Abstract
Ligusticum sinense is a popular herb in Chinese medicine. The circular double-stranded complete chloroplast genome of L. sinense was 146,342 bp in length, exhibiting a typical quadripartite structure. It contained a large single-copy region (LSC) of 91,788 bp, a small single-copy region (SSC) of 17,618 bp and two identical inverted repeat (IR) regions of 18,468 bp each. The overall nucleotide composition of chloroplast genome sequence is: A (30.8%), T (31.6%), C (19.2%), G (19.4%) and the total G + C content of 38.6%. The chloroplast genome contained 127 genes, including 83 protein-coding genes, 36 transfer RNA genes and 8 ribosomal RNA genes were annotated. The total of 15 genes duplicated in one of the IR, including 6 tRNA, 4 rRNA, and 5 protein-coding genes. The ML phylogenetic tree indicated that L. sinense is closely related to L. tenuissimum in the phylogenetic relationship.
Keywords: Ligusticum sinense, phylogenetic analysis, chloroplast genome
Ligusticum sinense belongs to the Umbelliferae family that has been used to relieve toothache and treat other oral diseases as one of Chinese medicine in China (Wei et al. 2014). In China, its Chinese named Chuan-Xiong as the herb that was also used to flavor food and add fragrance to cosmetics because of its warm and spicy qualities (Wang et al. 2011). At present, the study of L. sinense is only on the chemical composition and the reports on genomics and molecular biology are rare, which affects the further research and scientific utilization of this species. In this study, we published the chloroplast genome of L. sinense, which can help to research the phylogenetic relationship information, also can be useful for traditional Chinese medicine utilization research for future.
The samples of L. sinense were obtained from the herb market near the Hunan University of Chinese Medicine (47.28 N, 112.90E) located at Changsha, Hunan, China, and also preserved in liquid nitrogen for further study. The total genomic DNA of L. sinense was stored in Hunan University of Chinese Medicine (No. HNUCM-01). Total genomic DNA of L. sinense was isolated using the Plant Tissues Genomic DNA Extraction Kit (TIANGEN Biotech., Beijing and China) and sequenced. Adapters and low-quality reads were removed and controlled using FastQC (Andrews 2015). The chloroplast genome of L. sinense was assembled using MitoZ (Meng et al. 2019) and annotated by Geneious 8.1.7 (Kearse et al. 2012). The genes in chloroplast genome were predicted using CPGAVAS (Liu et al. 2012) and corrected using DOGMA (Wyman et al. 2004).
The circular double-stranded complete chloroplast genome of L. sinense (GenBank Accession number: NK9214541) was 146,342 bp in length, exhibiting a typical quadripartite structure. It contained a large single-copy region (LSC) of 91,788 bp, a small single-copy region (SSC) of 17,618 bp, and two identical inverted repeat (IR) regions of 18,468 bp each. The overall nucleotide composition of chloroplast genome sequence is: A (30.8%), T (31.6%), C (19.2%), G (19.4%) and the total G + C content of 38.6%. The chloroplast genome contained 127 genes, including 83 protein-coding genes, 36 transfer RNA genes, and 8 ribosomal RNA genes were annotated. The total of 15 genes duplicated in one of the IR, including 6 tRNA, 4 rRNA, and 5 protein-coding genes.
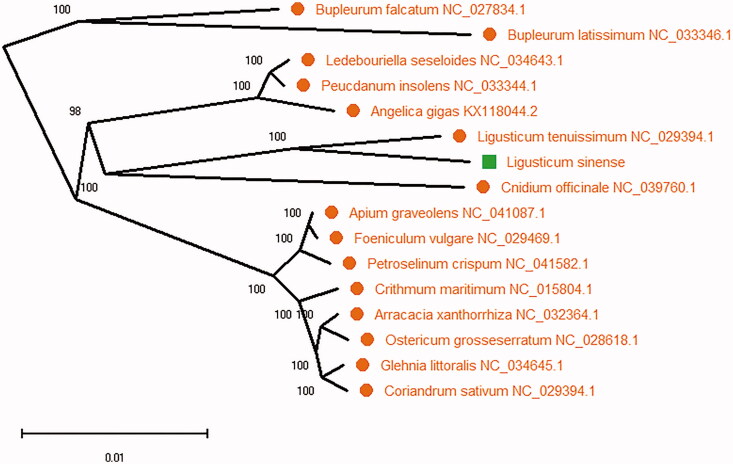
To confirm the phylogenetic relationship of L. sinense, 16 plant species complete chloroplast genome sequences were aligned using MAFFT (Katoh and Standley 2013) and maximum-likelihood (ML) analysis was conducted using MEGA X (Kumar et al. 2018) with 2000 bootstraps values at all the nodes under the substitution model. The phylogenetic tree was drawn and edited using MEGA X (Kumar et al. 2018). The ML phylogenetic tree (Figure 1) indicated that L. sinense is closely related to L. tenuissimum in the phylogenetic relationship (Figure 1). This complete chloroplast genome of L. sinense can provide useful information for phylogenetic studies and will be useful for traditional Chinese medicine utilization research for future.
Figure 1.
Phylogenetic maximum likelihood tree with L. sinense based on 16 plant species complete chloroplast genomes. The number on each node indicates bootstrap support value from 2000 replicates.
Disclosure statement
No potential conflict of interest was reported by the author(s). This study does not contain any studies with human participants or animals performed by any of the authors.
Data availability statement
The data that support the findings of this study are available from the corresponding author, upon reasonable request.
The data that support the findings of this study are openly available in Ligusticum sinense at NCBI, reference number [reference number NK9214541].
References
- Andrews S. 2015. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K.. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X.. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng GL, Li YY, Yang CT, Liu SL.. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. h, Xu L, Yang L, Liu ZL.. 2011. Composition, anti bacterial and antioxidant activities of essential oils from Ligusticum sinense and L. jeholense (Umbelliferae) from China. Rec Nat Prod. 5:314–318. [Google Scholar]
- Wei Q, Yang J, Ren J, Wang A, Ji T, Su Y.. 2014. Bioactive phthalides from Ligusticum sinense oliv cv. chaxiong. Fitoterapia. 93:226–232. [DOI] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL.. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, upon reasonable request.
The data that support the findings of this study are openly available in Ligusticum sinense at NCBI, reference number [reference number NK9214541].