Abstract
Aberration in microRNA expression or DNA methylation is a causal factor for polycystic ovarian syndrome. However, the epigenetic interactions between miRNA and DNA methylation remain unexplored in PCOS. We conducted a novel integrated analysis of RNA-seq, miRNA-seq, and methylated DNA-binding domain sequencing on ovarian granulosa cells to reveal the epigenetic interactions involved in the pathogenesis of PCOS. We identified 830 genes and 30 miRNAs that were expressed differently in PCOS, and seven miRNAs negatively regulated target mRNA expression. 130 miRNAs’ promoters were significantly differently methylated, while 13 were associated with miRNA expression. Furthermore, the hypermethylation of miR-429, miR-141-3p, and miR-126-3p′ promoter was found related to miRNA expression suppression and therefore their corresponding genes upregulation, including XIAP, BRD3, MAPK14, and SLC7A5. Our findings provide a novel insight in PCOS. The consequential reversal of genes silencing may participate in PCOS pathogenesis and served as potential molecular targets for PCOS.
Subject terms: Genetics research, DNA methylation
Introduction
Polycystic ovarian syndrome (PCOS) is one of the most prevalent endocrine disorders with the symptoms of hyperandrogenism, chronic anovulation, and polycystic ovaries [1]. It is also considered to be a common cause of body malfunction in women, with symptoms including hirsutism, acne, obesity, menstrual dysfunction, and infertility. It also appears to be associated with an increased risk of metabolic aberrations, including insulin resistance and hyperinsulinism, type 2 diabetes mellitus, dyslipidemia, cardiovascular disease, and endometrial carcinoma [2, 3].
Ovarian granulosa cells (GCs) have been demonstrated to play a major role in deciding the fate of follicles. In the early stage of follicle development, oocyte apoptosis results in follicular atresia-induced ambient GC death, which results in molecules that are essential for oocyte development and maintenance as well as self-renewal by apoptotic processes [4, 5]. Therefore, it is important to investigate the role of GCs in the pathogenesis of PCOS.
Current research has shown that the influence of multiple factors, including age, the environment/lifestyle, and the disease state environment, can modify the clinical presentation of PCOS via epigenetic modifications [6]. DNA methylation and microRNAs (miRNAs) are two main epigenetic modifications in the regulation of gene expression. MiRNAs are small noncoding RNAs acting as posttranscriptional negative regulators of gene expression, which are involved in the regulation of various diseases such as diabetes, insulin resistance, inflammatory disease, and cancer. Meanwhile, aberrant DNA methylation manifests in both global genome stability preservation and in localized gene promoter changes, which influences the transcription of disease-causing genes [7].
Recently, compelling evidence has indicated the roles of DNA methylation and miRNAs in PCOS, respectively. Previous studies detected significant alteration in genome-wide DNA methylation and transcriptional patterns in human ovaries, GCs, and the adipose tissue of PCOS involved in metabolic disturbances [8–10]. These studies indicated that DNA methylation may be responsible for the development of PCOS. Similarly, the altered miRNA expression detected in human adipose tissue, follicular fluid, GCs, serum, and peripheral blood leukocytes has been found to be associated with PCOS by targeting specific molecules and modulating various hormone-related and apoptosis-related pathways [11, 12].
Though the regulation of miRNAs and DNA methylation are both widely explored in PCOS, the epigenetic interaction mechanisms between miRNAs and DNA methylation remain to be explored. In various human disease including cancers, DNA methylation can regulate miRNA transcription by hyper-/hypo-methylating the promoter regions of miRNAs. Meanwhile, miRNAs can regulate genome-wide DNA methylation patterns by directly targeting DNA methyltransferases and methylation-related critical proteins [13, 14].
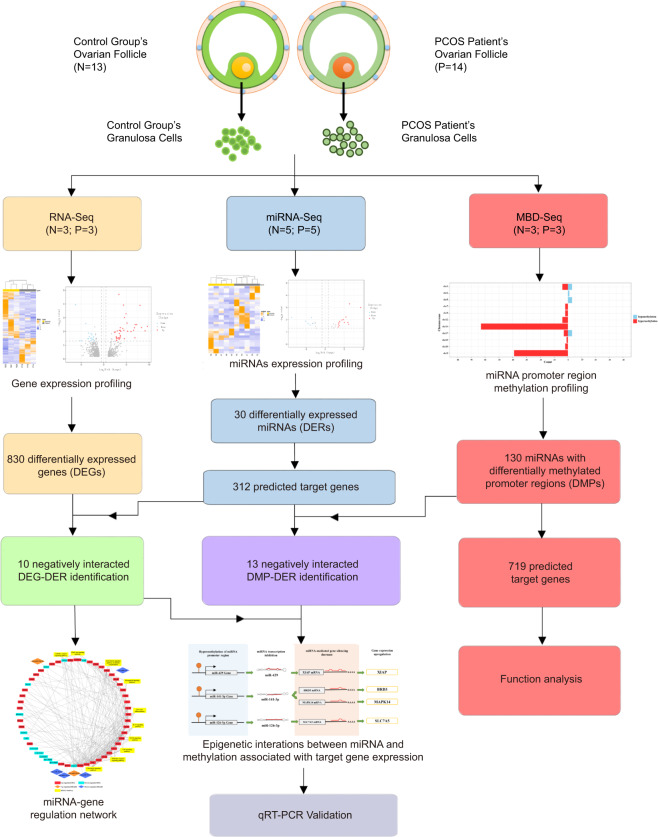
In this study, we identified differentially expressed genes (DEGs) and miRNA in GCs of PCOS patients by transcriptome and small RNA sequencing. Then the miRNA-gene networks were constructed to demonstrate their regulatory functions involved in biological processes and pathways. In addition, DNA methylation levels of miRNA promoter region were determined by methylated DNA-binding domain sequencing (MBD-seq). Furthermore, the correlations between differential miRNA expression and promoter methylation were analyzed. And the epigenetic interactions between miRNA and methylation associated with target gene expression of PCOS were identified and validated by quantitative real-time PCR (qRT-PCR) and methylation specific PCR (MSP). Our results revealed the complex regulatory interaction between miRNA and DNA methylation of their promoter regions in PCOS.
Materials and methods
Patient information and clinical characteristics
Twenty-seven participants (14 PCOS and 13 controls) for this study were recruited from Yuncheng center hospital. The study protocol was approved by the Ethics Committees on Human Research of Yuncheng center hospital (Approve ID: KYLL2019073). The diagnosis of PCOS patients proceeded by achieving at least two of the Rotterdam ESHRE/ASRM (2003) criteria [15]: (1) signs of clinical and/or biochemical hyperandrogenism, (2) ovulatory dysfunction, and (3) polycystic ovarian morphology. The control group were fertile women with male partners experiencing sterility or infertile women with tubal blockages and with normal menstrual cycles, no polycystic ovary morphology, no clinical signs of hyperandrogenism, and no history of endometriosis or other chronic diseases. All participants were non-related Han people age between 20 and 35 years old from the same geographical region undergoing in vitro fertilization (IVF) treatment, and all agreed to participate in this study and provided written informed consent. The preoperative baseline sex hormone levels were measured at 2–5 days after menstruation. The level of thyroid stimulating hormone, follicle stimulating hormone (FSH), luteinizing hormone (LH), testosterone (T), and prolactin (PRL) were measured with the full DXI800 automatic chemiluminescence immunoanalyzer (Beckman Coulter, CA, USA). The number of oocytes and follicles (>14 mm) on the day of oocyte retrieval, and cumulative dose of Gonadotropin (Gn) of the IVF protocol were also calculated.
Long agonist protocol for controlled ovarian hyperstimulation (COH) and isolation of GCs
All PCOS patients and control group underwent COH with long agonist protocol. All of the participators subcutaneously received the GnRH agonist (Diphereline, Ipsen Pharma Biotech, Paris, France) starting at the mid-luteal phase (7 days after ovulation) for 14–16 days. Once adequate pituitary downregulation was confirmed (serum FSH levels <5 U/L, serum LH levels <5 U/L, serum E2 levels <30 pmol/mL, and endometrial thickness <5 mm), the patients subcutaneously received Gn (Gonal-F, Merck, MN, USA; FSH-HP, Livzon, Zhuhai, China) for COH. The dosages of Gn were individualized according to serum hormone levels and transvaginal ultrasonic measurements of the follicles. When the dominant follicle was 18 mm or two follicles were at least 17 mm in diameter, all participators received 6000U hCG (Merck, MN, USA, or Fengyuan pharmaceutical Co, Anhui, China). Transvaginal oocyte aspiration was performed with ultrasound guidance 36 h after injection of hCG. And the GCs were obtained by follicular aspiration from oocyte aspiration process. To avoid blood cells pollution, only follicular aspirates which did not contain any visible blood contamination were used in this study. The GCs were then isolated from the cellular by centrifugation and washed twice with Dulbecco’s improved Eagle medium. And then the cells were stored at −80 °C for further experiments.
Total RNA and genomic DNA extraction
TRIzol (Invitrogen, Waltham, MA, USA) was used to extract total RNA from GC samples following the manufacturer’s protocol. The total RNA quantity was determined with Nanodrop 2000 (Thermo Scientific, Waltham, MA, USA) and quality was assessed by performing 1% agarose gel electrophoresis, stained using with 4S Red Plus Nucleic Acid Stain (Sango, Shanghai, China).
Genomic DNA from GCs of PCOS patients and control group was isolated by using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) as recommended by the manufacturer.
Transcriptome sequencing and data analysis
One microgram of total RNA was used to construct the RNA-seq libraries by the KAPA Stranded RNA-Seq Library Preparation Kit (KAPA Biosystem, Wilmington, MA, USA). The quality of libraries was assessed with Agilent 2100 Bioanalyzer (Agilent, United States).
Sequencing reads were aligned to the UCSC Human Genome Browser hg38 assembly with HISAT2 (version 2.0.5), after data preprocessing [16]. Stringtie (version 1.3.3) was then used to determine the read counts per gene based on Ensembl gene-level annotations (GRCh38.p12) [17]. The final unnormalized counts were assembled into a count-matrix with R (version 3.6.0), and this served as an input for DESeq2 (version 1.24.0) and edgeR (version 3.26.5) [18, 19]. To perform the clustering analysis, the R package ComplexHeatmap was used to calculate hierarchical clustering and visualize it [20]. In this package, the ‘Euclidean’ method was applied in distance calculation and the ‘complete’ method () was applied in combination. Next, the intersections of DEGs identified by DESeq2 and edgeR were used for further analysis, and expression changes with absolute value of |log2 fold changes| ≥ 1 and p values < 0.05 were considered significant.
To validate the DEGs, microarray datasets, GSE10946, GSE31681, and GSE40400, were downloaded from the Gene Expression Omnibus (GEO) database. The samples information of GEO datasets was shown in Table S4. Firstly, we used GEOquery package to obtain normalized data. Then highly correlated (Pearson’s r > 0.8) datasets were selected as validation datasets and combined as single dataset for PCOS and control groups respectively after log transformation. Finally, the DEGs of validation datasets were identified using limma package (p value < 0.05 and |logFC| > 1). In addition, the hypergeometric test p value was calculated after identifying the intersection of DEGs of experiment and validation datasets.
Small RNA sequencing and data analysis
One microgram of total RNA was used to construct the small RNA-seq libraries by the TruSeq® Small RNA Library Prep Kit (NEB, San Diego, CA, USA) following the manufacturer’s protocol. The quality of libraries was assessed with Agilent 2100 Bioanalyzer (Agilent, USA).
Briefly, after trimming the adapter sequences and filtering low-quality reads, all the clean reads were aligned against miRBase (version 22.1) using bowtie2 (version 2.1.0) [21, 22]. Read counts were then calculated through a customized script. In this study, the intersections of differentially expressed miRNA (DEmiRs) identified by DESeq2 and edgeR were considered significant, and expression changes with absolute values of |log2 fold changes| ≥ 1, RPM ≥ 0.1 and p values < 0.05 were considered significant. The target genes of DEmiRs were predicted with the miRTarbase database (Release 7.0) and used for further analysis [23]. Then, the functional analysis was performed by clusterProfiler as mentioned above [24].
PPI network analysis of DEGs and DEmiRs
Based on target genes of the previously predicted DEmiRs, the protein–protein interaction (PPI) network of DEG/DEmiR pairs was built with the STRING database (version 11.0) [25]. Cytoscape (version 3.6.0) was used to visualize the network [26].
MBD sequencing and bioinformatics analysis
Genomic DNA was fragmented with M220 Focused-ultrasonicator (Covaris, Woburn, MA, USA). The fragmented DNA with methylated CpGs was enriched using the MethylMiner™ Kit (Invitrogen, Waltham, MA, USA) as recommended by the manufacturer. The MBD-enriched fractions were used to generate indexed libraries with the NEBNext Ultra DNA Library Prep Kit for Illumina(NEB, San Diego, CA, USA) as recommended by the manufacturer. The quality of libraries was assessed with Agilent 2100 Bioanalyzer (Agilent, USA).
MBD-seq reads with low-quality data filtered out were mapped to the reference genome (Human hg38) using bowtie2 [21]. Then SAM files were converted to sorted BAM files under the constraint of MAPQ30. The MEDIPS package (version 1.24.0) was used for the analysis and comparison of DNA methylation datasets of PCOS and controls [27]. P values < 0.05 and |log2 fold changes| ≥ 1 were considered to show differentially methylated regions. Raw sequence data of MBD-seq were submitted to the GEO database (https://www.ncbi.nlm.nih.gov/geo/) and represented under one super-series along with transcriptome sequencing and small RNA sequencing data (Accession Number: GSE138575).
Methylation analysis of promoter region of miRNA associated with miRNA target gene expression
The transcription start site data of DEmiRs were extracted from the FANTOM5 database, and 100 bp upstream and downstream were considered to be promoter regions of DEmiRs [28]. The identified differentially methylated miRNA promoter regions of the MBD-seq data were defined as differentially methylated promoters (DMPs).
Validation of the DEGs and DEmiRs expression by qRT-PCR
Five hundred nanograms of total RNA was used for reverse transcription by using the PrimeScript RT Master Mix (TAKARA), which contained a mixture of random 6-mers and oligo dT primer. In addition, 500 ng of total RNA was used for reverse transcription using SuperScript II Reverse Transcription (Thermo Scientific, Waltham, MA, USA) with miRNA-specific stem loop primers. qRT-PCR was then carried out with a PowerUp SYBR Green Master Mix (Applied biosystems) at the StepOne Plus qPCR instrument (Applied Biosystems). Primer sequences are shown in Table S1.
Validation of the methylation level by MSP
EZ DNA Methylation-Gold™ Kit was used to conduct bisulfite conversion on the genomic DNA of GCs. And PCR was then performed by using primers designed for hypermethylated and hypomethylated DNA, respectively. Primer sequences are shown in Table S1.
Statistical analysis
Statistical analyses were conducted by using Statistical Package for Social Sciences version 20.0 software (IBM Corp., Armonk, NY, USA). The differences between groups were assessed by a two-sided Student’s t test, and p < 0.05 was considered statistically significant.
Results
Comparison of clinical data between PCOS patients and control group
The clinical data including age, height, weight, circumference, and serum levels of seven sex hormones—follicle-stimulating hormone (FSH), estrogen (E2), progesterone (P), PRL, LH, testosterone (T), human chorionic gonadotropin β (β-HCG), size of follicles, number of oocytes, and cumulative dose of Gn—are shown in Table 1. The PCOS patients showed significantly higher body mass indexes (BMI), increased levels of LH and increased number of follicles (>14 mm) compared to control group (p < 0.05) and lower FSH without statistic significant. Obesity, increased LH level and decreased FSH level are common features of PCOS patients, which are basically consistent with our observation [29]. No significant difference was found for other features. The GCs of the above-mentioned PCOS and control group were then obtained and used for subsequent experiments, as the graphic workflow in Fig. 1 showed.
Table. 1.
Clinical characteristics of polycystic ovarian syndrome (PCOS) and control subjects
| Clinical characteristics | PCOS (n = 14) | Control (n = 13) | p values |
|---|---|---|---|
| Age (years) | 29.3 ± 3.89 | 31.07 ± 4.79 | 0.293 |
| BMI (kg/m2) | 24.82 ± 3.36*a | 21.95 ± 2.58 | 0.005 |
| W (cm) | 75.2 ± 5.59 | 76.8 ± 6.27 | 0.596 |
| H (cm) | 92 ± 4.92 | 91.53 ± 5.26 | 0.166 |
| FSH (mlU/ml) | 5.01 ± 1.68 | 5.28 ± 2.13 | 0.742 |
| E2 (pg/ml) | 55.86 ± 44.08 | 55.67 ± 32.64 | 0.404 |
| P (ng/ml) | 0.78 ± 0.38 | 0.62 ± 0.2 | 0.184 |
| PRL (ng/ml) | 11.06 ± 6.62 | 11.31 ± 6.04 | 0.408 |
| LH (mIU/ml) | 7.76 ± 3.88*a | 3.54 ± 2.25 | 0.004 |
| T (ng/ml) | 1.69 ± 0.94 | 1.54 ± 0.46 | 0.659 |
| β-HCG (mIU/ml) | 0.98 ± 0.55 | 0.82 ± 0.62 | 0.217 |
| Fasting glucose (mmol/L) | 5.1 ± 0.31 | 5.11 ± 0.35 | 0.737 |
| Number of follicles ≥ 14 mm | 13.1 ± 4.53*a | 9.73 ± 2.94 | 0.033 |
| Number of oocytes | 12.44 ± 5 | 11.87 ± 5.04 | 0.787 |
| Gn administered (U) | 2046.5 ± 780.42 | 2192.5 ± 867.59 | 0.672 |
All data are presented as the mean ± SEM
BMI body mass index, W waist circumference, H hip circumference, FSH follicle stimulating hormone, E2 estrogen, P progesterone, PRL prolactin, LH luteinizing hormone, T testosterone, β-HCG human chorionic gonadotropin β, Gn Gonadotropin
*p < 0.05. The values that considered as statistically significant difference between two groups are shown in bold.
aIncreased
Fig. 1.
Workflow of experiment and data analysis
Identification of DEGs in GCs between PCOS patients and controls
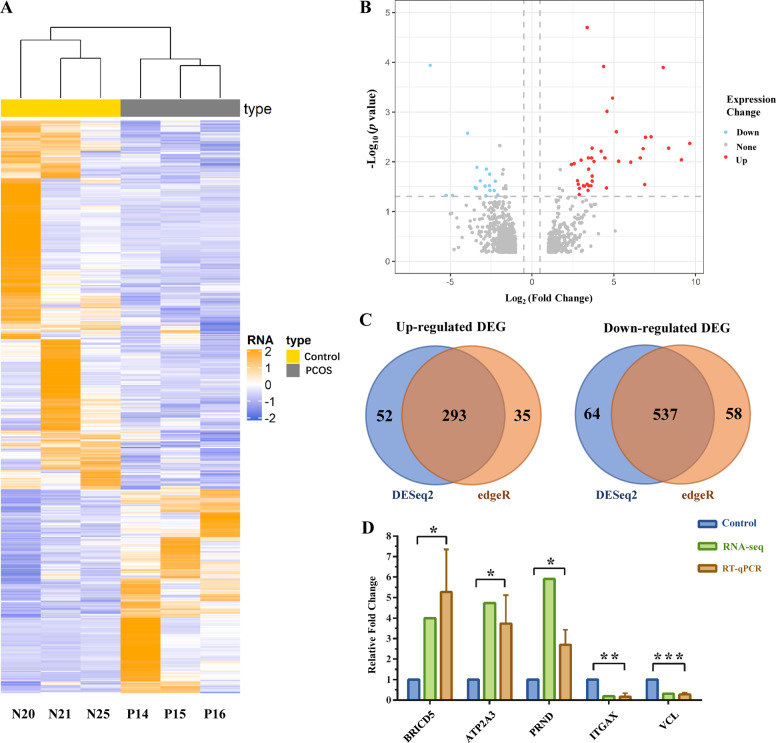
To determine the significant genes in PCOS patients, the transcriptome profiles of GCs from PCOS patients and control group women were sequenced and analyzed. Detailed information regarding the sequencing data is summarized in Table S2. In order to better identify DEGs, the intersection of two different identification methods, DESeq2 and edgeR, was used as the final dataset (Table S3). The hierarchical clustering revealed noticeable partition between GCs from PCOS and control group into two major opposite branches, indicating significantly different expression patterns between two groups (Fig. 2a). The results showed clustering of the downregulated DEGs in the GCs from the PCOS patients compared with controls. In total, 830 genes were found to be DEGs, including 538 upregulated genes and 292 downregulated genes in PCOS vs. control GCs (Fig. 2b, c). Moreover, we tested the reliability of transcriptome sequencing results by using qRT-PCR, and the results demonstrated a high consistency in the randomly selected five genes’ (three upregulated and two downregulated genes) expression level between RNA sequencing data and qRT-PCR (Fig. 2d).
Fig. 2.
Gene expression profile of PCOS vs. control granulosa cells (GCs) by RNA-seq. a Heat map of differentially expressed genes (DEGs) in GCs (P: PCOS GCs; N: Control GCs). b Volcano plot of DEGs in GCs. Upregulated genes are represented by red, and downregulated genes are represented by blue. c Venn diagram of identified DEGs in GCs between the DESeq2 method and edgeR method. Results of DESeq2 are shown as blue, and the edgeR method is orange. d Histogram of quantitative real-time PCR validation of the gene expression identified by RNA-seq. (*0.01 < p < 0.05; **p < 0.01). Color figure online
To validate the significant of identified DEGs, GSE10946, GSE31681, and GSE40400 including 27 control GCs samples and 18 PCOS GCs samples were downloaded from the GEO database. Then the quality control was conducted (Fig. S1) and highly correlated (Pearson’s r > 0.8) datasets were selected (21 Control vs. 18 PCOS) for further analysis. As a result, in total of 2853 DEGs were identified (p value < 0.05 and |logFC| > 1) and 103 of them were found both in experiment and validation pools, of which the hypergeometric test p value was <10−5 (Table S4). Consequently, the identified DEGs in this study were considered statistically significant for subsequent analysis.
Identification of DEmiRs in GCs between PCOS patients and controls
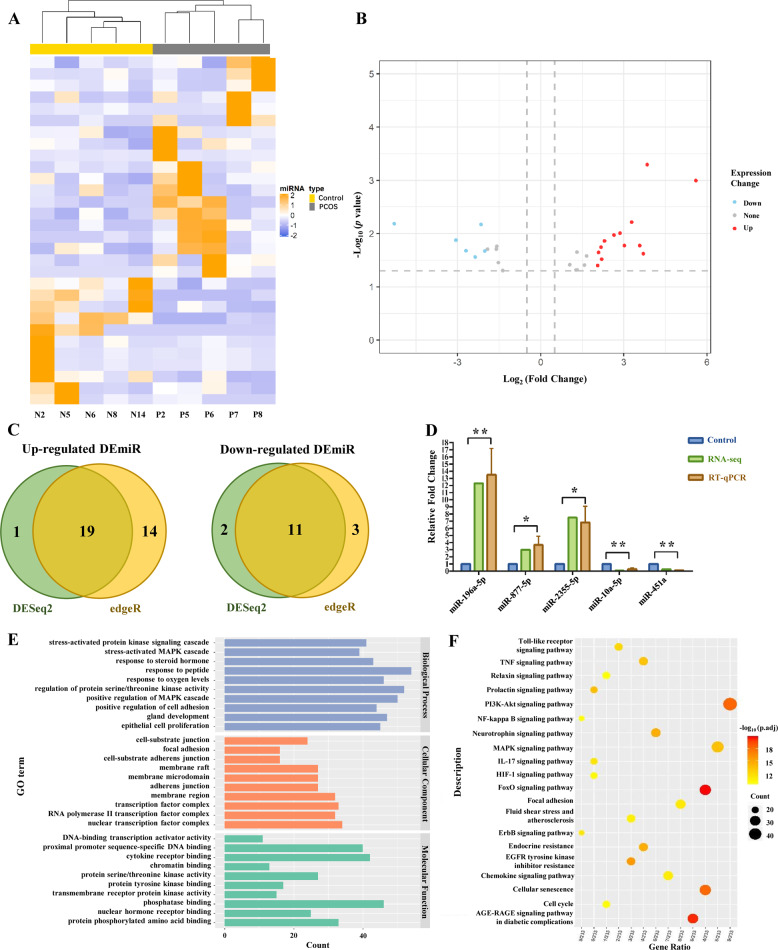
Small RNA libraries were constructed to identify miRNA expression differences between the GCs of PCOS and control group. A total of 23,447,261 and 21,642,858 clean reads which mapped to the genome were acquired from PCOS and control GCs, respectively (Table S2). We identified 30 significant DEmiRs (Table S5) and observed distinct signatures in the PCOS and control groups by performing hierarchical clustering (Fig. 3a). The results showed clustering of miRNAs overexpressed in the GCs from the PCOS patients compared with controls. Based on the intersection of two different identification methods, 19 miRNAs were upregulated and 11 were downregulated significantly in PCOS GCs (Fig. 3b, c). In addition, the results of small RNA sequencing (three upregulated and two downregulated miRNAs) were validated by qRT-PCR to prove the reliability of this method (Fig. 3d).
Fig. 3.
MiRNA profile of PCOS vs. Control GCs by small RNA-seq. a Heat map of DEmiRs in GCs (P: PCOS GCs; N: Control GCs). b Volcano plot of DEmiRs in GCs. Upregulated miRNAs are represented by red, and downregulated miRNAs are represented by blue. c Venn diagram of identified DEmiRs in GCs between DESeq2 method and edgeR method. Results of DESeq2 are shown as green, and the edgeR method is yellow. d Histogram of quantitative real-time PCR validation of the miRNA expression identified by miRNA-seq. (*0.01 < p < 0.05; **p < 0.01). e Gene Ontology (GO) terms of DEmiRs target genes. f Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of DEmiRs target genes. Color figure online
Then, to characterize the regulatory roles of DEmiRs, the potential miRNA target genes were predicted (Table S6). Among the 312 predicted target genes, PTEN, BCL2, HIF1A, CDKN1A, and DNMT1 had the highest count of potential regulatory miRNAs. Moreover, we analyzed the regulatory function of DEmiRs by using the Gene Ontology (GO) database and Kyoto Encyclopedia of Genes and Genomes (KEGG). Certain GO terms were significantly enriched in the biological process, such as cellular response to different stimuli, stress-activated protein kinase (SAPK) signaling cascade (GO: 0031098), stress-activated mitogen-activated protein kinase (MAPK) cascade (GO: 0051403), hormone response, response to steroid hormone (GO: 0048545), and response to peptide (GO: 1901652) (Fig. 3e). In addition, the results of the KEGG analysis indicated that the putative target genes of DEmiRs were involved in the FoxO signaling pathway, the AGE-RAGE signaling pathway in diabetic complications, PI3K-Akt signaling pathway, cellular senescence, and the MAPK signaling pathway (Fig. 3f).
Identification of the DEmiRs that negatively regulated targeted mRNA expression
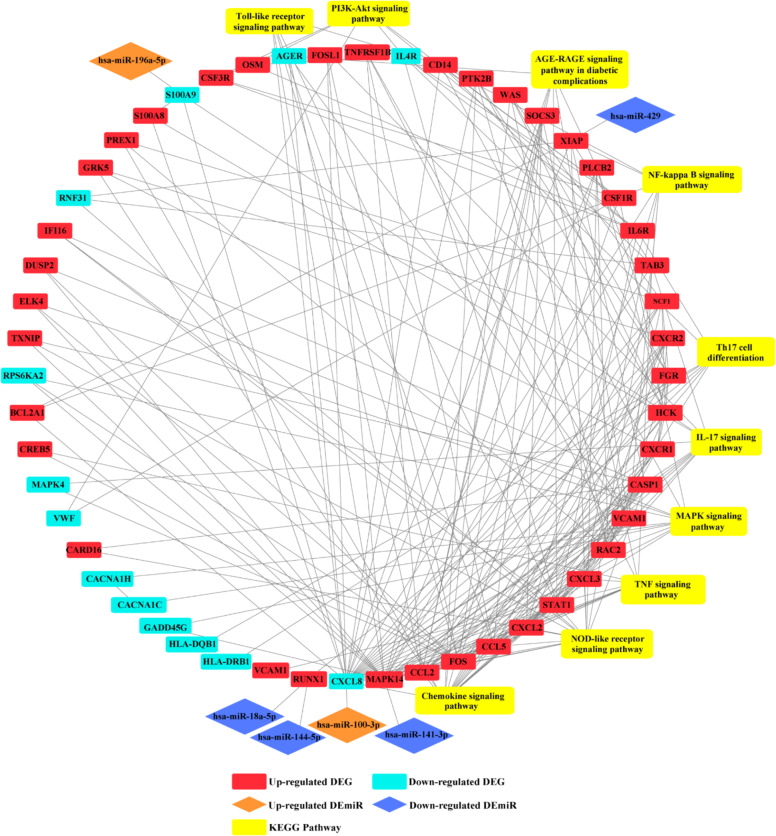
To illustrate the regulatory functions of the miRNAs in PCOS, we found ten pairs of miRNA and genes which had a significant negative correlation in expression, respectively, with the identified 830 DEGs and 30 DEmiRs (Table 2). In order to further investigate the role of miRNA–gene pairs in PCOS, a miRNA–gene interaction network was then constructed by Cytoscape (Fig. 4). The network consists of six paired miRNA gene and 47 DEGs genes that were highly aggregated in the interaction network. As demonstrated in Fig. 4, the DEGs in interaction with each other were associated with several downstream pathways, such as the NOD-like receptor signaling pathway, MAPK signaling pathway, interleukin (IL)-17 signaling pathway, tumor necrosis factor (TNF) signaling pathway, PI3K-Akt signaling pathway, nuclear factor (NF)-kappa B signaling pathway, AGE-RAGE signaling pathway in diabetic complications, Th17 cell differentiation, Toll-like receptor signaling pathway and the chemokine signaling pathway. Furthermore, the upregulation of miR-100-3p and miR-196a-5p was involved in the downstream pathways by inhibiting the expression of the respective target genes CXCL8 and S100A9. In addition, four downregulated miRNAs participated in the regulation of downstream pathways by reducing the transcription repression of target genes including miR-141-3p/MAPK14, miR-144-5/RUNX1, miR-18a-5p/RUNX1, and miR-429/XIAP.
Table. 2.
Negatively regulated miRNA–gene pairs in PCOS
| miRNA | Log2FC (miRNA) | miRNA expression change | Target gene | Log2FC (gene) | Gene expression change |
|---|---|---|---|---|---|
| hsa-miR-18a-5p | –2.15 | Down | RUNX1 | 1.47 | Up |
| hsa-miR-100-3p | 1.58 | Up | CXCL8 | –1.45 | Down |
| hsa-miR-126-3p | –1.92 | Down | SLC7A5 | 1.23 | Up |
| hsa-miR-141-3p | –3.06 | Down | BRD3 | 1.56 | Up |
| hsa-miR-141-3p | –3.06 | Down | MAPK14 | 1.36 | Up |
| hsa-miR-144-5p | –2.02 | Down | RUNX1 | 1.47 | Up |
| hsa-miR-196a-5p | 3.57 | Up | ANXA1 | –2.01 | Down |
| hsa-miR-196a-5p | 3.57 | Up | LSP1 | –1.75 | Down |
| hsa-miR-196a-5p | 3.57 | Up | S100A9 | –3.94 | Down |
| hsa-miR-429 | –1.60 | Down | XIAP | 2.20 | Up |
Log2FC (miRNA), miRNA expression change between GCs of PCOS patients and control group (PCOS GCs vs. Control GCs); Log2FC (Gene), gene expression change between GCs of PCOS patients and control group (PCOS GCs vs. Control GCs)
Fig. 4.
Integrated analysis of DEG–DEmiR pairs. (A red square represents an upregulated gene; a blue square represents a downregulated gene; an orange rhombus represents upregulated miRNA; a blue rhombus represents downregulated miRNA; a yellow square represents a related KEGG pathway). Color figure online
Identification of DMPs of miRNAs in GCs between PCOS patients and controls
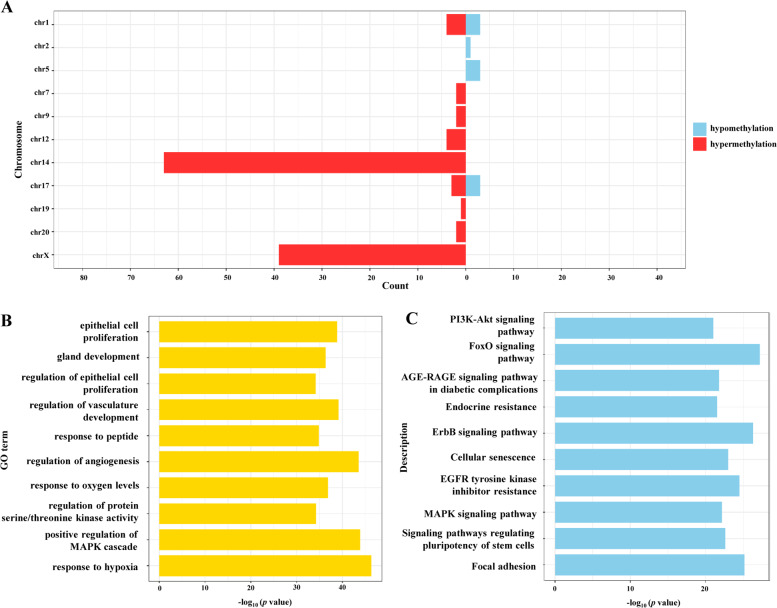
Aberrant DNA methylation of the miRNA promoter region directly regulates the expression of miRNA. To determine the global methylation pattern of miRNA promoter region in PCOS, we assessed the methylation levels and genomic distribution of both hypomethylated and hypermethylated miRNA promoter regions of 130 miRNA and found that they were significantly differently methylated (Table S7). As shown in Fig. 5a, we observed a significant hypermethylation of global miRNA promoter regions in PCOS GCs, as ten DMPs (7.69%) were hypomethylated and 120 DMPs (92.3%) were hypermethylated. In addition, the majority of hypermethylated DMPs were found to be located in chromosome 14 and chromosome X.
Fig. 5.
MiRNA promoter region methylation alternation of PCOS vs. Control GCs by methylated DNA-binding domain sequencing (MBD-seq). a Representation of the distribution of hypomethylated (blue) and hypermethylated (red) regions across genomes. b GO terms of promoter hypermethylated miRNA target genes. c KEGG analysis of promoter hypermethylated miRNA target genes. Color figure online
Then, a function analysis was performed on 719 predicted target genes of the aforementioned 130 miRNAs to understand the biological significance of the miRNAs with hypermethylated promoters in PCOS patients (Table S8). We found that the hypermethylated DEmiRs were mostly involved in the regulation of cell proliferation, development, and stimuli response, such as epithelial cell proliferation (GO: 0050673), gland development (GO: 0048732) and response to peptide (GO: 1901652). In addition, KEGG analysis results showed overrepresented biological processes including the PI3K-Akt signaling pathway, FoxO signaling pathway and MAPK signaling pathway.
Analysis of the regulatory effect of DNA methylation on miRNA and gene expression in GCs of PCOS patients
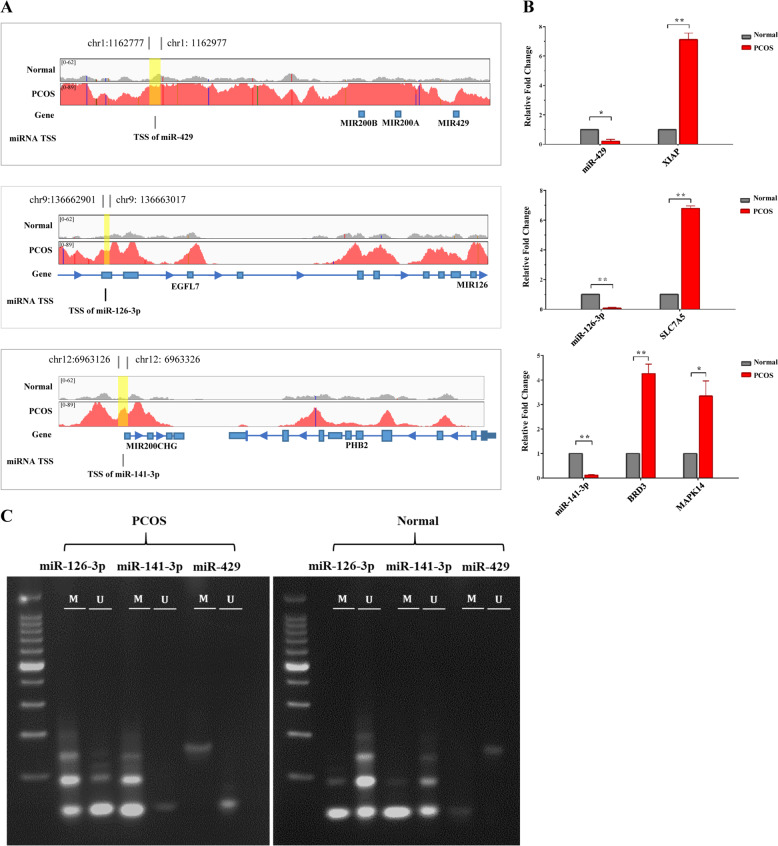
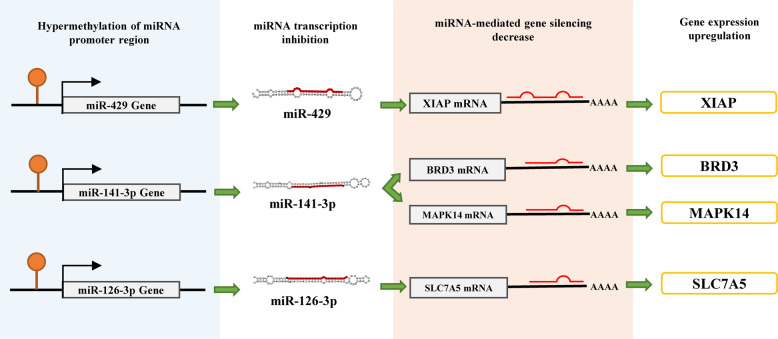
Next, we examined the interactions between miRNA and DNA methylation; 13 miRNA promoter methylation interactions that were significantly associated with expression were identified and are presented in Table 3. To further determine the effect of the interaction between DMPs and DEmiRs on gene expression, we investigated the expression change of promoter aberrant methylated DEmiRs target genes. We found three DEmiRs with DMPs were associated with target gene expression levels. As demonstrated in Fig. 6a, the promoter regions of miR-429, miR-141-3p, and miR-126-3p were significantly hypermethylated in PCOS GCs compared to controls. Subsequently, we validated the expression change of the three DEmiRs with DMPs and their target genes using qRT-PCR (Fig. 6b). The expression of miR-429 and its target gene XIAP were negatively correlated, while the expression patterns of miR-141-3p and its target genes BRD3 and MAPK14 and miR-126-3p and its target gene SLC7A5 were consistent with what we observed in miR-429 and XIAP. Then we also validated the methylation level of the promoter regions of the three DEmiRs between two groups of GCs by MSP (Fig. 6c). Compared with the control group, the promoter regions of miR-429, miR-141-3p, and miR-126-3p showed significant hypermethylation in GCs of the PCOS group. The schematic diagram of interactions between the above-mentioned miRNAs and DNA methylation associated with target-genes expression is presented in Fig. 7.
Table. 3.
The interaction between differentially methylated promoters (DMPs) and DEmiRs in PCOS
| miRNA | Chromosome | Log2FC (miRNA promoter region methylation) | Methylation level change | Log2FC (miRNA) | miRNA Expression change |
|---|---|---|---|---|---|
| hsa-miR-216a-5p | chr2 | −1.98 | − | 1.13 | Up |
| hsa-miR-429 | chr1 | 1.57 | + | −1.60 | Down |
| hsa-miR-126-3p | chr9 | 1.10 | + | −1.92 | Down |
| hsa-miR-141-3p | chr12 | 1.36 | + | −3.06 | Down |
| hsa-miR-1185-5p | chr14 | 1.22 | + | −2.21 | Down |
| hsa-miR-1197 | chr14 | 1.22 | + | −1.24 | Down |
| hsa-miR-412-5p | chr14 | 1.22 | + | −1.23 | Down |
| hsa-miR-431-3p | chr14 | 1.22 | + | −1.26 | Down |
| hsa-miR-431-5p | chr14 | 1.22 | + | −1.16 | Down |
| hsa-miR-433-3p | chr14 | 1.22 | + | −1.37 | Down |
| hsa-miR-485-5p | chr14 | 1.22 | + | −1.02 | Down |
| hsa-miR-487a-3p | chr14 | 1.22 | + | −1.07 | Down |
| hsa-miR-652-5p | chrX | 1.21 | + | −1.33 | Down |
Log2FC (miRNA promoter region methylation), methylation change located in promoter region of miRNAs between GCs of PCOS patients and control group (PCOS GCs vs. Control GCs); Log2FC (miRNA), miRNA expression change between GCs of PCOS patients and control group (PCOS GCs vs. Control GCs); +, hypermethylation; −, hypomethylation
Fig. 6.
The hypermethylation of miRNA promoters leads to miRNA silencing and the disinhibition of target genes. a A genomic view of miR-429, miR-141-3p, and miR-126-3p’s promoter region methylation in GCs of PCOS (red) and control (gray) groups in IGV browser. b Expression changes of miR-429, miR-141-3p, and miR-126-3p and target genes were verified by RT-qPCR (N = 3; P = 3). c Methylation level change of promoter region of miR-429, miR-141-3p, and miR-126-3p (N = 3; P = 3). Color figure online
Fig. 7.
Schematic diagram of interactions between miRNAs and methylation associated with target gene expression
Discussion
Epigenetic modification regulates gene expression during multiple critical physiological and pathological processes, including cell proliferation, apoptosis, inflammation, and carcinogenesis. Among all the epigenetic modifications, DNA methylation and miRNA are both major epigenetic regulators that are reportedly involved in the development of PCOS. Moreover, DNA methylation and miRNA interact with each other and, along with other epigenetic mechanisms, govern the gene expression levels of mRNA and proteins. However, the epigenetic interaction between miRNA and DNA methylation associated with mRNA transcription in PCOS remains unreported.
In the present study, significantly different expression of genes and miRNAs were found between GCs of PCOS patients and controls (Figs. 2a and 3a). We performed RNA-seq and found 830 DEGs in GCs of PCOS patients, with 538 (64.82%) upregulated and 292 (35.18%) downregulated (Fig. 2c). Using small RNA sequencing, 30 DEmiRs were identified, including eight (26.67%) upregulated and 22 (73.33%) downregulated miRNAs, and 312 target genes of DEmiRs were predicted using the miRTarbase database (Fig. 3c). The GO terms of DEmiRs target genes were significantly enriched in biological processes such as cellular response to different stimuli, the cascade of SAPK, and MAPK, which participate in modulating processes such as inflammation and immune activation, development, and apoptosis [30, 31].
Moreover, among the 312 predicted corresponding genes, 22 were identified as significantly differentially expressed based on transcriptome profiles, and the expression of nine genes was found negatively regulated by seven miRNAs by performing miRNA-seq (Table 2). For example, there was a negative correlation between miR-141-3p (downregulation) and two genes, BRD3 (upregulation) and MAPK14 (upregulation), respectively. Also, there were two miRNAs, miR-144-5p (downregulation), and miR-18a-5p (downregulation), targeting the same gene, RUNX1 (upregulation), in PCOS. To further illustrate the regulatory functions of negatively regulated PCOS-related DEGs and DEmiRs, we constructed a PPI network with Cytoscape (Fig. 4). The results illustrated that miR-100-3p (upregulation)/CXCL8 (downregulation), miR-196a-5p (upregulation)/S100A9 (downregulation), miR-141-3p (downregulation)/MAPK14 (upregulation), miR-144-5 (downregulation)/RUNX1 (upregulation), miR-18a-5p (downregulation)/RUNX1 (upregulation), and miR-429 (downregulation)/XIAP (upregulation) compactly participated in the regulation network consisting of 53 identified DEGs. Therefore, these were associated with the NOD-like receptor signaling pathway, MAPK signaling pathway, IL-17 signaling pathway, TNF signaling pathway, PI3K-Akt signaling pathway, NF-kappa B signaling pathway, AGE-RAGE signaling pathway in diabetic complications, Th17 cell differentiation, Toll-like receptor signaling pathway, and chemokine signaling pathway. Among these, the MAPK signaling pathway [32], PI3K-Akt signaling pathway [33], and Toll-like receptor signaling pathway [34] had been reported to be involved in PCOS development previously by performing apoptosis and cell proliferation related processes.
Furthermore, we performed MBD-seq to characterize the miRNA promoter methylation pattern in PCOS GCs. Compared to control group, the majority of DMPs in PCOS were hypermethylated (92.3%) and mainly located in chromosome 14 (48.46%) and chromosome X (30%). In various species, the miRNA distribution pattern is uneven, with miRNA concentrated into certain chromosomes and physically adjacent miRNA genes transcribed as clusters [35]. A previous study reported that certain chromosomes have relatively higher counts of miRNA associated with development and disease processes, such as chromosomes 1, 14, 19, and X [36]. In our study, we found 63 miRNAs with DMPs that are members of the human chromosome 14 microRNA cluster (C14MC), which was reported to control important biological functions in placental and embryonic development [37]. Surprisingly, all aforementioned members of C14MC shared the same hypermethylated promoter region, which is consistent with another study which showed that multiple pre-miRNA transcripts may be produced from the same pre-miRNA cluster. In contrast, 39 miRNAs localized in chromosome X corresponded to nine different hypermethylated promoter regions (Table S7). These observations suggest that underlying epigenetic mechanisms may play an important role in the chromosome 14 and chromosome X-regulated cellular processes in PCOS.
DNA methylation can result in the inhibition of miRNA transcription by hypermethylating the CpG islands in the promoter regions of miRNAs. In this study, we found 12 miRNA silencing events caused by the hypermethylation of the miRNA-coding gene promoter, and one hypomethylation of a miRNA promoter led to its upregulation. In addition, DNA methylation of the miRNA promoter region controlling its corresponding gene expression was observed in miR-429, miR-141-3p, and miR-126-3p. The miRNA-target interactions of miR-141-3p/BRD3 [38], miR-141-3p/MAPK14 [39], miR-126-3p/SLC7A5 [40], and miR-429/ XIAP [41] have been validated in previous study. In this study, all four target genes of these miRNAs, XIAP, BRD3, MAPK14, and SLC7A5, were upregulated due to the reduced suppression of miRNAs, which were caused by their transcription inhibition resulting from promoter hypermethylation (Fig. 6).
MAPK14 and miR-141-3p have both been identified as being related to PCOS. The coding protein of MAPK14 is a member of the mitogen-activated protein (MAP) kinase family and is involved in serval critical biological processes such as cell proliferation, differentiation, and development [31]. Cabergoline (Cb2), one of the dopamine receptor 2 agonizts, has been used to reduce the ovarian hyperstimulation risk associated with PCOS, and MAPK14 was shown to be a key gene implicated in these pathways in Cb2 response, which might be a potential biomarker for further studies of PCOS [42]. MAPK14 was also found to participate in the biosynthesis of human androgen, a hormone whose excessive production was mainly induced by PCOS [43]. Likewise, the differential expression of miR-141-3p was reported in rat models of PCOS induced by letrozole, and consistently with our findings, it showed a significantly decreased expression in the PCOS group and regulated the MAPK signaling pathway [44]. Our study further demonstrated the complicated epigenetic mechanism underlying miR-141-3p/MAPK14 regulation in GC function.
In addition, previous studies have also suggested that XIAP and miR-429 are involved in the mechanism of cell apoptosis and anovulation, respectively. Most of the GCs of polycystic ovaries had fundamental and significant differences in the rate of cell death and proliferation and were demonstrated to be more non-apoptotic than GCs of normal follicles [5]. It had previously been reported in another study that the X-linked inhibitor of the apoptosis protein encoded by XIAP was involved in blocking apoptosis by inhibiting caspase-3 activity, and the overexpression of XIAP has been reported in many diseases such as leukemia and breast cancer [45, 46]. On the other hand, miR-429 was found to play an essential role in mammal ovulation. The knockout of miR-429 led to increasing ZEB1 expression in the pituitary of mice, resulting in unsuccessful ovulation [47]. Our results suggested roles for miR-429 and its target gene XIAP in the regulation of the GCs of PCOS. Further investigation in a larger independent cohort is needed to determine their specifically regulatory function and potential as biological markers in PCOS.
DNA methylation can result in the inhibition or activation of miRNA transcription by hypermethylating or hypomethylating the CpG islands in the promoter regions of miRNAs. Conversely, miRNAs can also regulate DNA methylation by directly targeting DNA methyltransferases and methylation-related critical proteins, thereby affecting the whole-genome methylation pattern [14]. It is worth noting that in this study, DNMT1 was predicted to be a target gene of three DEmiRs including miR-126-3p, miR-18a-5p, and miR-429. However, the expression of DNMT1 was not determined in our transcriptome profile. It is worth further exploring to confirm that miRNAs modulate DNA methylation in PCOS by targeting DNMTs.
To better understand the role of epigenetic regulation in gene expression levels in PCOS, further investigation of how the individual epigenetic regulator interact and influence each other is required. Integrative analysis of the epigenetic interactions between miRNA and DNA methylation can aid in determining the pathogenesis of PCOS and the regulation function of GCs in it. It might also facilitate the foundation of novel therapeutic strategies and identification of diagnostic and prognostic markers.
In conclusion, this work represents the first multi-omics study exploring the alteration of miRNA expression and their promoters’ methylation in GCs of PCOS. We found abundant genes and miRNAs that are affected in GCs of PCOS. We also identified aberrant miRNAs promoters’ methylation that may participate in regulation of miRNA expression, and consequently of target gene expression. Together, these novel findings indicate epigenetic interactions between miRNA and DNA methylation in GCs that are related to PCOS.
Supplementary information
Acknowledgements
This study was supported by the Natural Science Foundation of Shanghai (19ZR1476100), National Infrastructures for Translational Medicine (Shanghai) (TMSK-2020-109), Interdisciplinary Program of Medical Engineering Cross Fund (YG2019GD02, YG2019QNB23, YG2019QNA49, and YG2019QNA52) and Laboratory Innovative Research Program of Shanghai Jiao Tong University (JCZXSJB2019002).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s10038-020-0819-6) contains supplementary material, which is available to authorized users.
References
- 1.Escobar-Morreale HF. Polycystic ovary syndrome: definition, aetiology, diagnosis and treatment. Nat Rev Endocrinol. 2018;14:270–84. doi: 10.1038/nrendo.2018.24. [DOI] [PubMed] [Google Scholar]
- 2.Azziz R, et al. The prevalence and features of the polycystic ovary syndrome in an unselected population. J Clin Endocrinol Metab. 2004;89:2745–9. doi: 10.1210/jc.2003-032046. [DOI] [PubMed] [Google Scholar]
- 3.Norman RJ, Dewailly D, Legro RS, Hickey TE. Polycystic ovary syndrome. Lancet. 2007;370:685–97. doi: 10.1016/S0140-6736(07)61345-2. [DOI] [PubMed] [Google Scholar]
- 4.Matsuda F, Inoue N, Manabe N, Ohkura S. Follicular growth and atresia in mammalian ovaries: regulation by survival and death of granulosa cells. J Reprod Dev. 2012;58:44–50. doi: 10.1262/jrd.2011-012. [DOI] [PubMed] [Google Scholar]
- 5.Das M, et al. Granulosa cell survival and proliferation are altered in polycystic ovary syndrome. J Clin Endocrinol Metab. 2008;93:881–7. doi: 10.1210/jc.2007-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li S, Zhu D, Duan H, Tan Q. The epigenomics of polycystic ovarian syndrome: from pathogenesis to clinical manifestations. Gynecol Endocrinol. 2016;32:942–6. doi: 10.1080/09513590.2016.1203409. [DOI] [PubMed] [Google Scholar]
- 7.Holubekova V, et al. Epigenetic regulation by DNA methylation and miRNA molecules in cancer. Future Oncol. 2017;13:2217–22. doi: 10.2217/fon-2017-0363. [DOI] [PubMed] [Google Scholar]
- 8.Yu Y, et al. Genome-wide screen of ovary-specific DNA methylation in polycystic ovary syndrome. Fertil Steril. 2015;104:145–53. doi: 10.1016/j.fertnstert.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 9.Kokosar M, et al. Epigenetic and transcriptional alterations in human adipose tissue of polycystic ovary syndrome. Sci Rep. 2016;6:1–18. [DOI] [PMC free article] [PubMed]
- 10.Xu J, et al. Comprehensive analysis of genome-wide DNA methylation across human polycystic ovary syndrome ovary granulosa cell. Oncotarget. 2016;7:27899–909. doi: 10.18632/oncotarget.8544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tu J, Cheung AH, Chan CL & Chan W. The role of microRNAs in ovarian granulosa cells in health and disease. Front Endocrinol. 2019;10:174. [DOI] [PMC free article] [PubMed]
- 12.Chen B, Xu P, Wang J, Zhang C. The role of MiRNA in polycystic ovary syndrome (PCOS) Gene. 2019;706:91–6. doi: 10.1016/j.gene.2019.04.082. [DOI] [PubMed] [Google Scholar]
- 13.Chhabra R. miRNA and methylation: a multifaceted liaison. Chembiochem. 2015;16:195–203. doi: 10.1002/cbic.201402449. [DOI] [PubMed] [Google Scholar]
- 14.Wang S, Wu W, Claret FX. Mutual regulation of microRNAs and DNA methylation in human cancers. Epigenetics. 2017;12:187–97. doi: 10.1080/15592294.2016.1273308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Group, T.R.E.A. Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome (PCOS) Hum Reprod. 2004;19:41–7. doi: 10.1093/humrep/deh098. [DOI] [PubMed] [Google Scholar]
- 16.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37:907–15. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11:1650–67. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Love MI, Huber W & Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed]
- 19.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–9. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 21.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47:D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chou C, et al. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46:D296–D302. doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu G, Wang L, Han Y, He Q. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS J Integr Biol. 2012;16:284–7. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szklarczyk D, et al. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lienhard M, Grimm C, Morkel M, Herwig R, Chavez L. MEDIPS: genome-wide differential coverage analysis of sequencing data derived from DNA enrichment experiments. Bioinformatics. 2014;30:284–6. doi: 10.1093/bioinformatics/btt650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lizio M, et al. Update of the FANTOM web resource: expansion to provide additional transcriptome atlases. Nucleic Acids Res. 2019;47:D752–D758. doi: 10.1093/nar/gky1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Azziz R, et al. Polycystic ovary syndrome. Nat Rev Dis Primers. 2016;2:1–18. [DOI] [PubMed]
- 30.Tibbles LA, Woodgett JR. The stress-activated protein kinase pathways. Cell Mol Life Sci. 1999;55:1230–54. doi: 10.1007/s000180050369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.LiuHT Z. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 2002;12:9–18. doi: 10.1038/sj.cr.7290105. [DOI] [PubMed] [Google Scholar]
- 32.Aydos A, et al. Identification of polycystic ovary syndrome (PCOS) specific genes in cumulus and mural granulosa cells. PLoS ONE. 2016;11:e168875. doi: 10.1371/journal.pone.0168875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao H, et al. Beneficial effects of Heqi san on rat model of polycystic ovary syndrome through the PI3K/AKT pathway. DARU. 2017;25:21. doi: 10.1186/s40199-017-0188-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bi X, Zhai Z, Wang S. Identification of the key pathways and genes related to polycystic ovary syndrome using bioinformatics analysis. Gen Physiol Biophys. 2019;38:205–14. doi: 10.4149/gpb_2018049. [DOI] [PubMed] [Google Scholar]
- 35.Chan W, et al. MetaMirClust: Discovery of miRNA cluster patterns using a data-mining approach. Genomics. 2012;100:141–8. doi: 10.1016/j.ygeno.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 36.Ghorai A & Ghosh U. miRNA gene counts in chromosomes vary widely in a species and biogenesis of miRNA largely depends on transcription or post-transcriptional processing of coding genes. Front Genet. 2014;5:100. [DOI] [PMC free article] [PubMed]
- 37.Dini P, et al. Kinetics of the chromosome 14 microRNA cluster ortholog and its potential role during placental development in the pregnant mare. BMC Genomics. 2018;19:954. [DOI] [PMC free article] [PubMed]
- 38.Zhang L, et al. microRNA-141 is involved in a nasopharyngeal carcinoma-related genes network. Carcinogenesis. 2010;31:559–66. doi: 10.1093/carcin/bgp335. [DOI] [PubMed] [Google Scholar]
- 39.Mateescu B, et al. miR-141 and miR-200a act on ovarian tumorigenesis by controlling oxidative stress response. Nat Med. 2011;17:1627–35. doi: 10.1038/nm.2512. [DOI] [PubMed] [Google Scholar]
- 40.Miko E, et al. miR-126 inhibits proliferation of small cell lung cancer cells by targeting SLC7A5. FEBS Lett. 2011;585:1191–6. doi: 10.1016/j.febslet.2011.03.039. [DOI] [PubMed] [Google Scholar]
- 41.Zhu W, et al. miR-200bc/429 cluster modulates multidrug resistance of human cancer cell lines by targeting BCL2 and XIAP. Cancer Chemoth Pharmacol. 2012;69:723–31. doi: 10.1007/s00280-011-1752-3. [DOI] [PubMed] [Google Scholar]
- 42.Ferrero H, et al. Dysregulated genes and their functional pathways in luteinized granulosa cells from PCOS patients after cabergoline treatment. Reproduction. 2018;155:373–81. doi: 10.1530/REP-18-0027. [DOI] [PubMed] [Google Scholar]
- 43.Udhane SS, Flück CE. Regulation of human (adrenal) androgen biosynthesis—new insights from novel throughput technology studies. Biochem Pharmacol. 2016;102:20–33. doi: 10.1016/j.bcp.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 44.Li D, et al. Differential expression of microRNAs in the ovaries from letrozole-induced rat model of polycystic ovary syndrome. DNA Cell Biol. 2016;35:177–83. doi: 10.1089/dna.2015.3145. [DOI] [PubMed] [Google Scholar]
- 45.Saraei R, et al. The role of XIAP in resistance to TNF-related apoptosis-inducing ligand (TRAIL) in Leukemia. Biomed Pharmacother. 2018;107:1010–9. doi: 10.1016/j.biopha.2018.08.065. [DOI] [PubMed] [Google Scholar]
- 46.Hussain AR, et al. XIAP over-expression is an independent poor prognostic marker in Middle Eastern breast cancer and can be targeted to induce efficient apoptosis. BMC Cancer. 2017;17:640. [DOI] [PMC free article] [PubMed]
- 47.Hasuwa H, Ueda J, Ikawa M, Okabe M. MiR-200b and miR-429 function in mouse ovulation and are essential for female fertility. Science. 2013;341:71–3. doi: 10.1126/science.1237999. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.