Abstract
Iris loczyi is a perennial rhizomatous herb distributed in Central Asia. We examined genomic architecture of the complete chloroplast genome in I. loczyi by assembling the Illumina MiSeq reads using de novo strategy. The chloroplast genome is 150,940 bp in length harboring 79 protein-coding genes, 30 tRNA genes, and four rRNA genes. It exhibits typical quadripartite structure comprising LSC (80,907 bp), SSC (17,853 bp), and a pair of IRs (26,090 bp). Phylogenetic analysis of 20 chloroplast genomes from Asparagales revealed that Iridaceae is a monophyletic group and the I. loczyi is clustered together with the congener, I. sanguinea.
Keywords: Iris loczyi, complete chloroplast genome, Iridaceae
Iris loczyi Kanitz (Iridaceae), is a rhizomatous perennial herb distributed in Central Asia including Afghanistan, Iran, Kazakhstan, Kyrgyzstan, Pakistan, Tadzhikistan, Uzbekistan (Khassanov and Rakhimova 2012). Like most plants in Iris L., I. loczyi is a well- recognized plant for its economical values in pharmaceutical and horticultural practices (Crisan and Cantor 2016). The plant has recently drawn much attention as it contains various secondary metabolites that might have the potential to manage diabetes (Mosihuzzman et al. 2013). However, the genomic information applicable for breeding program and other biological studies is scarce. In the present study, we investigated the genomic architecture in the whole chloroplast genome of I. loczyi using whole genome shotgun sequencing.
We collected young leaves of I. loczyi from Issyk kul, Kyrgyzstan (N42°47’0.8″, E77°31′41.9″). The voucher specimen was prepared and deposited at the Herbarium of Korea National Arboretum (KH) with the accession number KHB1544459. The total genomic DNA was extracted followed by manufacturer’s protocol (Quiagen, Hilden, Germany). After library preparation, the prepared libraries were sequenced on Illumina MiSeq platform (Illumina, San Diego, CA). Eight million high-quality 300 bp paired-end reads were obtained. We assembled 2.85 GB reads with de novo strategy using CLC Assembly Cell package (ver. 4.2.1) followed by Kim et al. (2015). The genes were predicted with GeSeq (Tillich et al. 2017) and manually curated based on Blast search result. The simple sequence repeats were investigated with MISA (Beier et al. 2017).
The complete chloroplast genome of I. loczyi (MT254070) is 150,940 bp in length with the typical quadripartite structure comprising LSC (80,907 bp), SSC (17,853 bp), and a pair of IRs (26,090 bp). The cp genome contained 113 genes including 79 protein-coding genes, 30 tRNA genes, and four rRNA genes. 463 simple sequence repeats were identified in the cp genome, most of which was penta-nucleotide.
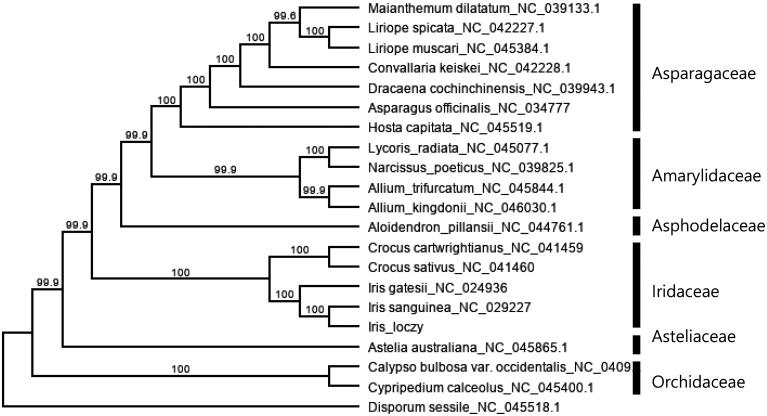
To investigate its phylogenetic relationship, the entire chloroplast genome sequences of 20 Asparagales taxa were aligned in MAFT (Katoh et al. 2019). All sequences other than I. loczyi were downloaded from NCBI Genebank. We assigned Disporum sessile D.Don (Colchicaceae) as an outgroup following phylogenetic relationships based on APG system (Stevens 2017). We inferred the phylogeny using Maximum-likelihood algorithm implemented in RAxML v. 4.0 with GTR GAMMA model. For the clade support, 1000 bootstrap replicates were used. The five species of Iridaceae formed a monophyletic group (BP = 100) with strong support on ML tree (Figure 1). In ML tree, Iridaceae grouped together with Asparagaceae, Amarylidaceae and Asphodelaceae, while Orchidaceae and Asteliaceae formed separate clades respectively. The ML tree also indicated that I. loczyi is closely related with I. sanguinea which is consistent with the previous subgeneric classification (Wilson 2004).
Figure 1.
Maximum-likelihood (ML) tree based on chloroplast genome sequences of 20 species of Asparagales, numbers on the nodes indicated the bootstrap support value (>50%).
Funding Statement
This work was supported by the grant ‘Central Asia Green Road Project II. Conservation of Plant Diversity and Ethnobotanical Research’ [KNA1-1-26, 20-1], financed by the Korea National Arboretum.
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The data that support the findings of this study are openly available in NCBI GenBank at https://www.ncbi.nlm.nih.gov/genbank/, accession number MT254070.
References
- Beier S, Thiel T, Münch T, Scholz U, Mascher M.. 2017. MISA-web: a web server for microsatellite prediction. Bioinformatics. 33(16):2583–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisan I, Cantor M.. 2016. New perspectives on medicinal properties and uses of Iris sp. Hop Med Plants. 24:24–36. [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD.. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinformatics. 20(4):1160–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khassanov FO, Rakhimova N.. 2012. Taxonomic revision of the genus Iris L. (Iridaceae Juss.) for the flora of Central Asia. Stapfia. 97:174–179. [Google Scholar]
- Kim K, Lee SC, Lee J, Lee HO, Joh HJ, Kim NH, Park HS, Yang TJ.. 2015. Comprehensive survey of genetic diversity in chloroplast genomes and 45S nrDNAs within Panax ginseng species. PLoS One. 10(6):e0117159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosihuzzman M, Naheed S, Hareem S, Talib S, Abbas G, Khan SN, Choudhary MI, Sener B, Tareen RB, Israr M.. 2013. Studies on α-glucosidase inhibition and anti-glycation potential of Iris loczyi and Iris unguicularis. Life Sci. 92(3):187–192. [DOI] [PubMed] [Google Scholar]
- Stevens PF. 2017. Angiosperm Phylogeny Website. Version 14.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S.. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CA. 2004. Phylogeny of Iris based on chloroplast matK gene and trnK intron sequence data. Mol Phylogenet Evol. 33(2):402–412. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are openly available in NCBI GenBank at https://www.ncbi.nlm.nih.gov/genbank/, accession number MT254070.