ABSTRACT
Plant resilience to oxidative stress possibly operates through the restoration of intracellular redox milieu and the activity of various posttranslationally modified proteins. Among various modes of redox regulation operative in plants cys oxPTMs are brought about by the activity of reactive oxygen species (ROS), reactive nitrogen species (RNS), and hydrogen peroxide. Cysteine oxPTMs are capable of transducing ROS-mediated long-distance hormone signaling (ABA, JA, SA) in plants. S-sulphenylation is an intermediary modification en route to other oxidative states of cysteine. In silico analysis have revealed evolutionary conservation of certain S-sulphenylated proteins across human and plants. Further analysis of protein sulphenylation in plants should be extended to the functional follow-up studies followed by site-specific characterization and case-by-case validation of protein activity. The repertoire of physiological methods (fluorescent conjugates (dimedone) and yeast AP-1 (YAP1)-based genetic probes) in the recent past has been successful in the detection of sulphenylated proteins and other cysteine-based modifications in plants. In view of a better understanding of the sulfur-based redoxome it is necessary to update our timely progress on the methodological advancements for the detection of cysteine-based oxPTM. This substantiative information can extend our investigations on plant–environment interaction thus improving crop manipulation strategies. The simulation-based computational approach has emerged as a new method to determine the directive mechanism of cysteine oxidation in plants. Thus, sulfenome analysis in various plant systems might reflect as a pinnacle of plant redox biology in the future.
KEYWORDS: Cysteine modifications, oxidative stress, redox function, sulfenylation, sulfenome
1. Introduction
Plant resilience to oxidative stress is attained through a tight regulation of reactive oxygen species (ROS) and reactive nitrogen species (RNS) homeostasis which is mediated by various enzymatic and non-enzymatic mechanisms. The high amount of dissolved oxygen in the cells is associated with low potential electron carrier thus leading to the formation of an oxidizing environment. Reduction of molecular oxygen results in the formation of superoxide radicals which are readily metabolized by the activity of superoxide dismutase (SOD) to form hydrogen peroxide.1 A subtle change in the redox milieu of plant cells triggers a series of signaling responses which are mediated by ROS and RNS activity. Nitric oxide is produced by enzymatic and non-enzymatic pathways and can stoichiometrically react with superoxide anions and oxygen to produce RNS molecules like peroxynitrite (ONOO_) and dinitrogen trioxide (N2O3).2 Pertubation of ROS homeostasis has been reported to be associated with phytohormone signaling, wound response, osmotic adjustment, and various developmental processes like root growth (gravitropic) and pollen tube elongation.3–8
Among various mechanisms of ROS and RNS sensing in cells, their reactivity with protein cysteine residues appears to be a major oxidative post-translational modification (oxPTM) among living kingdoms.9,10 The differential state of oxidation of sulfur-containing amino acids in proteins result in the formation of a wide range of oxPTMs like S-nitrosylation (-SNO), S-glutathionylation (-SSG), sulfhydration, sulfenylation (-SO2H), and sulfonylation (-SO3H) (Figure 1). The protein microenvironment and pKa value of the cysteine-based sulfur atom determine the reactivity of their thiol groups. The thiolate form of cysteine side chains is highly prone to oxidation by ROS. An interesting review by Ref.11 states the chronological advancements in the concept of reactive sulfur species (RSS) since 2001. The author addresses various questions on the nature and function of RSS. These molecules have been stated to constitute a group of redox-active inorganic (sulfenic acid, sulfinic acid) and organic (disulfane) species. Organic RSS has been reported to be formed by reaction with H2S or oxidative thiols (RSH) which in turn participate in various signaling processes in plant and animal cells. Cysteinyl-sulfenic acids are the primary molecules associated with redox signaling which further undergo various reversible and irreversible secondary modifications.
Figure 1.
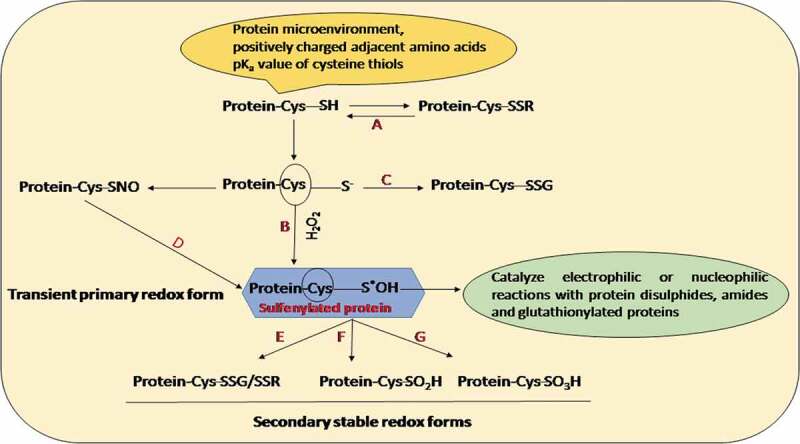
Cysteine modifications in plants. ROS-mediated reversible oxidation of cysteine residue results in the formation of sulfenic acid (R-SOH]. This oxidative modification may be unstable in the protein microenvironment thus leading to its oxidation into sulfinic acid (R-SO2H) and subsequent irreversible oxidation into sulfonic acid (R-SO3H). S-nitrosylation is a reversible cys oxPTM where NO is covalently linked to the reactive cysteine thiol (-SNO) (a-g)
Various investigations across the last decade have enhanced our fundamental knowledge on the physiological role of cys oxPTM and protein sulfenylation in plants. Various qualitative and quantitative methodologies have been devised to perform the detection of sulphenylated proteins and other cysteine-based modifications in plants. In view of a better understanding of the sulfur-based redoxome it is necessary to update our timely progress on the methodological advancements for the detection of cysteine-based oxPTM. Medicago truncatula and Arabidopsis thaliana have been analyzed for deciphering the spatio-temporal regulation of sulfenylated proteins and sulfenome expression, respectively.12,13 Interesting updates in the recent couple of years reveal the conserved homology of sulfenylated proteins which constitute a significant part of the plant and human sulfenomes.
Although a number of reviews has been published on the aspect of protein sulfenylation in plants, it has been necessary to critically summarize the recent developments on sulfenome analysis in plants. The review briefly analyzes the physiological role of cysteine-modifications and recent advancements in sulfenome analysis (in reference to certain plant systems – Arabidopsis and Medicago). This shall enable researchers to undertake future investigations for deciphering sulfenome of various plant systems during developmental and stress response.
2. Cysteine modifications (oxPTM) and their role in plant signaling
ROS and RNS in plant cells regulate various physiological processes associated with stress sensing, pathogen signaling, wound response, and developmental changes.3–8 ROS and RNS interaction and their role in redox regulation have been deciphered in the last decade of investigations in plants.14,15 Among the various mechanisms of ROS and RNS-mediated sensing of redox stimuli protein modification at the cysteine and methionine residue lead to a diverse set of oxPTMs in cells.16,17 NO and H2S-mediated oxPTM of proteins are manifested mostly in the form of S-nitrosylation and S-sulfhydration, respectively.18–21 Among the various proteins undergoing ROS-mediated oxPTM (protein sulfenylation, sulfinylation and sulfonylation) enzymes associated with calvin cycle, sulfur, and starch metabolism have been extensively studied in plants.15,22,23 Cys-mediated PTMs in plants have been associated with ROS-phytohormone crosstalk during stressful environments. Extensive reviews on the aspect of plant development and redox regulation have been evident by various authors.24–26 ROS levels in cells are preferentially kept under the tight control of enzymatic and nonenzymatic antioxidative systems.27–30 A critical analysis of the physiological state of plant cells has been supplemented with our current knowledge of the redox nature of cysteine residues present in various proteins. Cysteine-mediated oxPTM largely depends upon the redox environment of the protein.
Since all cysteine residues in proteins are not prone to ROS, RNS, or H2S-mediated modifications, it is likely that the local redox environment is an important determinant of cysteine reactivity in proteins. The pKa value of each cysteine residues in proteins exerts a strong influence on the reactivity of cysteine molecules. Furthermore, electrostatic environment, steric accessibility, hydrogen bonding (between thiols and thiolates) and nucleophilicity of cysteine residues are some of the important factors which regulate cysteine OxPTMs in plants.31 The reduced form of the thiol group (R-SH) in cysteine-oxPTMs is represented by the −2 oxidation state of the sulfur atom. The anionic (thiolate) form of sulfur present in cysteine is prone to oxidation, which in turn, is regulated by its pKa value.32 According to Ref.9 an increased number of hydrogen bonds associated with cysteine-sulfur lowers the pKa value and stabilizes the thiolate form. Most importantly, it has been evident that the three-dimensional conformation of the protein and the steric accessibility of cysteine are important regulators of its reactivity with ROS and other biomolecules.33 ROS-mediated reversible oxidation of cysteine residue results in the formation of sulfenic acid (R-SOH). This oxidative modification may be unstable in the protein microenvironment thus leading to its oxidation into sulfinic acid (R-SO2H) and subsequent irreversible oxidation into sulfonic acid (R-SO3H).17 The formation of sulfenic acid (R-SOH) has been reported to be catalyzed by the activity of sulfiredoxin (Srx) which reversibly reduces R-SO2H to R-SOH in Arabidopsis.13 This mechanism has been known to be operative in the presence of two known substrates, namely,- chloroplast-2cys Prx and mitochondrial PrxIIF.13,34 Sulfenic acid residues are capable of further reaction with free protein thiols which lead to the formation of intra/intermolecular disulfide bonds (R-SS-R/R-SS-R’) or may produce low molecular weight thiols (GSH) thus leading to cys S-glutathionylation in plants. Recent investigations reveal the role of S-glutathionylation in the regulation of stress tolerance, prevention of cysteine oxidation at the active site of proteins, and regulation of redox signaling. Glutredoxins (Grxs) and thioredoxins (Trxs) exert regulatory roles in the reduction of disulfide bonds and deglutathionylation of proteins. Unlike in prokaryotes and animal systems, the Trx/Grx system is known to exhibit much complex regulation in plants. Chloroplast has been known to be a major site of Trx and Grx activity where the enzymes are energized by the presence of NADPH.15,35
Hydrogen sulfide-mediated modification of cysteine residues is mostly manifested by the formation of protein S-sulfhydration which further enhances the catalytic activity of targeted proteins in animal systems.20 Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (38 kDa), β-tubulin (55 kDa), and actin (43 kDa) exhibit a higher degree of sufhydration.19 Proteomic analysis of H2S-mediated endogenous S-sulfhydration in Arabidopsis has been reported by Ref.36 The authors describe the implication of the biotin-switch method in the identification of 176 S-sulfhydrated proteins in Arabidopsis leaf (ecotype Col 0). The S-sulfhydrated proteins were mostly related to primary metabolic pathways (TCA cycle, glycolysis) and cytoskeletal components. Functional analysis of S-sulfhydrated proteins reveals the significance of H2S-mediated sulfide signaling in regulating metabolism during stress tolerance. Furthermore, S-sulfhydrated proteins in Arabidopsis appear similar to those in the mammalian system. According to Aroca et al.37,38 H2S-mediated cysteine signaling, therefore, primarily operates through protein sulfhydration in plants. This phenomenon involves modification in the activity of major enzymes like ascorbate peroxidase, glyceraldehyde-3-phosphate dehydrogenase, and glutamine synthetase in Arabidopsis. A recent review by Ref.39 summarizes the role of H2S in triggering persulfidation and sulfenylation of protein cysteine residues. Among various proteins undergoing persulfidation and sulfenylation, ABA receptor (PYL1), calcium-dependent protein kinase (CPK 3, 5, 6, 9), SNF1-related protein kinases (2.1, 2.2, 2.3 2.4), and protein phosphatases exert regulatory functions in ABA signaling of guard cells.
Last decade of investigations has revealed the multiple roles of S-nitrosylation in regulating plant growth and development.40 S-nitrosylation is a reversible cys oxPTM where NO is covalently linked to the reactive cysteine thiol (-SH) to form S-nitrosothiol. S-nitrosylation is also associated with denitrosylation and transnitrosylation.41 This cys oxPTM has been considered to be a nonenzymatic process which is mediated by NO or nitrogen oxides (NOx), or through metal-NO intermediates, SNOs, or ONOO.42 The redox potential difference between the NO donors regulates the extent of S-nitrosylation in cells. Detailed structural analysis of the S-nitrosylated proteins has been investigated in plants and other organisms.43–46 Recent investigations suggest that transnitrosylation involves the transfer of NO bearing moiety from one protein to another, thus nitrosylating the latter. Transnitrosylases are capable of transferring the nitroso group between the cysteine residues of the two proteins.41 Unlike in bacteria and mammals, no transnitrosylase has yet been reported in plants. Denitrosylation of proteins is mediated by the activity of thioredoxins and thioredoxin reductases.
The chloroplast localized 2-cysteine peroxiredoxin (2-CysPrx) essentially serves as an electron sink and is capable of oxidizing reductively activated proteins (fructose-1,6-bisphosphatase and NADPH-dependent malate dehydrogenase).47 This finding provides important insights into cysteine peroxiredoxin-mediated thioredoxin signaling and regulation of metabolic proteins in chloroplast. Recent investigation by,48 in seeds of Arabidopsis report tandem mass tag [iodoTMT)-based thiol redox proteomic approach to monitor protein thiol switches which are characterized by redox-sensitive cysteine residues in organellar proteins. Seed metabolism and efficient energy usage is crucial for maintaining germination vigor in seeds. An investigation by Ref.48 emphasizes on the role of operational protein thiol switches in regulating seed metabolism in mitochondria and cytosol. Cysteine containing peptides (412] were observed to undergo redox switching and were mostly related to the tricarboxylic acid (TCA) cycle, glutathione reductase 2, NADPH-thioredoxin reductase a/b, and thioredoxin-o1. Thus, proteins with functional redox cysteine residues were mostly associated with improved energy efficiency in mitochondria, increased ATP accumulation, and oxygen uptake in germinating seeds. Plant cysteine oxidases (PCOs) in Arabidopsis thaliana have been emphasized to function as oxygen sensors during hypoxia and stress tolerance.49 Redox-post translational modifications precisely control the process of protein compartmentation and regulate the activity of various antioxidant or redox-associated enzymes.50
2.1. Crosstalk among phytohormones, ROS, and metabolic enzymes is orchestrated by the formation of cysteine oxPTMs
2.1.1. Plant growth and immune response
Various S-nitrosylated proteins have been characterized in Arabidopsis which are mostly associated with growth and development, hormone signaling, ROS homeostasis, and abiotic stress tolerance. ABI5 transcription factor is an important regulator of seed germination and seedling growth in response to ABA and NO. S-nitrosylation of ABI5 results in its proteosomal degradation and subsequent promotion of seed germination.51 Furthermore, NO-mediated S-nitrosylation of SnRK2.6/OST1 components can negatively regulate ABA signaling during abiotic stress. VASCULAR-RELATED NAC-DOMAIN (VND), and a small family of NAM/ATAF/CUC (NAC) transcription factors are important regulators of xylem element differentiation. S-nitrosylation in the cys 264 and cys 320 of VND7 has been detected by biotin-switch assay.52 Thus, cys oxPTM of this protein is crucial for modulation of xylem differentiation. S-nitrosylation has been reported to regulate auxin signaling by mediating proteolytic degradation of Aux/IAA repressors.53 Auxin dependent-interaction with TIR1/AFBs elements lead to proteolytic degradation of AUX/IAA complex. S-nitrosylation of the TIR 1 receptor at cys 140 and cys 37 result in enhanced degradation of AUX/IAA complex. Interestingly, cytokinin signaling is negatively regulated by S-nitrosylation. The two component system of cytokinin signaling comprises of a histidine phosphotransfer protein which is S-nitrosylated at the cys 115 residue thus repressing its phosphorylation.40 The chloroplast-localized peroxiredoxin II E (PrxII) is an important component of the antioxidant machinery of cells that detoxifies H2O2. S-nitrosylation at the cys 121 residue of PrxII results in its decreased activity. This is further accompanied by tyrosine nitration and lipid peroxidation which elicits immune response during pathogen attack.54 S-nitrosylation of RBOHD at Cys-890 also reduces ROS generation in plants subjected to oxidative stress.55 Investigations therefore reveal the regulatory role of S-nitrosylation in biotic and abiotic defense in plants. Chlorophyll metabolism is regulated by S-nitrosylation, where NO plays an important role in regulation of GSNOR activity. Various component proteins associated with light and dark reaction have been reported to undergo S-nitrosylation. NAB1 is a cytosolic RNA-binding protein that represses the translation of light-harvesting protein at (LHCBM) PSII in Chlamydomonas reinhardtii. During low light intensity S-nitrosylation of NAB1 at cys 226 results in increased translation of LHCBM thus optimizing light harvest.56
2.1.2. ABA and Ca2+ signaling
Among the various plant hormones, abscisic acid (ABA) and salicylic acid signaling has been known to be mediated by cys oxPTMs. During abiotic and biotic stress increased ABA concentrations impart drought tolerance, regulate stomatal closure, and facilitate nutrient homeostasis in various plant organs. ABA can preferably bind to PYRABACTIN RESISTANCE/PYR1-LIKE/REGULATORY COMPONENT OF ABA RECEPTOR PYR/PYL/RCAR thus activating SnRK2/OST1 kinases.57 The ABA-mediated downstream signal cascade is further transduced by the activation of NADPH oxidases and RbohF thus promoting ROS formation in cells. In this context glutathione peroxidase 3 (GPX3) mediated ROS signaling is accompanied by thiol-disulfide exchange with the negative regulatory protein of ABA signaling named- ABI2 (PROTEIN PHOSPHATASE 2 C – PPC family of ABA insensitive 2).58 Similar mechanism of thiol-disulfide exchange has been reported in yeast and mammalian cells subjected to oxidative stress.59 Thiol peroxidases are widely known to function as redox sensors within the plant and animal kingdoms (Margis et al 2008). H2O2 sensor oxidant receptor Peroxidase 1 (ORP1/Gpx3) is also involved in thiol-disulfide signaling in yeast.60 ANNEXIN 1 (At ANN1) has been reported to mediate ROS-dependent Ca2+ flux in Arabidopsis roots. ANNEXIN 1 undergoes S-glutathionylation on cys111 and cys239 residues thus decreasing its Ca2+ affinity. This mechanism is effective in the prevention of ROS-mediated Ca2+ flux in plant cells subjected to biotic stress.61
2.1.3. Modulation of SA and JA signaling
Salicylic acid (SA) exerts regulatory effects on local and systemic acquired resistance to pathogen infection. NONEXPRESSOR of PR GENES 1 (NPR1) is a transcriptional coactivator that regulates SA-governed transcriptional response in cells. Interestingly, NPR1 (cytoplasm localized) undergoes oligomerization in cys82 and cys216 residues formed by the virtue of intramolecular bonds (R-SS-R).62 SA-signaling further triggers Trxh3/h5-dependent reduction of disulfide bonds. This leads to the monomerization of NPR1 proteins. S-nitrosylation at the cys 156 residue of NPR1 competes with oligomerization of the protein. Thus, cys modification of NPR1 is a precise switch-off mechanism for NPR-1-dependent signaling operative between cytoplasm and nuclei.63 Furthermore, NPR1-TGA2 (transcription factor) interaction is regulated by the modifications at cys521 and cys529 residues. TGA undergoes cys modifications at residues cys260 and cys266. This modification is an important determinant of TGA-NPR1 interaction which leads to downstream activation of PR genes during biotic stress.64 Similarly, S-nitrosylation has been known to play a pivotal role in SA biosynthesis and modulate its signaling.65 NPR1 has also been reported to undergo S-nitrosylation at the cys 156 residue thus modulating SA-induced gene expressions. Furthermore, TGA and SRG transcription factors are also regulated by S-nitrosylation which in turn regulates SA-mediated defense response.
JA signaling in plants is known to be modulated by cys oxPTMs. JA precursor 12-oxo-phytodienoic acid (OPDA) exerts transcriptional changes during the surge in JA biosynthesis.66 OPDA signaling is mediated by chloroplastic receptor CYCLOPHYLIN 20–3 (CYP20-3) in the chloroplast.67 The CYP20-3 receptor undergoes oxidative inhibition brought about by the formation of R-SS-R (intramolecular) bonds between cys53-cys170 and cys128-cys175 residues.68 CYP-20-3 is a redox-sensitive protein that exerts regulatory influence on JA and OPDA signaling. In Arabidopsis, CYP-20-3 binds to its target protein chloroplast SERINE ACETYL-TRANSFERASE1 (SAT1). The CYP-20-3 and SAT1 interaction is crucial for the regulation of cysteine biosynthesis in chloroplasts. The OPDA-CYP20-3 binding is crucial for JA-mediated stimulation of antioxidants.67
2.1.4. MAPK signaling and oxidative stress
MAPK signaling is associated with various aspects of plant growth, development and stress tolerance.69–72 MAPK functions as a three component signal cascade which is initiated by MAPK kinase kinases (MAPKKKs) that phosphorylates MAPK kinases (MAPKKs) which in turn phosphorlaytes specific MAPKs. Apart from plant MAPKs, yeast and mammalian MAPKs have also been reported to undergo cys oxPTM.73–75 Plant MAPKs are often known to be activated during oxidative stress. Among various MAPKs present in plants MAPK12 is stimulated both by the activity of H2O2 and ABA. MAPK3 and MAPK 6 are activated by OXIDATIVE SIGNALINDUCIBLE1 (OXI1) kinase which is also regulated by ROS levels in the cells. Tomato and Arabidopsis plants exhibit evolutionary conserved redox-regulation of MAPK signaling cascades (MAPK1/2 and MAPK6).76 Rice MAPKs-osMPK3 and osMPK6 also exhibit precise redox control. Rice thioredoxin (osTrx 23) negatively regulates the activity of rice MAPKs.77 Cys179 and cys210 are important regulatory sites of both the rice MAPKs (osMPK3 and osMPK6). Oxidative stress is known to induce sulfenylation of both the MAPKs in rice plants. Furthermore, subsequent reduction of the sulfenylated forms of MAPK produces S-glutathionylated forms which render the MAPKs inactive. MAP triple kinase 1 (OMTK1)-MMK3 pathway in alfalfa is known to regulate H2O2-dependent cell death during oxidative stress.78 MAPK 2, 4, and 7 in Arabidopsis have been reported to undergo H2O2-dependent sulfenylation. Mutation in cys32 of AtMAPK4 orthologue in B. napus (BnMPK4) also reveals a similar mode of cys modification. In this context, it is worth mentioning that Protein Tyrosine Phosphatase (PTPs) and Protein phosphatase Dual specificity phosphatase (DsPTPs) have been considered to be important regulators of MAPK signaling. AtDsPTP1 and AtPTPi exhibit sequence similarity in cys265 residue and their activity is negatively regulated by H2O2.79 Contrastingly, soybean GmPTP exhibits low sensitivity to H2O2-induced inhibition but undergoes GSSG induced S-glutathionylation at cys 78 and cys176.80 MAPK signaling and regulation of ROS generation provide control to the cys oxPTM of proteins during oxidative stress.
2.1.5. Modulation of metabolic enzymes and regulation of root hair growth
S-sulfhydration is an important H2S-mediated cys ox PTM which is known to modulate the activity of various metabolic and antioxidative enzymes.37,38 Biotin-switch method in Arabidopsis has been implied to detect sulfhydrated proteins namelychloroplastic glyceraldehyde-3-phosphate dehydrogenase (GAPA; 42 kD), Glyceraldehyde-3-phosphate dehydrogenase B (GAPB; 48 kD), cytosolic isoform of glyceraldehyde-3-phosphate dehydrogenase (GAPDH; 37 kD), chloroplastic Glutamine synthetase (GS2 isoform 43 kD) and cytosolic ascorbate peroxidase (Apx; 27 kD).38 Interestingly, functional analysis of the enzymes revealed that GS and APx/GAPDH exhibited contrasting nature in the modulation of their activity in response to NaHS (1 nM)-induced S-sulfhydration. GS exhibited a reduction in activity while APx and GAPDH showed a 40–60% enhancement in the activity after S-sulfhydration.38
Investigations have revealed that cytoskeletal proteins are likely to be regulated by S-sulfhydration and S-nitrosylation in plants.80,81 H2S-mediated inhibition of root hair growth in Arabidopsis involves S-sulfhydration of ACTIN-2 filaments. Increased H2S content in the L-CYSTEINE DESULFHYDRASE/O-acetylserine (thiol) lyase isoform a1 (LCD/oasa1) mutant results in enhanced S-sulfhydration in cys 287 residue of F-ACTIN filaments thus imparing root hair growth.82
2.1.6. Modulation of antioxidative defense and regulation of plant receptor kinase
Apart from its role in the protection of oxidative stress, S-glutathionylation has been suggested to be involved in the signaling process and modulates various target proteins in plants. Glutaredoxins invariably control the extent of protein S-glutathionylation in the cells. Plant cells subjected to oxidative stress are followed by a decrease in GSH/GSSG ratio thus triggering increased S-glutathionylation of proteins. Glutathione-mediated cellular redox homeostasis is further regulated by the S-glutathionylation of major metabolic and antioxidative enzymes. Although various proteins have been identified to undergo S-glutathionylation in plants, fewer reports depict the functional or structural modulation of proteins brought about by this cys oxPTM. GAPDH has been extensively studied for S-glutathionylation in plants.83 The modulation of GAPDH activity (inhibition) due to S-glutathionylation has been known to exert regulatory influence on certain primary metabolic pathways namely calvin cycle, glycolysis, and gluconeogenesis. Oxidative stress in Arabidopsis leads to S-glutathionylation of enzymes associated with ascorbate-glutathione (Asa-GSH cycle) namely monodehydroascorbate reductase (MDAR), dehydroascorbate reductase (DHAR 1), NADP-malic enzyme, and methionine sulfoxide reductase B1 (MSRB1).80). DHAR1 ACTIVITY is known to be inhibited by S-glutathionylation at the catalytic cys 20 residue.84 MSRB activity and its cys modification is likely to regulate methionine generation in cells. Similar to S-nitrosylation, S-glutathionylation also regulates the Asa-GSH cycle during environmental stress conditions. MSRB activity is crucial to prevent methionine oxidation in cells which might otherwise affect the function of various proteins. Treatment of Arabidopsis with GSH leads to downregulation of MSRB1 and MSRB2.85 Thus, S-glutathionylation is an important cys oxPTM which regulates redox homeostasis and abiotic stress tolerance in plants.86
S-glutathionylation causes functional deactivation of Arabidopsis thaliana kinase – (BRASSINOSTEROID-INSENSITIVE 1-ASSOCIATED RECEPTOR LIKE KINASE; (BAK1).86 Reports from the animal system also reveal S-glutathionylation-mediated reduction in enzyme activity in mouse cDPK and other kinases.87 BAK1 is known to interact with glutaredoxin (GRXC2), thus catalyzing S-glutathionylation of BAK1 at cys353, 374, and cys 408 residues. The conserved cysteine residues at Arabidopsis kinome exhibit S-glutathionylation during oxidative stress in cells. Investigations by Moffett et al. (2015)88 reveal that S-glutathionylation at the cys 408 site of BAK1 exerts allosteric control over the activity of this protein kinase.
The summary of various proteins undergoing cys oxPTM and their functional significance have been summarized in Table 1.
Table 1.
Various cys (oxPTM) modifications aand their functional relevance in plants
| Protein | Cys oxPTM | Functional significance | References |
|---|---|---|---|
| ABI2 [PROTEIN PHOSPHATASE 2 C – PPC family of ABA insensitive 2) | thiol-disulfide exchange | Inhibition of ABI2 and activation of ABA signaling cascade | 58 |
| H2O2 sensor oxidant receptor Peroxidase 1 (ORP1/Gpx3 | thiol-disulfide exchange | Redox regulation | 60 |
| ANNEXIN 1 [At ANN1] | S-glutathionylation | Decrease in its Ca2+ affinity | 61 |
| NONEXPRESSOR of PR GENES 1 [NPR1] | Oligomerization R-SS-R | Inhibition of NPR1 activity | 63, Mou et al. [2013] |
| TGA | Oligomerization R-SS-R | Regulates TGA-NPR1 interaction | 64 |
| CYCLOPHYLIN 20–3 (CYP20-3] | formation of R-SS-R (intramolecular) bonds | Regulates its binding to JA precursor 12-oxo-phytodienoic acid [OPDA) | 67,68 |
| (MAPK1/2 and MAPK6] in rice- osMPK3 and osMPK6 | S- sulfenylation and S-glutathionylation | Regulation of down- stream signal cascade during oxidative stress | 77 |
| AtDsPTP1 [protein tyrosine phosphatase) and AtPTPi | GSSG induced S-glutathionylation | Regulation of MAPK signaling | 79 |
| ]chloroplastic GAPA | S-sulfhydration | Regulation of oxidative stress | 38 |
| GAPDH | S-sulfhydration | Primary metabolism | 38 |
| ascorbate peroxidase | S-sulfhydration | Regulation of oxidative stress | 38 |
| chloroplastic Glutamine synthetase | S-sulfhydration | Nitrogen metabolism | 38 |
| ACTIN-2 filaments | S-sulfhydration | Inhibition of root hair growth | 82 |
| BRASSINOSTEROID- INSENSITIVE 1- ASSOCIATED RECEPTOR LIKE KINASE- BAK 1 | S-glutathionylation | Allosteric control of BAK1 | Moffetl et al. (2015) |
| MDAR, DHAR 1, NADP-malic enzyme, methionine sulfoxide reductase B1 (MSRB1) | S-glutathionylation | Abiotic stress tolerance, ROS homeostasis | Beger-Morales et al. (2016) |
| AtMAPK4 | S-sulfenylation | Abiotic stress tolerance, ROS homeostasis | 86,87 |
| Glyceraldehyde-3- phosphate dehydrogenase [GAPDH] | S-sulfenylation | Metabolic regulation of calvin cycle, regulation of regulate glycolysis and gluconeogenesis | 88 |
| GLUTATHIONE PEROXIDASE 3 (GPX3 | S-sulfenylation | Mediates the oxidation equivalent to AB12 [PP2C family ABA INSENSITIVE 1-negative regulator of ABA signaling] | 58,89 |
| MAPK9 and MAPK12 | S-sulfenylation | Regulates ABA-dependnet stomatal closure in Arabidopsis | 90 |
| SNF1-related protein kinase2 family proteins | S-sulfenylation | Regulates downstream signaling components (respiratory burst oxidase homolog RbohF and TFs] | 91 |
| SALT OVERLY SENSITIVE 1 [SOS1) | S-sulfenylation | Increased stability of SOS 1 and ion homeostasis | 92 |
| PLASTIDIC TYPE I SIGNAL PEPTIDASE 1 (PLSP1 | S-sulfenylation | Regulates proteolysis | 93 |
| Monodehydroascorbate reductase 1; glutathione-disulfide reductase; Acyl-CoA oxidase 1 and glyoxalase 1 | S-sulfenylation | redox signaling and β-oxidation of fatty acids | 94 |
| peroxidase | S-sulfenylation | Redox signaling | 95,96 |
| REGULATORY PARTICLE NON-ATPASE 12A (cytoplasmic thioredoxin h (Trxh] [RPN12A) | S-sulfenylation | Regulates cytokinin signaling | 97,98 |
| SnRK2.6/OST1 | S-nitrosylation | Negative regulation of ABA signaling | 51 |
| VASCULAR-RELATED NAC-DOMAIN [VND] | S-nitrosylation | Regulation of xylem differentiation | 52 |
| TIR 1 auxin receptor and Arabidopsis SKP1-like [ASK1] protein | S-nitrosylation | Enhances proteolytic degradation of AUX/IAA elements | 53 |
| Chloroplast-localized peroxiredoxin II E [PrxII] | S-nitrosylation | Reduced activity of PrxII triggers defense response by tyrosine nitration and lipid peroxidation | 54 |
| NAB1 (cytosolic RNA-binding protein] | S-nitrosylation | Repressed NAB1 activity increases the translation of light-harvesting protein at (LHCBM) PSII | 56 |
3. Mechanism of protein sulfenylation in plants
Primary redox modifications occurring in the cysteine residue of proteins mostly form sulfenic acid molecules. The oxidation state of sulfur in the sulfenic acid residue is represented with a value of 0. This renders it to function both as an electrophile and a nucleophile thus exchanging electron among several other biomolecules like ROS, H2O2, and thiol groups.115 Apart from forming sulfinic and sulfonic acid residues, the protein cystenyl sulfenic groups are also capable of reacting with protein disulfides, glutathionylated proteins, or amide backbone of proteins to produce cyclic sulphenamyde. The secondary redox forms of sulfenic acid are more stable in nature. The acidity of the cysteine thiols is largely regulated by multiple factors prevalent in the microenvironment of the protein (Figure 1). The presence of positively charged amino acids and likeliness of forming a hydrogen bond with cysteine thiolate of proteins modulate the biochemical property of cysteine thiols.10,99,100 The position of cysteine in the N-terminal end of the α-helix essentially creates a macro-dipole. Furthermore, the decrease of the dielectric constant of the cysteine group increases its overall electrostatic interactions.101–103 Precise investigations are therefore conclusive of the fact that the pKa of acidic cysteine thiols in certain proteins are usually lower than that of free cysteine. According to,104 the molecular environment of the cysteine thiol is capable of modulating the stability of sulfenic acid residues. Multiple cysteine residues at more than one site may undergo sulfenylation in wide classes of functional proteins.105 The formation and stability of sulfenic acid increases in the absence of neighboring cysteine thiols.
Although fewer plant-proteins with sulfenic residues have been characterized in crystallized form, our understanding of the formation of sulfenic acid residues has been assimilated from instances in human and animal systems. Certain instances of stabilization of sulfenic acid residues by adjacent hydrogen bonding in functional enzymes have also been reported by,106 and.107 Interestingly, the sulfenic acid residues have been observed to associate with polar and positively charged amino acids (serine, threonine, or argentine) in its vicinity.105 Apart from the acidic nature of cysteine, various extrinsic factors regulate the process of sulfenylation in the microenvironment of the protein. According to,35cysteine involves in prominent redox reactions with H2O2. However, the lower pKa value of cysteine in acidic pH reduces its chances of being oxidized.101,108 In order to react with H2O2, thiolates of strongly acidic cysteine require to overcome higher energy barriers.101,109 Investigations in the last decade have deciphered the role of sulfenic acid residues in imposing regulatory and catalytic activity in various enzymes during oxidative stress.110,111
4. Methodological approaches for functional characterization of sulfenylated proteins in plants
Proteomic approaches have been implied for the analysis of oxidative stress-induced modulation of sulfenylated proteins in plant systems (Table 2). The methods commonly involve tagging of the cysteine-based oxidative modification followed by isolation and enrichment of modified proteins which are then identified by mass spectrometry (LC-MS-MS). According to some significant advancements obtained for sulfenome analysis in plants, it appears that, although effective, in-vivo sulfenome analysis imposes certain problems arising due to various reasons. Excessive ROS formation may overoxidize the unstable transient sulfenic acid residues which prevents the sulfenylated form to be detected. In general, due to the transient nature, the identification of SOH is very challenging. Furthermore, tissue homogenization and extraction procedures may lead to the disruption of the compartmentalization of the redox state in various organelles. This results in the artificial oxidation of various proteins in their non-native state.116,117
Table 2.
Methodological approaches to analyze sulfenylated proteins in plants
| Methodological approach | Specific protein targeting | Isolation and identification of sulfenylated proteins | References |
|---|---|---|---|
| Differential alkylation based indirect proteomics approach | Blocking of de novo cysteine oxidation using NEM and IAM, followed by arsenite reduction and labeling with biotin-NEM | Streptavidin affinity mediated isolation of proteins followed by 2DE gel separation and detection by MS | 112 |
| Direct proteomics based approach | chemical and genetic probe method | ||
| Chemical probe | DCP-Bio1labelling [in vitro) | Streptavidin affinity mediated isolation followed by detection by MS | 12 |
| DAz-2 and DYn-2 [in vivo] | Protein biotinylation by click reaction and streptavidin affinity followed by detection by MS | 113 | |
| β-ketoesters and BCN[in vivo] | Protein biotinylation by click reaction and streptavidin affinity followed by 2DE gel separation and detection by MS | 114,115 | |
| Genetic probe | YAP-1-based method [in vivo] | Tandem affinity purification followed by detection by MS | 12 |
4.1. Alkylation-based proteomic method
Certain investigations report the implications of the differential alkylation method to detect cysteine modifications in proteins. In this process the free thiols in the sample are blocked by using iodoacetamide (IAM) or N-ethylmaleimide (NEM) which is followed by reduction and isolation of cys-based oxPTM. The commonly used reductants capable of detecting cys-based modifications are usually tris 2-carboxyethyl phosphine (TCEP) or dithiothreitol (DTT). Followed by reduction, the modified thiols are separated by affinity chromatographic methods118 or labeled with various tagged alkylating agents – namely IAM/NEM/maleimide/HDPD, iodoacetamidofluorescein (IAF), mBBr, Cys tandem mass tag (cysTMT), isobaric tag for relative and absolute quantification (iTRAQ), or isotope-coded affinity tag (ICAT). Followed by labeling, further analysis of cys-oxPTM is performed by western blot, avidin affinity tagging or fluorescent 2-D gel electrophoresis followed by mass spectrometry.119–123
Alkylation-based proteomics enable to detect protein sulfenylation by reversibly modified cysteine modifications. Arsenite is used as a reductant in this process.124,125 According to Ref. 126 this method has been used to detect peroxide-mediated sulfenylation in papain. However, fewer reports are available for this method being implied in plant investigations. The alkylation method of cys-oxPTM possesses certain limitations due to the reactive and transient nature of sulfenic acids. Sample preparation steps from the cell lysates increase the chance of non-native proteins being sulfenylated, thus providing false-positive results. Furthermore, the sulfenylated proteins are often not detected due to the presence of -SSG group or because of over-oxidation.117,127
4.2. Direct proteomic approach (probe-based)
Proteomic approaches implied for detecting cys-SOH mostly involve chemical probes used for labeling or direct trapping of sulfenylated proteins. This approach involves the use of affinity enrichment of the targets which enable better detection through mass spectrometry. Direct detection methods for sulfenylated proteins are based on the principle of electrophilic substitution undergone by sulfenylated proteins which react with nucleophilic probe called dimedone (5,5-dimethyl-1,3-cyclohexanedione). This reaction has been widely used as a dimedon-modified sulfenic acid detection method which is attempted through mass spectrometry.128 Dimedone-modified sulphenic residues, however, possesses a limitation in being detected at cellular levels. This pertains to the lack of functional groups which would have otherwise enriched the signal associated with dimedone-modified sulphenylated proteins. Signal enhancement for enriched dimedone-modified sulphenylated proteins has been possible by using dimedone-biotin/fluorophores conjugates.128–130 The dimedone-biotin/fluorophores conjugate is membrane impermeable and hence this method could not be used for in-vivo cellular localization of sulfenylated proteins.117 Substitutes of dimedone have been reported in various investigations which are namely DAz-1, 131,132 DAz-2,133 DYn-1, and DYn-2.109 These substitutes preferably tag and trap the modified cysteine groups in the proteins.
Recent developments suggest the application of a chemical probe (DYn-2) specific for sulphenylated proteins. The chemical probe is composed of a dimedone unit attached with an alkyne group which helps in the enrichment of labeled proteins. This chemical probe helps in tagging the sulfenic acid residue of proteins which can further undergo biotinylation by the principle of copper (I)-catalyzed azide-alkyne huisgen cyclo-addition reaction [click reaction,134,135]. This modified method facilitates enriched sulfenome analysis by mass spectrometry. Interestingly, DYn-2 is a nontoxic and membrane-permeable probe which does not alter the intracellular-redox state. Furthermore, this probe has been successful in wide detection of sulphenylated residues in proteins of human epidermoid carcinoma cell lines (epidermal growth factor- EGF), budding yeast (recombinant glutathione peroxidase 3 – Gpx3), and Arabidopsis.134,136 According to Ref.113 this method has been successful in the identification of 226 sulfenylated proteins in Arabidopsis cell suspension. The authors reported the identification of novel sulfenylated proteins in sub-cellular compartments arising from oxidative stress (H2O2 treatment). The extent of the formation of sulfenylated proteins was observed in the presence of oxidative stress induced by variable concentrations of H2O2 [0.1–20 mM). The authors obtained conclusive results from LC-MS/MS analysis which depicts the DYn-2 probe method to be sensitive in the traping and visualization of sulfenylated proteins in plants. According to,134 the in-vivo labeling of sulfenylated proteins might affect the basic oxidation levels of cysteine. In this report the authors conclusively presented the functional characterization and subcellular location of 226 sulphenylated proteins resulting from H2O2- induced oxidative stress. A novel generation benzothiazine-based chemoselective probe [BTD] and mass spectrometry assisted chemoproteomic approach has been recently reported by Ref.137 The methodology briefly describes the process of global site-specific identification and quantification of S-sulfenylayted proteins. In another report by Ref.138, the authors report the creation of a novel library of diverse carbon nucleophiles that are capable of reacting with cysteine sulfenic acid residues of proteins. The method described by,138 explains its advantages over fragment based-ligand methods that misses target cysteine residues due to their redox-sensitive nature.
4.3. Yeast AP-1 [YAP1]-based genetic probe approach
The authors of Ref87have reported YAP1-based sulfenic acid trapping method which involves the preparation of a genetic construct of the C-terminal domain of the yeast (Saccharomyces cerevisiae] AP-1–like (YAP1) transcription factor followed by a tandem affinity purification tag. The YAP1-based probe functions on the basis of H2O2-induced signaling response obtained from thiol-peroxidases. The peroxidase1 (ORP1/glutathione peroxidase 3) oxidant receptor in yeast is a H2O2 sensor which along with transcription factor YAP1 (yeast AP-1–like) regulates redox signaling mediated by sulphenic acid thiol-disulfide crosstalk mechanism.60 The authors87 reported a total of 100 sulphenylated proteins in Arabidopsis which comprises of H2O2-mediated sulfenome in various subcellular compartments. Novel findings by Ref.87 could identify 66 novel proteins undergoing oxPTM. A similar investigation using the YAP1 probe has been undertaken in the Medicago truncatula which reported an approximate of 90 sulfenylated proteins.12 Some other chemical probes like 1,3-cyclopentadione139 and the linear β-ketoester140 have also been implied to attempt the detection of sulphenylated proteins. However, according to Ref.115, the β-ketoester probe possesses properties of nonspecific recognition of proteins. An important aspect of using chemical probes for – SOH detection has been discussed by Ref.113 The chemical probes are likely to interfere with various signaling pathways of the cell which limit the efficacy of the probe-based in situ Cys-SOH trapping method. According to Ref. 109, the second-order rate constant for the reaction of dimedone with sulphenic acid is 2.7 × 10–2 M–1 S–1 which is sufficiently low to detect transient changes in cys-SOH.
A combination of the DCP-Bio1 and YAP method has been successful in probing of sulphenylated proteins associated with the symbiotic interaction of the Medicago truncatula and Sinorhizobium meliloti.12 Although the YAP-1 probe method has been successful in detecting orgnaellar cys-SOH, it imposes certain limitations in terms of its nonspecific reactivity, steric effect, and substrate biasness in cells.87 Furthermore, glutathione or redox-enzyme mediated reduction of disulfide bonds between the YAP1 and target proteins may underrepresent the sulfenome pool of the cells.,141 reported the schematic path of proteomic workflow which depicts the identification of 1000 sulfenylation sites present in 700 proteins. Recent report by Ref.142 using the YAP1-C approach reveals an in-vivo analysis of plastidial sufenome in response to light and H2O2-mediated oxidative stress. A major pool of all the sufenylated proteins [132) detected in plastids was associated with amino acid metabolism. In a modified method adopted by Ref.143 the identification of in situ sulfenylated cysteines by using a transgenic YAP-1 probe has been accomplished in Arabidopsis. This method is a noninvasive approach to detect sulfenylated cysteines in all organisms with genetic modifications (Figure 2).
Figure 2.
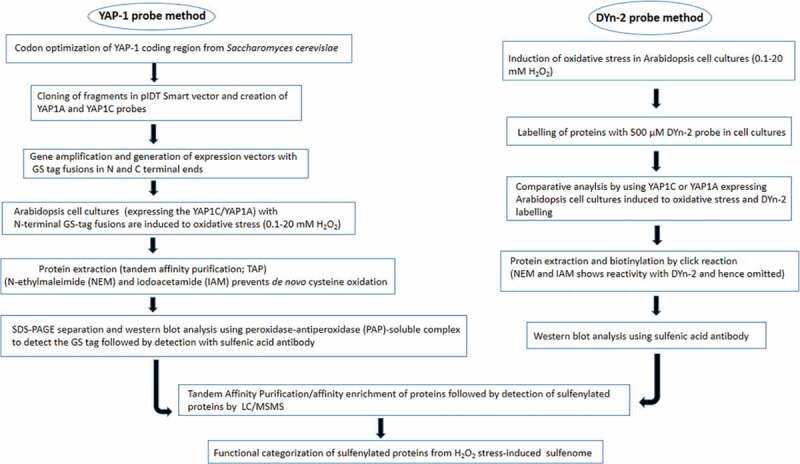
Methodology of sulfenome analysis in plants (YAP-1 and DYn- probe based approach)
5. Spatio-temporal regulation of sulfenome orchestrates primary metabolism, stress signaling, and redox homeostasis in plants
Redox ox-PTM is an important physiological phenomenon associated with the catalytic activity of various proteins that modulate signaling response and protein–protein interactions in cells. Proteins contain cysteine residues that are prone to reaction with different biomolecules such as H2O2, NO, or H2S. This results in the formation of sulfenyl-amide and glutathione conjugates which alter the activity of the enzyme. The altered biological activity of various proteins at the subcellular level regulate the downstream cascade of various signaling events. Protein sulfenylation is an important redox-oxPTM regulated by thioredoxin (Trx) and glutredoxin [Grx) family of proteins. Interestingly, the regulatory cysteines also impose changes in the active site or conformation of protein which modulates DNA–protein interaction. For instance,144 report the role of S-sulfenylation, S-nitrosylation and S-glutathionylation in altering the DNA-binding affinity of TGA1 protein (transcription factor]. Functional categorization of H2O2-dependent sulfenome in Arabidopsis has revealed a total of 67 novel proteins that undergo sulfenylation.87 Various sulfenylated proteins identified were mainly associated with signal transduction (13), proteolytic acitivity (19), RNA-binidng and translation (9), hormone homeostasis (4), protein transport (5), amino acid metabolism (5), and redox-associated enzymes (7). Interestingly, the time dependent-response of H2O2 treatment on sulfenome expression revealed that signal transduction and redox related proteins exhibited sulfenylation as an early response (10 min of oxidative stress) while the other classes of sulfenylated proteins could be detected as late responsive elements.87 Thus, the early response of sulfenylation is likely to operate through pathways of signal transduction and redox homeostasis within the cells. Sulfenylation in the redox proteins could also be detected at a later stage of oxidative stress (1 h) thus suggesting its vital role in redox regulation.87 Recent investigations by Ref.86 have implied a chemoproteomic approach to decipher the cysteine map of sulfenylated proteins in Arabidopsis. The Arabidopsis MITOGEN-ACTIVATED PROTEIN KINASE4 (AtMAPK4) undergoes sulfenylation at the cys181 residue, which corresponds to cys161 of human MAPK1. The authors concluded that the cysteine based-sulfenylation sites of the proteins are highly redox-sensitive. The next subsections of the review shall highlight the detailed role of protein sulfenylation in regulating plant stress tolerance, antioxidative defense system, transcriptional regulation, signal transduction, and redox signaling events.115,145,146,147
5.1. Protein sulphenylation regulates signaling and primary metabolism
Glyceraldehyde-3- phosphate dehydrogenase (GAPDH) in plants is represented by different isoforms located in chloroplasts and cytoplasm which are also sensitive to H2O2-mediated sulphenylation.88 Plastidial GAPDH is involved in catalyzing calvin cycle reactions and the cytoplasmic isoform regulates glycolysis and gluconeogenesis. Mammalian glyceraldehyde-3phosphate dehydrogenase (GAPDH), PTP1B, or p65subunit of the NF-kB transcription factor exhibit multiple regulation mediated by cysteine.19,20,148 Extensive investigations reveal that cys-oxPTM is capable of inducing signaling crosstalk among various biomolecules like ROS, ABA, jasmonic acid, and MAPK. GLUTATHIONE PEROXIDASE 3 (GPX3) is a major redox sensor protein capable of mediating the oxidation equivalent to AB12 (PP2C family ABA INSENSITIVE 1-negative regulator of ABA signaling) by thiol-disulfide exchange ROS activated guard cells.58,89 Protein kinases (MAPKs) impart precise regulation in various aspects like plant growth, development, and stress signaling.69–72,149 Plants (tomato and Arabidopsis) have been reported to possess conserved mechanism of MAPK-mediated redox signaling.76 MAPKs are essentially activated by the process of sulphenylation which occurs at the redox-sensitive cysteine residues.150 ABA dependent-stomatal closure during abiotic stress conditions is associated with the differential activity of various MAPKs (MAPK9 and MAPK12) which are in turn modulated by sulphenylation. The kinase activity of MAPK12 is positively regulated by ABA and H2O2.90 According to,87 the Arabidopsis MAPK family members (MAPK2, −4, and −7) exhibit H2O2-dependent sulphenylation. Arabidopsis MAPK family proteins – MAPK2, 4, and 7 have also been detected to undergo sulphenylation. Interestingly, the MAPKs are preferably activated by H2O2.76,151 Sulphenylation of MAPKs appears as an early signal response to oxidative stress which presumably acts downstream in the ROS-mediated signaling pathway. Furthermore, dephosphorylation by redox-sensitive phosphatases regulates the activity of MAPKs.152 The Arabidopsis protein tyrosine phosphatase (AtPTP1) undergoes sulphenylation which in turn negatively regulates MAPK activity.79 Another instance of the regulation of stress signaling appears from evidences of sulphenylation in plant-specific SNF1-related protein kinase2 family proteins. This protein phosphorylates various downstream signaling components (respiratory burst oxidase homolog RbohF and TFs) which in turn activate various stress-induced genes.91
Brassica napus ortholog of AtMAPK4 has been reported to undergo H2O2-dependent activity modulation which can be reversed by mutation at cysteine 232. However, according to,153 H2O2-mediated aggregation of the protein did not affect the protein kinase activity. Protein sulphenylation is also likely to regulate the process of protein translation. The stability of SALT OVERLY SENSITIVE 1 (SOS1) transcripts has been reported to be increased by H2O2 treatment. SOS1 encodes for a plasma membrane-bound Na+/H+ antiporter and the mRNA is highly unstable at normal conditions. However, salinity-induced surge in the endogenous H2O2 levels stabilizes the transcripts, thus leading to protein expression.92 According to Ref.93, the redox-regulated RNA-binding proteins are likely to undergo cys-oxPTM which regulates the stability of the SOS1 mRNA. The proteolytic control of PLASTIDIC TYPE I SIGNAL PEPTIDASE 1 (PLSP1) which removes thylakoid transfer signal protein is also regulated by the modifications at Cys166 and Cys286.154 Recent review by Ref.94 summarizes the role of sulphenylation in regulating peroxisomal proteins – (Monodehydroascorbate reductase 1; glutathione-disulfide reductase; Acyl-CoA oxidase 1 and glyoxalase 1) associated with redox signaling and β-oxidation of fatty acids.
5.2. Sulphenylated proteins exhibit precise subcellular compartmentalization and perform redox-balance and proteolytic activity
Peroxidases are one of the various enzymes which exhibit cysteine modifications at multiple sites during its reaction with H2O2. The formation of sulphenic acid residue in the cysteines of Prx is the first catalytic step of oxPTM. Certain isoforms of Prx possess cysteine groups where the sulphenic acid modification is reduced by the glutathione-Grx components.155 Interestingly, overoxidation of peroxidase isoforms may lead to its inactivation followed by H2O2 accumulation and subsequent downstream signaling response.95,96 Certain reports from microbial systems reveal that peroxide-mediated sulphenic acid formation modulates the DNA-binding ability of various transcription factors (OxyR, OhrR, AP1, orCrtJ) in bacteria and fungi.96 DYn-2-based detection of sulfenome in Arabidopsis113 revealed spatial compartmentalization of sulphenylated proteins in various organelles namely plastids, nucleus, mitochondria, endoplamic reticulum, golgi body, plasma membrane, and peroxisome. The details of all subcellular or organellar – sulfenylated proteins analyzed by Dyn-2 chemical probe and YAP1-c genetic probe approaches87,113 are being mentioned in Table 3. Major pool of sulphenylated proteins detected was associated with various pathways of primary metabolism (pentose phosphate pathway, glycolysis, TCA cycle, shikimate, amino acid, and lipid biosynthesis). Furthermore, the other proteins could be functionally categorized into hormonal signal pathway, redox homeostasis, and regulators of transcription and translation. Evidences, therefore, reveal that a similar class of proteins is likely subjected to sulphenylation in cytoplasm and organelles which infers its role in primary metabolism, signaling system, and redox management. The authors also compared sulfenome pool of Arabidopsis and Medicago trunculata by using another chemical probe Bio-DCP1. Some 30 sulphenylated proteins were also identified to exhibit similar mechanisms of redox function and signaling in Medicago trunculata. An important finding appears in the paper reported by Akter et al.113 where they compared the set of sulphenylated proteins separately detected by YAP1C and DYn-2 methods. Alarmingly, except for 16 proteins in common, the other set of sulphenylated proteins was found to be different in the two detection methods. The authors attributed the reaction kinetics of dimedone, and the internal concentration of the protein to be some of the essential factors which contribute to variable results (Figure 3).
Table 3.
Functional categories of various sulfenylated proteins identified from Arabidopsis and Medicago sulfenome
| Functional categorization | Protein | Subcellular location/number of cysteines | Method of detection | References |
|---|---|---|---|---|
| Arabidopsis sulfenome | Functional categorization of major types of sulphenylated proteins identified as components of Arabidopsis and Medicago sulfenome in response to oxidative stress (H2O2]. The sulfenome analysis was performed by Dyn-2 chemical probe and YAP1-c genetic probe approaches [87,113]. | |||
| Type 1- Protein degradation | OVARIAN TUMOR DOMAIN (OTU)-CONTAINING DUB (DEUBIQUITILATING ENZYME) 5 |
Cytosol (3) | DYn-2 chemical probe method and TAIR | 113 |
| Alpha/beta-Hydrolases superfamily protein | Cytosol, chloroplast [1) | DYn-2 chemical probe method and TAIR | 157,158 | |
| Peptidase M1 family protein | Cytosol, chloroplast [7) | DYn-2 chemical probe method and TAIR | 87 | |
| Zn-dependent exopeptidases superfamily protein | Cytosol, chloroplast [11] | DYn-2 chemical probe method and TAIR | 159 | |
| Cytosol aminopeptidase family protein | Cytosol, chloroplast (5] | DYn-2 chemical probe method and TAIR | 80 | |
| UBIQUITIN-ACTIVATING ENZYME 1 | Cytosol, nucleus and plasma membrane (18) | DYn-2 chemical probe method and TAIR | 113 | |
| COP9 SIGNALOSOME 5A | Cytosol, nucleus [2) | DYn-2 chemical probe method and TAIR | 87 | |
| 20S PROTEASOME BETA SUBUNIT G1 and 26S PROTEASOME REGULATORY COMPLEX | Cytosol (1] and chloroplast (8) | DYn-2 chemical probe method and TAIR | 80 | |
| CALRETICULIN 1A, 1B | Endoplasmic reticulum | DYn-2 chemical probe method and TAIR | 12 | |
| Ubiquitin-specific protease 12 (UBP12] and Ubiquitin-specific protease 13 (UBP13) | Cytoplasm [11) | YAP1-c | 87 | |
| Type 2- Primary metabolism | Pyruvate kinase family protein | Cytoplasm, plasma membrane (11] | 80 | |
| CYTOSOLIC-NAD-DEPENDENT MALATE DEHYDROGENASE 2 |
Cytoplasm, plasma membrane [6) | 160 | ||
| PHOSPHOENOLPYRUVATE CARBOXYKINASE | Cytoplasm, nucleus (12] | 113 | ||
| ARABINOSE KINASE | Cytoplasm, plasma membrane (22) | |||
| NADP-DEPENDENT MALATE DEHYDROGENASE |
Cytoplasm, apoplast [9) | 161,162 | ||
| S-FORMYLGLUTATHIONE HYDROLASE | Cytoplasm, apoplast [5] | 159 | ||
| GLYOXALASE I HOMOLOG | Cytoplasm, peroxisome, plastids, plasma membrane (1] | 113 | ||
| ACONITASE 1 | Cytoplasm, mitochondria plastids, plasma membrane, vacuoles (13) | |||
| IAA-CONJUGATE-RESISTANT 4 | Mitochondria [5) | 160 | ||
| ATP SYNTHASE ALPHA/BETA FAMILY | Mitochondria [3] | 12 | ||
| SUCCINYL-COA LIGASE, ALPHA SUBUNIT | Mitochondria, cell wall [8] | 158 | ||
| CYTOCHROME C-1 | Mitochondria, cytosol (2] | 113 | ||
| LONG-CHAIN ACYL-COA SYNTHETASE 4/ AMP-DEPENDENT SYNTHETASE AND LIGASE FAMILY PROTEIN |
Golgi apparatus, nucleus, plasma membrane (13) | |||
| 6-phosphogluconate dehydrogenase family | Plastid, mitochondria (6) | |||
| HEAT SHOCK COGNATE PROTEIN 70–1 | Plastid (4) | |||
| PLASTID ISOFORM TRIOSE PHOSPHATE ISOMERASE, |
Plastid [2) | 159 | ||
| Vacuolar ATP synthase subunit A (VHA-A] | Vacuolar membrane [6) | YAP1-c | 87 | |
| NADP-malic enzyme 2 (NADP-ME2] | Cytoplasm (7) | YAP1-c | ||
| Hormone homeostasis | RECEPTOR FOR ACTIVATED C KINASE 1B | Cytoplasm, ribosome and nucleus (2) | ||
| ACC OXIDASE 2 | Cytoplasm, endoplasmic reticulum | 113 | ||
| Amino acid metabolism | ARABIDOPSIS CYSTEINE SYNTHASE 1 | Plastid (5) | ||
| O-ACETYLSERINE (THIOL) LYASE ISOFORM C | Chloroplast, mitochondria (3) | |||
| PHOSPHOSERINE AMINOTRANSFERASE | Plastid (8) | |||
| ANTHRANILATE SYNTHASE 2 | Plastid (4) | |||
| ASPARTATE AMINOTRANSFERASE | Plastid (6) | |||
| VALINE-TOLERANT 1 | Plastid, cytosol (2) | |||
| SERINE-HYDROXYMETHYLTRANSFERASE 3 | Plastid (7) | |||
| Pyridoxine biosynthesis 1.3 (PDX1.3) | Plastid [7) | YAP1-c | 87 | |
| Redox-activity | STROMAL ASCORBATE PEROXIDASE | Plastid [4] | 163 | |
| MONODEHYDROASCORBATE REDUCTASE 6 | Plastid (5] | 113 | ||
| DIHYDROLIPOYL DEHYDROGENASES | Plastid [9) |
87 113 113 113 113 |
||
| GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE A SUBUNIT |
Plastid (5) | |||
| ELECTRON TRANSFER FLAVOPROTEIN BETA PHOSPHATE TRANSPORTER 3 (MPT3) |
Mitochondria (3) | |||
| THIOREDOXIN-DEPENDENT PEROXIDASE 1 | Cytosol, chloroplast, plasmamembrane | |||
| ATNRX1, NRX1, NUCLEOREDOXIN 1/DC1 domain-containing protein |
Cytosol (12) | |||
| AMINO ACID DEHYDROGENASE FAMILY | Cytosol (6) | |||
| Protein folding and transport | IMPORTIN ALPHA ISOFORM 1 | Cytosol, nuclear envelope, nulcleus (11) | ||
| HEAT SHOCK PROTEIN 81–3 | Cytosol, golgi, plasma membrane (5) | |||
| HEAT SHOCK COGNATE PROTEIN 70–1 | Cytosol, golgi, plasma membrane (7) | |||
| TCP-1/cpn60 chaperonin family protein | Cytosol, golgi, plasma membrane (9) | |||
| CHAPERONIN-60BETA3 | Cytoplasm, nucleus (6) | |||
| ERBB-3 BINDING PROTEIN 1 | Nucleus, plasma membrane (6) | |||
| LUMINAL BINDING PROTEIN | Endoplasmic reticulum (5) | |||
| γ-Glutamyl peptidase 3 (GGP3) | Cytoplasm (8) | YAP1-c | ||
| RNA-binding translation | ATP binding*leucine-tRNA ligases*aminoacyltRNA ligases*nucleotide binding*ATP |
Cytosol (20) | ||
| EUKARYOTIC TRANSLATION INITIATION FACTOR 3B-2 |
Cytosol, nucleus (3) | |||
| Nucleic acid-binding, OB-fold-like protein | Cytosol, plasma membrane (4) | |||
| CYCLOPHILIN 40 | Cytosol (7) | |||
| TRANSLATION INITIATION FACTOR 3 SUBUNIT H1 | Cytosol (7) | |||
| EUKARYOTIC TRANSLATION INITIATION FACTOR ISOFORM 4G1 |
Cytosol, nucleus (7) | |||
| Signal transduction | PEROXISOMAL 3-KETOACYL-COA THIOLASE 3 | Peroxysome (9) | ||
| CO-FACTOR FOR NITRATE REDUCTASE AND XANTHINE DEHYDROGENASE | Cytoplasm (9) | |||
| PHOSPHOLIPASE D ALPHA 1 | Cytoplasm (8) | |||
| INOSITOL 1,3,4-TRISPHOSPHATE 5/6-KINASE 4 (ITPK4) |
Cytosol, nucleus [9) | 87 | ||
| Map kinase 2, 4 and 7 (MAPK] | Cytosol [8) | YAP1-C | 87 | |
Figure 3.
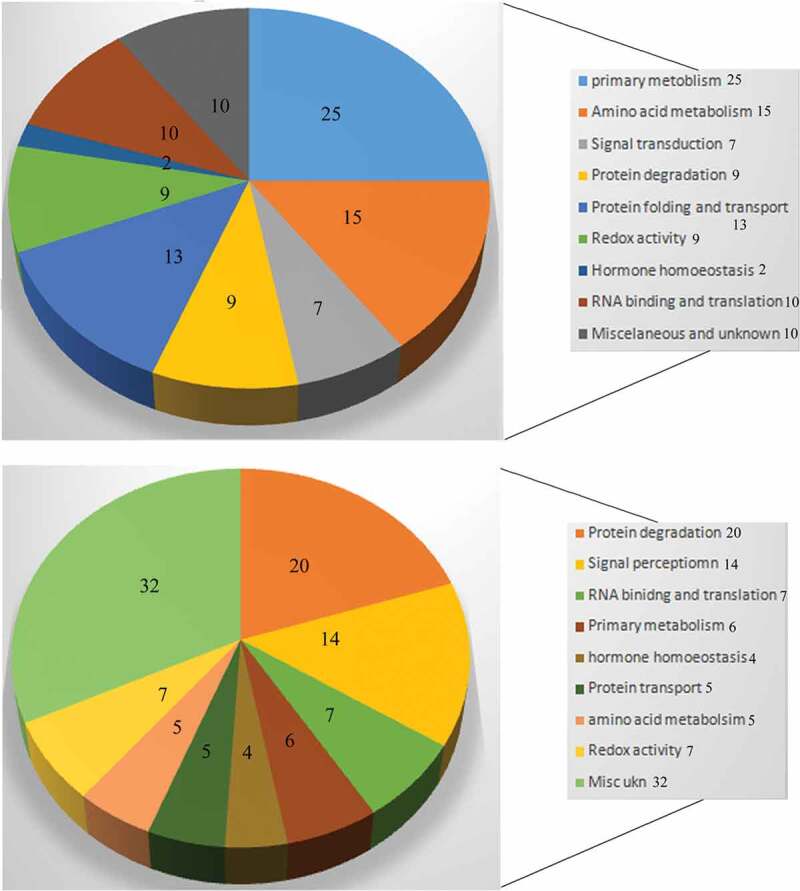
Comparative account of the functional categories of Arabidopsis sulfenome detected by Dyn-2 probe-based method113 (a) and YAP-1 genetic probe method87 (b). Numbers represent percentage representation of each class of sulfenylated proteins
The authors87 precisely report that 20% of the oxidative-stress–induced sulfenome in Arabidopsis has been associated with proteolytic function. Five proteins (UBIQUITINCONJUGATING ENZYME27 (UBC27), two subunits of the Skp/Cullin/F-box (SCF) E3-ubiquitin ligase complex (ASK1 and ASK2), 3 and 5A subunits of the constitute photomorphogenic9 (COP9) signalosome has been identified to be associated with ubiquitination. Among other proteins REGULATORY PARTICLE NON-ATPASE 12A (cytoplasmic thioredoxin h (Trxh) (RPN12A) is an important component of 26S proteasome which is also associated with the cytokinin signal pathway.97,98 Present findings provide insights to the fact that the sulphenylation of proteolytic components facilitates the degradation of oxidatively damaged proteins and imparts physiological stress tolerance (Figure 4). Similar to Arabidopsis, a similar mechanism of oxidative modification of 20S proteolytic proteins has been reported in response to sugar starvation in Maize.156
Figure 4.
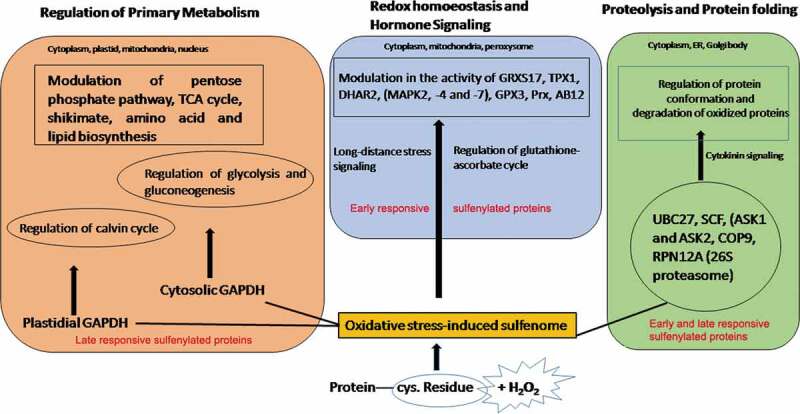
Signaling role of sulfenylated proteins in plants
The redox sulphenylated proteins detected among the components of sulfenome namely included MONOTHIOL GLUTAREDOXIN17 (GRXS17), THIOREDOXIN-DEPENDENT PEROXIDASE1 (TPX1), GLUTAREDOXIN C2 (GRXC2), and DHAR2. Certain enzymes (glutathione peroxidase, peroxiredoxins, and methionine sulfoxide reductases) with specialized redox mechanisms were not detected by the YAP1C method possibly because of the transient form of sulphenic acid formed by the cysteine residues. Certain proteins like TPX1 and DHAR exist among early and late responsive proteins. Investigations with dimedon-tagged sulphenylation in DHAR in presence of H2O2 revealed a remarkably lower rate of sulphenylation in comparison with the control [without H2O2). The authors discussed the findings to be resulting due to the fast conversion of sulphenic residue to sulfininc or sulfonic forms, respectively. Thus, the function of DHAR in relation to oxidative-stress-induced sulphenylation has been associated with important reactions of the glutathione-ascorbate cycle. Thus, sulphenylated DHAR appears to be an essential enzyme necessary to maintain an ascorbate pool in cells subjected to oxidative stress. Recent report by,142 involves the use of the YAP1-C approach for in-vivo detection of plastidial sufenome in response to light and H2O2-mediated oxidative stress. A major pool of all the sufenylated proteins (132) detected in plastids was associated with amino acid metabolism.
6. Homology of sulphenylatyed proteins in humans and plants: a conserved set of sulfenome
The comprehensive data set of sulphenylated proteins obtained from an organism is referred to as sulfenome (Table 3). Recent advancements in the methodological approaches have been successful to decipher the pool of sulphenylated proteins in human and plant system, preferably Arabidopsis. Proteomic approach mediated by mass spectrometric analysis has been beneficial in identifying proteins with cysteine sulphenic acid residues. Proteomic analysis reveals a better characterization of sulphenylated proteins in mammalian systems. Various reports provide validation of cysteine sulphenylation in mammals and plants. Although mammalian systems involve cysteine sulphenylation to be associated with a wide range of proteins namely oxidoreductase, dehydrogenase, kinase, phosphatase, transferase, and adaptor proteins, plants systems have been reported to involve mostly oxidoreductase and dehydrogenase protein family. Certain plant-based orthologs of animal proteins exhibit a similar mechanism of sulphenylation. Functional conservation of sulphenylation and its regulatory effect have been observed for the methionine sulfoxide reductase (MSR) family of proteins.164–166 The MSRB1 and MSRB2 isoforms from Arabidopsis and MSRA from human and mouse have been reported to undergo sulphenylation. Interestingly, the proteins are structurally unlike yet share some similarities in their catalytic nature and thus undergo nucleophilic substitution in the cysteine residue. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is another protein that possesses conserved sites for cysteine-sulphenylation in plants and humans.167,168
Functional categorization of sulphenylated proteins in mammals and plants reveal two mechanistic categories namely an intermediate to overoxidation or disulfide formation and as a direct regulator of protein function. The intermediate forms of cysteine-SOH are transient and they reside in the catalytic site of the proteins. MSR, GAPDH, dehydroascorbate reductase 2 (DHAR2; Arabidopsis), and peroxiredoxin (Prx) are such proteins with evolutionary conserved sulphenic acid residues.164–168 Alternatively, -SOH group not being associated with the catalytic activity of the protein has been suggested to mediate disulfide formation to prevent overoxidation. Mammalian reductase family proteins (glutathione reductase (GR), aldose reductase (AR) exhibit such mechanism oLDf sulphenic acid-mediated regulation of protein activity. Crystal form of GR exhibit a stable – SOH present in the cysteine 63 which inhibits the activity of the enzyme.169 Conversely the stable form of – SOH present in Cys298 and Cys303 of AR has been reported to activate its function.170,171 Sulphenylation-induced activation of kinase activity has been evident in mammalian epidermal growth factor receptor (EGFR).136 Recent interesting report by Ref.172 has evaluated the homology of sulfenome landscapes in human and plant systems. Using uniform annotation with current Uni-ProtKB accessions a total of 1684 sulphenylated proteins was compared from humans, Arabidopsis and Medicago. Around 29% and 5.8% of the total sulphenylatyed proteins were reported in multiple studies in humans and Arabidopsis, respectively. A comprehensive account of the comparative landscape of mammalian and plant sulfenome was obtained by reciprocal BLAST analysis. Proteins with 60% query coverage and 40% amino acid identity were considered to be homologous in nature. The authors reported that homologous identity exists among sulphenylated proteins reported from humans (31%], Arabidopsis and Medicago (42%) (Figure 5). GAPDH, cMDH1, and dihydrolipyl dehydrogenase (DLDH) are the evolutionary conserved sulphenylated proteins that have been characterized. Certain enzymes exhibiting sulphenylation in humans and Arabidopsis exhibit evolutionary conserved nature in terms of their biological activity. They are mostly associated with the primary metabolic pathways such as purine biosynthesis, pyruvate metabolism, and glycolysis pathway. Sulphenylation can essentially regulate the process of glycolysis and pentose phosphate pathway.173, 174, 175, 176 Thus, our current understanding of the mechanism of protein sulphenylation and sulfenome appears to possess homology across diverse groups of biological systems.
Figure 5.
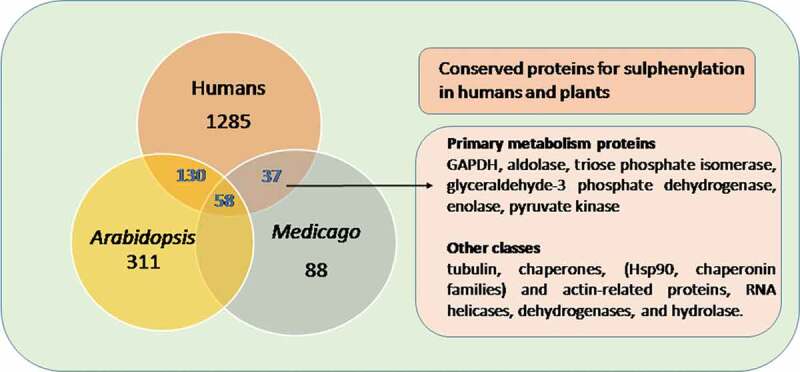
Conserved homologous protein targets for sulfenylation in humans, Arabidopsis and Medicago. Overlapping numbers represent the conserved pool of sulfenylated proteins among the three organisms
7. Deciphering the signaling mechanisms of cys oxPTM-mediated molecular crosstalk and bioengineering of plants to endure stress: future perspectives
The increasing number of investigations in the last decade has led to the identification of a large number of proteins that undergo cys oxPTM. Plant endurance to biotic and abiotic stress is largely regulated by the activity of antioxidant enzymes, regulation of primary metabolism, and modulation in the activity of receptor kinases. In this context, a plethora of effects are mediated by crosstalk among phytohormones, Ca2+ and ROS. Thus, cys oxPTMs of proteins modulate physiological pathways thus imparting stress tolerance to plants.
To date, our current knowledge on the physiological significance of cysteine oxPTM and protein sulphenylation provides insights into its role in stress signaling, metabolism, and redox homeostasis. Recent advancements have identified novel proteins associated with cys-mediated oxPTM among which sulphenylation possesses immense functional significance. Oxidative stress-induced sulfenome identification substantiates the fact that sulphenylated proteins are important components in the ROS-generated signal cascade. However, future investigations are necessary to perform validation and functional characterization of the sulphenylated proteins. Comparative proteomic analysis of sulfenome in some plant systems explains that differences exist among the sulphenylted proteins identified by different techniques. Thus, it is necessary to collate data from various methods with variable sensitivity. Modification of non-native proteins during sample preparation in lysates imposes hurdles of false-positive results. Plant sulfenome analyzed through in vitro stress-induced responses should also be essentially validated by in vivo experimental analysis. Application of a UV-cleavable dimedone probe is expected to provide better results in sulfenome identification in the future. Unlike mammalian systems, investigations on plant sulfenome require further advancements. The evolutionary conserved set of sulfenomes detected in humans and plants shall facilitate the functional interpretation of various sulphenylated proteins in plants. It is essential to overcome the current technical limitations in methodologies adapted to detect sulphenylated proteins in vivo. The kinetics of chemical probes implied in various investigations appears to be an important factor for sulfenome detection in plants. Thus, future investigations with novel modified protocols are necessary for detecting transient forms of sulphenic acid residues in plants. Long-distance abiotic-stress signaling, photobiomodulation and hormonal crosstalk during plant development are some of the potential areas to be investigated in the context of protein sulfenylation. Individual protein targets should be considered for more detailed analysis through functional characterization and case to case validation. Insights to S-nitrosylation mediated negative regulation of NO signaling in plants is likely to be addressed in the upcoming years.
Acknowledgments
The author (SM) is thankful to Ms. Piyali Mukherjee for her sincere assistance in the preparation of schemes and figures.
References
- 1.Del Río A. ROS and RNS in plant physiology: an overview. J Exp Bot. 2015;66:1–19. [DOI] [PubMed] [Google Scholar]
- 2.Zaffagnini M, DeMia M, Morisse S, Di Giacinto N, Marchand CH, Maes A, Trost P. Protein S-nitrosylation in photosynthetic organisms: A comprehensive overview with future perspectives. Biochim Biophys Acta. 2016a;1864:952–966. [DOI] [PubMed] [Google Scholar]
- 3.Apostol I, Heinstein PF, Low PS. Rapid stimulation of an oxidative burst during elicitation of cultured plant cells. Role in defense and signal transduction. Plant Physiol. 1989;90(1):109–116. doi: 10.1104/pp.90.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duan Q, Kita D, Johnson EA, Aggarwal M, Gates L, Wu H-M, Cheung AY. Reactive oxygen species mediate pollen tube rupture to release sperm for fertilization in Arabidopsis. Nat Commun. 2014;5:3129. [DOI] [PubMed] [Google Scholar]
- 5.Foreman J, Demidchik V, Bothwell JHF, Mylona P, Miedema H, Torres MA, Linstead P, Costa S, Brownlee C, Jones JDG, et al. Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature. 2003;422:442–446. [DOI] [PubMed] [Google Scholar]
- 6.Fryer MJ, Ball L, Oxborough K, Karpinski S, Mullineaux PM, Baker NR. Control of Ascorbate Peroxidase 2 expression by hydrogen peroxide and leaf water status during excess light stress reveals a functional organisation of Arabidopsis leaves. Plant J. 2003;33:691–705. [DOI] [PubMed] [Google Scholar]
- 7.Levine A, Tenhaken R, Dixon R, Lamb C. H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell. 1994;79:583–593. [DOI] [PubMed] [Google Scholar]
- 8.Wise RR, Naylor AW. Chilling-enhanced photooxidation: evidence for the role of singlet oxygen and superoxide in the breakdown of pigments and endogenous antioxidants. Plant Physiol. 1987;83:278–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roos G, Foloppe N, Messens J. Understanding the pK(a) of redox cysteines: the key role of hydrogen bonding. Antioxidant Redox Signal. 2013;18:94–127. [DOI] [PubMed] [Google Scholar]
- 10.Zaffagnini M, Fermani S, Calvaresi M, Orrù R, Iommarini L, Sparla F, Trost P. Tuning Cysteine Reactivity and SulfenicAcid Stability by Protein Microenvironment in Glyceraldehyde-3-Phosphate Dehydrogenases of Arabidopsis thaliana. Antioxidant Redox Signal. 2016b;24:502–517. [DOI] [PubMed] [Google Scholar]
- 11.Giles G, Nasim M, Ali W, Jacob C. The reactive sulfur species concept: 15 years on. Antioxidants. 2017;6:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oger E, Marino D, Guigonis JM, Pauly N, Puppo A. Sulfenylated proteins in the Medicago truncatula–Sinorhizobium melilotis symbiosis. J Proteomics. 2012;75:4102–4113. [DOI] [PubMed] [Google Scholar]
- 13.Rey P, Bécuwe N, Barrault M-B, Rumeau D, Havaux M, Biteau B, Toledano MB. The Arabidopsis thaliana sulfiredoxin is a plastidic cysteine-sulfinic acid reductase involved in the photooxidative stress response. Plant J. 2007;49:505–514. [DOI] [PubMed] [Google Scholar]
- 14.Montrichard F, Alkhalfioui F, Yano H, Vensel WH, Hurkman WJ, Buchanan BB. Thioredoxin targets in plants: the first 30 years. J Proteomics. 2009;72:452–474. [DOI] [PubMed] [Google Scholar]
- 15.Schürmann P, Buchanan BB. The ferredoxin/thioredoxin system of oxygenic photosynthesis. Antioxidant Redox Signal. 2008;10:1235–1273. [DOI] [PubMed] [Google Scholar]
- 16.Jacques S, Ghesquière B, Van Breusegem F, Gevaert K. Plant proteins under oxidative attack. Proteomics. 2013;13:932–940. [DOI] [PubMed] [Google Scholar]
- 17.Roos G, Messens J. Protein sulfenic acid formation: from cellular damage to redox regulation. Free Radic Biol Med. 2011;51:314–326. [DOI] [PubMed] [Google Scholar]
- 18.Álvarez C, García I, Moreno I, Pérez-Pérez ME, Crespo JL, Romero LC, Gotor C. Cysteine-generated sulfide in the cytosol negatively regulates autophagy and modulates the transcriptional profile in Arabidopsis. Plant Cell. 2012c;24:4621–4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mustafa AK, Gadalla MM, Sen N, Kim S, Mu W, Gazi SK, Barrow RK, Yang G, Wang R, Snyder SH. H2S signals through protein S-sulfhydration. Sci Signal. 2009;2:ra72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paul BD, Snyder SH. H2S signaling through protein sulfhydration and beyond. Nat Rev Mol Cell Biol. 2012;13:499–507. [DOI] [PubMed] [Google Scholar]
- 21.Yu M, Lamattina L, Spoel SH, Loake GJ. Nitric oxide function in plant biology: a redox cue in deconvolution. New Phytol. 2014;202:1142–1156. [DOI] [PubMed] [Google Scholar]
- 22.Glaring MA, Skryhan K, Kötting O, Zeeman SC, Blennow A. Comprehensive survey of redox sensitive starch metabolising enzymes inArabidopsis thaliana. Plant Physiol Biochem. 2012;58:89–97. [DOI] [PubMed] [Google Scholar]
- 23.Kopriva S, Mugford SG, Baraniecka P, Lee BR, Matthewman CA, Koprivova A. Control of sulfur partitioning between primary and secondary metabolism in Arabidopsis. Front Plant Sci. 2012;3:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Considine MJ, Foyer CH. Redox regulation of plant development. Antioxidant Redox Signal. 2014;21:1305–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dietz KJ. Redox regulation of transcription factors in plant stress acclimation and development. Antioxidant Redox Signal. 2014;21:1356–1372. [DOI] [PubMed] [Google Scholar]
- 26.Schmidt R, Schippers JHM. ROS-mediated redox signalling during cell differentiation in plants. Biochim Biophys Acta—Gen Subjects. 2014;1850:1497–1508. [DOI] [PubMed] [Google Scholar]
- 27.Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annual Rev Plant Biol. 2004;55:373–399. [DOI] [PubMed] [Google Scholar]
- 28.Del Río LA. Redox pioneer: professor Christine Helen Foyer. Antioxidant Redox Signal. 2011;15:2383–2391. [DOI] [PubMed] [Google Scholar]
- 29.Hernández I, Alegre L, Van Breusegem F, Munné-Bosch S. How relevant are flavonoids as antioxidants in plants?. Trend Plant Sci. 2009;14:125–132. [DOI] [PubMed] [Google Scholar]
- 30.Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, Vandepoele K, Gollery M, Shulaev V, Van Breusegem F. ROS signaling: the new wave?. Trends Plant Sci. 2011;16:300–309. [DOI] [PubMed] [Google Scholar]
- 31.Waszczak C, Akter S, Jacques S, Huang J, Messens J, Van Breusegem F. Oxidative post-translational modifications of cysteine residues in plant signal transduction. J Exp Bot. 2015;66:2923–2934. [DOI] [PubMed] [Google Scholar]
- 32.Harris TK, Turner GJ. Structural basis of perturbed pKa values of catalytic groups in enzyme active sites. Intern Union Biochem Mol Biol Life. 2002;53:85–98. [DOI] [PubMed] [Google Scholar]
- 33.Marino SM, Gladyshev VN. Cysteine function governs its conservation and degeneration and restricts its utilization on protein surfaces. J Mol Biol. 2010;404:902–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iglesias-Baena I, Barranco-Medina S, Lázaro-Payo A, López- Jaramillo FJ, Sevilla F, Lázaro JJ. Characterization of plant sulfiredoxin and role of sulphinic form of 2-Cys peroxiredoxin. J Exp Bot. 2010;61:1509–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zaffagnini M, Bedhomme M, Marchand CH, Morisse S, Trost P, Lemaire SD. Redox regulation in photosynthetic organisms: focus on glutathionylation. Antioxidant Redox Signal. 2012;16:567–586. [DOI] [PubMed] [Google Scholar]
- 36.Romero LC, MÁ A, Serna A, Gotor C. Proteomic analysis of endogenous S-sulfhydration in Arabidopsis thaliana. Nitric Oxide. 2013;31:S23. [Google Scholar]
- 37.Aroca A, Schneider M, Scheibe R, Gotor C, Romero LC. Hydrogen sulfide regulates the cytosolic/nuclear partitioning of glyceraldehyde-3-phosphate dehydrogenase by enhancing its nuclear localization. Plant Cell Physiol. 2017;58(6):983–992. doi: 10.1093/pcp/pcx056. [DOI] [PubMed] [Google Scholar]
- 38.Aroca Á, Serna A, Gotor C, Romero LC. S-sulfhydration: a cysteine posttranslational modification in plant systems. Plant Physiol. 2015;168:334–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou H, Zhang J, Shen J, Zhou M, Yuan X, Xie Y. Redox-based protein persulfidation in guard cell ABA signaling. Plant Signal Behav. 2020:1741987. doi: 10.1080/15592324.2020.1741987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feng J, Chen L, Zuo J. Protein S-nitrosylation in plants: current progresses and challenges. J Integr Plant Biol. 2019;12:1206–1223. [DOI] [PubMed] [Google Scholar]
- 41.Stomberski CT, Hess DT, Stamler JS. Protein S-nitrosylation: determinants of specificity and enzymatic regulation of S-nitrosothiol-based signaling. Antioxid Redox Signal. 2019;30:1331–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y, Yun BW, Kwon E, Hong JK, Yoon J, Loake GJ. Snitrosylation: an emerging redox-based post-translational modification in plants. J Exp Bot. 2006;57:1777–1784. [DOI] [PubMed] [Google Scholar]
- 43.Chen YJ, Lu CT, Su MG, Huang KY, Ching WC, Yang HH, Liao YC, Chen YJ, Lee TY. dbSNO 2.0: A resource for exploring structural environment, functional and disease association and regulatory network of protein S-nitrosylation. Nucleic Acids Res. 2015;43:D503–D511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu J, Huang X, Chen L, Sun X, Lu C, Zhang L, Wang Y, Zuo J. Site-specific nitrosoproteomic identification of endogenously S-nitrosylated proteins in Arabidopsis. Plant Physiol. 2015;167:1731–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu J, Yang H, Mu J, Lu T, Peng J, Deng X, Kong Z, Bao S, Cao X, Zuo J. Nitric oxide regulates protein methylation during stress responses in plants. Mol Cell. 2018;67:702–710 e704. [DOI] [PubMed] [Google Scholar]
- 46.Puyaubert J, Fares A, Reze N, Peltier J-B BE. Identification of endogenously S-nitrosylated proteins in Arabidopsis plantlets: effect of cold stress on cysteine nitrosylation level. Plant Sci. 2014;215–216:150–156. [DOI] [PubMed] [Google Scholar]
- 47.Vaseghi MJ, Chibani K, Telman W, Liebthal MF, Gerken M, Mueller SM, Dietz KJ. The chloroplast 2-cysteine peroxiredoxin functions as thioredoxin oxidase in redox regulation of chloroplast metabolism. BioRxiv. 2018;7:e38194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nietzel T, Mostertz J, Ruberti C, Née G, Fuchs P, Wagner S, Moseler A, Müller-Schüssele SJ, Benamar A, Poschet G, et al. Redox-mediated kick-start of mitochondrial energy metabolism drives resource-efficient seed germination. Proc Nat Acad Sci. 2020;117:741–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.White M, Kamps J, Samuel K, Flashman E. The Plant Cysteine Oxidases from Arabidopsis thaliana are kinetically tailored to act as oxygen sensors. J Biol Chem. 2018;293:11786–11795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Foyer CH, Baker A, Wright M, Sparkes IA, Mhamdi A, Schippers JHM, Breusegem FV. On the move: redox-dependent protein relocation in plants. J Exp Bot. 2020;71:620–631. [DOI] [PubMed] [Google Scholar]
- 51.Albertos P, Romero-Puertas MC, Tatematsu K, Mateos I, Sanchez-Vicente I, Nambara E, Lorenzo O. S-nitrosylation triggers ABI5 degradation to promote seed germination and seedling growth. Nat Commun. 2015;6:8669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kawabe H, Ohtani M, Kurata T, Sakamoto T, Demura T. Protein S-nitrosylation regulates xylem vessel cell differentiation in Arabidopsis. Plant Cell Physiol. 2018;59:17–29. [DOI] [PubMed] [Google Scholar]
- 53.Iglesias MJ, Terrile MC, Correa-Aragunde N, Colman SL, Izquierdo-Alvarez A, Fiol DF, Paris R, Sanchez-Lopez N, Marina A, Calderon Villalobos LIA, et al. Regulation of SCF(TIR1/AFBs) E3 ligase assembly by S-nitrosylation of Arabidopsis SKP1-like1 impacts on auxin signaling. Redox Biol. 2018;18:200–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Romero-Puertas MC, Laxa M, Matte A, Zaninotto F, Finkemeier I, Jones AM, Perazzolli M, Vandelle E, Dietz KJ, Delledonne M. S-nitrosylation of peroxiredoxin II E promotes peroxynitrite-mediated tyrosine nitration. Plant Cell. 2007;19:4120–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yun BW, Feechan A, Yin M, Saidi NB, Le Bihan T, Yu M, Moore JW, Kang JG, Kwon E, Spoel SH, et al. S-nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature. 2011;478:264–268. [DOI] [PubMed] [Google Scholar]
- 56.Berger H, De Mia M, Morisse S, Marchand CH, Lemaire SD, Wobbe L, Kruse O. A light switch based on protein Snitrosylation fine-tunes photosynthetic light harvesting in Chlamydomonas. Plant Physiol. 2016;171:821–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signaling network. Annual Rev Plant Biol. 2010;61:651–679. [DOI] [PubMed] [Google Scholar]
- 58.Miao Y, Lv D, Wang P, Wang X-C, Chen J, Miao C, Song CP. An Arabidopsis glutathione peroxidase functions as both a redox transducer and a scavenger in abscisic acid and drought stress responses. Plant Cell. 2006;18:2749–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gutscher M, Sobotta MC, Wabnitz GH, Ballikaya S, Meyer AJ, Samstag Y, Dick TP. Proximity-based protein thiol oxidation by H2O2-scavenging peroxidases. J Biol Chem. 2009;284:31532–31540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Delaunay A, Pflieger D, Barrault MB, Vinh J, Toledano MB. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell. 2002;111:471–481. [DOI] [PubMed] [Google Scholar]
- 61.Konopka-Postupolska D, Clark G, Goch G, Debski J, Floras K, Cantero A, Fijolek B, Roux S, Hennig J. The role of annexin 1 in drought stress in Arabidopsis. Plant Physiol. 2009;150:1394–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mou Z, Fan W, Dong X. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell. 2003;113:935–944. [DOI] [PubMed] [Google Scholar]
- 63.Tada Y, Spoel SH, Pajerowska-Mukhtar K, Mou Z, Song J, Wang C, Zuo J, Dong X. Plant immunity requires conformational charges of NPR1 via S-nitrosylation and thioredoxins. Science. 2008;321:952–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Després C, Chubak C, Rochon A, Clark R, Bethune T, Desveaux D, Fobert PR. The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell. 2003;15:2181–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Feechan A, Kwon E, Yun BW, Wang Y, Pallas JA, Loake GJ. A central role for S-nitrosothiols in plant disease resistance. Proc Natl Acad Sci USA. 2005;102:8054–8059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Taki N, Sasaki-Sekimoto Y, Obayashi T, Kikuta A, Kobayashi K, Ainai T, Yagi K, Sakurai N, Suzuki H, Masuda T, et al. 12-oxophytodienoic acid triggers expression of a distinct set of genes and plays a role in wound-induced gene expression in Arabidopsis. Plant Physiol. 2005;139:1268–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Park SW, Li W, Viehhauser A, He B, Kim S, Nilsson AK, Andersson MX, Kittle JD, Ambavaram MMR, Luan S, et al. Cyclophilin 20-3 relays a 12-oxo-phytodienoic acid signal during stress responsive regulation of cellular redox homeostasis. Proc Nat Acad Sci USA. 2013;110:9559–9564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Motohashi K, Koyama F, Nakanishi Y, Ueoka-Nakanishi H, Hisabori T. Chloroplast cyclophilin is a target protein of thioredoxin—thiol modulation of the peptidyl-prolyl cis-trans isomerase activity. J Biol Chem. 2003;278:31848–31852. [DOI] [PubMed] [Google Scholar]
- 69.Beckers GJM, Jaskiewicz M, Liu Y, Underwood WR, He SY, Zhang S, Conrath U. Mitogen-activated protein kinases 3 and 6 are required for full priming of stress responses in Arabidopsis thaliana. Plant Cell. 2009;21:944–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kosetsu K, Matsunaga S, Nakagami H, Colcombet J, Sasabe M, Soyano T, Takahashi Y, Hirt H, Machida Y. The MAP kinase MPK4 is required for cytokinesis in Arabidopsis thaliana. Plant Cell. 2010;22:3778–3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pitzschke A, Djamei A, Bitton F, Hirt H. A major role of the MEKK1–MKK1/2–MPK4 pathway in ROS signalling. Mol Plant. 2009;2:120–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Takahashi F, Yoshida R, Ichimura K, Mizoguchi T, Seo S, Yonezawa M, Maruyama K, Yamaguchi-Shinozaki K, Shinozaki K. The mitogen-activated protein kinase cascade MKK3–MPK6 is an important part of the jasmonate signal transduction pathway in Arabidopsis. Plant Cell. 2007;19:805–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cross JV, Templeton DJ. Oxidative stress inhibits MEKK1 by site specific glutathionylation in the ATP-binding domain. Biochem J. 2004;381:675–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Day AM, Veal EA. Hydrogen peroxide-sensitive cysteines in the Sty1 MAPK regulate the transcriptional response to oxidative stress. J Biol Chem. 2010;285:7505–7516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Templeton DJ, Aye M-S, Rady J, Xu F, Cross JV. Purification of reversibly oxidized proteins (PROP) reveals a redox switch controlling p38 MAP kinase activity. PLoS ONE. 2010;5:e15012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou J, Xia XJ, Zhou Y-H, Shi K, Chen Z, Yu JQ. RBOH1- dependent H2O2 production and subsequent activation of MPK1/2 play an important role in acclimation-induced cross-tolerance in tomato. J Exp Bot. 2014;65:595–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xie G, Kato H, Sasaki K, Imai R. A cold-induced thioredoxin h of rice, OsTrx23, negatively regulates kinase activities of OsMPK3 and OsMPK6 in vitro. FEBS Lett. 2009;583:2734–2738. [DOI] [PubMed] [Google Scholar]
- 78.Nakagami H, Kiegerl S, Hirt H. OMTK1, a novel MAPKKK, channels oxidative stress signaling through direct MAPK interaction. J Biol Chem. 2004;279:26959–26966. [DOI] [PubMed] [Google Scholar]
- 79.Gupta R, Luan S. Redox control of protein tyrosine phosphatases and mitogen activated protein kinases in plants. Plant Physiol. 2003;132:1149–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dixon DP, Skipsey M, Grundy NM, Edwards R. . Stress induced protein S-glutathionylation in Arabidopsis. Plant Physiol. 2005; 138:2233–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lam YW, Yuan Y, Isaac J, Babu CV, Meller J, Ho SM. Comprehensive identification and modified-site mapping of S-nitrosylated targets in prostate epithelial cells. PLoS ONE. 2010;5:e9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yemets AI, Krasylenko YA, Lytvyn DI, Sheremet YA, Blume YB. Nitric oxide signalling via cytoskeleton in plants. Plant Sci. 2011;181:545–554. [DOI] [PubMed] [Google Scholar]
- 83.Li JS, Chen SS, Wang XF, Shi C, Liu HX, Yang J, Shi W, Guo JK, Jia HL. Hydrogen sulfide disturbs actin polymerization via S-sulfhydration resulting in stunted root hair growth. Plant Physiol. 2018;178:936–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zaffagnini M, Michelet L, Marchand C, Sparla F, Decottignies P, Le Maréchal P, Miginiac-Maslow M, Noctor G, Trost P, Lemaire SD. The thioredoxin-independent isoform of chloroplastic glyceraldehyde-3-phosphate dehydrogenase is selectively regulated by glutathionylation. FEBS Lett. 2007;274:212–226. [DOI] [PubMed] [Google Scholar]
- 85.Dixon DP, Davis BG, Edwards R. Functional divergence in the glutathione transferase superfamily in plants identification of two classes with putative functions in redox homeostasis in Arabidopsis thaliana. J Biol Chem. 2002;277:30859–30869. [DOI] [PubMed] [Google Scholar]
- 86.Bender KW, Wang X, Cheng GB, Kim HS, Zielinski RE, Huber SC . Glutaredoxin AtGRXC2 catalyses inhibitory glutathionylation of Arabidopsis BRI1-associated receptor-like kinase 1 (BAK1) in vitro. Biochem J. 2015. May 1;467(3):399-413 [DOI] [PubMed] [Google Scholar]
- 87.Moffett AS, Bender KW, Huber SC, Shukla, D.. Allosteric Control of a Plant Receptor Kinase through S-Glutathionylation. Biophys J. 2017a;113:2354–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Moffett AS, Bender KW, Huber SC, Shukla D.. Molecular dynamics simulations reveal the conformational dynamics of arabidopsis thaliana bri1 and bak1 receptor-like kinases. J Biol Chem. 2017b;292:12643–12652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Waszczak C, Akter S, Eeckhout D, Persiau G, Wahni K, Bodra N, Molle IV, Smet B, Vertommen D, Gevaert K, et al. Sulfenome mining in Arabidopsis thaliana. Proc Nat Acad Sci USA. 2014;111:11545–11550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Trost P, Fermani S, Calvaresi M, Zaffagnini M. Biochemical basis of sulphenomics: how protein sulphenic acids may be stabilized by the protein microenvironment. Plant Cell Environ. 2017;40:483–490. [DOI] [PubMed] [Google Scholar]
- 91.Merlot S, Gosti F, Guerrier D, Vavasseur A, Giraudat J. The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 2001;25:295–303. [DOI] [PubMed] [Google Scholar]
- 92.Jammes F, Song C, Shin D, Munemasa S, Takeda K, Gu D, Cho D, Lee S, Giordo R, Sritubtim S, et al. MAP kinases MPK9 and MPK12 are preferentially expressed in guard cells and positively regulate ROS-mediated ABA signaling. Proceed Nat Acad Sci USA. 2009;106:20520–20525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kulik A, Wawer I, Krzywinska E, Bucholc M, Dobrowolska G. SnRK2 protein kinases—Key regulators of plant response to abiotic stresses. OMICS. 2011;15:859–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chung JS, Zhu JK, Bressan RA, Hasegawa PM, Shi H. Reactive oxygen species mediate Na+-induced SOS1 mRNA stability in Arabidopsis. Plant J. 2008;53:554–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lorković ZJ. Role of plant RNA-binding proteins in development stress response and genome organization. Trend Plant Sci. 2009;14:229–236. [DOI] [PubMed] [Google Scholar]
- 96.Sandalio LM, Gotor C, Romero LC, Romero-Puertas MC. Multilevel Regulation of Peroxisomal Proteome by Post-Translational Modifications. Int J Mol Sci. 2019;20:4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wood ZA, Poole LB, Karplus PA. Peroxiredoxinevolution and theregulation of hydrogen peroxide signaling. Science. 2003;300:650–653. [DOI] [PubMed] [Google Scholar]
- 98.D’Autreaux B, Toledano MB. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat Rev Mol Cell Biol. 2007;8:813–824. [DOI] [PubMed] [Google Scholar]
- 99.Yamazaki D, Motohashi K, Kasama T, Hara Y, Hisabori T. Target proteins of the cytosolic thioredoxins in Arabidopsis thaliana. Plant Cell Physiol. 2004;45:18–27. [DOI] [PubMed] [Google Scholar]
- 100.Ryu MY, Cho SK, Kim WT. RNAi suppression of RPN12a decreases the expression of type-A ARRs, negative regulators of cytokininsignaling pathway, in Arabidopsis. Mol Cells. 2009;28:375–382. [DOI] [PubMed] [Google Scholar]
- 101.Fermani S, Sparla F, Marri L, Thumiger A, Pupillo P, Falini G, Trost P. Structure of photosynthetic glyceraldehyde-3-phosphate dehydrogenase (isoform A4) from Arabidopsis thaliana in complex with NAD. ActaCrystallographica. Section F. Struct Biol Crystall Commun. 2010;66:621–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nelson KJ, Parsonage D, Hall A, Karplus PA, Poole LB. Cysteine pK(a) values for the bacterial peroxiredoxinAhpC. Biochem. 2008;47:12860–12868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ferrer-Sueta G, Manta B, Botti H, Radi R, Trujillo M, Denicola A. Factors affecting protein thiol reactivity and specificity in peroxide reduction. Chem Res Toxicol. 2011;24:434–450. [DOI] [PubMed] [Google Scholar]
- 104.Iqbalsyah TM, Moutevelis E, Warwicker J, Errington N, Doig AJ. The CXXC motif at the N terminus of an alpha-helical peptide. Protein Sci. 2006;15:1945–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kortemme T, Creighton TE. Ionisation of cysteine residues at the termini of model alpha-helical peptides. Relevance to unusual thiolpKa values in proteins of the thioredoxin family. J Mol Biol. 1995;253:799–812. [DOI] [PubMed] [Google Scholar]
- 106.Carballal S, Alvarez B, Turell L, Botti H, Freeman BA, Radi R. Sulfenic acid in human serum albumin. Amino Acids. 2007;32:543–551. [DOI] [PubMed] [Google Scholar]
- 107.Salsbury FR Jr, Knutson ST, Poole LB, Fetrow JS. Functional site profiling and electrostatic analysis of cysteines modifiable to cysteine sulfenic acid. Protein Sci. 2008;17:299–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Race PR, Bentley ML, Melvin JA, Crow A, Hughes RK, Smith WD, Banfield MJ. Crystal structure of Streptococcus pyogenessortase A: implications for sortase mechanism. J Biol Chem. 2009;284:6924–6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kulathu Y, Garcia FJ, Mevissen TE, Busch M, Arnaudo N, Carroll KS, Komander D. Regulation of A20 and other OTU deubiquitinases by reversible oxidation. Nat Commun. 2013;4:1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Winterbourn CC. Reconciling the chemistry and biology of reactive oxygen species. Nat Chem Biol. 2008;4:278–286. [DOI] [PubMed] [Google Scholar]
- 111.Paulsen CE, Carroll KS. Cysteine-mediated redox signaling: chemistry, biology, and tools for discovery. Chem Rev. 2013;113:4633–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Claiborne A, Mallett TC, Yeh JI, Luba J, Parsonage D. Structural, redox, and mechanistic parameters for cysteine-sulfenic acid function in catalysis and regulation. Advan Prot Chem. 2001;58:215–276. [DOI] [PubMed] [Google Scholar]
- 113.Lim JC, Choi HI, Park YS, Nam HW, Woo HA, Kwon KS, Kim YS, Rhee SG, Kim K, Chae HZ. Irreversible oxidation of the active-site cysteine of peroxiredoxin to cysteine sulfonic acid for enhanced molecular chaperone activity. J Biol Chem. 2008;283:28873–28880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hansen RE, Roth D, Winther JR. Quantifying the global cellular thiol-disulfide status. Proc Nat Acad Sci USA. 2009;106:422–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gupta V, Carroll KS DYn-2 Based Identification of Arabidopsis Sulfenomes. Sulfenic acid chemistry, detection and cellular lifetime. Biochim Biophys Acta—Gen Subjects. 2014;1840:847–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Akter S, Huang J, Bodra N, De Smet B, Wahni K, Rombaut D, Pauwels J, Gevaert K, Carroll K, Van Breusegem F, et al. DYn-2 Based Identification of Arabidopsis Sulfenomes. Mol Cell Proteom. 2015;14:1183–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Poole TH, Reisz JA, Zhao W, Poole LB, Furdui CM, King SB. Strained cycloalkynes as new protein sulfenic acid traps. J Am Chem Soc. 2014;136:6167–6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gupta V, Carroll KS. Sulfenic acid chemistry, detection and cellular lifetime. Biochim Biophys Acta—Gen Subjects. 2014;1840:847–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Alvarez S, Zhu MK. Chen S. Proteomics of Arabidopsis redox 1450 proteins in response to methyl jasmonate. J Proteome.. 2019;73:30–40. [DOI] [PubMed] [Google Scholar]
- 120.Go YM, Jones DP. Redox compartmentalization in eukaryotic cells. Biochim Biophys Acta. 2008;1780:1273–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Leonard SE, Carroll KS. Chemical “omics” approaches for understanding protein cysteine oxidation in biology. Curr Opin Chem Biol. 2011;15:88–102. [DOI] [PubMed] [Google Scholar]
- 122.Lee K, Lee J, Kim Y, Bae D, Kang KY, Yoon SC, Lim D. Defining the plant disulfide proteome. Electrophoresis. 2004;25:532–541. [DOI] [PubMed] [Google Scholar]
- 123.Alvarez S, Zhu M, Chen S. Proteomics of Arabidopsis redox proteins in response to methyl jasmonate. J Proteome. 2009;73:30–40. [DOI] [PubMed] [Google Scholar]
- 124.Bykova NV, Hoehn B, Rampitsch C, Banks T, Stebbing JA, Fan T, Knox R. Redox-sensitive proteome and antioxidant strategies in wheat seed dormancy control. Proteomics. 2011;11:865–882. [DOI] [PubMed] [Google Scholar]
- 125.Liu P, Zhang H, Wang H, Xia Y. Identification of redox-sensitive cysteines in the Arabidopsis proteome using OxiTRAQ, a quantitative redox proteomics method. Proteomics. 2014;14:750–762. [DOI] [PubMed] [Google Scholar]
- 126.Parker J, Zhu N, Zhu M, Chen S. Profiling thiol redox proteome using isotope tagging mass spectrometry. J Visual Exp. 2012;61:e3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang H, Wang S, Lu Y, Alvarez S, Hicks LM, Ge X, Xia Y. Proteomic analysis of early-responsive redox-sensitive proteins in Arabidopsis. J Proteome Res. 2012;11:412–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Carballal S, Alvarez B, Turell L, Botti H, Freeman BA, Radi R . Sulfenic acid in human serum albumin. Amino Acids. 2004; 32:543–551. [DOI] [PubMed] [Google Scholar]
- 129.Saurin AT, Neubert H, Brennan JP, Eaton P. Widespread sulfenic acid formation in tissues in response to hydrogen peroxide. Proc Nat Acad Sci USA. 2004;101:17982–17987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tyther R, Ahmeda A, Johns E, McDonagh B, Sheehan D. Proteomic profiling of perturbed protein sulfenation in renal medulla of the spontaneously hypertensive rat. J Proteome Res. 2010;9:2678–2687. [DOI] [PubMed] [Google Scholar]
- 131.Lin WS, Armstrong DA, Gaucher GM. Formation and repair of papain sulfenic acid. Canad J Biochem Physiol. 1975;53:298–307. [DOI] [PubMed] [Google Scholar]
- 132.Couturier J, Chibani K, Jacquot JP, Rouhier N. Cysteine based redox regulation and signaling in plants. Front Plant Sci. 2013;4:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Charles RL, Schro¨ der E, May G, Free P, Gaffney PR, Wait R, Begum S, Heads RJ, Eaton P. Protein sulfenation as a redox sensor – proteomics studies using a novel biotinylateddimedone analogue. Mol Cell Proteom. 2007;6:1473–1484. [DOI] [PubMed] [Google Scholar]
- 134.Poole LB, Klomsiri C, Knaggs SA, Furdui CM, Nelson KJ, Thomas MJ, Fetrow JS, Daniel LW, King SB. Fluorescent and affinity-based tools to detect cysteine sulfenic acid formation in proteins. Bioconjug Chem. 2007;18:2004–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Poole LB, Zeng BB, Knaggs SA, Yakubu M, King SB. Synthesis of chemical probes to map sulfenic acid modifications on proteins. Bioconjug Chem. 2005;16:1624–1628. [DOI] [PubMed] [Google Scholar]
- 136.Reddie KG, Seo YH, Muse III WB, Leonard SE, Carroll KS. A chemical approach for detecting sulfenic acid-modified proteins in living cells. Mol Biosyst. 2008;4:521–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Seo YH, Carroll KS. Profiling protein thiol oxidation in tumor cells using sulfenic acid-specific antibodies. Proc Nat Acad Sci USA. 2009;106:16163–16168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Leonard SE, Reddie KG, Carroll KS. Mining the thiol proteome for sulfenic acid modifications reveals new targets for oxidation in cells. ACS Chem Biol. 2009;4:783–799. [DOI] [PubMed] [Google Scholar]
- 139.Truong TH, Carroll KS. Bioorthogonal chemical reporters for analyzing protein sulfenylation in cells. Curr Prot Chem Biol. 2012;4:101–122. [Google Scholar]
- 140.Wang W, Hong S, Tran A, Jiang H, Triano R, Liu Y, Chen X, Wu P. Sulfated ligands for the copper (I)-catalyzedazide-alkyne cycloaddition. Chem Asian J. 2011;6:2796–2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Paulsen CE, Truong TH, Garcia FJ, Homann A, Gupta V, Leonard SE, Carroll KS. Peroxide-dependent sulfenylation of the EGFR catalytic site enhances kinase activity. Nat Chem Biol. 2012;8:57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Fu L, Liu K, Ferreira RB, Carroll KS, Yang J. Proteome-wide analysis of cysteine s-sulfenylation using a benzothiazine-based probe. Curr Prot Prot Sci. 2019;95:e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gupta V, Yang J, Daniel C, Carroll KS. Diverse Redoxome Reactivity Profiles of Carbon Nucleophiles. J Am Chem Soc. 2017;139:5588–5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Qian J, Klomsiri C, Wright MW, King SB, Tsang AW, Poole LB, Furdui CM. Simple synthesis of 1, 3-cyclopentanedione derived probes for labelling sulfenic acid proteins. Chemical Commun. 2011;47:9203–9205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Qian J, Wani R, Klomsiri C, Poole LB, Tsang AW, Furdui CM. A simple and effective strategy for labeling cysteine sulfenic acid in proteins by utilization of β-ketoesters as cleavable probes. Chem Commun. 2012;48:4091–4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yang J, Gupta V, Carroll KS, Liebler DC. Site-specific mapping and quantification of protein S-sulphenylation in cells. Nat Commun. 2014;5:4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.De Smet B, Willems P, Fernandez-Fernandez AD, Alseekh S, Fernie AR, Messens J, Van Breusegem F. In vivo detection of protein cysteine sulfenylation in plastids. Plant J. 2018;97:4. [DOI] [PubMed] [Google Scholar]
- 148.Wei B, Willems P, Huang J, Tian C, Yang J, Messens J, Breusegem FV. Identification of sulfenylated cysteines in Arabidopsis thaliana proteins using a disulfide-linked peptide reporter. bioRxiv. 2020;11:777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lindermayr C, Sell S, Müller B, Leister D, Durner J. Redox regulation of the NPR1-TGA1 system of Arabidopsis thaliana by nitric oxide. Plant Cell. 2010;22:2894–2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.De Smet B. Protein Cysteine Sulfenylation in Plant Stress Responses : A Journey through the Organelles. Ghent University. Faculty of Sciences; Vrije Universiteit Brussel. Faculty of Sciences and Bioengineering Sciences; 2020. [Google Scholar]
- 151.Paulsen CE, Carroll KS. Orchestrating redox signaling networks through regulatory cysteine switches. American Chem Soc Chem Biol. 2010;5:47–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Poole LB, Karplus PA, Claiborne A. Protein sulfenic acids in redox signaling. Annual Rev Pharmacol Toxicol. 2004;44:325–347. [DOI] [PubMed] [Google Scholar]
- 153.Krishnan N, Fu C, Pappin DJ, Tonks NK. H2S-induced sulfhydration of the phosphatase PTP1B and its role in the endoplasmic reticulum stress response. Sci Signal. 2011;4:ra86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Betsuyaku S, Takahashi F, Kinoshita A, Miwa H, Shinozaki K, Fukuda H, Sawa S. Mitogen-activated protein kinase regulated by the CLAVATA receptors contributes to shoot apical meristem homeostasis. Plant Cell Physiol. 2011;52:14–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Xie G, Sasaki K, Imai R, Xie D. A redox-sensitive cysteine residue regulates the kinase activities of OsMPK3 and OsMPK6 in vitro. Plant Sci. 2014;227:69–75. [DOI] [PubMed] [Google Scholar]
- 156.Kovtun Y, Chiu WL, Tena G, Sheen J. Functional analysis of oxidative stress activated mitogen-activated protein kinase cascade in plants. Proc Nat Acad Sci USA. 2000;97:2940–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Truong TH, Carroll KS. Redox regulation of protein kinases. Crit Rev Biochem Mol Biol. 2013;48:332–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhang T, Zhu M, Song WY, Harmon AC, Chen S. Oxidation and phosphorylation of MAP kinase 4 cause protein aggregation. Biochim Biophys Acta Proteins Proteom. 2015;1854:156–165. [DOI] [PubMed] [Google Scholar]
- 159.Midorikawa T, Endow JK, Dufour J, Zhu J, Inoue K. Plastidic type I signal peptidase 1 is a redox-dependent thylakoidal processing peptidase. Plant J. 2014;80:592–603. [DOI] [PubMed] [Google Scholar]
- 160.Noguera-Mazon V, Lemoine J, Walker O, Rouhier N, Salvador A, Jacquot JP, Lancelin JM, Krimm I. Glutathionylation induces the dissociation of 1-CysD-peroxiredoxin non- covalent homodimer. J Biol Chem. 2006;281:31736–31742. [DOI] [PubMed] [Google Scholar]
- 161.Basset G, Raymond P, Malek L, Brouquisse R. Changes in the expression and the enzymic properties of the 20S proteasome in sugar-starved maize roots. Evidence for an in vivo oxidation of the proteasome. Plant Physiol. 2002;128(3):1149–1162. doi: 10.1104/pp.010612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tanou G, Job C, Rajjou L, Arc E, Belghazi M, Diamantidis G, Molassiotis A, Job D. Proteomics reveals the overlapping roles of hydrogen peroxide and nitric oxide in the acclimation of citrus plants to salinity. Plant Journal. 2009;60:795–804. [DOI] [PubMed] [Google Scholar]
- 163.Balmer Y, Koller A, Val GD, Schu¨rmann P, Buchanan BB. Proteomics uncovers proteins interacting electrostatically with thioredoxinin chloroplasts. Photosynthesis Res. 2004;79:275–280. doi: 10.1023/B:PRES.0000017207.88257.d4. [DOI] [PubMed] [Google Scholar]
- 164.Marchand CH, Vanacker H, Collin V, Issakidis-Bourguet E, Le Mare´ Chal P, Decottignies P. Thioredoxin targets in Arabidopsis roots. Proteomics. 2010;10:2418–2428. [DOI] [PubMed] [Google Scholar]
- 165.Rouhier N, Villarejo A, Srivastava M, Gelhaye E, Keech O, Droux M, Finkemeier I, Samuelsson G, Dietz KJ, Jacquot JP, et al. Identification of plant glutaredoxin targets. Antioxidant Redox Signal. 2005;7:919–929. [DOI] [PubMed] [Google Scholar]
- 166.Marchand C, Le Mare´ Chal P, Meyer Y, Miginiac-Maslow M, Issakidis-Bourguet E, Decottignies P. New targets of Arabidopsis thioredoxins revealed by proteomic analysis. Proteomics. 2004;4:2696–2706. [DOI] [PubMed] [Google Scholar]
- 167.Marchand CH, Le Mare´chal P, Meyer Y, Decottignies P. Comparative proteomic approaches for the isolation of proteins interacting with thioredoxin. Proteomics. 2006;6:6528–6537. [DOI] [PubMed] [Google Scholar]
- 168.Stro¨her E, Dietz KJ. The dynamic thiol-disulphide redox proteome of the Arabidopsis thaliana chloroplast as revealed by differential electrophoretic mobility. Physiol Plant. 2008;133:566–583. [DOI] [PubMed] [Google Scholar]
- 169.Kim G, Levine A. Methionine residue promotes hyperoxidation of the catalytic cysteine of mouse methionine sulfoxidereductase A. Biochemistry. 2016;55:3586–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Tarrago L, Laugier E, Zaffagnini M, Marchand C, Le Marechal P, Lemaire SD, Rey P. Plant thioredoxin CDSP32 regenerates 1-cys methionine sulfoxidereductase B activity through the direct reduction of sulfenic acid. J Biol Chem. 2010;285:14964–14972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Tarrago L, Laugier E, Zaffagnini M, Marchand C, Le Marechal P, Rouhier N, Lemaire SD, Rey P. Regeneration mechanisms of Arabidopsis thaliana methionine sulfoxidereductases B by glutaredoxins and thioredoxins. J Biol Chem. 2009;284:18963–18971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Bedhomme M, Adamo M, Marchand CH, Couturier J, Rouhier N, Lemaire SD, Zaffagnini P, Trost M. Glutathionylation of cytosolic glyceraldehyde-3-phosphate dehydrogenase from the model plant Arabidopsis thaliana is reversed by both glutaredoxins and thioredoxins in vitro. Biochem J. 2012;445:337–347. doi: 10.1042/BJ20120505. [DOI] [PubMed] [Google Scholar]
- 173.Peralta D, Bronowska AK, Morgan B, Doka E, Van Laer K, Nagy P, Grater F, Dick TP. A proton relay enhances H2O2 sensitivity of GAPDH to facilitate metabolic adaptation. Nat Chem Biol. 2015;11:156–163. [DOI] [PubMed] [Google Scholar]
- 174.Becker K, Savvides SN, Keese M, Schirmer RH, Karplus PA. Enzyme inactivation through sulfhydryl oxidation by physiologic NO-carriers. Nat Struct Biol. 1998;5(4):267–271. doi: 10.1038/nsb0498-267. [DOI] [PubMed] [Google Scholar]
- 175.Kaiserova K, Srivastava S, Hoetker JD, Awe SO, Tang XL, Cai J, Bhatnagar A. Redox activation of aldose reductase in the ischemic heart. J Biol Chem. 2006;281:15110–15120. [DOI] [PubMed] [Google Scholar]
- 176.Wetzelberger K, Baba SP, Thirunavukkarasu M, Ho YS, Maulik N, Barski OA, Conklin DJ, Bhatnagar A. Postischemic deactivation of cardiac aldose reductase: role of glutathione S-transferase P and glutaredoxin in regeneration of reduced thiols from sulfenic acids. J Biol Chem. 2010;285:26135–26148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Huang J, Willems P, Van Breusegem F, Messens J. Pathways crossing mammalian and plant sulfenomic landscapes. Free Rad Biol Med. 2018;122:193–201. [DOI] [PubMed] [Google Scholar]
- 178.Ralser M, Wamelink MM, Kowald A, Gerisch B, Heeren G, Struys EA, Klipp E, Jakobs C, Breitenbach M, Lehrach H, et al. Dynamic rerouting of the carbohydrate flux is key to counteracting oxidative stress. J Biol. 2007;6:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Claiborne A, Yeh JI, Mallett TC, Luba J, Crane EJ, Charrier V, Parsonage D. Protein-sulfenic acids: diverse roles for an unlikely player in enzyme catalysis and redox regulation. Biochem. 2001;38:15407–15416. [DOI] [PubMed] [Google Scholar]
- 180.Margis R, Dunand C, Teixeira FK, Margis-Pinheiro M. Glutathione peroxidase family—an evolutionary overview. FEBS J. 2008;275:3959–3970. [DOI] [PubMed] [Google Scholar]
- 181.Moffett AS, Bender KW, Huber SC, Shukla, D. Allosteric Control of a Plant Receptor Kinase through S-Glutathionylation. Biophys J. 2017a;113:2354–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]


