Abstract
The complete mitochondrial genome of Anax parthenope was assembled from next-generation sequencing data. The circularized mitochondrial genome spans 15,306 bp with a high A + T content of 74.7% and consists of 13 protein-coding genes, 22 tRNA genes, and 2 rRNA genes. Its gene composition and order were similar to Anax imperator. The overall base composition is 36.6% for A, 34.8% for T, 16.8% for C, and 11.8% for G. A phylogeny of 31 dragonfly species clustered this species within Odonata.
Keywords: Aeshnidae, gene order, mitogenome, phylogeny
Odonata (Insecta: Pterygota) are a kind of ancient, medium and small winged insects, consisting of three suborders, namely Anisoptera, Zygoptera, and Anisozygoptera. It takes an important position in the evolution of winged insects and as sensitive indicator organisms for freshwater ecosystems. Currently, a total of 6322 species were recorded in the world (Schorr and Paulson 2020). However, in the recent 30 years, the increasing water pollution degraded the habitat quality and resulted in the loss of dragonfly biodiversity. Besides, species relationships within Odonata are poorly understood. Anax Leach in Brewster, 1815, a genus of family Aeshnidae, is a cosmopolitan genus of the 32 species which are distributed in the temperature and tropical zones (Schorr and Paulson 2020). To date, only a few mitochondrial and ribosomal sequences have been reported for Aeshnidae (NCBI Reference Sequence, accessed 2020 Apr 19). To explore its evolution in Odonata, we report the complete mitochondrial genome of Anax parthenope julis Brauer.
In this study, the voucher specimen of A. parthenope was collected from Nanjing, Jiangsu Province, China (31.580°N, 119.176°E; NCBI BioSample accession SAMN14733969; specimen voucher number 33m-3). Total genomic DNA was extracted from one leg of a single individual using the QIAamp DNA Micro Kit (Qiagen GmbH, Hilden, Germany) and sequenced by NovaSeq 6000. The voucher specimen and extracted DNA were preserved at −20 °C in the Nanjing Agricultural University, China. The mitogenome was assembled with NOVOPlasty v3.8.2 (Dierckxsens et al. 2017), annotated with MitoZ v2.4-alpha (Meng et al. 2019), and deposited in GenBank with the accession number MT408029.
The circularized mitochondrial genome spans 15,306 bp with a high A + T content of 74.7% and consists of 13 protein-coding genes, 22 tRNA genes, and two rRNA genes. Its gene composition and order were similar to Anax imperator (NC_031821.1). The overall base composition is 36.6% for A, 34.8% for T, 16.8% for C, and 11.8% for G, and the G-C skew is −11.15. Nine PCGs and 14 tRNA genes are located on the positive strand, while four PCGs (ND1, ND4, ND4L, and ND5), eight tRNA genes (tRNALeu, tRNAVal, tRNAGln, tRNACys, tRNATyr, tRNAPhe, tRNAHis, and tRNAPro), and two rRNA genes (l-rRNA and s-rRNA) are located on the reverse strand. For the 13 PCGs, 12 of them are terminated with TAA stop codon, while ND5 is ended with the incomplete TA stop codon.
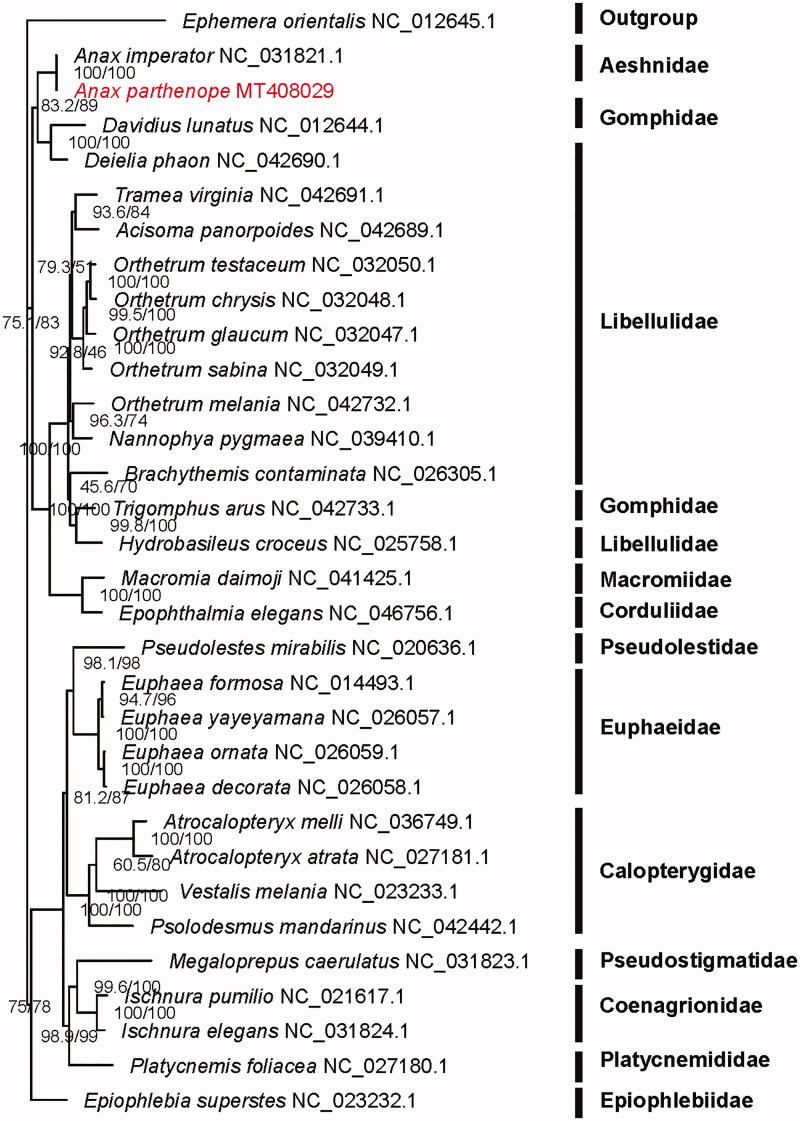
Amino acid sequences were aligned using MAFFT v7.407 (Katoh and Standley 2013) and trimmed with trimAl v1.4.1 (Capella-Gutiérrez et al. 2009) with the heuristic method ‘automated1’. The phylogeny was reconstructed using IQ-TREE v2.0 (Nguyen et al. 2015) with the partitioning method. Monophyly of Aeshnidae was recovered because A. parthenope was clustered within Aeshnidae (Figure 1).
Figure 1.
Maximum-likelihood phylogenetic tree inferred from 13 PCGs. SH-aLRT and UFBoot support values are given on nodes.
Funding Statement
This work was supported by the Programme of Biodiversity Survey, Monitoring and Assessment of Ministry of Ecology and Environment of China.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The data that support the findings of this study are openly available in NCBI GenBank database at https://www.ncbi.nlm.nih.gov with the accession number MT408029, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
References
- Schorr M, Paulson D.. 2020. World list of Odonata; [accessed 2020 Apr 20]. Available from: https://www.pugetsound.edu/academics/academic–resources/slater–museum/biodiversity–resources/dragonflies/world–odonata–list.
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldon T.. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G.. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng G, Li Y, Yang C, Liu S.. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are openly available in NCBI GenBank database at https://www.ncbi.nlm.nih.gov with the accession number MT408029, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.