Abstract
Hand, foot and mouth disease is a common viral infectious disease caused by enteroviruses, including coxsackie A16 (CVA16) and enterovirus 71 (EV71). HFMD can cause severe symptoms in children which can be fatal. Human scavenger receptor class B member 2 (SCARB2) is a cellular receptor for EV71 and CVA16, providing a potential approach for preventing EV71 infection and transmission. In this present study, we constructed and assessed the potential of recombinant SCARB2, using E. coli expression system. To generate this construct, scarb2 gene was cloned into pET22b vector and expressed in E. coli BL21 (DE3). The expression of SCARB2 was induced by 0.1 mM IPTG and analyzed using SDS-PAGE, followed by Western blot. Expressed SCARB2 was in inclusion bodies and refolded to obtain the soluble form with recovery efficacy of 100%. This recombinant protein was then validated for binding with EV71 via indirect ELISA in two different pHs (7.4 and 5.5), which partially revealed the mechanism of virus–receptor interaction. These results envisaged potential applications for utilizing recombinant SCARB2 in preventing the virus transmission.
Keywords: EV71, HFMD, Inclusion bodies, Refolding, SCARB2
Introduction
Hand, foot and mouth disease (HFMD) is a common viral infectious disease characterized by common symptoms including fever, painful sores in the mouth, and a rash with blisters on hands, feet and buttocks. In most cases, the disease is mild and self-limiting, occurring mainly in children under 10 years old, but it is most commonly seen in children under 5 years of age. It can also appear in adolescents and occasionally in adults. Viruses causing HFMD are spread by direct contact with saliva, mucus, fluid from blisters and stool of infected people. Adults infected with HFMD may shed the virus and have no symptoms (WHO). The dominant cause of HFMD is a group of enteroviruses, including coxsackie A16 (CVA16) and enterovirus 71 (EV71) (Yamayoshi et al. 2012a). Although complications are uncommon, EV71 infection can lead to severe symptoms in children and has been associated with neurological diseases, including acute flaccid paralysis, aseptic meningitis, and brain stem encephalitis with neurogenic pulmonary edema, which can be fatal (Lee 2016). There are no specific antiviral drugs or vaccine commercially available against enteroviruses causing HFMD hitherto (WHO). EV71 belongs to the human enterovirus A species of the enterovirus genus within the family Picornaviridae (King et al. 2000). Virion includes a non-enveloped capsid surrounding a core of single-stranded, positive-polarity RNA approximately 7.5 kb in size (Brown and Pallansch 1995). Since its discovery in 1969, EV71 has been recognized as a frequent cause of epidemics of HFMD associated with severe neurological sequelae in a small proportion of cases (WHO 2011).
Approaches have been intensively evaluated to prevent the spread of the virus, in which vaccines are the most priority (Yi et al. 2017). Several significant milestones have been archived recently in vaccine development of China, which leads to its phase 3 testing on trial (Li et al. 2014; Zhu et al. 2013, 2014). In December 2015 and January 2016, two HFMD vaccines approved by the China Food and Drug Administration were commercially produced and began to apply on Chinese children. However, these vaccines required long duration to demonstrate their efficacy and safety before becoming commercially available for application (Yi et al. 2017). In addition, the outreach efforts toward preventing the virus from entering its target cells also achieved certain results. Many studies were conducted to identify receptors on the surface of human cells that play a role in facilitating the infection of EV71. In particular, SCARB2 receptor presents in many cell types in the body and has been shown to be an important factor in the infection of EV71, which can be bound by all existing EV71 strains (Yamayoshi et al. 2009). Moreover, SCARB2 is also a co-receptor for CVA16, one of the main causative agents of HFMD besides EV71 (Yamayoshi et al. 2012b). SCARB2 has been confirmed to have ten potential N-glycosyl binding sites, considered to be responsible for its proper folding (Yamayoshi and Koike 2011).
Recently, recombinant receptor produced in E. coli has been shown to interact with EV71 in vitro (Xu et al. 2018). However, the receptor was used in insoluble form, so it is not suitable for related or further research, as well as putting into applications. On the other hand, the role of the receptor’s glycosylation remained unclear in interaction with EV71. In this study, we created a SCARB2-expressing plasmid to produce recombinant receptor in E. coli along with a refolding procedure. Then, the interaction between recombinant SCARB2 and EV71 was evaluated by indirect ELISA.
Materials and Methods
Bacterial Strains, Plasmids, Reagents and Growth Conditions
The scarb2 gene was amplified from pPHAGE-C-TAP-SCARB2 (Harvard PlasmID Database) (Huttlin et al. 2015). E. coli DH5α and E. coli BL21 (DE3) were employed as host strains for cloning and protein expression, respectively. Both were routinely grown in Luria–Bertani (LB) medium containing 100 μg/mL of ampicillin at 37 °C. The pET22b plasmid was used for both cloning and expression for scarb2 gene, and protein expression was controlled by T7 promoter via IPTG (isopropyl ß-D-1-thiogalactopyranoside) (Biobasic) inducer. DNA marker (Bioline, 33,053), protein maker (GE, 17,044,601) and pre-stained protein marker (Thermo Scientific, 26,612) were utilized for band size marking. Recombinant protein was constructed, expressed and stored at −200C.
Construction of Recombinant pET-SCARB2 (Fig. 1)
Fig. 1.
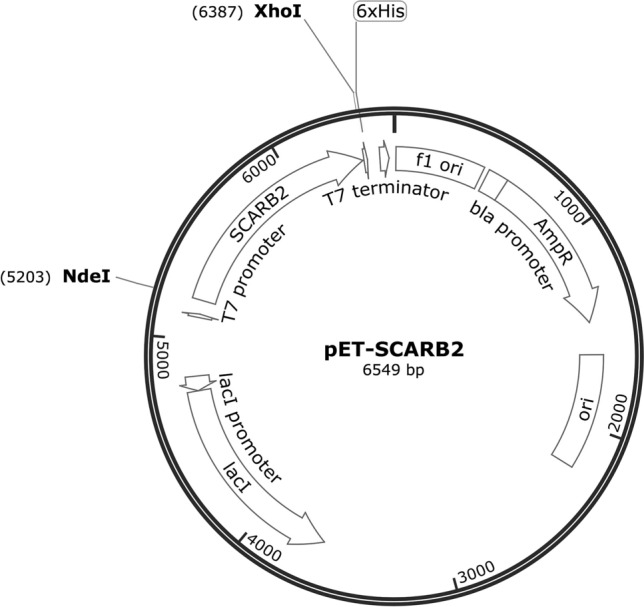
pET-SCARB2 map
pPHAGE-C-TAP-SCARB2 plasmid was used as a template for amplification of scarb2 gene by PCR with specific primers (scarb2F: CATATGatcgagaagaaaattgtg and scarb2R: CTCGAGaatcatagacttcagtcgac). scarb2 gene and pET22b plasmid were digested with NdeI and XhoI (Thermo Scientific). The ligation reaction containing the double-digested insert and expression vector was performed in the presence of T4 DNA ligase (Thermo Scientific), and the resulting mixture was transformed into competent E. coli DH5α. The transformants were initially screened on ampicillin-containing LB agar plate and then re-screened by PCR with specific primers and T7pro/T7ter. Plasmids derived from positive colonies confirmed by PCR colonies were sent to the Macrogen, Korea, for sequencing.
Expression of Recombinant SCARB2 in E. coli BL21 (DE3)
The expression of recombinant SCARB2 was conducted as described with some modifications (Seidmoradei et al. 2020). pET-SCARB2 obtained from previous steps was transformed into competent E. coli BL21 (DE3) strain. Positive colonies were inoculated in LB media shaking tubes supplemented with ampicillin and allowed to grow at 37 °C in 16 h. The cultures were then sub-cultured at 1:10 (v/v) and inoculated at 37 °C until OD600 reached 0.8–1.0. At this point, the cultures were induced with the final concentration of 0.1 mM IPTG, and the protein expressions were performed at 37 °C in 4 h. Harvested cells proceeded for sonication on ice in PBS buffer (pH 7.4) to obtain proteins in total, soluble and insoluble fractions. A sample of the bacterial culture was taken as negative control with non-induced E. coli BL21 (DE3)/pET-SCARB2. Recombinant protein expression was analyzed by SDS-PAGE and Coomassie Brilliant Blue stain, followed by Western blot and probed with His-probe (Santa Cruz) and goat anti-mouse IgG-HRP antibody (Proteintech). The membrane plot was developed using 100 µL of ready-to-use 3, 3′, 5, 5′-tetra methyl benzidine (TMB) (Thermo Scientific, 37,574) and incubated at room temperature. The reaction was stopped with PBS buffer and digitized documented.
Refolding of SCARB2 Inclusion Bodies
The refolding of recombinant SCARB2 was conducted as described with some modifications (Tat Truong Dang et al. 2014). Harvested cells proceeded for sonication on ice in lysis buffer (20 mM Tris–HCl pH 8.0, 1 M NaCl, 5 mM EDTA). The inclusion bodies were obtained by centrifugation at 13,000 rpm for 20 min, then washed five times in washing buffer (20 mM Tris–HCl pH 8.0, 1% Triton X-100) and re-washed in non-Triton washing buffer. Obtained pellet was re-suspended in dissolving buffer (20 mM Tris–HCl pH 8.0, 4 M Gu-HCl), well-vortexed and put still for one hour at 4°C. Supernatant was then harvested using centrifugation at 13,000 rpm for 20 min and suspended in refolding buffer (20 mM Tris–HCl pH 8.0, 2 M Urea, 10% Sucrose, 0.1% Tween 80). Mixture was then well-stirred at 40C for 14 h, and the solubilized cell pellet comprising inclusion bodies was centrifuged at 13,000 rpm for 20 min to remove insoluble particulates. Finally, the refolding result was verified using SDS-PAGE and silver-stained, followed by Western blot and probed with anti-His (Santa Cruz) and goat anti-mouse IgG-HRP antibody (Proteintech). The membrane plot was developed using 100 µL of ready-to-use TMB (Thermo Scientific) and incubated at room temperature. The reaction was stopped with PBS buffer and digitized documented.
The supernatant containing soluble SCARB2 obtained from previous steps was dialyzed against PBS buffer (pH 7.4) to remove refolding buffer, which could interfere with later experiments. The dialyzed SCARB2 protein was concentrated by 10 MWCO Amicon® Ultra centrifugal filter (Thermo Fisher). The obtained SCARB2 concentration was determined by using Bradford assay (Sigma).
Enzyme Immunoassay
The EV71 concentration was determined using TCID50 as previously described (Quynh-Anh Nguyen-Ngoc 2015; Thao et al. 2010). For UV inactivation, the conditions were followed as described (Zou et al. 2013). Briefly, EV71 stock was spread on a Petri dish and then treated with UV irradiation in a Biosafety Cabinet for 30 min or longer at room temperature. Interaction of recombinant SCARB2 protein with EV71 was evaluated by the indirect enzyme-linked immunosorbent assay (ELISA) conducted as described with some modifications (Ding et al. 2015). The 96-well microtiter plates were coated with 105 UV-inactivated EV71 virions in 100 µL carbonate buffer (pH 9.6) at 4°C overnight and then blocked with 100 μL of 3% skim milk in PBS-T for an hour at room temperature. After being washed three times with PBS-T, 100 μL of recombinant SCARB2 with concentration at 109, 1010, 1011 molecules (diluted in PBS pH 7.4 and PBS pH 5.5) was added and incubated at 37 °C for 2 h. Solutions were removed, and 100 μL/well of anti-His-tag-HRP antibody (Proteintech) at 1/20,000 dilution was added and incubated for 1 h at room temperature. The plate was developed using 100 µL of TMB (Sigma, T0440) and incubated at room temperature. The reaction was stopped with 100 μL of HCl 2 N after 30 min, and the absorbance at 450 nm was measured on a microplate reader (Thermo). Negative controls were created using no virions coating or no SCARB2 incubating. The cutoff value was calculated as negative control value plus 3 times of SD. The values higher than cutoff value were indicated as positive results.
Results and Discussion
Amplification of scarb2 Gene and Verification of Recombinant pET-SCARB2
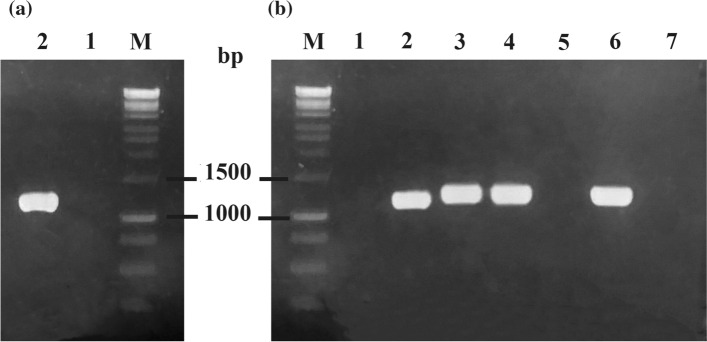
The scarb2 gene was amplified from pPHAGE-C-TAP-SCARB2 plasmid using specific primers. The length of the amplified product with about 1179 bp correlated with scarb2 gene (Fig. 2a). Under the PCR conditions performed in our experiments, the scarb2 gene was successfully isolated, which was then cloned into the pET expression vector. PCR colonies were performed to screen for recombinant pET22b-SCARB2 from transformed E. coli DH5α. Amplicons with sizes of approximately 1200 bp were obtained by using reversed primer of scarb2 and T7pro (Fig. 2b). The results showed that scarb2 gene was inserted with correct orientation. Sequencing results confirmed the amplified product was in 100% homologous with SCARB2 gene from GenBank (data not shown). Collectively, the recombinant vector of desire was successfully constructed.
Fig. 2.
Amplification of scarb2 gene a and verification of recombinant plasmid by PCR colonies of E. coli DH5α/pET22b-SCARB2 b in 1.5% agarose gel electrophoresis. M, DNA marker 1 kb; 1, negative control; 2, amplified scarb2 gene by specific primers; 3–7, screened colonies by scarb2R primer and T7pro
Expression of Recombinant SCARB2
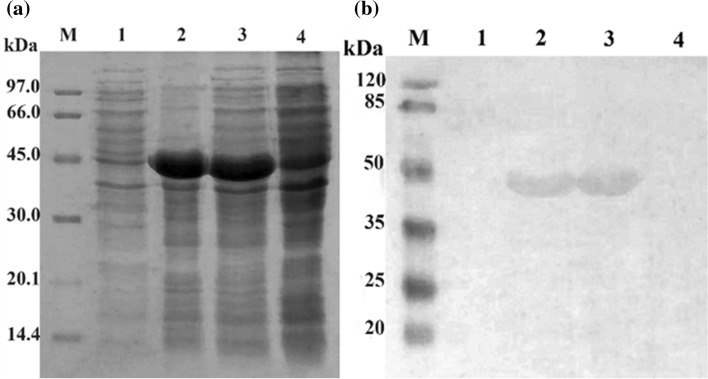
The pET22b-SCARB2 vector was transformed into E. coli BL21 (DE3) cells. Positive clones were induced by IPTG to produce recombinant proteins. To verify the recombinant SCARB2 proteins, the E. coli cells were lysed, subjected to 12.5% SDS-PAGE and stained with Coomassie Blue. The separated bands on gel showed overexpression of one band at about 45 kDa (lane 2, 3; Fig. 3a), which were exact the predicted sizes of SCARB2. There was no overexpression band in the negative control (lane 4, Fig. 3a). In addition, the SCARB2 was designed to fuse with His-tag at the C-terminal, so the presence of SCARB2 was confirmed using His-probe and goat anti-mouse IgG-HRP antibody in Western blot. The results indicated that the protein excessively expressed in the SDS-PAGE gels was SCARB2 (lane 2, 3; Fig. 3b), and this protein expressed mainly in the insoluble fraction. Thus, recombinant SCARB2 fused to His-tag was successfully expressed in E. coli BL21 (DE3) in inclusion bodies.
Fig. 3.
Coomassie brilliant blue staining of expressed SCARB2 analyzed by SDS-PAGE on 12.5% gel a and confirmed by Western blot probed with anti-His, b M, protein maker a and pre-stained protein marker b; 1–3, E. coli BL21 (DE3)/pET22b-SCARB2 (+ IPTG); 1, soluble phase; 2, insoluble phase; 3, total phase; 4, E. coli BL21 (DE3)/pET22b-SCARB2 (-IPTG)
Human SCARB2 has a theoretical molecular weight of 54.3 kDa (COPaKB 2015). The weight discrepancy between its theoretical (54.3 kDa) and obtained in this study (45 kDa) was due to the deletion of transmembrane domain, which anchors the receptor onto mammalian cell membrane. The extracellular domain was chosen to express since the function of capturing or interacting with viruses solely related to this region. This truncated transmembrane protein could help enhance the solubilization of expressed protein. It could also reduce the chance of transmembrane domain, which contains mostly hydrophobic amino acids, to interact with nascent polypeptides, thereby increasing the solubility of target protein. SCARB2′s structure has been confirmed as it carries 10 high mannose-type N-linked oligosaccharide chains and two disulfide bridges, which contribute to the receptor in its proper folding (Dang et al. 2014; Yamayoshi and Koike 2011). Glycosylation and disulfide bridges could have facilitated the ability of interacting with virus capsid. Therefore, elimination of these factors to study the dependence of receptor function was performed. As E. coli expression system does not facilitate post-translation modifications like glycosylation, which has resulted in accumulation of recombinant product in the form of insoluble inclusion bodies, this obstacle might lead to difficulties in further experiments; therefore, a refolding procedure was necessary.
Solubilization and Refolding of Recombinant SCARB2 from Inclusion Bodies
SCARB2 obtained from E. coli BL21 (DE3)/pET22b-SCARB2 formed inclusion bodies. The suspension of inclusion bodies in the washing buffer containing 1% Triton X-100 aided for the inclusion bodies cleaning process. Dissolving in high molarity urea, Gu-HCl containing buffer, 10% sucrose and 0.1% tween 80 contributed to complete denaturation and refolding of targeted protein. After extraction from the inclusion bodies with a refolding buffer, the final product was dialyzed and concentrated by 10 MWCO. Finally eliminating the molarity of urea, Gu-HCl concentration by dialysis contributed to increase proper folded protein, as well as ensured the ability of SCARB2 in later experiments.
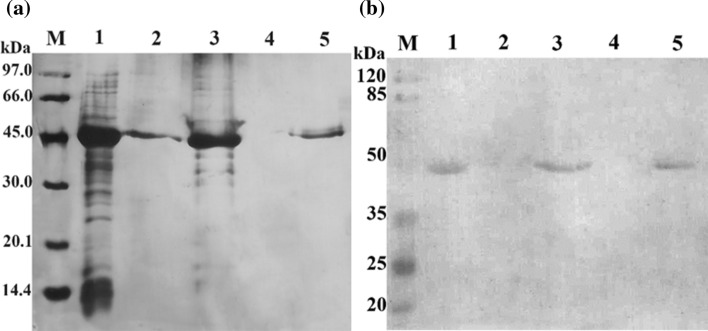
As can be observed from the electrophoresis gel, the targeted protein bands in lane 1, lane 3 and lane 5 were at corresponding molecular weight (Fig. 4a). Simultaneously, the targeted bands that appeared in lane 2 indicated that the dissolution step was not fully completed. However, the targeted bands disappeared in lane 4, which showed that the protein of interest was successfully refolded. The refolding efficiency was about 100%. As fused with His-tag at the C-terminal, the refolding result was confirmed by Western blot using anti-His (Fig. 4b). The corresponding bands on the membrane plot proved the success of our refolding method. This was the first report of obtaining the soluble form of SCARB2 from inclusion bodies (Xu et al. 2018).
Fig. 4.
Silver staining of dissolution and refolding results of recombinant SCARB2 by SDS-PAGE on 12.5% gel a and validated by Western blot probed with anti-His b. M, protein maker a and pre-stained protein marker b; 1, inclusion bodies; 2, precipitated phase after dissolving; 3, supernatant phase after dissolving; 4, precipitated phase after refolding; 5, supernatant phase after refolding
Evaluation of the Interaction between Recombinant SCARB2 and EV71
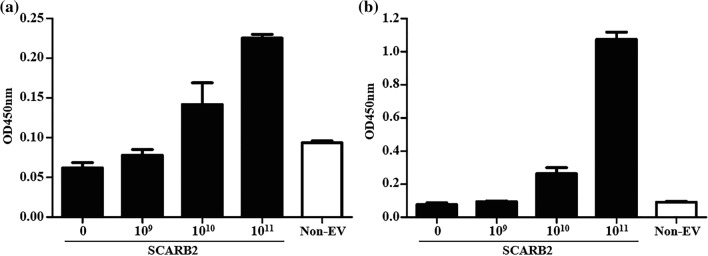
Indirect ELISA was conducted to determine the interaction between recombinant SCARB2 and EV71 at two different pHs (5.5 and 7.4). In order to investigate the ratio of interaction between EV17 and SCARB2, molecule was utilized instead of concentration. As revealed in the previous study, a conformational change was observed in SCARB2 induced by pH, which led to a noticeable dissimilarity in a specific region between structures at two pH levels as an implication for the receptor’s function (Dang et al. 2014). Our result showed that recombinant SCARB2 and EV71 did have interaction in dose-dependent manner at both pH levels, but at lower pH SCARB2 (at 1011 dilution) exhibited a significantly higher interaction, which was approximately 5 times higher compared to that of pH 7.4 (Fig. 5). Firstly, this result was in line with Yamayoshi et al. (2011), supporting the finding that SCARB2 was an EV71 main receptor. Secondly, this result was obtained under soluble form of SCARB2, which was different from Xu et al. (2018) as their insoluble protein product. Moreover, critical region of SCARB2 accountable for binding to EV71 was identified (Chen et al. 2012; Yamayoshi and Koike 2011). Zhao et al. (2014) stated that pH 5.5 was the point at which the binding region showed significant flexibility, whereas at higher pH level, this region was more compact and steadier. This crucial observation contributed to the explanation of differences between the two pH-induced interactions.
Fig. 5.
The interaction of recombinant SCARB2 and EV71 determined by indirect ELISA at pH 7.4 a and pH 5.5 b
There have been considerations about the role of glycosylation in SCARB2. Yamayoshi et al. (2011) stated that the N-linked carbohydrate chains of human SCARB2 were not necessary for the EV71–SCARB2 interaction. Nevertheless, the study of Dang et al. (2014) indicated that the attachment of SCARB2 to EV71 decreased when glycosylation was reduced. In agreement with this opinion, Zhao et al. (2014) confirmed the role of glycan as an attachment point for interaction between SCARB2 and EV71 due to the finding of partial cover of glycan to the residues responsible for viral attachment region. The discrepancy was partly revealed in this study. Our soluble and non-disulfide, non-glycosylated receptor had showed the ability to attach with EV71 in vitro, despite low signals. Upon acidification, the binding signals increased. It could be that SCARB2 underwent confirmation changes, thereby exposing viral attachment region though we could not exclude the role of post-translation modifications. This could indicate that conformational change played a more important role in virus binding than post-translation modifications. Therefore, further experiments need to be conducted for fully understanding the principal mechanism. On the other hand, SCARB2-soluble form could facilitate the initial step for cell and in vivo experiments, as the expression system using E. coli BL21 (DE3) could not provide needed post-translation modifications like glycosylation and disulfide bridges, which might affect the attachment efficacy. Consequently, to clarify the unclear role of glycosylation, higher expression systems should be employed. The E. coli Origami strain and yeast expression system could be potential candidates for further investigations.
Conclusion
In this study, the recombinant vector carrying the gene scarb2 (pET22b-SCARB2) encoding for SCARB2 protein was successfully constructed. Transformed E. coli BL21 (DE3) with recombinant vector pET22b-SCARB2 was successfully created. Extraction of SCARB2 from inclusion bodies was first time documented with a yield of 100%. Interaction of EV71 and recombinant SCARB2 was confirmed via indirect ELISA. To our knowledge, our present study documented for the first time a procedure for obtaining soluble form of E. coli-derived SCARB2 using refolding method. Because early steps of viral infection require crucial interactions between host cell receptors and viruses, blocking or interfering with the interactions could have potential applications. Due to its wide distribution in human cells and organs (Kuronita et al. 2002), entrance through one of the main EV71′s receptors SCARB2 could bring more severe consequences to patients. On that account, appropriate strategies, such as receptor-displaying probiotics or generally recognized as safe organisms, for utilizing recombinant SCARB2 should be performed to help reduce the harm of EV71 on children during the epidemic seasons.
Author contributions
All authors contributed to the study conception and design. All authors read and approved the final manuscript.
Funding
Hai-Vy Vo-Nguyen was funded by Vingroup Joint Stock Company and supported by the Domestic Master/PhD Scholarship Programme of Vingroup Innovation Foundation (VINIF), Vingroup Big Data Institute (VINBIGDATA).
Compliance with ethical standards
Conflicts of interest
The authors declare that they have no conflict of interest.
References
- Brown BA, Pallansch MA. Complete nucleotide sequence of enterovirus 71 is distinct from poliovirus. Virus Res. 1995;39:195–205. doi: 10.1016/0168-1702(95)00087-9. [DOI] [PubMed] [Google Scholar]
- Chen P, et al. Molecular determinants of enterovirus 71 viral entry cleft around Gln-172 on VP1 protein interacts with variable region on scavenge receptor B 2. J Biol Chem. 2012;287:6406–6420. doi: 10.1074/jbc.M111.301622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COPaKB (2015) Protein sequence of human SCARB2 (Uniprot ID: Q14108).
- Dang M, et al. Molecular mechanism of SCARB2-mediated attachment and uncoating of EV71. Protein cell. 2014;5:692–703. doi: 10.1007/s13238-014-0087-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, et al. Characterization of the antibody response against EV71 capsid proteins in Chinese individuals by NEIBM-ELISA. Scientific reports. 2015;5:1–11. doi: 10.9734/JSRR/2015/14076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin EL, et al. The BioPlex network: a systematic exploration of the human interactome. Cell. 2015;162:425–440. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King A, et al. Picornaviridae. New York and San Diego, CA: Academic Press; 2000. Virus taxonomy: Seventh report of the International Committee for the Taxonomy of Viruses; pp. 657–673. [Google Scholar]
- Kuronita T, Eskelinen E-L, Fujita H, Saftig P, Himeno M, Tanaka Y. A role for the lysosomal membrane protein LGP85 in the biogenesis and maintenance of endosomal and lysosomal morphology. J cell sci. 2002;115:4117–4131. doi: 10.1242/jcs.00075. [DOI] [PubMed] [Google Scholar]
- Lee KY. Enterovirus 71 infection and neurological complications Korean. J pediatrics. 2016;59:395. doi: 10.3345/kjp.2016.59.10.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, et al. An inactivated enterovirus 71 vaccine in healthy children New England. J Med. 2014;370:829–837. doi: 10.1056/NEJMoa1303224. [DOI] [PubMed] [Google Scholar]
- Quynh-Anh Nguyen-Ngoc T-TTN, Dang-Trinh M-A, Van Nguyen K, Tran B-P, Nguyen H-L, Le-Phan K-N, Le-Ha T-D, Hoang-Ngoc K-Q, Le LT, Nguyen Y-N, Nguyen T-T, Cao B-V. Enterovirus 71 (EV71) neutralization effect of IgY antibodies obtained from egg yolk Vietnam. Journal of Preventive Medicine. 2015;5:144. [Google Scholar]
- Seidmoradei R, Zeinoddini M, Saeedinia AR, Xhodadadi N. Intein-mediated fusion soluble expression of recombinant human interferon beta (rhIFN-β) Iranian. J Sci Technol Trans A: Sci. 2020;44:371–377. [Google Scholar]
- Dang TT, Trinh MT, Tran-Van H. Cloning, expression and purification of human fibroblast growth factor 1 (FGF-1) in Escherichia coli. J Biotechnol. 2014;12:615–621. [Google Scholar]
- Thao NTT, Ngoc NTK, Tú PV, Thúy TT, Cardosa MJ, McMinn PC, Phuektes P. Development of a multiplex polymerase chain reaction assay for simultaneous identification of human enterovirus 71 and coxsackievirus A16. J virol methods. 2010;170:134–139. doi: 10.1016/j.jviromet.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO The WHO Regional Office for the Western Pacific's Institutional Repository for Information Sharing. https://iris.wpro.who.int/.
- WHO . A guide to clinical management and public health response for hand, foot and mouth disease (HFMD) Manila: WHO Regional Office for the Western Pacific; 2011. [Google Scholar]
- Xu T, Lin Z, Wang C, Li Y, Zhao M, Hua L, Zhu B. Prokaryotic expression and identification of scavenger receptor B2. Acta Virol. 2018;62:50–57. doi: 10.4149/av_2018_106. [DOI] [PubMed] [Google Scholar]
- Yamayoshi S, Fujii K, Koike S. Scavenger receptor B2 as a receptor for hand, foot, and mouth disease and severe neurological diseases. Frontiers in microbiology. 2012;3:1–6. doi: 10.3389/fmicb.2012.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamayoshi S, et al. Human SCARB2-dependent infection by coxsackievirus A7, A14, and A16 and enterovirus 71. J virol. 2012;86:5686–5696. doi: 10.1128/JVI.00020-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamayoshi S, Koike S. Identification of a human SCARB2 region that is important for enterovirus 71 binding and infection. J virol. 2011;85:4937–4946. doi: 10.1128/JVI.02358-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamayoshi S, Yamashita Y, Li J, Hanagata N, Minowa T, Takemura T, Koike S. Scavenger receptor B2 is a cellular receptor for enterovirus 71. Nat med. 2009;15:798–801. doi: 10.1038/nm.1992. [DOI] [PubMed] [Google Scholar]
- Yi E-J, Shin Y-J, Kim J-H, Kim T-G, Chang S-Y. Enterovirus 71 infection and vaccines. Clin exp vaccine res. 2017;6:4–14. doi: 10.7774/cevr.2017.6.1.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F-C, et al. Efficacy, safety, and immunology of an inactivated alum-adjuvant enterovirus 71 vaccine in children in China: a multicentre, randomised, double-blind, placebo-controlled, phase 3 trial. The Lancet. 2013;381:2024–2032. doi: 10.1016/S0140-6736(13)61049-1. [DOI] [PubMed] [Google Scholar]
- Zhu F, et al. Efficacy, safety, and immunogenicity of an enterovirus 71 vaccine in China New England. J Med. 2014;370:818–828. doi: 10.1056/NEJMoa1304923. [DOI] [PubMed] [Google Scholar]
- Zou S, et al. Inactivation of the novel avian influenza A (H7N9) virus under physical conditions or chemical agents treatment. Virol J. 2013;10:289. doi: 10.1186/1743-422X-10-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Ren J, Padilla-Parra S, Fry EE, Stuart DI. Lysosome sorting of β-glucocerebrosidase by LIMP-2 is targeted by the mannose 6-phosphate receptor. Nature Communications. 2014;5(1):4321. doi: 10.1038/ncomms5321. [DOI] [PMC free article] [PubMed] [Google Scholar]