has been used for the lean tissue, and the Dvo:w (obtained using vegetable oil) instead of Po:w has been used for the adipose tissue. The mechanistic basis of their equations is that the drug is distributed in each tissue and plasma according to their composition of lipids, water, and plasma proteins. The lipophilicity of each tissue, which is determined by its ratio of hydrophilic and lipophilic components and the lipophilicity of the drug (Po:w or Dvo:w values at pH 7.4), rules the distribution of drugs into each tissue. Besides, the binding to proteins commonly existing in the plasma and interstitial tissue also influences tissue distribution. The contribution of lipophilic (neutral lipid contents), hydrophilic (water contents), and the mixed-natured components (phospholipid contents) are also incorporated into the equations. In phospholipid contents, 30% of the volume was put in the lipophilic side and the rest in the hydrophilic side [6,7].
In summary, equations 5 and 6 demonstrate the contribution of solubility and free concentration to the tissue distribution of drugs. The maximum achievable free concentration of drugs in a tissue will be identical to its solubility. Thus the free concentration ratio of (tissue/plasma) will be identical to the solubility ratio of (tissue/plasma). The general notion “unbound plasma concentration and unbound tissue concetration are equal at equilibrium,” mentioned in the previous section in this tutorial, is in fact confusing. It should have been rewritten as “Unbound drug concentrations in plasma water and tissue water are equal at equilibrium”. It may be further clarified into four sequential sentences if influences by ionization or transporters are ignored.
1) Unbound drug concentrations in plasma water and tissue water are equal at equilibrium.
2) Unbound drug concentrations in plasma lipid and tissue lipid are equal at equilibrium.
3) The lipid: water compositions of the plasma and tissue are different, and water solubility and lipid solubility of the drug are different.
4) Thus, the averaged unbound concentration in the tissue (water and lipid) differs from that in the plasma.
Because equations 5 and 6 does not consider the distribution of ionized forms of the acidic, basic or neutral drugs, Rodgers and Rowland [8] proposed modified forms of the Berezhkovsky's equations by adding pKa of the drug and pH of plasma and tissues (equations not shown in this tutorial).
HOW ACCURATE ARE THE PBPK-PREDICTED Vss?
According to the report on the modified method by Rodgers and Rowland [8], 84% of their predicted partition coefficients were within 0.3 to 3 times the experimental values measured in 13 rat tissues for 49 structurally unrelated drugs, which outperformed Berezhkovsky's method (61%). When 24 different allometric, semi-mechanistic, and mechanistic (PBPK) models predicting Vss were compared using 18 drugs' human i.v. PK data [9], performance differences were not conclusive. However, in the three PBPK models (Poulin & Theil, Berezhkovsky, and Rodger & Rowland), Berezhkovsky's model worked best (61% of predicted Vss were within 0.5–2 times the human PK-estimated Vss; 39% by Poulin and Theil, 50% by Rodger and Rowland) [9]). Graham [10] has also reviewed the performance of the three PBPK methods to predict Vss in her Ph.D. dissertation. Predictions by the Poulin & Theil model were within 5-fold experimental values, and all three were within 10-fold when the experimentally obtained rat Vss for 35 drugs were tested. There are no defined cutoff criteria for the performance of Vss prediction methods. However, the five or 10-fold range seems too broad to use as a reliable method to design first-in-human clinical trials. As discussed in this tutorial, the three PBPK methods are sophisticated tools that incorporate all kinds of in vitro information. Notwithstanding this sophistication, the performance is not satisfactory. What can we do more? The older method may be worth trying - the allometric method.
ALLOMETRIC METHOD TO PREDICT Vd
Because describing the allometric method from A to Z is not in this tutorial's scope, we briefly introduce current routines to predict Vd using allometric methods. To perform allometric analysis, PK data from at least three species (e.g., mouse, rat, dog) are necessary. For the prediction of human Vd, PK parameters obtained from i.v. PK studies are necessary. When Vss is the only volume parameter to predict, the Vss estimated using NCA (non-compartmental analysis) is enough for allometric analysis. However, to predict compartmental PK parameters (e.g., Vc, Vp, Q: inter-compartmental clearance) in humans, the compartmental analysis should be done for animal data before allometric analysis. Because the compartmental volume parameters should be accordant between species, it is recommended that the same compartmental structure is chosen regardless of species. For example, when mice and rat PK data are modeled using a 2-compartment model and dog PK data are modeled using a 3-compartment model, it is impossible to apply the allometric method to predict human Vd parameters. In this case, dog PK data should also be analyzed using a 2-compartmental model, although it may be best described by a 3-compartment model. There are several different allometric methods: simple allometry using bodyweight only (Y = a(BW)b); brain weight added (Y × brain weight = a(BW)b); maximum life span (MLP) added (Y × MLP = a(BW)b). In the prediction of Vd, simple allometry using bodyweight only may be good enough in many cases. However, there is no rule on the choice of covariates or the exponents (fixed at 1 or 0.75 or best fit). Moreover, the allometric model chosen for the Vd terms may not be identical to that used for Q.
Results of the allometric prediction may significantly differ by researchers because the researchers may choose their favorite among several different ways in fitting the exponent or incorporating the covariate (BW, brain weight, MLP) for each of Vss, Vc, Vp, or Q. It is the researchers' discretion which allometric method to use. There is no rule for such choices, but the rationale or logic of model selection should be recorded. When the rationale is unclear, selecting the best-fitting model after trying several different allometric models would work.
WHICH METHOD SHOULD BE CHOSEN TO PREDICT HUMAN Vd?
There are a few reports on the performance comparison among several different PBPK methods and allometric methods in predicting Vd [11,12]. However, the kinds of PBPK and allometric methods used in their comparison process were different than those discussed in this tutorial, and their allometric method used was the simplest one.
The quantity or quality of in vitro and in vivo data available when performing prediction differs by the molecules' development stages or the companies' policies. When there is no animal PK data, you must use PBPK methods to predict animal or human Vd. When animal i.v. PK data are available in ≥ 3 species, allometric methods should also be tried, and its best fit result may be compared with the Vss predicted by the PBPK method. Even when the difference between Vd's predicted by PBPK and allometry is vast (this is not uncommon), you don't have to be embarrassed. Selecting one over the other after a thorough review of physiological plausibility is what may be done. The r2-value of the allometric curve may also be considered. Or you may use both of them (scenario of using two different kinds of Vd's). It should be remembered that uncertainty is the nature of preclinical to the clinical prediction of small molecule drugs.
Glossary
- Vd
volume of distribution
- Vp
plasma volume
- VT
tissue volume
- ktp
tissue plasma partition coefficient
- Vc
central volume of distribution
- Vp
peripheral volume of distribution
- Q
inter-compartmental clearance
- Vss
steady-state volume of distribution
- pKa
the symbol for the acid dissociation constant at logarithmic scale
- Ctotal, T
total concentration in tissue
- Ctotal, p
total concentration in plasma
- fut
unbound fraction in tissue
- fup
unbound fraction in plasma
- R
macromoleculalr concentration ratio of the tissue interstitial fluid and plasma
- Po:w
octanol:buffer partition coefficient of the non-ionized species
- Dvo:w
olive oil (vegetable oil):buffer partition coefficient of the non-ionized and ionized species
- Vnl
fractional tissue volume content of neutral lipid
- Vph
fractional tissue volume content of phopholipid
- Vw
fractional tissue volume content of water
- NCA
non-compartmental analysis
- BW
bodyweight
- MLP
maximum life span
Footnotes
Funding: This research is part of the EDISON (EDucation-research Integration through Simulation On the Net) Program of the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (grant number: NRF-2016M3C1A6936614).
Reviewer: This article was invited.
Conflict of Interest: - Authors: Nothing to declare
- Editors: Nothing to declare
- Conceptualization: Yim DS, Choi S.
- Data curation: Choi S.
- Formal analysis: Yim DS.
- Methodology: Choi S.
- Supervision: Yim DS.
- Writing - original draft: Yim DS.
- Writing - review & editing: Yim DS, Choi S.
References
- 1.Peters SA. Physiologically-based pharmacokinetic (PBPK) modeling and simulations: principles, methods, and applications in the pharmaceutical industry. Hoboken (NJ): John Wiley & Sons; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poulin P, Theil FP. A priori prediction of tissue:plasma partition coefficients of drugs to facilitate the use of physiologically-based pharmacokinetic models in drug discovery. J Pharm Sci. 2000;89:16–35. doi: 10.1002/(SICI)1520-6017(200001)89:1<16::AID-JPS3>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 3.Sawada Y, Hanano M, Sugiyama Y, Harashima H, Iga T. Prediction of the volumes of distribution of basic drugs in humans based on data from animals. J Pharmacokinet Biopharm. 1984;12:587–596. doi: 10.1007/BF01059554. [DOI] [PubMed] [Google Scholar]
- 4.Ritschel WA, Hammer GV. Prediction of the volume of distribution from in vitro data and use for estimating the absolute extent of absorption. Int J Clin Pharmacol Ther Toxicol. 1980;18:298–316. [PubMed] [Google Scholar]
- 5.Poulin P, Theil FP. Development of a novel method for predicting human volume of distribution at steady-state of basic drugs and comparative assessment with existing methods. J Pharm Sci. 2009;98:4941–4961. doi: 10.1002/jps.21759. [DOI] [PubMed] [Google Scholar]
- 6.Poulin P, Krishnan K. Molecular structure-based prediction of the partition coefficients of organic chemicals for physiological pharmacokinetic models. Toxicol Methods. 1996;6:117–137. [Google Scholar]
- 7.Poulin P, Krishnan K. A biologically-based algorithm for predicting human tissue: blood partition coefficients of organic chemicals. Hum Exp Toxicol. 1995;14:273–280. doi: 10.1177/096032719501400307. [DOI] [PubMed] [Google Scholar]
- 8.Rodgers T, Rowland M. Physiologically based pharmacokinetic modelling 2: predicting the tissue distribution of acids, very weak bases, neutrals and zwitterions. J Pharm Sci. 2006;95:1238–1257. doi: 10.1002/jps.20502. [DOI] [PubMed] [Google Scholar]
- 9.Jones RD, Jones HM, Rowland M, Gibson CR, Yates JW, Chien JY, et al. PhRMA CPCDC initiative on predictive models of human pharmacokinetics, part 2: comparative assessment of prediction methods of human volume of distribution. J Pharm Sci. 2011;100:4074–4089. doi: 10.1002/jps.22553. [DOI] [PubMed] [Google Scholar]
- 10.Graham HS. Prediction of drug distribution in rat and human. Dissertation, The University of Manchester; 2012. [Google Scholar]
- 11.Petersson C, Papasouliotis O, Lecomte M, Badolo L, Dolgos H. Prediction of volume of distribution in humans: analysis of eight methods and their application in drug discovery. Xenobiotica. 2020;50:270–279. doi: 10.1080/00498254.2019.1625084. [DOI] [PubMed] [Google Scholar]
- 12.Fagerholm U. Prediction of human pharmacokinetics--evaluation of methods for prediction of volume of distribution. J Pharm Pharmacol. 2007;59:1181–1190. doi: 10.1211/jpp.59.9.0001. [DOI] [PubMed] [Google Scholar]