ABSTRACT
Eosinophils are bone marrow-derived granulocytes that display key effector functions in allergic diseases. Nonetheless, recent data highlight important roles for eosinophils in the tumor microenvironment (TME). Eosinophils have been attributed with pleiotropic and perhaps conflicting functions, which may be attributed at least in part to variations in eosinophil quantitation in the TME. Thus, a reliable, quantitative, and robust method for the assessment of eosinophilic infiltration in the TME is required. This type of methodology could standardize the identification of these cells and promote the subsequent generation of hypothesis-driven mechanistic studies. To this end, we conducted a comprehensive analysis of multiple primary tumors from distinct anatomical sites using a standardized method. Bioinformatics analysis of 10,469 genomically profiled primary tumors revealed that eosinophil abundance within different tumors can be categorized into three groups representing tumors with high, intermediate, and low eosinophil levels. Consequently, eosinophil abundance, as well as spatial distribution, was determined in tissue tumor arrays of six tumors representing all three classifications (colon and esophagus – high; lung – intermediate; cervix, ovary, and breast – low). With the exception of breast cancer, eosinophils were mainly localized in the tumor stroma. Importantly, the tumor anatomical site was identified as the primary predictive factor of eosinophil stromal density highlighting a distinction between mucosal-barrier organs versus non-mucosal barrier organs. These findings enhance our understanding of eosinophil diversity in the TME and provide a compelling rationale for future experiments assessing the activity of these cells.
KEYWORDS: Eosinophils, tumor microenvironment, mucosal barrier tissues, allergy
Introduction
Eosinophils are bone marrow-derived granulocytes that are traditionally studied in the context of allergic diseases and parasite infections.1 Under steady-state conditions, eosinophils accumulate in mucosal surfaces such as the gastrointestinal tract, which serves as the largest eosinophil reservoir in the body.2
Recently, unforeseen roles have been discovered for eosinophils in various settings that are far and beyond allergic inflammation, including cancer.3,4 In support of this notion, eosinophils have been shown to infiltrate multiple tumors and are, in fact, an integral part of the tumor microenvironment (TME). For example, eosinophils have been described in various tumors including bladder,5 gastric,6,7 colorectal,8–10 esophageal,11,12 head and neck,13,14 lung,15 liver,16 breast,17,18 ovary,19 and uterine cervix,20,21 as well as in malignant pleural effusions.22 In addition, eosinophils can accumulate in response to various therapeutic strategies.3
Standard methods of immune profiling in tumors have lacked thorough and systematic measures of eosinophils. This is likely due to several reasons such as technical difficulties to evaluate eosinophil numbers in situ and the underappreciated role of eosinophils in tumors as more than bystander cells.23 Identification and characterization of eosinophils in situ in human tissues is particularly complex. The majority of association studies assessing eosinophil levels with tumor stage/grade or disease prognosis, originated from either association of blood eosinophil counts or tumor tissue cytologic assessment with acid dye labeling of the granule proteins within intact eosinophils. Using the latter method, it is often difficult to discern eosinophils in highly degranulated structures of tumor tissues. This may explain, in part, why eosinophil numbers and contribution to tumor growth are quite variable and contradictory. More recently, unique RNA signatures have been identified for various immune cells.24 Nonetheless, methods for identifying eosinophils by mRNA transcript expression through microarrays or by RNA sequencing have been difficult, likelydue to instability of eosinophil RNA for analysis by array platform. Thus, standardized methods for quantitation of eosinophils in tumors are required. This is specifically important since tumor-infiltrating eosinophils were associated with favorable prognosis in several tumors (e.g. oral,14,25 esophageal,11,12 gastric,6,7 and colon8–10) and poor prognosis in others (bladder,5 cervical20,21 tumors, and Hodgkin’s lymphoma26,27). While there is no physiological reason to assume that eosinophils within a given microenvironment will be instructed toward a single effector function, this controversy can also be explained by variability in the identification of and quantitation of tumor-infiltrating eosinophils.
In this study, we used an established RNA profiling algorithm to measure the abundance of eosinophils in over ten thousand primary tumors from distinct anatomical sites. Subsequently, we confirmed our bioinformatics findings by eosinophil-specific immunohistochemical staining of 2,890 primary tumors. Using these two unique approaches we identified that the anatomical location of the tumor was the primary predictive factor of eosinophil stromal density highlighting a clear distinction between mucosal-barrier organs versus non-mucosal barrier organs. Our study establishes a reliable and quantitative method for the detection and analysis of eosinophils in the TME. The method and the findings that presented in this study provide a compelling rationale for future hypothesis-driven mechanistic studies, which will assess the role of eosinophils in tumors displaying eosinophilic infiltration.
Materials and methods
Total cancer care database and tumor gene profile analysis
Patients were consented to the Total Cancer CareTM (TCC) protocol (IRB-approved, Liberty IRB #12.11.0023).28 Pathology quality control evaluation of tumors was performed as part of the TCC tissue collection protocol, which includes percent malignant cellularity, necrosis, and stromal cell presence. Tumor samples were assayed using the custom Rosetta/Merck HuRSTA_2a520709 Affymetrix gene expression microarray platform (GEO: GPL15048). CEL files (which store the results of the intensity calculations) were normalized against the median CEL file using IRON,29 which yields Log2 intensity values per probeset. Principle component analysis (PCA) of all samples revealed that the first principle component was highly correlated to RNA integrity number (RIN), suggesting an RNA quality difference among samples. A partial least squares (PLS) model was trained upon the fresh frozen samples for which RIN was available and used to re-estimate the RNA quality of all samples. Then, the first principle component was removed to correct the signals for RNA quality. In total, 10,469 fresh frozen macro dissected primary tumors were identified for the related analyses.
Eosinophils normalized content abundance
Eosinophil abundance in the TCC patient cohort was evaluated using the ‘Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data’ (ESTIMATE) algorithm (Estimate R package v1.0.13).30 Normalized gene expression probeset identifiers were mapped to Entrez GeneIDs. As defined in the ESTIMATE package, the data were filtered to only the common genes. A single probeset was then selected per gene by choosing the probeset having the highest median expression across all tumors. This resulting dataset was used in the ESTIMATE function to produce stromal and immune scores for each cohort. Next, the Human RSTA array was reduced to the LM22 signature genes (Supplementary Table 1) as defined by CIBERSORT31 by choosing a representative probeset that detects the gene and has the highest median expression among matching probesets. LM22 is a validated leukocyte signature matrix generated from microarray data, which contains 547 genes that distinguish 22 human hematopoietic cell phenotypes.31 The CIBERSORT web tool (https://cibersort.stanford.edu/index.php) was accessed on 2017–05-19 to generate signature scores (using quantile normalization).
Immune cell infiltrate composition proportions from CIBERSORT31 were extracted in relative mode, where the proportions are relative to the total immune cell fraction of the tumor. To normalize the content across tumors, we scaled the ESTIMATE30 immune scores such that the lowest immune score was 0 (rather than negative) and analyzed multiple immune cell infiltrate fractions by this adjusted immune score, to yield the Eosinophil Normalized Content Abundance.30 Eosinophil levels were compared to other immune cell types that were previously estimated from CIBERSORT. The normalized content abundance (NCA) for eosinophils was correlated to the additional immune cells using Spearman’s correlation within each site of origin. The correlation coefficients were visualized using ComplexHeatmap in R4.0.3.
Tissue microarrays
Tissue microarrays were purchased from US Biomax Inc and were composed of paraffin-embedded primary tumor tissues collected prior to therapy from breast, colon, esophagus, lung, ovary, and uterine cervix. The following arrays were obtained: Breast- BR20810, BR20811, BR20812, BR20813, BR20814, BR486, BR804b, BR90; Colon- CO601, CO602, CO992, CO702b, CO703, CO952, CO953, T054b, T055; Esophagus- ES1021, ES2001a, ES2082; Lung- LC20812, LC20813, LC20814, LC20815, LC20816; Ovary- OV20810, OV20811, OV20812; Uterine cervix- CR2089, CR2088. Tumor samples from the aforementioned arrays were conjoined with the following clinical parameters: age, sex, pathological grade, clinical stage, tumor size, lymph node involvement, and receptor expression (progesterone receptor, estrogen receptor, and Human Epidermal Growth Factor Receptor 2-HER2). The sample size was calculated using G*power32 with an effect size of 0.1, 80% power, and 1% significance due to multiple comparisons.
Immunohistochemistry and analysis of stained tumor samples
Tumor arrays were stained with anti-eosinophil peroxidase (EPX), as previously described33 (kindly provided by the Lee laboratories Mayo clinic, Scottsdale, AZ). Anti-EPX stained slides were scanned at X20 magnification by means of the Leica© Aperio digital pathology slide scanner with a resolution of 0.32 μm/pixel. Thereafter, the scanned photomicrographs were analyzed using Qupath© bioimaging analysis software.34 Tumor cores on each array were annotated to regions of tumor and stroma based on differential cell appearance using the Nuclear Fast Red staining, and their relative area was calculated. Eosinophils were detected using color deconvolution, and cells were assigned to as positive or negative cells based upon the smoothed DAB channel information of mean intensity in the cell compartment (nucleus and cytoplasm). Extra-cellular staining (EPX degranulation) was not measured. The number of DAB-positive cells (i.e. eosinophils) and the area of tumor or stroma were used to calculate the density of eosinophils within the given region. Finally, eosinophil density data (i.e. the number of DAB positive cells per mm2) was exported along with marked images showing the detected cells, for visual verification. The detection and export steps were fully automated using a batch processing script.
Statistical analysis
Categorical variables were described as frequency and percentage. Continuous variables were evaluated for normal distribution using histogram and Q-Q plot and reported as mean and standard deviation (SD) or median and interquartile range (IQR). Spearman’s rank correlation coefficient was used to evaluate the association between eosinophil density, patient age, pathological grade, clinical stage, and tumor size. Classification and regression trees (CART) algorithm was used to identify a subgroup of patients/malignancies according to eosinophil density. Kruskal–Wallis test and Mann–Whitney test were used to compare eosinophil density between the different types of malignancy. Wilcoxon signed-rank test was used to compare eosinophil density in stroma and intra-tumoral. All statistical tests were two-sided and p-value < 0.05 was considered statistically significant. Statistical analysis was performed with SPSS statistical software (IBM SPSS Statistics, version 25, IBM Corp., Armonk, NY, USA, 2017).
Results
Bioinformatics analysis predict that eosinophils are differentially distributed in distinct primary tumors according to their anatomical location
Tumor-infiltrating eosinophils have been described in multiple tumors.35 Nonetheless, their relative distribution within tumors is largely unknown. Thus, we first aimed to determine whether eosinophils are differentially distributed within primary tumors from distinct anatomical locations. To this end, we used the TCC database, which profiled 10,469 genomically distinct solid, primary, non-metastatic tumor samples representing 22 tumor types. To specifically assess the abundance of eosinophils in the tumors, the CIBERSORT deconvolution algorithm was used.31 This algorithm is a versatile computational method for quantifying cell fractions from bulk tissue gene expression profiles using a signature matrix and was recently utilized to estimate the immune composition of tumor biopsies.24 The normalized content of eosinophils (defined using LM22 signature genes,31 see Supplementary Table 1), which represents their abundance, significantly differed between tumors from distinct anatomical locations (p-value <0.001). Further analysis employing this data for tree classification using classification and regression tree (CART) algorithm36 revealed that eosinophil distribution within different tumors can be categorized into three groups representing tumors with high (38.89 ± 75.45), intermediate (18.59 ± 47.88), and low (11.02 ± 34.88) eosinophil levels (Figure 1, Supplementary table 2). Interestingly, gastrointestinal tumors including stomach, rectum, large bowel, pancreas, and esophageal were among the tumors, which were highly infiltrated with eosinophils (Figure 1).
Figure 1.
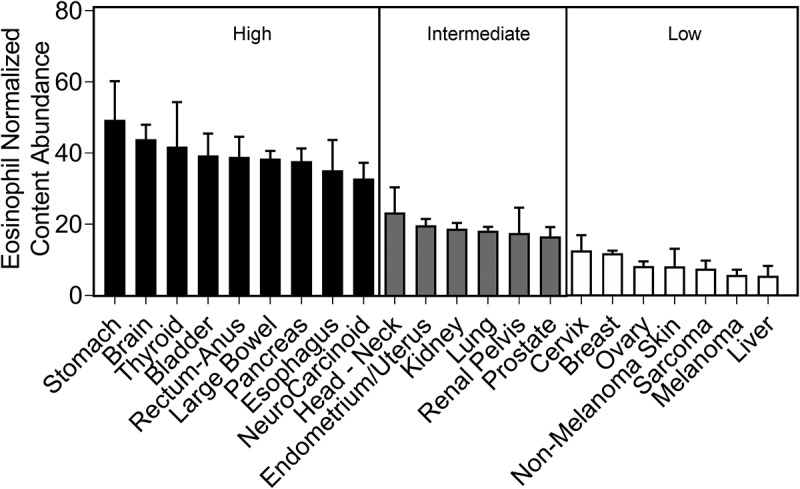
CIBERSORT analysis predicts differential eosinophil distribution according to the tumor anatomical location. Eosinophil abundance was assessed using the CIBERSORT deconvolution algorithm on 10,469 primary tumors of the TCC genomic database. Eosinophil normalized content abundance significantly differed between different anatomical sites (p-value < 0.001). Classification and regression tree (CART) algorithm categorized tumor types into three groups representing tumors with high (seen in black), intermediate (seen in gray) and low (seen in white) eosinophil levels
Anti-EPX staining reveals that eosinophils are differentially distributed within distinct tumor anatomical sites
Next, we obtained tissue tumor arrays of six tumor types representing all three classifications attained from the TCC data: namely, colon and esophagus – representing highly infiltrated tumors; lung – representing tumors infiltrated with intermediate levels; and cervix, ovary, and breast – representing tumors with low infiltration. The slides were subjected to anti-EPX immunohistochemical staining. We specifically chose to use this methodology since anti-EPX staining has been demonstrated as an efficient tool to identify and quantify eosinophils in pathological settings such as allergic diseases.33,37 Furthermore, using tumor arrays enables a side-by-side comparison of eosinophils in multiple specimens using the same methodology, thus reducing technical variability. Our cohort included 2,890 patients (Table 1), mostly female (66.8%), with a mean age of 53.4 ± 11.45 (ranging from 18 to 95). The cohort contained patients with six different primary tumors; 576 breast cancer (20%), 399 cervical cancer (13.8%), 837 lung cancer (29%), 586 ovarian cancer (20.3%), 306 esophageal cancer (10.5%), and 186 colon cancer (6.4%).
Table 1.
Patient clinical characteristics
| Total | Breast | Uterine cervix | Lung | Ovary | Esophagus | Colon | ||
|---|---|---|---|---|---|---|---|---|
| N | 2890 | 576 | 399 | 837 | 586 | 306 | 186 | |
| Age (years), mean (SD) | 53.4 (11.45) | 50.42 (11.26) | 48.25 (10.33) | 58.28 (9.51) | 49.65 (11.8) | 58.11 (8.73) | 56.25 (13.21) | |
| Female sex, n(%) | 1930 | 576 (100%) | 399 (100%) | 206 (24.6%) | 586 (100%) | 92 (30%) | 71 (38.2%) | |
| Male sex, n(%) | 960 | 0 (0%) | 0 (0%) | 631 (75.4%) | 0 (0%) | 214 (70%) | 115 (61.8%) | |
| Stage, n(%) | ||||||||
| 1 | 1490 | 81 (14%) | 280 (70.2%) | 602 (71.9%) | 469 (80%) | 6 (2%) | 52 (28%) | |
| 2 | 1060 | 429 (74.5%) | 103 (25.8%) | 124 (14.8%) | 80 (13.7%) | 240 (78.4%) | 84 (45.2%) | |
| 3 | 298 | 64 (11.1%) | 14 (3.5%) | 106 (12.7%) | 16 (2.7%) | 59 (19.3%) | 39 (20.9%) | |
| 4 | 42 | 2 (0.35%) | 2 (0.5%) | 5 (0.6%) | 21 (3.6%) | 1 (0.3%) | 11 (5.9%) | |
| Grade, n(%) | ||||||||
| 1 | 450 | 27 (4.7%) | 62 (15.5%) | 124 (14.8%) | 103 (17.6%) | 93 (30.4%) | 41 (22%) | |
| 2 | 1285 | 404 (70.1%) | 244 (61.2%) | 317 (37.9%) | 138 (23.5%) | 105 (34.3%) | 77 (41.4%) | |
| 3 | 601 | 58 (10.1%) | 76 (19%) | 190 (22.7%) | 196 (33.5%) | 61 (20%) | 20 (10.8%) | |
| NA* | 554 | 87 (15.1%) | 17 (4.3%) | 206 (24.6%) | 149 (25.4%) | 47 (15.3%) | 48 (25.8%) | |
| T, n(%) | ||||||||
| 1 | 878 | 84 (14.6%) | 287 (72%) | 60 (7.2%) | 438 (74.7%) | 8 (2.6%) | 1 (0.6%) | |
| 2 | 1390 | 381 (66.1%) | 105 (26.3%) | 684 (81.7%) | 97 (16.6%) | 85 (27.8%) | 38 (20.4%) | |
| 3 | 466 | 61 (10.6%) | 7 (1.7%) | 49 (5.8%) | 51 (8.7%) | 209 (68.3%) | 89 (47.8%) | |
| 4 | 156 | 50 (8.7%) | 0 (0%) | 44 (5.3%) | 0 (0%) | 4 (1.3%) | 58 (31.2%) | |
| N, n(%) | ||||||||
| 0 | 2455 | 502 (87.2%) | 388 (97.2%) | 653 (78%) | 551 (94%) | 237 (77.4%) | 124 (66.7%) | |
| 1 | 361 | 57 (9.9%) | 11 (2.8%) | 152 (18.2%) | 34 (5.8%) | 69 (22.6%) | 38 (20.4%) | |
| 2 | 74 | 17 (2.9%) | 0 (0%) | 32 (3.8%) | 1 (0.2%) | 0 (0%) | 24 (12.9%) | |
| M, n(%) | ||||||||
| 0 | 2848 | 575 (99.8%) | 399 (100%) | 832 (99.4%) | 566 (96.6%) | 305 (99.7%) | 171 (92%) | |
| 1 | 42 | 1 (0.2%) | 0 (0%) | 5 (0.6%) | 20 (3.4%) | 1 (0.3%) | 15 (8%) | |
*NA; specific classification was not available
Eosinophils were identified within the TME of tumors located in the colon (Figure 2(a)), lung (Figure 2(b)), uterine cervix (Figure 2(c)), esophagus (Figure 2(d)), ovary (Figure 2(e)) and breast (figure 2(f)). To accurately quantify eosinophil levels, Qupath© software was used,34 and anti-EPX stained eosinophils were automatically detected and quantified in each tissue sample. Thereafter, the tissue area was determined, and eosinophil density was calculated by assessing eosinophils per tissue mm2 (Figure 2(g)). The uniform handling and staining of all samples facilitated the comparison of eosinophil density between anatomical sites. Analysis of eosinophil density revealed significant differences (p-value <0.01) between anatomical sites (Figure 2(I), Supplementary table 3). Our immunohistochemical staining resulted in different eosinophil densities in esophageal, lung, and uterine cervix eosinophil accumulation in comparison to the CIBERSORT prediction (Figure 1). The histological analysis revealed breast tumors had significantly fewer infiltrating eosinophils compared to each of the other anatomical sites (p-value <0.01), whereas colon tumors had significantly increased numbers of infiltrating eosinophils compared to each of the other anatomical sites (p-value <0.05). Eosinophil accumulation was significantly distinct between most anatomical site comparisons, yet no significant difference was present between cervical and esophageal tumors. Importantly, although not quantitated, eosinophil degranulation was observed in several tumor samples (Supplementary Figure 1).
Figure 2.
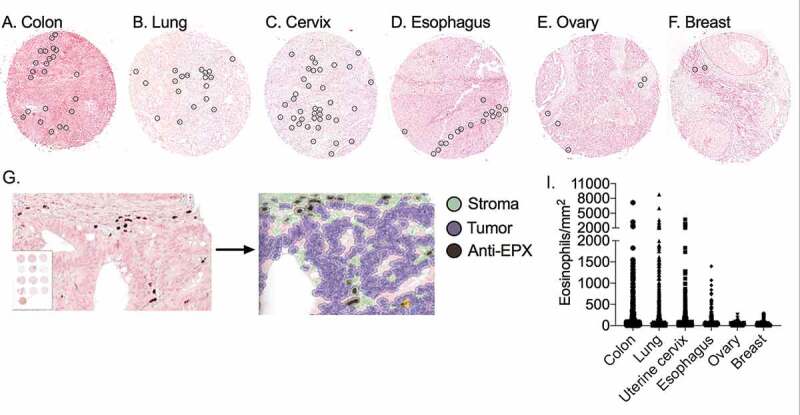
Anti-EPX immunohistochemical staining shows differential eosinophil distribution within distinct tumor anatomical sites. Tissue tumor arrays consisting of 2,890 individual patients were stained with anti-EPX. Representative photomicrographs of infiltrating eosinophils (stained brown with DAB and circled) in colon (a), lung (b), uterine cervix (c), esophagus (d), ovary (e) and breast (f) tumors. Qupath© software annotations of tumor cores to regions of tumor (purple), stroma (green) and DAB positive cells (black) eosinophils (g). Eosinophil density differed (p-value < 0.001) between anatomical sites (i)
Eosinophils are predominantly present in the stroma of the tumor microenvironment
We further assessed the distribution of eosinophils within the different tumors with emphasis on their location (i.e. stroma vs. intra-tumoral). This analysis (Supplementary Table 3) revealed that in colon (Figure 3(a)), lung (Figure 3(b)), cervical (Figure 3(c)), esophageal (Figure 3(d)), and ovarian tumors (Figure 3(e)) eosinophils were predominantly localized in the stroma (p-value <0.01). In contrast, in breast tumors (figure 3(f)), eosinophils localized primarily in intra-tumoral areas (p-value <0.01).
Figure 3.
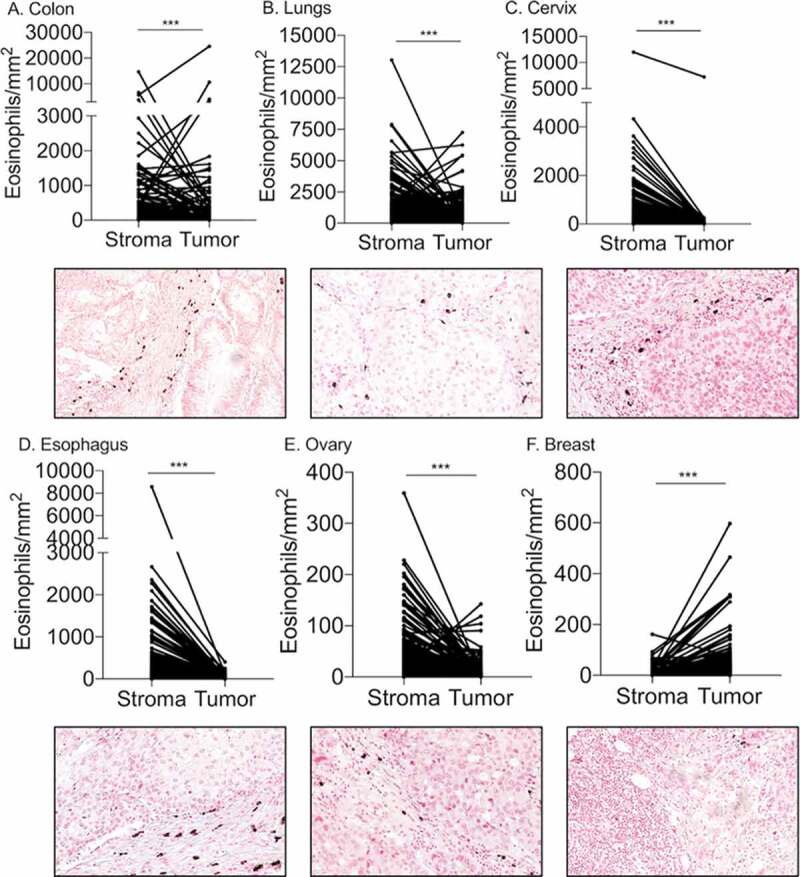
Localization analysis of eosinophils in the TME reveals predominant stromal infiltration. Paired analysis of eosinophil density in the tumor stroma versus intra-tumoral regions was conducted in colon (a), lung (b), cervical (c), esophageal (d) and ovarian (e) and breast (f) tumors. Representative photomicrographs of each tumor type are located bellow the corresponding graph. *** p- value < 0.001
Eosinophil stromal density is primarily determined by the tumor anatomical site
Eosinophil density could potentially be influenced by multiple parameters including age, sex, tumor stage, grade, size, lymph node involvement, and tumor anatomical site. To determine the primary factor, which is predictive of eosinophil stromal density we used an empiric approach employing the CART algorithm.36 This analysis revealed that the density of stromal eosinophils could be primarily classified according to the tumor anatomical site (Figure 4(a)) in respect to age, sex, tumor stage, grade, size, and lymph node involvement. Analysis of 2,890 samples, which represent 100% of the cohort (see Node 0, Figure 4(a)) in combination with all of the aforementioned clinical and pathological parameters demonstrated the tumor anatomical site to be the best predictor of eosinophil stromal density. Interestingly, this classification divided eosinophil stromal density into two major subsets collectively termed Node 1 and Node 2. Node 1 was comprised of colon, lung, esophageal, and cervical tumors, whereas Node 2 was comprised of breast and ovarian tumors (Figure 4(a)). This classification suggested that mucosal barrier oriented (Node 1) vs. non-mucosal barrier tumors (Node 2) may serve as a distinctive marker for stromal eosinophil density (p-value <0.01). Next, stromal eosinophil density of each sample was assessed with respect to the classification of tumors into mucosal vs. non-mucosal barrier. This analysis demonstrated that the percentage of samples, which displayed increased stromal eosinophil density was higher in mucosal barrier tumors (blue) in comparison with non-mucosal barrier tumors (red) (Figure 4(b)). We further performed a cross-validation analysis on the TCC cohort (Figure 4(c)) by assessing the eosinophil normalized content abundance in mucosal versus non-mucosal barrier tumors (see classification in Supplementary Table 4). This analysis recapitulated our findings demonstrating that mucosal barrier tumors had increased eosinophil abundance (26.38 ± 61.31) in comparison with non-mucosal tumors (17.24 ± 47.4, p-value<0.001). Collectively, these data suggest that mucosal barrier tumors will be characterized by abundant eosinophilia.
Figure 4.
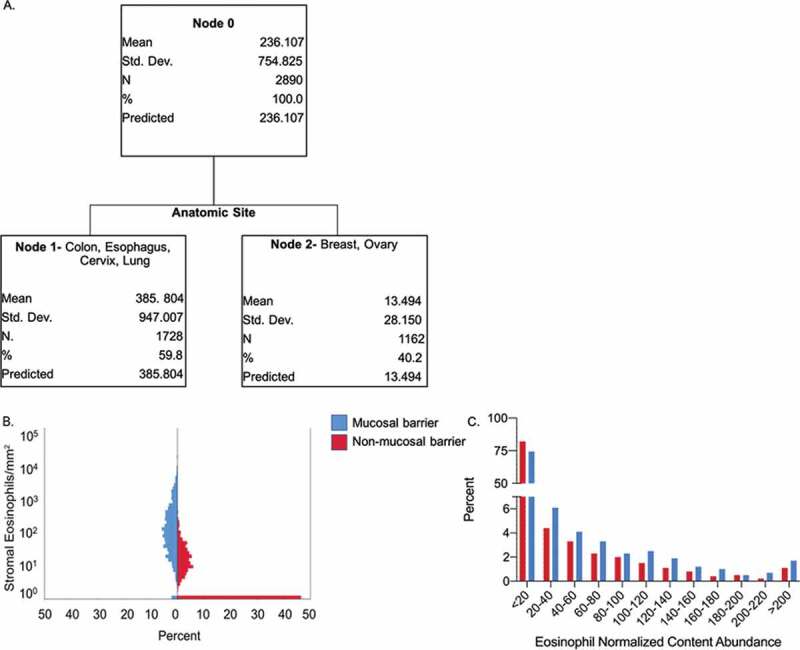
CART algorithm determines tumor anatomic site as the primary predictive factor of eosinophil stromal density. CART algorithm analyzed stromal eosinophil density in all 2,890 cohort samples (A, Node 0) in respect to age, sex, tumor stage, grade, size, lymph node involvement and tumor anatomical site. Tumor anatomical site was the best predictor of eosinophil stromal density classifying eosinophil stromal density into two subsets. One subset comprised of mucosal barrier tumors; colon, lung, esophageal and cervical (A, Node 1), and another comprised of non-mucosal barrier tumors; breast and ovarian tumors (A, Node 2). Analysis of stromal eosinophil density assessing the percentage of patients demonstrating a particular eosinophil density in the mucosal barrier (B, in blue) versus non-mucosal barrier (B, in red) subsets, showed significantly increased infiltration in mucosal barrier tumors (p-value <0.001). Similar analysis conducted on the TCC cohort (c), showed significantly increased eosinophil abundance in mucosal barrier tumors (p-value<0.001)
Age- or sex-specific differences in eosinophil infiltration
To assess age- and sex-specific patterns of eosinophil infiltration, eosinophil density was correlated to patient age (Supplementary Figure 2a-f) and associated with patient sex (Supplementary Figure 2g, Supplementary table 5). No significant correlation between eosinophil density and patient age was seen in colon (Supplementary Figure 2a), lung (Supplementary Figure 2b), uterine cervix (Supplementary Figure 2c), esophageal (Supplementary Figure 2d), ovarian (Supplementary Figure 2e) and breast (Supplementary figure 2f) tumors. Evaluation of sex-related differences was performed in colon, lung, and esophageal tumors (Supplementary Figure 2g). Only lung tumors showed significant sex differences (p-value = 0.048), with females (mean eosinophil density 314 ± 782) showing higher eosinophil infiltration than males (228 ± 492).
Association of eosinophil density with hormone receptors and HER2 expression
Considering the roles that hormone receptors (i.e. estrogen and progesterone receptors) and HER2 expression have on breast cancer prognosis and therapeutics,38 we evaluated eosinophil density in breast cancer with respect to the expression of these receptors. Stromal eosinophils were decreased (p-value = 0.009) in high estrogen receptor expression tumors, while eosinophil density was not associated with progesterone receptor or HER2 protein expression (Figure 5).
Figure 5.
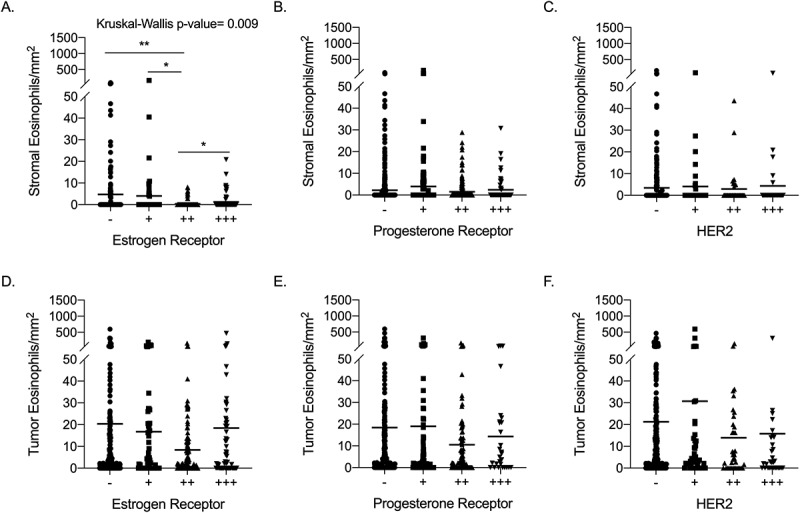
Association of eosinophil density with breast cancer hormone receptors and HER2 expression Eosinophil density in breast tumor stroma was evaluated in respect to estrogen receptor (a), progesterone receptor (b) and HER2 (c) expression using Kruskal-Wallis test followed by Mann-Whitney test for comparisons. Similarly, Intra-tumoral eosinophil density was evaluated in breast tumors in respect to estrogen receptor (d), progesterone receptor (e) and HER2 (f) expression. * p value <.05, ** p value < .01 of Mann-Whitney test
Association of eosinophils with tumor grade, size and clinical stage
Clinical and pathological evaluation of a given tumor involves multiple parameters including the tumor spread, cellular differentiation, and factors such as the TNM classification system, which considers the tumor size (T), involvement of regional lymph nodes (N), and metastasis (M). We were interested to determine whether eosinophil density correlated with any of the following parameters: a) tumor pathological grade; b) tumor stage; and c) tumor size in our sample cohort (Figure 6, Supplementary table 6).
Figure 6.
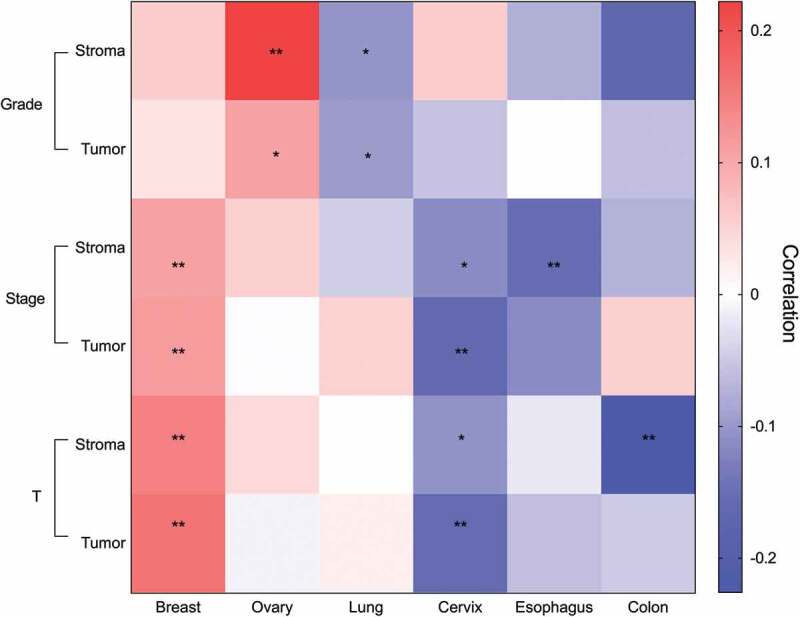
Unique clinical association patterns for eosinophils in distinct primary tumors. Heatmap describing eosinophil stromal and intra-tumoral density correlation with pathological grade, clinical stage and tumor size. Correlation coefficient presented in red to blue color, as indicated in the right color bar. Pathological grade was categorized as grade 1–3, clinical stage was categorized by stages 1–4, tumor size (termed T) was classified according to TNM staging system. * p value < .05, ** p value < .01
In colon tumors, stromal eosinophil density was inversely correlated to tumor size (p-value = 0.002) but not tumor grade or stage. In esophageal tumors, stromal eosinophil density was inversely correlated to tumor stage (p-value = 0.007) but not tumor grade or size. Inspection of cervical tumors revealed significant inverse correlation between stromal and intra-tumoral eosinophil density in regard to tumor stage (stroma p-value = 0.024, intra-tumoral p-value = 0.001) and size (stroma p-value = 0.035, intra-tumoral p-value = 0.002). Nonetheless, no correlations were observed in eosinophil density in respect to tumor grade.
In contrast to cervical tumors, lung tumors revealed a significant inverse correlation in stromal (p-value = 0.01) and intra-tumoral (p-value = 0.016) eosinophils with tumor pathological grade, while eosinophil density was not correlated with lung tumor stage or size.
Review of ovarian tumors revealed that stromal (p-value = 0.001) and intra-tumoral (p-value = 0.025) eosinophil density was significantly positively correlated with tumor grade. No significant correlation was observed between eosinophil density and ovarian tumor stage and size. Finally, breast tumors displayed significantly positive correlation between stromal and intra-tumoral eosinophil density in regard to tumor stage (stroma p-value = 0.009, intra-tumoral p-value = 0.007) and size (stroma p-value = 0.001, intra-tumoral p-value = 0.001). Eosinophil density in breast tumors was not associated with pathological grade.
Collectively, these data demonstrate unique clinical association patterns for eosinophils in distinct primary tumors.
Correlation between eosinophils and other immune cells in the TME
In the TME, eosinophils have been attributed with immunomodulatory functions since they can interact with various cell types including T cells,39,40 NK cells41,42 and macrophages.39,43 To provide insights into the potential cross-talk between eosinophils and additional immune cell in a given TME, we utilized the TCC database to assess the correlation between the abundance of eosinophils with additional immune cells including CD8+, CD4+ T cells, NK cells and macrophages in a given TME (Figure 7 and Supplementary table 7). A positive and significant correlation was identified between eosinophil abundance and resting memory CD4+ T cells in 11 of 22 tumor types. In contrast, a significant inverse correlation was identified in 9 of 22 tumor types between eosinophil abundance and CD8+ T cells. Associations between eosinophils and additional immune cells were diverse and displayed distinct correlation patterns depending on the specific type of tumor assessed (Figure 7).
Figure 7.
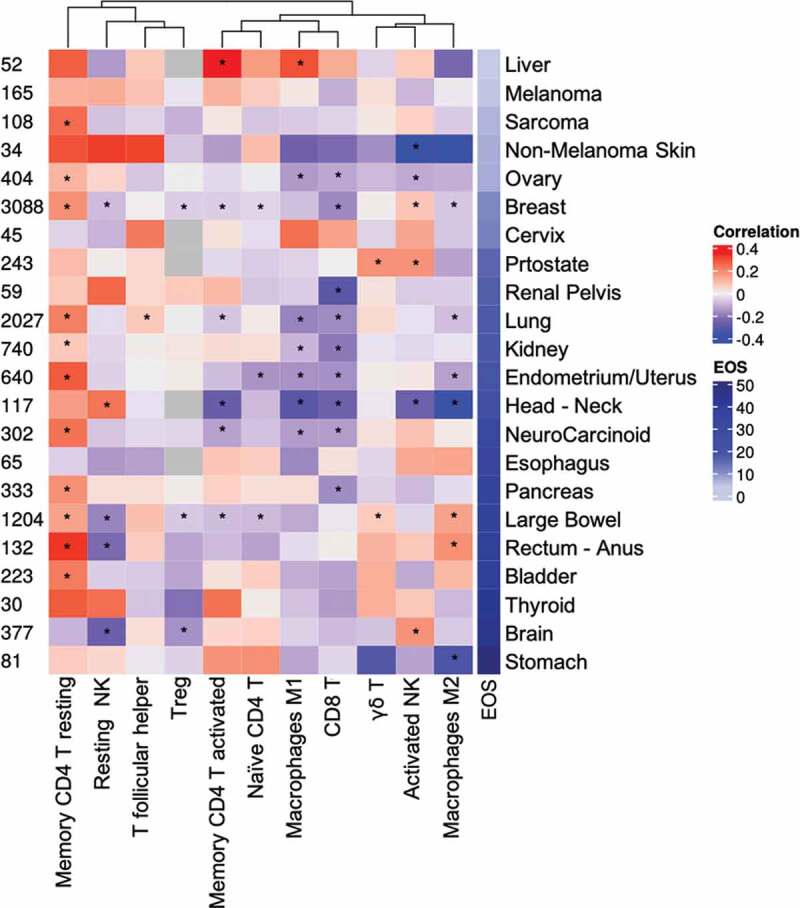
Eosinophil correlation with immune cell populations in the tumor microenvironment. A Heatmap of correlation coefficients between eosinophil estimates and immune cell types for the TCC database is shown. For each cancer type, the Spearman correlation of normalized content abundance (NCA) eosinophil levels and corresponding immune cell population was measured. The mean eosinophil estimate per cancer type is shown at the right and sample numbers at the left. * p value< .05
Discussion
Eosinophils have been primarily studied in the context of allergic inflammatory diseases.1 Nonetheless, numerous clinical and experimental studies have shown that they can infiltrate various tumors.3 Despite the fact that the eosinophils infiltrate multiple tumors, several caveats exist, which limit our interpretation of previous data. For example, the fact that there is no standardized method for eosinophil detection and quantitation prevents the ability to compile data from different experiments and to accurately define the level of eosinophil density within a given TME.37,44 In this study, we provide a comprehensive high-throughput evaluation of eosinophils in various TMEs combining bioinformatics and standardized pathological methods. We demonstrate that the abundance, localization, and clinical association of tumor-infiltrating eosinophils are primarily dictated by the anatomical site of the tumor with a clear distinction between mucosal barrier and non-mucosal barrier organs. Herein, we used an unbiased bioinformatics approach followed by corroborating experimental data. First, we analyzed 10,469 primary tumors in the TCC database, which represent 22 tumor types, by employing the CIBERSORT algorithm to infer the presence of eosinophils in tumors. This method of quantification was able to categorize eosinophil distribution within different tumors into three groups representing tumors with high, intermediate, and low eosinophil levels. Next, using an experimental approach, we analyzed the presence of eosinophils in independent tumor tissue arrays of the representative tumor types from each derived classification using standardized immunohistochemical staining methods. The algorithm predictions were partially recapitulated and highlighted the presence of eosinophils in mucosal barrier tumors. The differences between the bioinformatics and experimental approach may be explained by the fact that our staining was conducted on a different patient cohort from the one which was analyzed in the TCC database. Nonetheless, since immunohistochemical staining identifies eosinophils more accurately than the use of RNA-sequencing signature, such signatures can be only used as hypothesis-generating tools rather than definitive quantification. Collectively, these data suggest that specific environmental cues provide distinct signals for eosinophil recruitment and likely dictate the different activities eosinophils display in different TMEs.
One of the major outcomes of our study is the standardized side-by-side comparison of tissue eosinophils in multiple specimens from different tumor types. Using artificial intelligence, we mapped the localization of eosinophils within the different tumors and assessed the density of eosinophils in the stromal vs. intra-tumoral compartments. We demonstrate that tumor-infiltrating eosinophils were predominantly present in the tumor stroma with the exception of breast cancer where intra-tumoral eosinophilia was more prominent than stromal eosinophils. These findings are consistent with previous studies observing prominent stromal eosinophilia in bladder and colorectal cancer.5,8 Interestingly, employing the CART algorithm on our stained tumor microarrays revealed that the tumor anatomical site was the primary predictive variable for the stromal density of eosinophils. This analysis further demonstrated that mucosal barrier tumors differ from non-mucosal barrier tumors in stromal eosinophil density. Indeed, mucosal barrier tumors were characterized by increased stromal eosinophilia. This finding is likely explained by the fact that tissues with mucosal barriers are natural niches for eosinophil migration under homeostatic conditions and in various diseases.45 For example, the gastrointestinal tract is the largest reservoir of eosinophils under baseline conditions and eosinophils migrate to the gastrointestinal tract in allergic GI disorders and colitis under the direction of the eotaxin family of chemokines.2,46 In addition, eosinophils are resident cells in the lungs47 and their levels may be markedly increased in subsets of patients with asthma45 or even chronic obstructive pulmonary disease.48 We speculate that the difference in eosinophil density between mucosal barrier and non-mucosal barrier tumors is due to the ability of mucosal cells, such as epithelial cells or myeloid cells, to secrete eosinophil chemotactic factors such as CCL11, CCL24, and/or CCL26.23,45,49–51 Nonetheless, it is likely that additional factors facilitate eosinophil homing to these sites. Moreover, it is possible that lack of exposure to appropriate survival cytokines such as IL-5 in a given TME regulates eosinophil levels as well.52
The differential localization of eosinophils in distinct tumors resulted in unique clinical associations. This could be exemplified by comparing the clinical association of eosinophils in colon versus breast tumors, where eosinophils were primarily located in the stromal vs. intra-tumoral compartments, respectively. In colon tumors, stromal eosinophil density was inversely correlated with tumor size. Unfortunately, the cohorts used for this study do not have applicable survival data. Thus, survival analyses should be performed to associate the effect on patient outcome. These findings are consistent with previous studies showing a high density of eosinophils in the stroma of colon tumors; in those studies, eosinophilia was associated with a favorable outcome.8,53 One suggested mechanism for the eosinophil effect on colorectal tumors may be direct cytotoxic activities of eosinophils toward colon tumor cells. In fact, direct eosinophil-mediated cytotoxicity has been observed in co-culture studies of mouse or human eosinophils with colorectal cancer cells (MC38, CT26, SW480, Caco-2, Colo-205).51,54 In addition, mouse models of colorectal cancer demonstrated eosinophil recruitment, prolonged survival, and degranulation in the TME. Eosinophil-deficient mice had increased tumor burden compared to wild-type mice and cell depletion experiments indicated that eosinophil anti-tumorigenic activities were independent of CD8+ T cells.51 We have previously shown that in colorectal cancer, eosinophil tumor infiltration is inversely correlated with tumor stage,51 whereas no correlation was identified in this study. The difference between these data is likely explained by the fact that in our previous study the stained biopsies were divided into four different groups based on the numbers of intra-tumoral eosinophils and we did not account for the localization of eosinophils (i.e. intra-tumoral versus stromal localization). Thus, the comparative statistical analysis in our previous study was conducted only within the different groups of colorectal cancer patients and with no consideration of eosinophil localization. In this study, we compared multiple tumor types side by side and therefore did not use the aforementioned scoring system.
In contrast to colon cancer, our study revealed that eosinophil density in breast cancer was positively correlated with clinical stage and tumor size. These data suggest tumor-promoting activities for eosinophils in primary breast cancer. Moreover, stromal eosinophils were decreased in high estrogen receptor expression tumors. A proposed mechanism for the pro-tumorigenic activities of eosinophils in primary breast cancer was described using the 4T1 syngeneic murine orthotopic breast cancer model.55 Eosinophil peroxidase increased mammary tumor growth and enhanced lung metastases.55 Furthermore, in-vitro, peroxidase treatment stimulated robust migration of human mammary fibroblasts by inducing transcription of pro-tumorigenic and metastatic MMP1, MMP3, and COX-2 genes.55 In support of a tumor-promoting role for eosinophils in primary breast tumors, previous epidemiological studies suggested a link between increased expression of IL-5, a key eosinophil survival, and priming factor,56 in breast carcinomas and higher rates of distant metastasis and recurrence.57 Additionally, evaluation of human breast cancer revealed that a high proportion of tumors contained extensive, occult deposition of eosinophil peroxidase.58 Collectively, we set forth the notion that the opposing roles of eosinophils in distinct tumors is a result of functional heterogeneity determined by the physiological microenvironment as opposed to technical variance in detection methods.
One possible mechanism of action for eosinophils in the TME is by interacting with diverse cells of the immune system. To define potential interactions between eosinophils and additional immune cells we conducted a correlation analysis between eosinophils, T cell subsets, NK cells, and polarized macrophages using the TCC database. This analysis revealed a positive correlation between eosinophils and memory resting CD4+ T cells and an inverse correlation with CD8+ T-cells across multiple tumor types. Recent data demonstrated an active cross-talk exists between eosinophils and CD8+ T cells, where eosinophils promote the influx of CD8+ T cells into the TME, especially following immunotherapy.39,59 Thus, the finding of an inverse correlation between eosinophils and CD8+ T cells is of great interest. Since the associations between eosinophils and CD8+ T cells were observed primarily in settings of immunotherapy regimes or depletion of T regulatory cells. We hypothesize that under different non-immunosuppressed conditions, eosinophils may still associate with CD8+ T cells. Further studies should be done regarding this potential crosstalk.
The use of tissue microarrays enabled the high-throughput uniform detection of eosinophils in 2,890 tumor samples, thus reducing technical variability and empowering the conduction of more rigorous research. Nonetheless, several limitations should be considered in the interpretation of our study results. First, eosinophil density in the examined fraction of each analyzed tumor may not adequately represent the heterogenous infiltrate in the whole tumor area. Despite this limitation, previous studies demonstrated strong correlations between tumor microarray histospots and whole-tissue sections.60,61 Second, degranulation of eosinophils may play important roles in the TME.51,62,63 Although eosinophil degranulation was observed in multiple specimens, our study did not assess the extent of tissue degranulation and therefore we cannot conclude regarding the “activation status” of eosinophils and any clinical or pathological outcome. Moreover, since the quantitation of eosinophils was based on a positive intra-cellular DAB stain, only intact eosinophils were counted thus, introducing a potential bias in areas of extensive degranulation. Currently, the available methodologies, which are used to evaluate eosinophil degranulation in the field of cancer,8,59 are based on pathologist observations such as the scoring originally developed for eosinophilic esophagitis.33 This type of analysis cannot be scaled up to the number of slides/samples, which have been assessed in our study. Further development is required to provide an automated unbiased quantification method of eosinophil tissue degranulation.
In summary, our data contribute to the spatial characterization of eosinophils in the TME and set forward the use of artificial intelligence-based methods to dissect their tissue quantitation and localization. Furthermore, our data highlight the involvement of eosinophils in tumors from mucosal barrier sites, which should be further explored in experimental settings. Finally, our data suggest that differential infiltration of eosinophils into anatomically distinct tumors contributes to their distinct activities in each TME. We believe that subsequent studies including our own will utilize this method and findings, to focus on gaining mechanistic insights regarding the roles of eosinophils in primary and metastatic tumors in mucosal barrier organs.
Supplementary Material
Acknowledgments
A.M. wishes to thank the Munitz lab for their excellent input and discussions over the past years.
Funding Statement
A.M. is supported by the US-Israel Bi-national Science Foundation (grant no. 2015163), by the Israel Science Foundation (grant no. 886/15), the Israel Cancer Research Fund, the Israel Cancer Association Avraham Rotstein Donation, the Cancer Biology Research Center, Tel Aviv University and the Emerson Collective, the Israel Science Foundation (grant no.542/20).
Conflicts of Interest
The authors declare no conflict of interest.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website
References
- 1.Rosenberg HF, Dyer KD, Foster PS.. Eosinophils: changing perspectives in health and disease. Nat Rev Immunol. 2013;13:9–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Piazuelo, M. B. & Correa, P. in Eosinophils: Structure, Biological Properties and Role in Disease 73–96 (Nova Science Publishers, Inc., 2012)
- 3.Grisaru-Tal S, Itan M, Klion AD, Munitz A. A new dawn for eosinophils in the tumour microenvironment. Nat Rev Cancer. 2020;2:594–607. [DOI] [PubMed] [Google Scholar]
- 4.Spencer PFW. Functions of tissue-resident eosinophils. Nat Rev Immunol. 2017;17:746–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Popov H, I S Donev, Ghenev P (September 10, 2018) Quantitative Analysis of Tumor-associated Tissue Eosinophilia in Recurring Bladder Cancer. Cureus 10(9): e3279. doi: 10.7759/cureus.3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iwasaki K, Torisu M, Fujimura T. Malignant tumor and eosinophils - prognostic significance in gastric cancer. Cancer. 1986;58:1321–1327. [DOI] [PubMed] [Google Scholar]
- 7.Songun I, Van De Velde CJH, Hermans J, Pals ST, Verspaget HW, Vis AN, Menon AG, Litvinov SV, Van Krieken JHJM. Expression of oncoproteins and the amount of eosinophilic and lymphocytic infiltrates can be used as prognostic factors in gastric cancer. Br J Cancer. 1996;74:1783–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prizment AE, Vierkant RA, Smyrk TC, Tillmans LS, Lee JJ, Sriramarao P, Nelson HH, Lynch CF, Thibodeau SN, Church TR, et al. Tumor eosinophil infiltration and improved survival of colorectal cancer patients: Iowa Women's Health Study. Mod Pathol. 2016. May;29(5):516–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pretlow TP, Keith EF, Cryar AK, Bartolucci AA, Pitts AM, Pretlow TG 2nd, Kimball PM, Boohaker EA. Eosinophil infiltration of human colonic carcinomas as a prognostic indicator. Cancer Res. 1983. Jun;43(6):2997–3000. [PubMed] [Google Scholar]
- 10.Fernández-Aceñero MJ, Galindo-Gallego M, Sanz J, Aljama A. Prognostic influence of tumor-associated eosinophilic infiltrate in colorectal carcinoma. Cancer. 2000;88:1544–1548. [PubMed] [Google Scholar]
- 11.Zhang Y, Ren H, Wang L, Ning Z, Zhuang Y, Gan J, Chen S, Zhou D, Zhu H, Tan D, et al. (2014). Clinical impact of tumor-infiltrating inflammatory cells in primary small cell esophageal carcinoma. International journal of molecular sciences, 15(6), 9718–9734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyazaki S, Prefectural I, Hospital C, Moriya T, Satomi S. Tumor-associated tissue eosinophilia in human esophageal squamous cell carcinoma. (2006). [PubMed]
- 13.Said M, Wiseman S, Yang J, Alrawi S, Douglas W, Cheney R, Hicks W, Rigual N, Loree T, Spiegel G, et al. Tissue eosinophilia: a morphologic marker for assessing stromal invasion in laryngeal squamous neoplasms. BMC Clin Pathol. 2005. Jan 7;5(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dorta RG, Landman G, Kowalski LP, Lauris JR, Latorre MR, Oliveira DT. Tumour-associated tissue eosinophilia as a prognostic factor in oral squamous cell carcinomas. Histopathology. 2002. Aug;41(2):152–157. [DOI] [PubMed] [Google Scholar]
- 15.Tataroǧlu C, Kargi A, Özkal S, Eşrefoǧlu N, Akkoçlu A. Association of macrophages, mast cells and eosinophil leukocytes with angiogenesis and tumor stage in non-small cell lung carcinomas (NSCLC). Lung Cancer. 2004;43:47–54. [DOI] [PubMed] [Google Scholar]
- 16.Steel JL, Kim KH, Dew MA, Unruh ML, Antoni MH, Olek MC, Geller DA, Carr BI, Butterfield LH, Gamblin TC. Cancer-related symptom clusters, eosinophils, and survival in hepatobiliary cancer: an exploratory study. Journal of pain and symptom management, 2010;39(5), 859–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ownby EH, Roi DL, Isenberg RR, Bernnan JM. Peripheral lymphocyte and eosinophil counts as indicators of prognosis in primary breast cancer. Cancer. 1983;52:126–130. [DOI] [PubMed] [Google Scholar]
- 18.Onesti CE, Josse C, Poncin A, Frères P. Predictive and prognostic role of peripheral blood eosinophil count in triple-negative and hormone receptor-negative/HER2-positive breast cancer patients undergoing neoadjuvant treatment. 9, 33719–33733 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bishara S, Griffin M, Cargill A, Bali A, Gore ME, Kaye SB, Shepherd JH, Van Trappen PO. Pre-treatment white blood cell subtypes as prognostic indicators in ovarian cancer. Eur J Obstet Gynecol Reprod Biol. 2008. May;138(1):71–5. [DOI] [PubMed] [Google Scholar]
- 20.Spiegel GW, Ph D, Ashraf M, Brooks JJS Eosinophils as a marker for invasion in cervical squamous neoplastic lesions. 2002;117–124 [DOI] [PubMed]
- 21.van Driel WJ, Hogendoorn PC, Jansen FW, Zwinderman AH, Trimbos JB, Fleuren GJ. Tumor-associated eosinophilic infiltrate of cervical cancer is indicative for a less effective immune response. Hum Pathol. 1996. Sep;27(9):904–911. [DOI] [PubMed] [Google Scholar]
- 22.Stathopoulos GT, Sherrill TP, Karabela SP, Goleniewska K, Kalomenidis I, Roussos C, Fingleton B, Yull FE, Peebles RS Jr, Blackwell TS. Host-derived interleukin-5 promotes adenocarcinoma-induced malignant pleural effusion. Am J Respir Crit Care Med. 2010;182:1273–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reichman H, Karo-atar D, Munitz A. Emerging roles for eosinophils in the tumor microenvironment. Trends Cancer. 2016;2:664–675. [DOI] [PubMed] [Google Scholar]
- 24.Chen B, Khodadoust MS, Liu CL, Newman AM. Profiling tumor infiltrating immune cells with CIBERSORT Binbin. Methods Mol Biol. 2018;1711:243–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jain M, Kasetty, S., Sudheendra, U. S., Tijare, M., Khan, S., & Desai, A. Assessment of tissue eosinophilia as a prognosticator in oral epithelial dysplasia and oral squamous cell carcinoma - An image analysis study. Patholog Res Int. 2014;2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elnblad G, Sundstrom C. Infiltration of eosinophils in Hodgkin’s disease involved lymph nodes predicts prognosis. Hematol Oncol. 1993;11:187–193. [DOI] [PubMed] [Google Scholar]
- 27.Von Wasielewski R, Seth S, Franklin J, Fischer R, Hübner K, Hansmann ML, Diehl V, Georgii A. Tissue eosinophilia correlates strongly with poor prognosis in nodular sclerosing Hodgkin’s disease, allowing for known prognostic factors. Blood. 2000;95:1207–1213. [PubMed] [Google Scholar]
- 28.Fenstermacher DA, Wenham RM, Rollison DE, Dalton WS. Implementing personalized medicine in a cancer center. Cancer J. 2011;17:528–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Welsh EA, Eschrich SA, Berglund AE, Fenstermacher DA. Iterative rank-order normalization of gene expression microarray data. BMC Bioinform. 2013;14:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW, Levine DA et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newman A, AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen J. Statistical power analysis for the behavioural sciences. second. Hillsdale (New Jersey): Lawrence Earlbaum Associates; 1988. [Google Scholar]
- 33.Protheroe C, Woodruff SA, de Petris G, Mukkada V, Ochkur SI, Janarthanan S, Lewis JC, Pasha S, Lunsford T, Harris L, et al. A novel histological scoring system to evaluate mucosal biopsies from patients with eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2009;7:749–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, McQuaid S, Gray RT, Murray LJ, Coleman HG, et al. QuPath: open source software for digital pathology image analysis. Sci Rep. 2017;7:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Varricchi G, Galdiero MR, Loffredo S, Lucarini V, Marone G, Mattei F, Marone G, & Schiavoni G. Eosinophils: the unsung heroes in cancer? Oncoimmunol. 2017;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Breiman L, Friedman J, Olshen RA, Stone CJ. Classification And Regression Trees (1st ed.). Classification and regression trees. Wadsworth Int Gr. (1st ed.) 1984. [Google Scholar]
- 37.Willetts L, Parker K, Wesselius LJ, Protheroe CA, Jaben E, Graziano P, Moqbel R, Leslie KO, Lee NA, Lee, JJ. Immunodetection of occult eosinophils in lung tissue biopsies may help predict survival in acute lung injury. Respir Res. 2011;12:8–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Onitilo AA, Engel JM, Greenlee RT, Mukesh BN. Breast cancer subtypes based on ER/PR and Her2 expression: comparison of clinicopathologic features and survival. Clin Med Res. 2009;7:4–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carretero R, Sektioglu IM, Garbi N, Salgado OC, Beckhove P, Hämmerling GJ. Eosinophils orchestrate cancer rejection by normalizing tumor vessels and enhancing infiltration of CD8 + T cells. Nat Immunol. 2015;16:609–617. [DOI] [PubMed] [Google Scholar]
- 40.Odemuyiwa SO, Ghahary A, Li Y, Puttagunta L, Lee JE, Musat-Marcu S, Ghahary A, Moqbel R. Cutting edge: human eosinophils regulate t cell subset selection through indoleamine 2,3-dioxygenase. J Immunol. 2004;173:5909–5913. [DOI] [PubMed] [Google Scholar]
- 41.Schuijs MJ, Png S, Richard AC, Tsyben A, Hamm G, Stockis J, Garcia C, Pinaud S, Nicholls A, Ros XR, et al. ILC2-driven innate immune checkpoint mechanism antagonizes NK cell antimetastatic function in the lung. Nat Immunol. 2020;21:998–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pesce S, Thoren FB, Cantoni C, Prato C, Moretta L, Moretta A, Marcenaro E. The innate immune cross talk between NK cells and eosinophils is regulated by the interaction of natural cytotoxicity receptors with eosinophil surface ligands. Front Immunol. 2017;8:510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kratochvill F, Neale G, Haverkamp JM, Van de Velde LA, Smith AM, Kawauchi D, McEvoy J, Roussel MF, Dyer MA, Qualls JE, et al. TNF counterbalances the emergence of M2 tumor macrophages. Cell Rep. 2015;12:1902–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meyerholz DK, Griffin MA, Castilow EM, Varga SM. Comparison of histochemical methods for murine eosinophil detection in a RSV vaccine-enhanced inflammation model. Toxicol Pathol. 2009;37:249–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Travers J, Rothenberg ME. Eosinophils in mucosal immune responses. Mucosal Immunol. 2015;8:464–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waddel A, Ahrens R, Steinbrecher K, Donovan B, Rothenberg ME, Munitz A. Colonic eosinophilic inflammation in experimental colitis is mediated by Ly6Chigh CCR2+ inflammatory monocyte/macrophage-derived CCL11. J Immunol. 2011;23:5993–6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mesnil C, Raulier S, Paulissen G, Xiao X, Birrell MA, Pirottin D, Janss T, Starkl P, Ramery E, Henket M, et al. Lung-resident eosinophils represent a distinct regulatory eosinophil subset Find the latest version: lung-resident eosinophils represent a distinct regulatory eosinophil subset. J Clin Invest. 2016;126:3279–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brightling CE, Monteiro W, Ward R, Parker D, Morgan MD, Wardlaw AJ, Pavord ID. Sputum eosinophilia and short-term response to prednisolone in chronic obstructive pulmonary disease: A randomised controlled trial. Lancet. 2000;356:1480–1485. [DOI] [PubMed] [Google Scholar]
- 49.Cho H, Lim SJ, Won KY, Bae GE, Kim GY, Min JW, Noh BJ. Eosinophils in colorectal neoplasms associated with expression of CCL11 and CCL24. J Pathol Transl Med. 2016;50:45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lorena SCM, Oliveira DT, Dorta RG, Landman G, Kowalski LP. Eotaxin expression in oral squamous cell carcinomas with and without tumour associated tissue eosinophilia. Oral Dis. 2003;9:279–283. [DOI] [PubMed] [Google Scholar]
- 51.Reichman H, Itan M, Rozenberg P, Yarmolovski T, Brazowski E, Varol C, Gluck N, Shapira S, Arber N, Qimron U, et al. Activated eosinophils exert antitumorigenic activities in colorectal cancer. Cancer Immunol Res. 2019. doi: 10.1158/2326-6066.CIR-18-0494. [DOI] [PubMed] [Google Scholar]
- 52.Steven J, Ackerman BSB. Mechanisms of eosinophilia in the pathogenesis of hypereosinophilic disorders. Immunol Allergy Clin North Am. 2007;27:257–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lau MC Lau MC, Haruki K, Väyrynen SA, Dias Costa A, Borowsky J, Zhao M, Fujiyoshi K, Arima K, Twombly TS, et al. Prognostic significance of immune cell populations identified by machine learning in colorectal cancer using routine hematoxylin & eosin stained sections. (2020). doi: 10.1158/1078-0432.CCR-20-0071 [DOI] [PMC free article] [PubMed]
- 54.Gatault S, Delbeke M, Driss V, Sarazin A, Dendooven A, Kahn JE, Lefèvre G, Capron M. IL-18 is involved in eosinophil-mediated tumoricidal activity against a colon carcinoma cell line by upregulating LFA-1 and ICAM-1. J Immunol. 2015;195:2483–2492. [DOI] [PubMed] [Google Scholar]
- 55.Panagopoulos V, Leach DA, Zinonos I, Ponomarev V, Licari G, Liapis V, Ingman WV, Anderson P, DeNichilo MO, Evdokiou A. Inflammatory peroxidases promote breast cancer progression in mice via regulation of the tumour microenvironment. Int J Oncol. 2017;1191–1200. doi: 10.3892/ijo.2017.3883. [DOI] [PubMed] [Google Scholar]
- 56.Rothenberg ME, Hogan SP. The Eosinophil. Annu Rev Immunol. 2006;24:147–174. [DOI] [PubMed] [Google Scholar]
- 57.Eiró N, González L, González LO, Fernandez-Garcia B, Lamelas ML, Marín L, González-Reyes S, del Casar JM, Vizoso FJ. Relationship between the inflammatory molecular profile of breast carcinomas and distant metastasis development. PLoS One. 2012;7:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Samoszuk MK, Nguyen V, Gluzman I, Pham JH. Short communication occult deposition of eosinophil peroxidase in a subset of human breast carcinomas. 148, 701–706 (1996). [PMC free article] [PubMed] [Google Scholar]
- 59.Simon SCS, Hu X, Panten J, Grees M, Renders S, Thomas D, Weber R, Schulze TJ, Utikal J, Umansky V. Eosinophil accumulation predicts response to melanoma treatment with immune checkpoint inhibitors. Oncoimmunol. 2020;9:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Camp RL, Charette LA, Rimm DL. Validation of tissue microarray technology in breast carcinoma. 80, 1943–1950 (2000). [DOI] [PubMed] [Google Scholar]
- 61.Torhorst J Bucher C, Kononen J, Haas P, Zuber M, Köchli OR, Mross F, Dieterich H, Moch H, Mihatsch M, et al. Tissue microarrays for rapid linking of molecular changes to clinical endpoints. 159, 2249–2256 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Legrand F, Driss V, Delbeke M, Loiseau S, Hermann E, Dombrowicz D, Capron M. Human eosinophils exert TNF-α and granzyme a-mediated tumoricidal activity toward colon carcinoma cells. J Immunol. 2010;185:7443–7451. [DOI] [PubMed] [Google Scholar]
- 63.Andreone S, Spadaro F, Buccione C, Mancini J, Tinari A, Sestili P, Gambardella AR, Lucarini V, Ziccheddu G, Parolini I, et al. IL-33 promotes CD11b/CD18-mediated adhesion of eosinophils to cancer cells and synapse-polarized degranulation leading to tumor cell killing. Cancers (Basel). 2019;11:1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


