Abstract
The COVID-19 pandemic caused by SARS-CoV-2 is responsible for the global health emergency. Here, we explore the diverse mechanisms of SARS-CoV-induced inflammation. We presume that SARS-CoV-2 likely contributes analogous inflammatory responses. Possible therapeutic mechanisms for reducing SARS-CoV-2-mediated inflammatory responses comprise FcR inactivation. Currently, there is no specific remedy available against the SARS-CoV-2. Consequently, recognizing efficacious antiviral leads to combat the virus is crucially desired. The coronavirus (CoV) main protease (Mpro also called 3CLpro), which plays an indispensable role in viral replication and transcription, is an interesting target for drug design. This review compiles the latest advances in biological and structural research, along with development of inhibitors targeting CoV Mpros. It is anticipated that inhibitors targeting CoV Mpros could be advanced into wide-spectrum antiviral drugs in case of COVID-19 and other CoV-related diseases. The crystal structural and docking results have shown that Ebselen, N3, TDZD-8 and α-ketoamide (13b) inhibitors can bind to the substrate-binding pocket of COVID-19 Mpro. α-ketoamide-based inhibitor 13b inhibits the replication of SARS-CoV-2 in human Calu3 lung cells. Quantitative real-time RT-PCR (qRT-PCR) showed that the treatment with Ebselen, TDZD-8 and N3 reduced the amounts of SARS-CoV-2, respectively, 20.3-, 10.19- and 8.4-fold compared to the treatment in the absence of inhibitor. Moreover, repurposing of already present drugs to treat COVID-19 serves as one of the competent and economic therapeutic strategies. Several anti-malarial, anti-HIV and anti-inflammatory drugs as mentioned in Table 2 were found effective for the COVID-19 treatment. Further, hydroxychloroquine (HCQ) was found more potent than chloroquine (CQ) in inhibiting SARS-CoV-2 in vitro. Furthermore, convalescent plasma from patients who have recuperated from viral infections can be employed as a therapy without the appearance of severe adverse events. Hence, it might be valuable to examine the safety and efficacy of convalescent plasma transfusion in SARS-CoV-2-infected patients.
Keywords: Inflammatory responses, SARS-CoV-2, COVID-19, Drug design, Drug repurposing
Introduction
A newly emerged pathogen, 2019 novel coronavirus (2019-nCoV), caused an outbreak of contagious respiratory disease in the last month of 2019 in Wuhan, Hubei Province, China [1–4]. On January 10, 2020, first full-length genome sequence of the virus was discharged, which helped in the quick identification of virus in patients by employing reverse transcription polymerase chain reaction (RT-PCR) [1]. On February 12, the World Health Organization (WHO) permanently named the virus as severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) and the disease as coronavirus disease 2019 (COVID-2019). Initially, the disease cluster was related to Huanan seafood market, probably because of animal proximity. Later on, human-to-human transmission was observed [5] and the disease rapidly spread globally [3, 4]. Owing to global expansion, the World Health Organization (WHO) proclaimed the outbreak an epidemic on March 11, 2020. The disease has spread to more than 100 countries, territories or areas with over 21,026,758 confirmed cases and over 755,786 confirmed deaths worldwide as of August 15, 2020 [6]. Coronaviruses (CoVs) can infect both humans and other animals, causing common cold to severe diseases, such as severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) [7]. However, bats which host a variety of coronaviruses were found resistant to coronavirus-induced disease [8]. The infected patients suffer from fever, fatigue, cough and dyspnea [9, 10]. Viruses are small obligate intracellular parasites which consist of nucleic acid (single- or double-stranded DNA or RNA) and a protein coat called capsid. The capsid protects the viral genome from nucleases and attaches the virion (a complete virus particle) to specific receptors exposed on the approaching host cell during infection. The capsid and its enclosed nucleic acid together constitute the nucleocapsid. The main purpose of the virion is to dispatch its RNA or DNA genome into the host cell so that the genome can be expressed (transcribed and translated) by the host cell. Though SARS-CoV-2 is categorized into the beta-coronaviruses cluster, it is distinct from MERS-CoV and SARS-CoV. Recent investigations demonstrated that SARS-CoV-2 genes contribute < 80% nucleotide identity and 89.10% nucleotide resemblance with SARS-CoV genes [3, 4]. CoVs are comparatively giant pleomorphic or spherical particles with bulbous surface extensions [11]. The diameter of the viral particle ranges from 80 to 120 nm [12]. CoVs possess a positive-sense single-stranded RNA genome enclosed within a membrane envelope. The viral envelope consists of a lipid bilayer where the structural proteins such as envelope (E), membrane (M) and spike (S) are anchored [13]. The spike protein embedded on the viral membrane gives them crown-like appearance (Fig. 1a). The size of genome ranges from roughly 27 to 34 kb, the largest amongst known RNA viruses [14].
Fig. 1.
a Coronavirus virion structure showing the presence of membrane (M), envelope (E), nucleocapsid (N) and spike (S) proteins (Reprinted with permission from Ref. [15], Copyright 2012 MDPI); b transmission electron microscope image of SARS-CoV-2, showing spikes on the outer edge of the virus; c scanning electron microscope image of SARS-CoV-2 (Yellow), exhibiting spherical or pleomorphic shape. Reprinted from Ref. [16], Credit: NIAID-RML
The SARS-CoV-2 genome is composed of ~ 30,000 nucleotides. Its replicase gene encodes pp1a and pp1ab, overlapping polyproteins that are essential for viral replication and transcription [3, 4]. Unlike SARS-CoV and MERS-CoV, the SARS-CoV-2 also influences gastrointestinal system, liver, kidney, heart and central nervous system along with the lower respiratory system, leading to multiple organ failure [1, 17]. Moreover, the recent research demonstrated higher infectious nature of SARSCoV-2 than SARS-CoV [18]. The S protein of the coronaviruses facilitates its entry into target cells. The entry of virus relies on binding of the S protein to a cellular receptor, which expedites viral attachment to the surface of target cells. The entry also entails S protein priming by cellular proteases, that entail S protein cleavage and permit fusion of viral and cellular membranes. SARS-S employs angiotensin-converting enzyme 2 (ACE2) as the entrance receptor [19] and engages the cellular serine protease TMPRSS2 for S protein priming [20–22]. SARS-S and SARS-2-S exhibit ~ 76% amino acid identity. However, it is anonymous whether SARS-2-S like SARS-S engages ACE2 and TMPRSS2 for host cell entry.
Several non-structural proteins such as main protease (Mpro also known as 3CLpro), papain-like protease (PLpro) and RNA-dependent RNA polymerase (RdRp) are also encoded by the viral genome [23, 24]. After reaching the host cells, the viral genome is discharged as a single-stranded positive RNA. Later on, it is translated into viral polyproteins employing host cell protein translation machinery, which are then divided into effector proteins by viral Mpro and PLpro [23, 24]. PLpro also acts as a deubiquitinase which may deubiquinate definite host cell proteins, such as interferon factor 3 and NF-κB, causing immune repression [24, 25]. The interaction between viral S protein and ACE2 on the host cell surface is of significant interest as it begins the infection process. Cryo-EM results showed 10–20 times higher binding affinity of SARS-CoV-2 S protein to ACE2 compared to SARS-CoV S protein [26, 27]. Alike SARS-CoV infection, SARS-CoV-2 disease also results in enlarged secretion of IL-1b, MCP-1, IFN-c, IP-10, IL-4 and IL-10 [9]. The anticipation also persists for discovery of therapeutic leads targeting the highly preserved proteins linked with both SARS-CoV and SARS-CoV-2. RdRp and Mpro of SARS-CoV-2 experience over 95% of sequence homology with those of SARS-CoV regardless of only 79% sequence homology at the genome level [27–30]. On the behalf of array arrangement and similar design, SARS-CoV and SARS-CoV-2 contribute to a huge protected receptor-binding domain (RBD), a domain of S protein, and 76% of array resemblance in their S proteins [27–30]. Moreover, the PLpro sequences of SARS-CoV-2 and SARS-CoV are only 83% similar; still, they share similar active sites [29]. So far, there have been no SARS-CoV-2-specified antiviral drugs. Previously, structure-based activity inspection and high-throughput investigations have described potential inhibitors for MERS-CoV and SARS-CoV 3CLpro [31–33]. Medicinal plants have attracted remarkable attention for developing formal drugs against several diseases with no or minimal side effects [34]. Recently, Qamar et al. acquired structural insight of SARS-CoV-2 Mpro and designed dominant anti-COVID-19 natural compounds [35].
In the first section of the review, we explore feasible mechanisms of SARS-CoV-2-mediated inflammatory responses and potential therapeutic interventions to overcome these responses. In the second section, we explore structure-assisted drug design strategies and inhibitors to target COVID-19 Mpro. In the last section, we explore potential drug targets and anti-COVID-19 drugs (by drug repurposing).
Mechanisms of COVID-19-mediated inflammatory responses
At present, there is no effective antiviral remedy for the newly emerged SARS-CoV-2, which is regularly causing fatal inflammatory responses and acute lung injury. Owing to similarity in inflammatory responses of COVID-19 and SARS-CoV infection, here, we explore the diverse mechanisms of SARS-CoV-mediated inflammation.
Inflammation led by rapid viral replication and cellular damage
The previous investigations have demonstrated that the SARS-CoV not only infects macrophages, vascular endothelial cells and alveolar epithelial cells, but also lymphocytes and monocytes [36]. According to the latest reports, both SARS-CoV-2 and SARS-CoV use angiotensin-converting enzyme 2 (ACE2) as the entry receptor for infection which recommends the possibility of the same type of cells being targeted and infected by the duo [3, 37]. According to Ming Yang, the early and quick viral replication may cause endothelial and epithelial cell apoptosis and vascular leakage, which in turn may cause the release of cytokines and chemokines [38]. Besides, SARS-CoV-2 infection may also result in pyroptosis of lymphocytes [38]. According to Guan et al., ~ 82% patients undergo peripheral blood lymphopenia due to COVID-19 [39], indicating viable pulmonary infiltration of lymphocytes via pyroptosis [9].
Inflammation led by virus-induced ACE2 downregulation and shedding
Foregoing studies showed ACE2-related lung injury in SARS-CoV disease [40, 41]. The spike (S) protein of SARS-CoV can downregulate ACE2 [42, 43] and prompt the shedding of catalytically active ACE2 ectodomain [44, 45]. Depletion of pulmonary ACE2 activity has been advised to be associated with severe lung injury [46, 47]. Furthermore, the decrease in ACE2 activity can lead to inhibition of the renin–angiotensin system (RAS) and increase inflammation and vascular permeability. The failure of ACE2 expression in a murine ARD model resulted in enhanced lung swelling, increased vascular permeability and reduced lung function [46]. ACE2 was also shed by the action of disintegrin and metalloprotease 17 in human airway epithelia to leak enzymatically active soluble ACE2 (sACE2) [45]. Furthermore, both SARS-CoV disease and inflammatory cytokines such as TNF-a and IL-1b can increase shedding of ACE2 [45, 48]. According to Haga et al., the S protein of SARS-CoV can induce ACE2 shedding, while the S protein of HNL63-CoV does not induce ACE2 shedding [44]. HNL63-CoV infection only causes the common cold, suggesting a potential pathogenic role of sACE2 in SARS-CoV infection. These findings demonstrate direct involvement of sACE2 in the inflammatory responses of SARS-CoV, and possibly SARS-CoV-2 as well.
Anti-spike IgG (anti-S-IgG)-prompted inflammatory responses
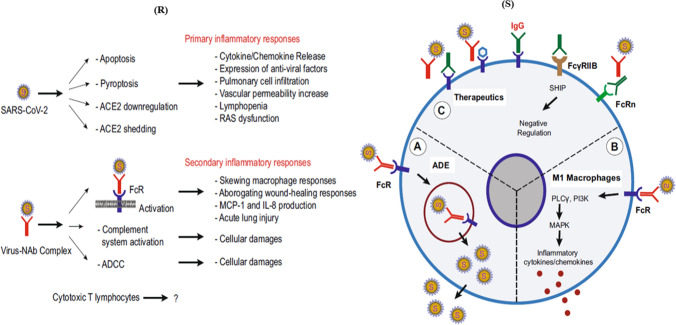
Antiviral neutralizing antibodies (Nab) perform a significant job in viral clearance. But, preceding investigations on SARS-CoV contamination in animal models demonstrated that corresponding anti-S-protein-neutralizing antibodies (anti-S-IgG) can also result in acute lung injury by changing inflammatory responses [49]. The study clearly indicated that despite viral repression, the presence of anti-spike protein antibody at the critical stage of SARS-CoV infection can actually result in acute lung injury which persists until the late stages. Close monitoring of SARS-CoV vaccine-induced pulmonary injury has also been reported in many animal models such as African green monkeys and mice [50–52]. Zhang et al. intimated that faster development of the anti-S-neutralizing antibody results in higher possibility of death of the patients from the disease [53]. Additionally, the anti-S-IgG can encourage proinflammatory macrophage or monocyte deposition and the generation of IL-8 and MCP-1 in the lungs. These proinflammatory responses were considered to be moderated through the binding of the virus–anti-S-IgG complex to the Fc receptors (FcR) existing on macrophages or monocytes [48]. Furthermore, antibody-dependent cell-mediated cytotoxicity (ADCC) may also be implicated. Fu et al. speculated that antibody-dependent enhancement (ADE) of viral infection may be the cause of persistent inflammation and non-clearance of virus in the patients in which neutralizing antibody is produced earlier (Fig. 2S) [54]. ADE advances viral cellular uptake of contagious virus–antibody complexes succeeding their association with FcR or more receptors, resulting in increased infection of target cells [55–58]. Hence, linkage of FcR with the virus–anti-S-IgG complex may ease both inflammatory responses and non-stop viral replication in the lungs of some patients (Fig. 2S). Fu et al. divided SARS-CoV-mediated inflammatory responses in two categories, i.e. primary and secondary responses (Fig. 2R) [54].
Fig. 2.
(R) Potential mechanisms of inflammatory responses caused by SARS-CoV-2. Based on recent investigations on SARS-CoV, the inflammatory responses in COVID-19 were divided into primary and secondary responses. Primary inflammatory responses exist early after viral infection, before the arrival of neutralizing antibodies (NAb). These responses are primarily led by active viral replication, viral-induced ACE2 downregulation and shedding, and host antiviral responses. Secondary inflammatory responses commence with the production of adaptive immunity and NAb. The virus–NAb complex can also cause FcR-mediated inflammatory responses; (S) antibody-dependent enhancement (ADE) of viral infection and inflammatory responses mediated by Fc receptor. A, ADE exists when antiviral NAb cannot completely neutralize the virus. Rather, the virus–NAb complex adheres to the Fc receptor (FcR), resulting in viral endocytosis and infection of the target cells. B, Binding of virus–NAb complex to FcR can also stimulate proinflammatory signalling, altering macrophage responses to the growth of proinflammatory (M1 or classically activated) macrophages in lungs. The M1 macrophages secrete inflammatory cytokines including IL-8 and MCP-1, resulting in lung injury. C, Possible therapeutics based on targeting the Fc receptors to prevent SARS-CoV-2-mediated inflammatory responses. From left to right, FcR can be blocked by employing anti-Fc specific antibodies, intravenous immunoglobulin (IVIG) or small molecules. The inhibitory FcR, FccRIIB, may also be selected to inhibit FcR activation. Reprinted with permission from Ref. [54]. Copyright 2020 Springer Nature
Primary inflammatory responses take place soon after viral infection, i.e. before the production of neutralizing antibodies (NAb). Such responses are mostly propelled by quick viral replication, viral-moderated ACE2 downregulation and antiviral responses of host cells. Although, as discussed earlier, the appearance of NAb can also activate FcR-mediated inflammatory responses and result in acute lung injury, Peiris et al. reported that the generation of antiviral IgG results in severe respiratory disease in 80% of SARS-CoV-infected patients [59]. Owing to non-lethal nature of primary inflammatory responses, this part of review primarily explores deadly secondary inflammatory responses. There are many promising therapeutic approaches that can be relevant or developed for the purpose (Fig. 2S). These approaches are mainly based on blocking of Fc receptor. Due to this, binding of virus–NAb complex to Fc receptor is prevented and inflammatory response is triggered off [60]. First, FcR deactivation can be achieved by developing small-molecule inhibitors that may interact with the Ig-binding domains of FcR for the purpose. Second, impeding FcR, FcγRIIB, may also be intended to inhibit FcR activation. Many FcγRIIB-specified antibodies are now being generated as novel immune repressors [61].
Third, FcR activation can also be hindered by targeting the newborn Fc receptor (FcRn), which is urgent for increasing the half-life of IgG. According to Nimmerjahn and Ravetch, antibody-mediated blockage of FcRn can inhibit the IgG interaction with FcRn, which can lessen circulating IgG levels [62]. Besides, intravenous immunoglobulin (IVIG) can also block the binding of antiviral NAb to other FcRs [63]. We also presume that SARS-CoV-2 also contributes similar inflammatory responses. Possible therapeutic techniques to reduce SARS-CoV-2-induced inflammatory responses comprise various methods to block FcR activation. In the absence of a tested clinical FcR blocker, the use of intravenous immunoglobulin to block FcR activation may be an applicable alternative for the crucial treatment of pulmonary inflammation to inhibit acute lung injury. Owing to lack of targeted therapeutics and effective treatment options, it becomes compulsory to rapidly discover new drugs for the treatment of COVID-19. One of the most distinguished drug targets included in coronaviruses is the main protease (Mpro, which is also called as 3CLpro) [64]. The Mpro is crucial for operating the polyproteins that are translated from the viral RNA [65]. Thus, this enzyme appears to be an attractive drug target for this virus [66]. Mpro breaks down the polyprotein at no less than 11 protecting sites. The practical significance of Mpro in the virus life cycle, as well as unavailability of related homologues in humans, makes the Mpro as an interesting target for antiviral drug formation [66]. The researchers are trying to employ structure-assisted drug design methodologies to repurpose existing drugs to target the Mpro.
The crystal structure of COVID-19 Mpro in complex with N3 inhibitor
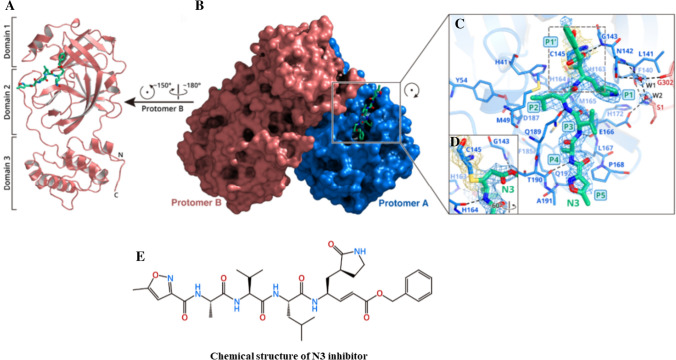
For understanding the inhibitory mechanism of N3 compound, Jin et al. established the crystal structure of COVID-19 Mpro in complex with this compound at 2.1 Å resolution (Fig. 3) [67]. Although the asymmetric unit possesses only one polypeptide, formation of dimer takes place between the two molecules and is labelled as protomers A and B (Fig. 3b). Each protomer is formed of three domains (Fig. 3a). Domains I and II possess antiparallel β-barrel structure. Domain III holds five α-helices ordered into a large antiparallel globular clump and is linked to domain II by means of a prolonged loop region. Mpro of COVID-19 has a Cys–His catalytic dyad. The substrate binding occurs in a cleft between domains I and II. These characteristics are close to those of other Mpros reported previously [64, 68–71]. The electron density map clearly manifests the binding of N3 in the substrate-binding pocket in an expanded form (Fig. 3c). However, atoms of the inhibitor backbone form an antiparallel sheet with residues of the long strand on the one side and with residues of the loop connecting domains II and III.
Fig. 3.
Crystal structure of COVID-19 Mpro in complex with N3 inhibitor; a one protomer of the dimeric Mpro–inhibitor complex, where green sticks represent N3 inhibitor; b depicts surface of the dimeric Mpro–inhibitor complex, where blue-coloured part represents protomer A, salmon-coloured part represents Protomer B, and green sticks indicate N3 inhibitor; c zoomed view of the substrate-binding region. The fundamental residues constituting the binding pocket are shown in sticks; W1 and W2 represent two water molecules and are shown as red spheres; the 2Fo-Fc density map formed at 1.2 σ is shown around N3 inhibitor (blue mesh), C145-A (yellow mesh), and the two waters (blue mesh); d C–S covalent bond; e chemical structure of N3 inhibitor. Reprinted with permission from Ref. [67]
Figure 3c demonstrates the specific interactions of N3 with Mpro in detail. The electron density manifests the formation of covalent bond between the Cβ of the vinyl group and Sγ atom of C145-A, confirming the Michael addition reaction (Fig. 3c, d). The side chains of H163-A, E166-A, H172-A, F140-A and S1-B (from protomer B) and main chains of L141- A and F140-A are used in the formation of S1 subsite, which also possesses two arranged water molecules (labelled W1 and W2). At P1 site, the lactam forms a hydrogen bond with H163-A. At P2 site, the side chain of Leu slips extremely into the hydrophobic S2 subsite. At P3 site, the side chain of Val gets exposed to solvent and exhibits tolerance of this site to a wide range of functional groups. At P4 site, the side chain of Ala is enclosed by the side chains of Q192- A, M165-A, F185-A and L167-A and the main chain of Q189-A, forming a small hydrophobic pocket. At P5 site, van der Waals interactions exist with P168-A and the residues 190–191. The benzyl group inserts into the S1′ site and establishes van der Waals relations with T24-A and T25-A. Moreover, N3 forms many hydrogen bonds with the main chain of the residues in the substrate-binding pocket, which also supports in locking the inhibitor interior the substrate-binding pocket (Fig. 3c). Thus, these interactions furnish an excellent model for quickly recognizing lead inhibitors to strike COVID-19 Mpro via in silico screening. Besides, Cinanserin has been clinically tested to inhibit SARS-CoV Mpro [72] in humans in the 1960s [73]. Hence, this can be further optimized as an antiviral drug lead. Similarly, Ebselen has been recognized as the strongest inhibitor of Mpro activity with an IC50 value of 0.48 μM.
The crystal structure of COVID-19 Mpro in complex with α-ketoamide inhibitors
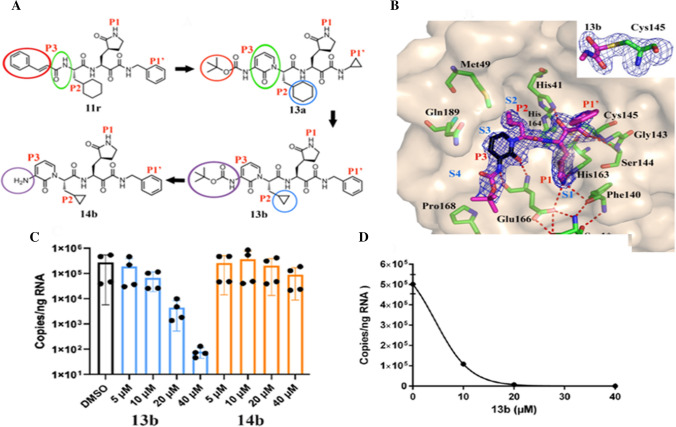
Alike N3, α-ketoamides have been considered as potent inhibitors of the Mpro of α-and β-coronaviruses in addition to 3C proteases of enteroviruses [74]. The finest of these inhibitors (11r; Fig. 4a) exhibited half maximal effective concentration (EC50) of 400 picomolar (pm) against MERS-CoV in Huh7 cells. For the purpose of increasing the half-life of this inhibitor in plasma, Zhang et al. altered the P3–P2 amide bond of this inhibitor with a pyridone ring (Fig. 4a, green circles) [75]. Moreover, for enhancing solubility and reducing binding to plasma proteins, hydrophobic cinnamoyl moiety was replaced with less hydrophobic Boc group (Fig. 4a, red circles) to give 13a inhibitor. To improve the antiviral activity against SARS-CoV and SARS-CoV-2, P2 cyclohexyl moiety of 13a was replaced by the smaller cyclopropyl in 13b (Fig. 4a, blue circles). Next, the compatibility of the modified inhibitor (13b) was determined with the three-dimensional structure of the target (Fig. 4b) for the purpose of designing antiviral activity. Figure 4b presents X-ray crystal structures of the complex between SARS-CoV-2Mpro and α-ketoamide (13b) in two different forms (in the space groups C2 and P212121) at the resolution of 1.95 and 2.20 Å. In C2 space group, both protomers of the Mpro dimer are bound to have identical conformations, whereas in P212121 space group the two protomers are independent to acquire different conformations.
Fig. 4.
a Chemical structures of α-ketoamide inhibitors 11r, 13a, 13b and 14b. Coloured circles feature the modifications from one development step to the next (see detail in text); b compound 13b in the substrate-binding cleft positioned between domains I and II of the Mpro. The Fo-Fc density is manifested for the inhibitor. Carbon atoms of the inhibitor are magenta coloured, but black in the pyridone ring; nitrogen atoms are blue, oxygens are red and sulphur yellow. S1, S2, S3 and S4 subunits (light blue) indicate the canonical binding pockets for P1, P2, P3 and P4 units (red) of the peptidomimetic inhibitor. Dashed red lines indicate hydrogen bonds. The interaction between the N-terminal residue of chain B, Ser1* and Glu166 of chain A is necessary for preserving the S1 pocket in the precise shape and the enzyme in the functional form. Inset represents the formation of thiohemiacetal by the nucleophilic attack of the catalytic cysteine onto the α-carbon of the inhibitor; c and d inhibition of SARS-CoV-2 replication by 13b inhibitor in human Calu3 lung cells; c SARS-CoV-2-infected Calu-3 cells stimulated with DMSO (black bar) and varied amounts (5, 10, 20 and 40 μM) of 13b (blue bars), and 14b (orange bars) evaluated at 24 h; d a dose–response curve for the estimation of the EC50 value of the inhibitor 13b against SARS-CoV-2. Reprinted from Ref. [75]. Copyright 2020 American Association for the Advancement of Science
In fact, in the latter crystal structure, the essential residue Glu166 acquires an inactive conformation in the protomer B (as indicated by its distance from His172 and the absence of H-bonding interlinkage with the P1 part of the inhibitor), despite the fact that compound 13b is bound in the same fashion as in molecule A. The same sensation has also been mentioned with the SARS-CoV Mpro [70] and is congruous with the half-site pursuit reported for this enzyme [76]. In all versions of the hindered Mpros of SARS-CoV-2, the inhibitor ties the facile substrate-binding site at the surface of every protomer, amid the domains I and II (Fig. 4b). Figure 4b inset clearly indicates the formation of a thiohemiacetal by the nucleophilic attack of the Cys145on the α-keto group of the inhibitor. Moreover, the warhead of the α-ketoamides can interact with the catalytic point of the target proteases via two hydrogen bonding interactions [74], rather than only one as in case of aldehydes [77] or Michael acceptors [78]. Further, the P1 γ-lactam part of the inhibitor, devised as a glutamine surrogate [78, 79], is extremely fixed in the S1 pocket of the Mpro, where the lactam nitrogen contributes a three-centre (bifurcated) hydrogen bond (HB) to the oxygen of Phe140 and to the Glu166 carboxylate, and the carbonyl oxygen undertakes a HB from the imidazole of His163. The P2 cyclopropyl methyl part inserts comfortably into the S2 subsite. Further, the P3 amide contributes a HB to the oxygen of Glu166. Compared to other protecting groups of the inhibitors, the Boc group on P3 does not employ the S4 subsite of the Mpro [80], but is located near Pro168 (Fig. 4b). That’s why removal of the Boc group as in compound 14b (Fig. 4a, purple circles) weakens the inhibitory power of this compound by a factor of nearby 2. The half maximal inhibitory concentration (IC50) value for inhibition of the SARS-CoV-2 Mpro has been reported as ~ 0.67 μM compared to ~ 0.90 μM and ~ 0.58 μM for the Mpro of SARS-CoV and the MERS-CoV, respectively. RNA replication was discouraged with EC50 of ~ 1.75 μM in a SARS-CoV replicon [81]. In human Calu3 cells infected with SARS-CoV-2, an EC50 of 4–5 μM was observed, whereas compound 14b missing the Boc part was reported almost inactive (Fig. 4c). The results demonstrate that the hydrophobic and bulky Boc portion is essential to cross the cellular membrane. Further, the highly hydrophobic portion might be excellent here, but it may again lead to enhanced plasma protein binding as seen for the cinnamoyl containing 11r.
Inhibitory activity of potential drugs
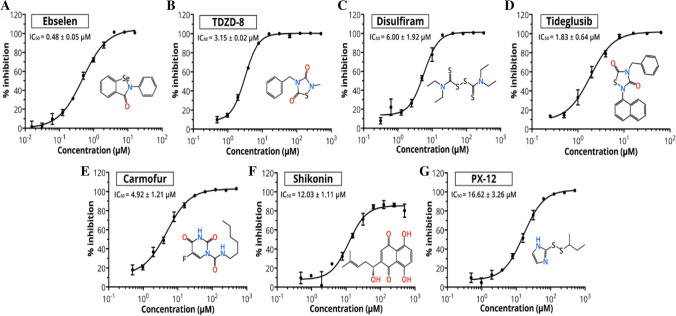
The timely development of operative antiviral drugs for clinical use is highly challenging. Conventional drug development methods normally require years of investigations and cost billions of dollars [82]. The repurposing of officially accepted pharmaceutical drugs and drug candidates suggests an alternative method to rapidly recognize potential drug leads for handling of rapidly emerging viral infections [83]. The convergence of structure-based ab initio drug design, virtual screening and high-throughput screening approaches suggests a new way to discover new drugs for treating COVID-19. Jin et al. employed FRET assay and screened a library of ~ 10,000 compounds including approved drugs, clinical trial drug candidates and natural products [67]. Some FDA-approved drugs or clinical/preclinical drugs with their IC50 values ranging from 0.48 to 16.62 μM are shown in Fig. 5. Amongst these, disulfiram and carmofur are FDA-approved drugs, whereas ebselen, TDZD-8, shikonin, tideglusib and PX-12 are currently in clinical trials or preclinical studies. Jin et al. also developed a similar model for COVID-19 Mpro [67]. Molecular docking was employed to investigate the targeting behaviour of N3 against COVID-19 Mpro. The results revealed the suitability of N3 inside the substrate-binding pocket. Kinetic analysis was also employed for assessing the efficacy of N3 against this virus [78]. The progress curve demonstrated time-dependent irreversible inhibitory behaviour of the N3 against the COVID-19 Mpro. The shape of the curve further assisted the mechanism of two-step irreversible inhibition. The inhibitor first links with Mpro of the virus, and then, a stable covalent bond is devised between the two. Thus, N3 exhibits very strong inhibition ofCOVID-19 Mpro.
Fig. 5.
Plots showing the inhibitory activity for the potential drug leads. The hydrolytic activity of SARS-CoV-2 Mpro was measured in the presence of different concentrations of drugs. Curves produce best fits for calculating the IC50 values. Reprinted with permission from Ref. [67]
For further supporting the in vitro results of the Mpro inhibition, the drug leads were further assessed for averting the cells from being infected by SARS-CoV-2. The results demonstrated that at concentration of 10 μM, Ebselen, N3 and TDZD-8 exhibited the most powerful antiviral effects. The drug leads completely inhibited the cytopathic effect (CPE) in SARS-CoV-2-infected Vero cells.
Quantitative real-time RT-PCR (qRT-PCR) showed that the treatment with Ebselen, TDZD-8 and N3 reduced the amounts of SARS-CoV-2, respectively, 20.3-, 10.19- and 8.4-fold compared to the treatment in the absence of inhibitor. Owing to antioxidant, cytoprotective and anti-inflammatory properties, Ebselen has also been used for the treatment of hearing loss and bipolar disorders. In humans, the toxicity of Ebselen has been judged in a number of clinical trials [84, 85]. TDZD-8 has been used in the treatment of Alzheimer's disease and found effective in various animal models [86]. Further, Cinanserin also showed moderate inhibition against SARS-CoV-2 with an IC50 value of 20.61 μM. These data firmly recommend the clinical potential of Ebselen, TDZD-8 and Cinanserin for COVID-19 treatment.
In another investigation, compound 13b constrained the recombinant SARS-CoV-2 Mpro with IC50 value of ~ 0.67 μM. The comparable IC50 values for restriction of the Mpros of SARS-CoV and the MERS-CoV were ~ 0.90 μM and ~ 0.58 μM, respectively. In a SARS-CoV replicon [81], RNA replication was prevented with EC50 value of ~ 1.75 μM. For further investigation of inhibitory/antiviral activity of 13b and 14b, Calu-3 cells were infected with SARS-CoV-2 using multiplicity of infection (MOI) of 0.05. The infected cells were then stimulated with DMSO (black bar) and varied amounts (5, 10, 20 and 40 μM) of 13b (blue bars) and 14b (orange bars) (Fig. 4c). The analysis was performed for 24 h. Total RNA was separated from cell lysates, and viral RNA content was analysed by qPCR. For the evaluation of the EC50 value of compound 13b against SARS-CoV-2, a dose–response curve was also framed (Fig. 4d). The results showed the EC50 value of 4–5 μM for the inhibitor 13b, whereas the inhibitor 14blacking the Boc group was found almost inactive (Fig. 4c). The results recommend that the hydrophobic and bulky Boc group is essential to cross the cellular membrane, but highly hydrophobic part may again lead to enlarged plasma protein binding as seen for the cinnamoyl comprising 11r. Thus, pyridone-containing α-ketoamide inhibitors can play a significant role in anti-coronaviral drugs.
Crucial proteins and their functions in viral disease
Recognition of drug targets is essential for repurposing of existing drugs for the treatment of the COVID-19. Therefore, it becomes necessary to investigate COVID-19-specific targets and their task in viral infection. Analogous to SARS-CoV, receptor-binding domain (RBD) in the S protein of SARS-CoV-2 binds to human ACE2 receptor for gaining access into the host cells [87]. In viral disease, the S protein, but not the other structural proteins, membrane glycoprotein (M), envelope (E) and nucleocapsid (N) in SARS-CoV, evokes an immune response [88]. The two viral proteases, Mpro and PLpro, cleavage the viral peptides into useful units for virus replication and loading within the host cells. Hence, drugs targeting these particular proteases in other viruses such as HIV drugs, ritonavir and lopinavir, were analysed [89]. RdRp or RNA replicase is an enzyme that is necessary for viral RNA generation. The existing antiviral drugs like remdesivir (RDV) [89] may block this RdRp. Perhaps, communication between viral S protein and its receptor ACE2 on host cells, and consequent viral endocytosis into the cells, may likewise be an applicable drug target. Arbidol (a broad-spectrum antiviral drug) acts as a virus–host cell fusion inhibitor. It inhibits viral entry into host cells against influenza virus [90]. That’s why it is under a clinical trial for curing COVID-19 [91, 92]. Transmembrane protease, serine 2 (TMPRSS2), is generated by the host cells, and it assists in proteolytic transformation of S protein mainly to the receptor ACE2 in human cells [93]. Camostat mesylate (CM) prevents the generation of TNF-α and monocyte chemoattractant protein-1 (MCP-1) by monocytes and is impressive in curing dibutyltin dichloride-prompted rat pancreatic fibrosis [94]. Hoffmann et al. further reported that CM is a clinically approved inhibitor of TMPRSS2. It can prevent entry of SARS-CoV-2 to the human cells. Thus, the CM may prove as a potential drug for treating COVID-19 [93].
Entanglement of ACE2 in CoV disease is of more significance because ACE2 is a powerful negative regulator and prevents over activation of the RAS. In addition to control of body fluid, electrolytes [43, 95] and blood pressure, RAS may be associated with elicitation of inflammatory lung infection. It leads to degradation of angiotensin II to angiotensin (1–7). The harmony between angiotensin II and angiotensin (1–7) elicits vasodilation conciliated by AT2 [46, 96, 97]. Owing to limited number of studies [43, 97], it is not clear how the functions of ACE2, AT1 and AT2 receptors are changed in coronavirus-induced infections. Hence, it is still a matter of discovery whether some drugs that target the proteins (such as L-163491 a partial antagonist of AT1 and partial agonist of AT2 receptor) may reduce coronavirus-prompted lung injury [98]. Table 1 presents some crucial proteins and their functions during the viral infection mechanism [99].
Table 1.
Crucial proteins and their functions during the viral infection process [99]
| Target candidate | Full name | Role during viral infection | Drug candidate |
|---|---|---|---|
| 3CLpro | Coronavirus main protease 3CLpro | A protease for the proteolysis of viral polyprotein into functional units | Lopinavir [89, 101] |
| PLpro | Papain-like protease PLpro | A protease for the proteolysis of viral polyprotein into functional units | Lopinavir [89, 101] |
| RdRp | RNA-dependent RNA polymerase | An RNA-dependent RNA polymerase for replicating viral genome | Remdesivir [89, 100, 103], ribavirin [28, 100, 102] |
| S protein | Viral spike glycoprotein | A viral surface protein for binding to host cell receptor ACE2 | Arbidol [90, 92, 104a] |
| TMPRSS2 | Transmembrane protease, serine 2 | A host cell-produced protease that primes S protein to facilitate its binding to ACE2 | Camostat mesylate [93] |
| ACE2 | Angiotensin-converting enzyme 2 | A viral receptor protein on the host cells which binds to viral S protein | Arbidol [90, 92, 104a] |
| AT2 | Angiotensin AT2 receptor | An important effector involved in the regulation of blood pressure and volume of the cardiovascular system | L-163491 [98] |
Reprinted from Ref. [99]; copyright American Chemical Society, 2020
aAn inhibitor of viral entry to host cells. Its direct action on S protein and ACE2 is yet to be confirmed
Existing drugs with potential therapeutic applications for COVID-19
As SARS-CoV-2 is a recently detected virus, no clear-cut drugs or vaccines have been declared so far. Further, the development of antiviral drugs is both time- and assets-consuming. Hence, repurposing of already present drugs to treat COVID-19 serves as one of the competent and economic therapeutic strategies. Based on genomic array data integrated with protein structure design, the scientists became capable of immediately responding to an implied list of existing drugs with therapeutic possibility for COVID-19. Table 2 lists some such drugs with their mechanism of action on COVID-19. Baricitinib works as an inhibitor of Janus kinase (JAK), clogging JAK1 and JAK2 subtypes. The drug has been approved in Europe, and the United States European Union approved it as a second-line remedy for rheumatoid arthritis in adults, either alone or in combination with methotrexate. Due to anti-inflammatory effect and viable capacity to inhibit viral entry [106], Richardson et al. proposed baricitinib as potential drug for COVID-19 treatment [106]. An established dose of the anti-HIV (lopinavir–ritonavir) combination along with Arbidol or ribavirin [92] is under clinical trials. Remdesivir has been recently identified as an auspicious antiviral drug against a wide progression of RNA viruses such as SARS and MERS-CoV [5] disease in cultured cells, mice and nonhuman primate (NHP) representations. It is presently under clinical testing for treating Ebola virus disease [107]. RDV is an adenosine analogue, which merges into nascent viral RNA chains and leads to premature termination [108]. The drug is presently being investigated in phase III clinical trials in both China and the USA. Favipiravir [109] (also known as T-705) is an antiviral drug developed by Toyama Chemical of Japan and is active against many RNA viruses. It has been found active against influenza virus, foot and mouth disease virus and flavivirus. It has also been approved for clinical trials to treat COVID-19 [101].
Table 2.
Existing drugs with therapeutic potentials for COVID-19 (drug repurposing) [99]
| Drug candidate | CAS RN | Target | Possible mechanism of action on COVID-19 | Disease indication |
|---|---|---|---|---|
| Baricitinib (C16H17N7O2S) [106] | 1187594-09-7 | JAK kinase | A JAK inhibitor that may interfere with the inflammatory processes | Approved drug for rheumatoid arthritis |
| Lopinavir (C37H48N4O5) [89a] | 192725-17-0 | Viral proteases: 3CLpro or PLpro | Protease inhibitors that may inhibit the viral proteases: 3CLpro or PLpro | Lopinavir and ritonavir are approved drug combination for HIV infection |
| Ritonavir (C37H48N6O5S2) [89a, 110c] | 155213-67-5 | |||
| Darunavir (C27H37N3O7S) [105] | 206361-99-1 | Approved drug for HIV infection | ||
| Favipiravir (Favilavir) (C5H4FN3O2) [100, 109] | 259793-96-9 | RdRp | A purine nucleoside that acts as an alternate substrate leading to inaccurate viral RNA synthesis | Viral infections |
| Remdesivir (C27H35N6O8P) [89a, 98, 103a] | 1809249-37-3 | A nucleotide analogue that may block viral nucleotide synthesis to stop viral replication | Ebola virus infection | |
| Ribavirin (C8H12N4O5) [28, 100–102] | 36791-04-5 | RSV infection, hepatitis C, some viral haemorrhagic fevers | ||
| Galidesivir (C11H15N5O3) [105b] | 249503-25-1 | Hepatitis C, Ebola virus, Marburg virus | ||
| BCX-4430 (salt form of galidesivir) [105b] | 222631-44-9 | Hepatitis C, Ebola virus, Marburg virus | ||
| Arbidol (C22H25BrN2O3S) [92, 104a] | 131707-23-8 | S protein/ACE2d | An inhibitor that may disrupt the binding of viral envelope protein to host cells and prevent viral entry to the target cell | Influenza antiviral drug |
| Chloroquine (C18H26ClN3) [99, 103] | 54-05-7 | Endosome/ACE2 | A drug that can elevate endosomal pH and interfere with ACE2 glycosylation | Malarial parasite infection |
| Nitazoxanide (C12H9N3O5S) [100] | 55981-09-4 | N/A | A drug that may inhibit viral protein expression | Various helminthic, protozoal and viral infection-caused diarrheea |
Reprinted from Ref. [99]; copyright American Chemical Society, 2020
aDrugs under clinical trials for treating COVID-19 (repurposing)
bDrugs under clinical trials for other virus-induced diseases
cRitonavir is a pharmacokinetic profile enhancer that may potentiate the effects of other protease inhibitors due to its ability to attenuate the degradation of those drugs by the liver enzyme CYP3A4 and thus is used in combination with antiviral lopinavir [109]. An inhibitor of viral entry to host cells. Its direct action on S protein and ACE2 is yet to be confirmed. The molecular formulae of the drugs have been provided in the parentheses below their chemical names
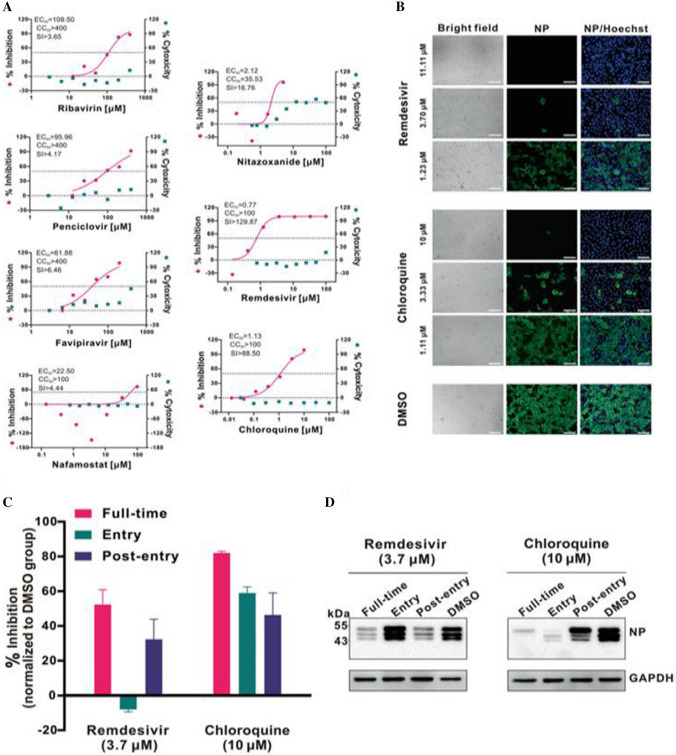
Chloroquine (CQ), a widely employed anti-malarial and autoimmune infection drug, has recently been reported as a potent broad-spectrum antiviral drug [111, 112]. CQ blocks viral disease by increasing endosomal pH along with interfering with the glycosylation of cellular receptors of SARS-CoV [113]. It has been reported effective in the treatment of coronavirus in China [31]. Previous studies demonstrated that alike CQ, hydroxychloroquine (HCQ) inhibited both the entry and the post-entry stages of SARS-CoV-2. Thus, both the drugs could be effective against SARS-CoV-2 [114, 115]. Another study showed more potency of HCQ than CQ in inhibiting SARS-CoV-2 in vitro [116, 117]. In addition to the aforementioned drugs, many other antiviral drugs are also listed in Table 2. Wang et al. evaluated in vitro antiviral efficiency of few FAD-authorized drugs such as penciclovir, ribavirin, nitazoxanide, nafamostat, chloroquine, favipiravir (T-705) and remdesivir (GS- 5734) against a clinical isolate of COVID-19. The results showed requirement of high concentration of ribavirin, penciclovir and favipiravir to curtail the viral infection (Fig. 6a), although favipiravir was found 100% effective in protecting mice against Ebola virus. Nafamostat and nitazoxanide were inhibitive against the COVID-19 with EC50 = 22.50 μM, CC50 > 100 μM, SI > 4.44 and EC50 = 2.12 μM, CC50 > 35.53 μM, SI > 16.76, respectively.
Fig. 6.
a In vitro antiviral activities of different doses of the tested antivirals for 48 h against SARS-CoV-2. The left and right Y-axes of the graphs show mean % inhibition of virus yield and cytotoxicity of the drugs, respectively; b immunofluorescence microscopy of viral disease upon treatment of chloroquine and remdesivir; c and d time-of-addition experiment of chloroquine and remdesivir. Chloroquine works at both entry, transport and at post-entry stages of the COVID-19 in Vero E6 cells. For all the experimental groups, cells were infected with SARS-CoV-2 at MOI of 0.05. Reprinted from Ref. [103]. Copyright 2020 Springer Nature
Notably, remdesivir (EC50 = 0.77 μM; CC50 > 100 μM; SI > 129.87) and chloroquine (EC50 = 1.13 μM; CC50 > 100 μM; SI > 88.50) potentially blocked viral disease at low micromolar concentration and exhibited high SI (Fig. 6a, b). The time-of-addition assay results demonstrated that RDV acted at post-virus entry stage, whereas chloroquine (CQ) performed at both entry and at post-entry stages of the COVID-19 in Vero E6 cells (Fig. 6c, d). Further, the EC90 value of RDV and CQ against SARS-CoV-2 in Vero E6 cells was 1.76 μM and 6.90 μM, respectively.
In addition to the above-mentioned commercial antiviral drugs, there are also many small molecular inhibitors that can inhibit viral enzymes inclusive of proteases and components for RdRp. As discussed earlier, viral Mpro carries a large sequence homology between SARS-CoV and SARS-CoV-2. The inhibitors applicable against SARS-CoV Mpro may also be pertinent to SARS-CoV-2 Mpro. The inhibitors such as benzopurpurin B, NSC-306711, N-65828 and C-473872, which may curb the action of viral NSP15 and poly(U)-specific endoribonuclease, were found to curtail SARS-CoV infections with IC50 of 0.2–40 μM [118]. It is also necessary to establish safe and efficient vaccines to check the COVID-19 pandemic, inhibit its spread and finally prevent its future recurrence. As mentioned earlier, owing to remarkable sequence homology of the SARS-CoV-2 with lethal SARS and MERS coronaviruses, the vaccines related to these two viruses can possibly expedite the drafting of anti-SARS-CoV-2 vaccines. Buchholz et al. demonstrated that viral S protein subunit vaccines generated higher NAb titers and increased complete protection as compared to full-length S protein, live-attenuated SARS-CoV and DNA-based S protein vaccines [119]. As a whole, S protein is the approved target site in MERS/SARS vaccine development, and the similar approach can be potentially convenient in developing SARS-CoV-2 vaccines. Furthermore, convalescent plasma from patients who have recuperated from viral infections can be employed as a therapy without the appearance of severe adverse events. Theoretically, it should be more effective to administer the convalescent plasma at the early stage of disease [120]. Hence, it might be valuable to examine the safety and efficacy of convalescent plasma transfusion in SARS-CoV-2-infected patients.
Conclusions
At present, there is no efficacious antiviral remedy for COVID-19, which generally causes fatal inflammatory responses and severe lung injury. Here, we scrutinize the different mechanisms of SARS-CoV-induced inflammation. We also presume that SARS-CoV-2 probably shares identical inflammatory responses. Possible therapeutic means to lessen SARS-CoV-2-mediated inflammatory responses comprise different methods to block FcR activation. Due to non-availability of a proven clinical FcR blocker, intravenous immunoglobulin may be used to block FcR activation and treat pulmonary inflammation. Although the proposed strategies remain to be clinically tested for efficacy, recognition of SARS-CoV-2 Mpro as a preserved target provides an opportunity for the development of broad-spectrum inhibitors against COVID-19 and other CoV-related diseases. The crystal structural and docking results have shown that Ebselen, N3, TDZD-8 and α-ketoamide (13b) inhibitors can bind to the substrate-binding pocket of COVID-19 Mpro. The inhibitor 13b inhibits the replication of SARS-CoV-2 in human Calu3 lung cells. Quantitative real-time RT-PCR (qRT-PCR) results showed that the treatment with Ebselen, TDZD-8 and N3 reduced the amounts of SARS-CoV-2, respectively, 20.3-, 10.19- and 8.4-fold compared to the treatment in the absence of inhibitor. Further, the development of antiviral drugs is both time- and assets-consuming. Hence, repurposing of already present drugs to treat COVID-19 serves as one of the competent and economic therapeutic strategies. Several anti-malarial, anti-HIV and anti-inflammatory drugs as mentioned in Table 2 were found effective for the COVID-19 treatment. Similar to chloroquine (CQ), hydroxychloroquine (HCQ) inhibited the entry, transport and the post-entry stages of SARS-CoV-2. Moreover, HCQ was found more potent than CQ in inhibiting SARS-CoV-2 in vitro. Furthermore, it might be valuable to examine the safety and efficacy of convalescent plasma transfusion in SARS-CoV-2-infected patients.
References
- 1.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W. A novel coronavirus from patients with pneumonia in China. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KSM, Lau EHY, Wong JY, Xing X, Xiang N, Wu Y, Li C, Chen Q, Li D, Liu T, Zhao J, Li M, Tu W, Chen C, Jin L, Yang R, Wang Q, Zhou S, Wang R, Liu H, Luo Y, Liu Y, Shao G, Li H, Tao Z, Yang Y, Deng Z, Liu B, Ma Z, ZhangY Shi G, Lam TTY, Wu JTK, Gao GF, Cowling BJ, Yang B, Leung GM, Feng Z. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu M-Q, Chen Y, Shen XR, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song Z-G, Hu Y, Tao Z-W, Tian J-H, Pei Y-Y, Yuan M-L, Zhang Y-L, Dai F-H, Liu Y, Wang Q-M, Zheng J-J, Xu L, Holmes EC, Zhang Y-Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan JF-W, Yuan S, Kok K-H, To KK-W, Chu H, Yang J, Xing F, Liu J, Yip CC-Y, Poon RW-S, Tsoi H-W, Lo SK-F, Chan K-H, Poon VK-M, Chan W-M, Ip JD, Cai J-P, Cheng VC-C, Chen H, Hui CK-M, Yuen K-YA, familial cluster of pneumonia associated with the, novel coronavirus indicating person-to-person transmission: a study of a family cluster. The Lancet. 2019;2020(395):514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coronavirus Disease (COVID-2019) (2020) Situation reports 1-97; World Health Organization. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situationreports
- 7.de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anthony SJ, Johnson CK, Greig DJ, Kramer S, Che X, Wells H, Hicks AL, Joly DO, Wolfe ND, Daszak P, Karesh W, Lipkin WI, Morse SS, Mazet JAK, Goldstein T. Global patterns in coronavirus diversity. Virus Evol. 2017;3(1):vex012. doi: 10.1093/ve/vex012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan XGJ, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xin X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xin J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H, Spitters C, Ericson K, Wilkerson S, Tural A, Diaz G, Cohn A, Fox L, Patel A, Gerber SI, Kim L, Tong S, Lu X, Lindstrom S, Pallansch MA, Weldon WC, Biggs HM, Uyeki TM, Pillai SK. First case of 2019 novel coronavirus in the United States for the Washington State. N Engl J Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldsmith CS, Tatti KM, Ksiazek TG, Rollin PE, Comer JA, Lee WW, RotaPA Bankamp B, Bellini WJ, Zaki SR. Ultrastructural characterization of SARS coronavirus. Emerg Infect Dis. 2004;10:320–326. doi: 10.3201/eid1002.030913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai MM, Cavanagh D. The molecular biology of coronaviruses. Adv Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sexton NR, Smith EC, Blanc H, Vignuzzi M, Peersen OB, Denison MR. Homology-based identification of a mutation in the coronavirus RNA-dependent RNA polymerase that confers resistance to multiple mutagens. J Virol. 2016;90:7415–7428. doi: 10.1128/JVI.00080-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belouzard S, Millet JK, Licitra BN, Whittaker GR. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.COVID-19 is an emerging, rapidly evolving situation. National Institute of Allergy and Infectious Disease (2020). https://www.niaid.nih.gov/news-events/novel-coronavirus-sarscov2-images
- 17.Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou J, Liu W, Bi Y, Gao GF. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang B, Bragazzi NL, Li Q, Tang S, Xiao Y, Wu J. An updated estimation of the risk of transmission of the novel coronavirus (2019-nCov) Infect Dis Model. 2020;5:248–255. doi: 10.1016/j.idm.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H, Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glowacka I, Bertram S, Müller MA, Allen P, Soilleux E, Pfefferle S, Steffen I, TsegayeTS He Y, Gnirss K, Niemeyer D, Schneider H, Drosten C, Pöhlmann S. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J Virol. 2011;85:4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuyama S, Nagata N, Shirato K, Kawase M, Takeda M, Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J Virol. 2010;84:12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shulla A, Heald-Sargent T, Subramanya G, Zhao J, Perlman S, Gallagher T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J Virol. 2011;85:873–882. doi: 10.1128/JVI.02062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ziebuhr J, Snijder EJ, Gorbalenya AE. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J Gen Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- 24.Baez-Santos YM, St John SE, Mesecar AD. The SARS coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antiviral Res. 2015;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee T-W, Cherney MM, Huitema C, Liu J, James KE, Powers JC, Eltis LD, JamesMNG, Crystal structures of the main peptidase from the SARS coronavirus inhibited by a substrate like aza-peptide epoxide. J Mol Biol. 2005;353:1137–1151. doi: 10.1016/j.jmb.2005.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh C-L, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV Spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. Genomic characterization and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/s0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morse JS, Lalonde T, Xu S, Liu WR. Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. ChemBioChem. 2020;21:730–738. doi: 10.1002/cbic.202000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chan JF-W, Kok K-H, Zhu Z, Chu H, To KK-W, Yuan S, Yuen K-Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong N, Yang X,Ye L, Chen K, Chan EW-C, Yang M, Chen S (2020) Genomic and protein structure modelling analysis depicts the origin and infectivity of 2019-nCoV, a new coronavirus which caused a pneumonia outbreak in Wuhan, China, bioRxiv. 10.1101/2020.01.20.913368v2
- 31.Gosh AK, Xi K, Ratia K, Santarsiero BD, Fu W, Harcourt BH, Rota PA, Baker SC, Johnson ME, Mesecar AD. Design and synthesis of peptidomimetic severe acute respiratory syndrome chymotrypsin-like protease inhibitors. J Med Chem. 2005;48:6767–6771. doi: 10.1021/jm050548m. [DOI] [PubMed] [Google Scholar]
- 32.Kumar V, Tan KP, Wang YM, Lin SW, Liang PH. Identification, synthesis and evaluation of SARS-CoV and MERS-CoV 3C-like protease inhibitors. Bioorg Med Chem. 2016;24:3035–3042. doi: 10.1016/j.bmc.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pillaiyar T, Manickam M, Namasivayam V, Hayashi Y, Jung S-H. An overview of severe acute respiratory syndrome-coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J Med Chem. 2016;59:6595–6628. doi: 10.1021/acs.jmedchem.5b01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qamar MTU, Maryam A, Muneer I, Xing F, Ashfaq UA, Khan FA, Anwar F, Geesi MH, Khalid RR, Rauf SA, Siddiqi AR. Computational screening of medicinal plant phytochemicals to discover potent pan-serotype inhibitors against dengue virus. Sci Rep. 2019;9:1–16. doi: 10.1038/s41598-018-38450-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qamar MTU, Alqahtani SM, Alamri MA, Chen L-L. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J Pharm Anal. 2020 doi: 10.1016/j.jpha.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gu J, Gong E, Zhang B, Zheng J, Gao Z, Zhong Y, Zou W, Zhan J, Wang S, Xie Z, Zhuang H, Wu B, Zhong H, Shao H, Fang W, Gao D, Pei F, Li X, He Z, Xu D, Shi X, Anderson VM, Leong AS. Multiple organ infection and the pathogenesis of SARS. J Exp Med. 2005;202:415–424. doi: 10.1084/jem.20050828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao Y, Zhao Z, Wang Y, Zhou Y, Ma Y, Zuo W. Single-Cell RNA expression profiling of ACE2, the putative receptor of Wuhan 2019-nCoV. BioRxiv. 2020 doi: 10.1101/2020.01.26.919985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang M (2020) Cell pyroptosis, a potential pathogenic mechanism of 2019-nCoV infection. 10.2139/ssrn.3527420
- 39.Guan W-j, Ni Z-y, Hu Y, Liang W-h, Ou C-q, He J-x, Liu L, Shan H, Lei C-l, Hui DSC, Du B, Li L-j, Zeng G, Yuen K-Y, Chen R-c, Tang C-l, Wang T, Chen P-y, Xiang J, Li S-y, Wang J-l, Liang Z-J, Peng Y-x, Wei L, Liu Y, Hu Y-h, Peng P, Wang J-m, Liu J-y, Chen Z, Li G, Zheng Z-j, Qiu S-q, Luo J, Ye C-j, Zhu S-y, Zhong N-S. Clinical characteristics of 2019 novel coronavirus infection in China. N Engl J Med. 2020 doi: 10.1056/NEJMoa2002032. [DOI] [PubMed] [Google Scholar]
- 40.Kuba K, Imai Y, Rao S, Gao H, Guo F, Guan B, Huan Y, Yang P, Zhang Y, Deng W, Bao L, Zhang B, Liu G, Wang Z, Chappell M, Liu Y, Zheng D, Leibbrandt A, Wada T, Slutsky AS, Liu D, Qin C, Jiang C, Penninger JM. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Imai Y, Kuba K, Penninger JM. The discovery of angiotensin-converting enzyme 2 and its role in acute lung injury in mice. Exp Physiol. 2008;93:543–548. doi: 10.1113/expphysiol.2007.040048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang S, Guo F, Liu K, Wang H, Rao S, Yang P, Jiang C. Endocytosis of the receptor-binding domain of SARS-CoV spike-protein together with virus receptor ACE2. Virus Res. 2008;136:8–15. doi: 10.1016/j.virusres.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Glowacka I, Bertram S, Herzog P, Pfefferle S, Steffen I, Muench MO, Simmons G, Hofmann H, Kuri T, Weber F, Eichler J, Drosten C, Pöhlmann S. Differential downregulation of ACE2 by the spike proteins of severe acute respiratory syndrome coronavirus and human coronavirus NL63. J Virol. 2010;84:1198–1205. doi: 10.1128/JVI.01248-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haga S, Yamamoto N, Nakai-Murakami C, Osawa Y, Tokunaga K, Sata T, Yamamoto N, Sasazuki T, Ishizaka Y. Modulation of TNF-α-converting enzyme by the spike protein of SARS-CoV and ACE2 induces TNF-α production and facilitates viral entry. Proc Natl Acad Sci USA. 2008;105:7809–7814. doi: 10.1073/pnas.0711241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lambert DW, Yarski M, Warner FJ, Thornhill P, Parkin ET, Smith AI, Hooper NM, Turner AJ. Tumor necrosis factor-alpha convertase (ADAM17) mediates regulated ectodomain shedding of the severe-acute respiratory syndrome-coronavirus (SARS-CoV) receptor, angiotensin-converting enzyme-2 (ACE2) J Biol Chem. 2005;280:30113–30119. doi: 10.1074/jbc.M505111200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Imai Y, Kuba K, Rao S, Huan Y, Guo F, Guan B, Yang P, Sarao R, Wada T, Leong-Poi H, Crackower MA, Fukamizu A, Hui CC, Hein L, Uhlig S, Slutsky AS, Jiang C, Penninger JM. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436:112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuba K, Imai Y, Penninger JM. Angiotensin-converting enzyme 2 in lung diseases. Curr Opin Pharmacol. 2006;6:271–276. doi: 10.1016/j.coph.2006.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jia HP, Look DC, Tan P, Shi L, Hickey M, Gakhar L, Chappell MC, Wohlford-Lenane C, McCray PB., Jr Ectodomain shedding of angiotensin converting enzyme 2 in human airway epithelia. Am J Physiol Lung Cell Mol Physiol. 2009;297:L84–L96. doi: 10.1152/ajplung.00071.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu L, Wei Q, Lin Q, Fang J, Wang H, Kwok H, Tang H, Nishiura K, Peng J, Tan Z, Wu T, Cheung KW, Chan KH, Alvarez X, Qin C, Lackner A, Perlman S, Yuen KY, Chen Z. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019;4:e123158. doi: 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bolles Deming D, Long K, Agnihothram S, Whitmore A, Ferris M, Funkhouser W, Gralinski L, Totura A, Heise M, Baric RS. A double-inactivated severe acute respiratory syndrome coronavirus vaccine provides incomplete protection in mice and induces increased eosinophilic proinflammatory pulmonary response upon challenge. J Virol. 2011;85:12201–12215. doi: 10.1128/JVI.06048-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clay C, Donart N, Fomukong N, Knight JB, Lei W, Price L, Hahn F, Van estrienen J, Harrod KS. Primary severe acute respiratory syndrome coronavirus infection limits replication but not lung inflammation upon homologous rechallenge. J Virol. 2012;86:4234–4244. doi: 10.1128/JVI.06791-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tseng CT, Sbrana E, Iwata-Yoshikawa N, Newman PC, Garron T, Atmar RL, Peters CJ, Couch RB. Immunization with SARS coronavirus vaccines leads to pulmonary immunopathology on challenge with the SARS virus. PLoS ONE. 2012;7:e35421. doi: 10.1371/journal.pone.0035421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang L, Zhang F, Yu W, He T, Yu J, Yi CE, Ba L, Li W, Farzan M, Chen Z, Yuen KY, Ho D. Antibody responses against SARS coronavirus are correlated with disease outcome of infected individuals. J Med Virol. 2006;78:1–8. doi: 10.1002/jmv.20499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fu Y, Cheng Y, Wu Y. Understanding SARS-CoV-2-mediated inflammatory responses: from mechanisms to potential therapeutic tools. Virol Sin. 2020 doi: 10.1007/s12250-020-00207-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Halstead SB, O’Rourke EJ. Antibody-enhanced dengue virus infection in primate leukocytes. Nature. 1977;265:739–741. doi: 10.1038/265739a0. [DOI] [PubMed] [Google Scholar]
- 56.Haslwanter D, Blaas D, Heinz FX, Stiasny K. A novel mechanism of antibody-mediated enhancement of flavivirus infection. PLoS Pathog. 2017;13:e1006643. doi: 10.1371/journal.ppat.1006643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ochiai H, Kurokawa M, Matsui S, Yamamoto T, Kuroki Y, Kishimoto C, Shiraki K. Infection enhancement of influenza A NWS virus in primary murine macrophages by anti-hemagglutinin monoclonal antibody. J Med Virol. 1992;36:217–221. doi: 10.1002/jmv.1890360312. [DOI] [PubMed] [Google Scholar]
- 58.Takada A, Kawaoka Y. Antibody-dependent enhancement of viral infection: molecular mechanisms and in vivo implications. Rev Med Virol. 2003;13:387–398. doi: 10.1002/rmv.405. [DOI] [PubMed] [Google Scholar]
- 59.Peiris JS, Chu CM, Cheng VC, Chan KS, Hung IF, Poon LL, Law KI, Tang BS, Hon TY, Chan CS, Chan KH, Ng JS, Zheng BJ, Ng WL, Lai RW, Guan Y, Yuen KY. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/s0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nimmerjahn F, Ravetch JV. Fcγ receptors as regulators of immune responses. Nat Rev Immunol. 2008;8:34–47. doi: 10.1038/nri2206. [DOI] [PubMed] [Google Scholar]
- 61.Veri MC, Gorlatov S, Li H, Burke S, Johnson S, Stavenhagen J, Stein KE, Bonvini E, Koenig S. Monoclonal antibodies capable of discriminating the human inhibitory Fcgamma-receptor IIB (CD32B) from the activating Fcgamma-receptor IIA (CD32A): biochemical, biological and functional characterization. Immunology. 2007;121:392–404. doi: 10.1111/j.1365-2567.2007.02588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nimmerjahn F, Ravetch JV. Anti-inflammatory actions of intravenous immunoglobulin. Annu Rev Immunol. 2008;26:513–533. doi: 10.1146/annurev.immunol.26.021607.090232. [DOI] [PubMed] [Google Scholar]
- 63.Kurlander RJ, Hall J. Comparison of intravenous gamma globulin and a monoclonal anti-Fc receptor antibody as inhibitors of immune clearance in vivo in mice. J Clin Invest. 1986;77:2010–2018. doi: 10.1172/JCI112530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anand K, Ziebuhr J, Wadhwani P, Mesters JR, Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: Basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 65.Hilgenfeld R. From SARS to MERS: Crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281:4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pillaiyar T, Manickam M, Namasivayam V, Hayashi Y, Jung SH. An overview of severe acute respiratory syndrome-coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J Med Chem. 2016;59:6595–6628. doi: 10.1021/acs.jmedchem.5b01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y, Zhang B, Li, Zhang L, Duan Y, Yu J, Wang L, Yang K, Liu F, You T, Liu X, Yang X, Bai F, Liu H, Liu X, Guddat LW, Xiao G, Qin C, Shi Z, Jiang H, Rao Z, Yang H (2020) Structure-based drug design, virtual screening and high-throughput screening rapidly identify antiviral leads targeting COVID-19. bioRxiv. 10.1101/2020.02.26.964882
- 68.Xue X, Yu H, Yang H, Xue F, Wu Z, Shen W, Li J, Zhou Z, Ding Y, Zhao Q, Zhang XC, Liao M, Bartlam M, Rao Z. Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design. J Virol. 2008;82:2515–2527. doi: 10.1128/JVI.02114-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang F, Chen C, Yang K, Xu Y, Lin X, Gao F, Liu H, Chen X, Zhao Q, Liu X, Cai Y, Yang H. Michael acceptor-based peptidomimetic inhibitor of main protease from porcine epidemic diarrhea virus. J. Med. Chem. 2017;60:3212–3216. doi: 10.1021/acs.jmedchem.7b00103. [DOI] [PubMed] [Google Scholar]
- 70.Yang H, Yang M, Ding Y, Liu Y, Lou Z, Zhou Z, Sun L, Mo L, Ye S, Pang H, Gao GF, Anand K, Bartlam M, Hilgenfeld R, Rao Z. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. PNAS. 2003;100:13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao Q, Li S, Xue F, Zou Y, Chen C, Bartlam M, Rao Z. Structure of the main protease from a global infectious human coronavirus, HCoV-HKU1. J Virol. 2008;82:8647–8655. doi: 10.1128/JVI.00298-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen L, Gui C, Luo X, Yang Q, Günther S, Scandella E, Drosten C, Bai D, He X, Ludewing B, Chen J, Luo H, Yang Y, Yang Y, Zou J, Thiel V, Chen K, Shen J, Shen X, Jiang H. Cinanserin is an inhibitor of the 3C-like proteinase of severe acute respiratory syndrome coronavirus and strongly reduces virus replication in vitro. J Virol. 2005;79:7095–7103. doi: 10.1128/JVI.79.11.70957103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rubin B, Waugh MH. Antiphlogistic effects of antiserotonin (SQ 10,643) and aminopyrine in rats versus endotoxin and other agents. Proc Soc Exp Biol Med. 1965;119:438–443. doi: 10.3181/00379727-119-30204. [DOI] [PubMed] [Google Scholar]
- 74.Zhang L, Lin D, Kusov Y, Nian Y, Ma Q, Wang J, von Brunn A, Leyssen P, Lanko K, Neyts J, de Wilde A, Snijder EJ, Liu H, Hilgenfeld R. α-Ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication: Structure-based design, synthesis, and activity assessment. J Med Chem. 2020 doi: 10.1021/acs.jmedchem.9b01828. [DOI] [PubMed] [Google Scholar]
- 75.Zhang L, Lin D, Sun X, Curth U, Drosten C, Sauerhering L, Becker S, Rox K, Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen H, Wei P, Huang C, Tan L, Liu Y, Lai L. Only one protomer is active in the dimer of SARS 3C-like proteinase. J Biol Chem. 2006;281:13894–13898. doi: 10.1074/jbc.M510745200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhu L, George S, Schmidt MF, Al-Gharabli SI, Rademann J, Hilgenfeld R. Peptide aldehyde inhibitors challenge the substrate specificity of the SARS-coronavirus main protease. Antiviral Res. 2011;92:204–212. doi: 10.1016/j.antiviral.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tan J, George S, Kusov Y, Perbandt M, Anemüller S, Mesters JR, Norder H, Coutard B, Lacroix C, Leyssen P, Neyts J, Hilgenfeld R. 3C protease of enterovirus 68: structure-based design of Michael acceptor inhibitors and their broad-spectrum antiviral effects against picornaviruses. J Virol. 2013;87:4339–4351. doi: 10.1128/JVI.01123-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dragovich PS, Zhou R, Skalitzky DJ, Fuhrman SA, Patick AK, Ford CE, Meador JW, Worland ST. Solid-phase synthesis of irreversible human rhinovirus 3C protease inhibitors. Part 1: optimization of tripeptides incorporating N-terminal amides. Bioorg Med Chem. 1999;7:589–598. doi: 10.1016/s0968-0896(99)00005-x. [DOI] [PubMed] [Google Scholar]
- 80.Zhu L, Hilgenfeld R (2012) Crystal structure of SARS coronavirus main protease complexed with an alpha, beta-unsaturated ethyl ester inhibitor SG85. PDB Entry-3TNT. 10.2210/pdb3TNT/pdb
- 81.Kusov Y, Tan J, Alvarez E, Enjuanes L, Hilgenfeld R. A G-quadruplex-binding macrodomain within the “SARS-unique domain” is essential for the activity of the SARS-coronavirus replication-transcription complex. Virology. 2015;484:313–322. doi: 10.1016/j.virol.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bedford J, Farrar J, Ihekweazu C, Kang G, Koopmans M, Nkenhasong J. A new twenty-first century science for effective epidemic response. Nature. 2019;575:130–136. doi: 10.1038/s41586-019-1717-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Farha MA, Brown ED. Drug repurposing for antimicrobial discovery. Nat Microbiol. 2019;4:565–577. doi: 10.1038/s41564-019-0357-1. [DOI] [PubMed] [Google Scholar]
- 84.Masaki C, Sharpley AL, Cooper CM, Godlewska BR, Singh N, Vasudevan SR, Harmer CJ, Churchill GC, Sharp T, Rogers RD, Cowen CJ. Effects of the potential lithium-mimetic, ebselen, on impulsivity and emotional processing. Psychopharmacology. 2016;233:2655–2661. doi: 10.1007/s00213-016-4319-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kil J, Lobarinas E, Spankovich C, Spankovich C, Griffiths SK, Antonelli PJ, Lynch E, Le Prell CG. Safety and efficacy of ebselen for the prevention of noise-induced hearing loss: randomised, double-blind, placebo-controlled, phase 2 trial. The Lancet. 2017;390:969–979. doi: 10.1016/S0140-6736(17)31791-9. [DOI] [PubMed] [Google Scholar]
- 86.Koehler D, Shah ZA, Williams FE. The GSK3β inhibitor, TDZD-8, rescues cognition in a zebrafish model of okadaic acid-induced Alzheimer's disease. Neurochem Int. 2019;122:31–37. doi: 10.1016/j.neuint.2018.10.022. [DOI] [PubMed] [Google Scholar]
- 87.Li F, Li W, Farzan M, Harrison SC. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 88.Bisht H, Roberts A, Vogel L, Subbarao K, Moss B. Neutralizing antibody and protective immunity to SARS coronavirus infection of mice induced by a soluble recombinant polypeptide containing an N-terminal segment of the spike glycoprotein. Virology. 2005;334:160–165. doi: 10.1016/j.virol.2005.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sheahan TP, Sims AC, Leist SR, Schafer A, Won J, Brown AJ, Montgomery SA, Hogg A, Babusis D, Clarke MO, Spahn JE, Bauer L, Sellers S, Porter D, Feng JY, Cihlar T, Jordan R, Denison MR, Baric RS. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat Commun. 2020;11:222. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kadam RU, Wilson IA. Structural basis of influenza virus fusion inhibition by the antiviral drug arbidol. Proc Natl Acad Sci USA. 2017;114:206–214. doi: 10.1073/pnas.1617020114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Therapeutic options for the 2019 novel coronavirus (2019-nCoV). https://www.nature.com/articles/d41573-020-00016-0 [DOI] [PubMed]
- 92.The Efficacy of Lopinavir Plus Ritonavir and Arbidol Against Novel Coronavirus Infection (ELACOI). https://clinicaltrials.gov/ct2/show/NCT04252885
- 93.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu N-H, Nitsche A, Müller MA, Drosten C, Pöhlmann S. SARS-CoV-2Cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:1–10. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Phillips JM, Gallagher T, Weiss SR. Neurovirulent murine coronavirus JHM.SD uses cellular zinc metalloproteases for virus entry and cell-cell fusion. J Virol. 2017;91:e01564–16. doi: 10.1128/JVI.01564-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wu Y. Compensation of ACE2 function for possible clinical managementof 2019-nCoV-induced acute lung injury. Virol Sin. 2020;7:1–3. doi: 10.1007/s12250-020-00205-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Donoghue M, Hsieh F, Baronas E, Godbout K, Gosselin M, Stagliano N, Donovan M, Woolf B, Robison K, Jeyaseelan R, Breitbart RE, Acton S. A novel angiotensin-converting enzyme related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1–9. Circ Res. 2000;87:e1–e9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- 97.Tipnis SR, Hooper NM, Hyde R, Karran E, Christie G, Turner AJ. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J Biol Chem. 2000;275:33238–33243. doi: 10.1074/jbc.M002615200. [DOI] [PubMed] [Google Scholar]
- 98.De Witt BJ, Garrison EA, Champion HC, Kadowitz PJ. L-163,491 is a partial angiotensin AT1 receptor agonist in the hindquarters vascular bed of the cat. Eur J Pharmacol. 2000;404:213–219. doi: 10.1016/s0014-2999(00)00612-9. [DOI] [PubMed] [Google Scholar]
- 99.Liu C, Zhou Q, Li Y, Garner LV, Watkins SP, Carter LJ, Smoot J, Gregg AC, Daniels AD, Jervey S, Albaiu D. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent Sci. 2020;6:315–331. doi: 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Guo D. Old weapon for new enemy: drug repurposing for treatment of newly emerging viral diseases. Virol Sin. 2020;11:1–3. doi: 10.1007/s12250-020-00204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Maxmen A. More than 80 clinical trials launch to test coronavirus treatments. Nature. 2020;578:347–348. doi: 10.1038/d41586-020-00444-3. [DOI] [PubMed] [Google Scholar]
- 102.Arabi YM, Shalhoub S, Mandourah Y, Al-Hameed F, Al-Omari A, Al Qasim E, Jose J, Alraddadi B, Almotairi A, Al Khatib K, Abdulmomen A, Qushmaq I, Sindi AA, Mady A, Solaiman O, Al-Raddadi R, Maghrabi K, Ragab A, Al Mekhlafi GA, Balkhy HH, Al Harthy A, Kharaba A, Gramish JA, Al-Aithan AM, Al-Dawood A, Merson L, Hayden FG, Fowler R. Ribavirin and interferon therapy for critically III patients with middle east respiratory syndrome: a multicenter observational study. Clin Infect Dis. 2019;70:1837–1844. doi: 10.1093/cid/ciz544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, Shi Z, Hu Z, Zhong W, Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.KangjiXinguangzhuangBingduFeiyanZhuanliXinxiYanbao. https://tech.sina.cn/2020-02-17/detail-iimxxstf2046715.d.html
- 105.Warren TK, Wells J, Panchal RG, Stuthman KS, Garza NL, Van Tongeren SA, Dong L, Retterer CJ, Eaton BP, Pegoraro G, Honnold S, Bantia S, Kotian P, Chen X, Taubenheim BR, Welch LS, Minning DM, Babu YS, Sheridan WP, Bavari S. Protection against filovirus diseases by a novel broad-spectrum nucleoside analogue BCX4430. Nature. 2014;508:402–405. doi: 10.1038/nature13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Richardson P, Griffin I, Tucker C, Smith D, Oechsle O, Phelan A, Stebbing J. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet. 2020;395:e30–e31. doi: 10.1016/S0140-6736(20)30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mulangu S, Dodd LE, Davey RT, Mbaya OT, Proschan M, Mukadi D, Manzo ML, Nzolo D, Oloma AT, Ibanda A, Ali R, Coulibaly S, Levine AC, Grais R, Diaz J, Lane HC, Muyembe-Tamfum J-J, PALM Writing Group, PALM Consortium Study Team (2019) A randomized, controlled trial of Ebola virus disease therapeutics. N Engl J Med 381:2293–2303. 10.1056/NEJMoa1910993 [DOI] [PMC free article] [PubMed]
- 108.Warren TK, Jordan R, Lo MK, Ray AS, Mackman RL, Soloveva V, Siegel D, Perron M, Bannister R, Hui HC, Larson N, Strickley R, Wells J, Stuthman KS, Van Tongeren SA, Garza NL, Donnelly G, Shurtleff AC, Rettere CJ, Gharaibeh D, Zamani R, Kenny T, Eaton BP, Grimes E, Welch LS, Gomba L, Wilhelmsen CL, Nichols DK, Nuss JE, Nagle ER, Kugelman JR, Palacios G, Doerffler E, Neville S, Carra E, Clarke MO, Zhang L, Lew W, Ross B, Wang Q, Chun K, Wolfe L, Babusis D, Park Y, Stray KM, Trancheva I, Feng JY, Barauskas O, Xu Y, Wong P, Braun MR, Flint M, McMullan LK, Chen SS, Fearns R, Swaminathan S, Mayers DL, Spiropoulou CF, Lee WA, Nichol ST, Cihlar T, Bavari S. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mifsud EJ, Hayden FG, Hurt AC. Antivirals targeting the polymerase complex of influenza viruses. Antiviral Res. 2019;169:104545. doi: 10.1016/j.antiviral.2019.104545. [DOI] [PubMed] [Google Scholar]
- 110.Zeldin RK, Petruschke RA. Pharmacological and therapeutic properties of ritonavir-boosted protease inhibitor therapy in HIV-infected patients. J Antimicrob Chemother. 2004;53:4–9. doi: 10.1093/jac/dkh029. [DOI] [PubMed] [Google Scholar]
- 111.Savarino A, Di Trani L, Donatelli I, Cauda R, Cassone A. New insights into the antiviral effects of chloroquine. Lancet Infect Dis. 2006;6:67–69. doi: 10.1016/S1473-3099(06)70361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yan Y, Zou Z, Sun Y, Li X, Xu K-F, Wei Y, Jin N, Jiang C. Anti-malaria drug chloroquine is highly effective in treating avian influenza A H5N1 virus infection in an animal model. Cell Res. 2013;23:300–302. doi: 10.1038/cr.2012.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vincent MJ, Bergeron E, Benjannet S, Erickson BR, Rollin PE, Ksiazek TG, Seidah NG, Nichol ST. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol J. 2005;2:69. doi: 10.1186/1743-422X-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Colson P, Rolain JM, Lagier JC, Brouqui P, Raoult D. Chloroquine and hydroxychloroquine as available weapons to fight COVID-19. Int J Antimicrob Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Biot C, Daher W, Chavain N, Fandeur T, Khalife J, Dive D, Clercq ED. Design and synthesis of hydroxyferroquine derivatives with antimalarial and antiviral activities. J Med Chem. 2006;49:2845–2849. doi: 10.1021/jm0601856. [DOI] [PubMed] [Google Scholar]
- 116.Liu J, Cao R, Xu M, Wang X, Zhang H, Hu H, Li Y, Hu Z, Zhong W, Wang M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020;6:16. doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yao X, Ye F, Zhang M, Cui C, Huang B, Niu P, Liu X, Zhao L, Dong E, Song C, Zhan S, Lu R, Li H, Tan W, Liu D. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ortiz-Alcantara J, Bhardwaj K, Palaninathan S, Frieman M, Baric RS, Kao CC. Small molecule inhibitors of the SARS-CoV Nsp15 endoribonuclease. Virus Adapt Treat. 2010;2:125–133. doi: 10.2147/VAAT.S12733. [DOI] [Google Scholar]
- 119.Buchholz UJ, Bukreyev A, Yang L, Lamirande EW, Murphy BR, Subbarao K, Collins PL. Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity. Proc Natl Acad Sci USA. 2004;101(26):9804–9809. doi: 10.1073/pnas.0403492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cheng Y, Wong R, Soo YOY, Wong WS, Lee CK, Ng MHL, Chan P, Wong KC, Leung CB, Cheng G. Use of convalescent plasma therapy in SARS patients in Hong Kong. Eur J Clin Microbiol Infect Dis. 2005;24:44–46. doi: 10.1007/s10096-004-1271-9. [DOI] [PMC free article] [PubMed] [Google Scholar]