Abstract
Crataegus kansuensis Wils. is an important wild eco economical species of the family Rosaceae. The complete chloroplast genome reported here is 159,865 bp in length, including two inverted repeats (IRs) of 26,384 bp, which are separated by a large single-copy (LSC) and a small single-copy (SSC) of 87,815 bp and 19,282 bp, respectively. The whole chloroplast genome of C. kansuensis contains 113 genes, including 79 protein-coding genes, 30 transfer RNA, and 4 ribosome RNA. Phylogenetic analysis indicated that C. kansuensis is closely related to that of C. chungtienensis and C. marshallii, and the genus Crataegus L. was sister to the genus Amelanchier Medik.
Keywords: Crataegus kansuensis, chloroplast genome, phylogenetic analysis
Gansu hawthorn (Crataegus kansuensis E.H.Wilson) is a fruit tree within the family Rosaceae, and is naturally distributed in the west and northwest of China. This species has very important ecological and economic values in the ecosystem of the shrub forest community. Hawthorn fruits are edible and used in traditional medicine, and there is a sizable market for natural health products (NHPs) made from hawthorn leaves, flowers, and fruits (Edwards et al. 2012). However, the genus Crataegus L. has long been recognized as one of the taxonomically challenging group due to the problems evoked by polyploidy, hybridization, and apomixis. In the present study, we determined the complete chloroplast genome (cpDNA) sequence of the Gansu hawthorn based on the next-generation sequencing method. The annotated cpDNA has been deposited into GenBank with the accession number MF784433. It is necessary to develop genomic resources for C. kansuensis for its utilization and to provide valuable genetic information for the phylogenetic studies.
We extracted the total genomic DNA from the fresh leaves of a single individual using the method of Li et al. (2013). And the sequencing library was constructed and quantified following the methods introduced by Dong et al. (2017). DNA sample and voucher specimen of C. kansuensis were deposited at the herbarium of the Institute of Chinese Materia Medica (Specimen accession number: PGP00008), China Academy of Chinese Medical Sciences (N39°56′18.96′′, E116°25′38.64′′). The whole-genome sequencing was conducted with 150 bp paired-end reads on the Illumina HiSeq X Ten platform. In all, 3.78 G raw reads were obtained, and after the quality-trimmed using the software CLC Genomics Workbench v7.5 (CLC bio, Aarhus, Denmark), the resultant 3.77 G reads were assembled with the program SPAdes 3.6.1 (Bankevich et al. 2012) (Kmer = 95). The chloroplast genome contigs selected by the Blast program, taken Pyrus pyrifolia (GenBank: AP012207) as the reference. The selected contigs were assembled using Sequencher 4.10. Gene annotation of C. kansuensis was performed using DOGMA annotation (Wyman et al. 2004) and manually corrected for codons and gene boundaries using BLAST searches.
The circular cpDNA of C. kansuensis was 159,865 bp in length, containing two short inverted repeat (IRa and IRb) regions of 26,384 bp, each, which was separated by a large single-copy (LSC) region of 87,815 bp and a small single-copy (SSC) region of 19,282 bp. The GC content of the whole chloroplast genome was 36.6%. The cpDNA of the Gansu hawthorn comprised 113 genes, including 79 protein-coding genes, 4 ribosomal RNA genes, and 30 transfer RNA genes. In these genes, 19 were duplicated in the IR regions and 19 genes contained one or two introns. 17 harbored a single intron, and 2 (ycf3 and clpP) contained double introns.
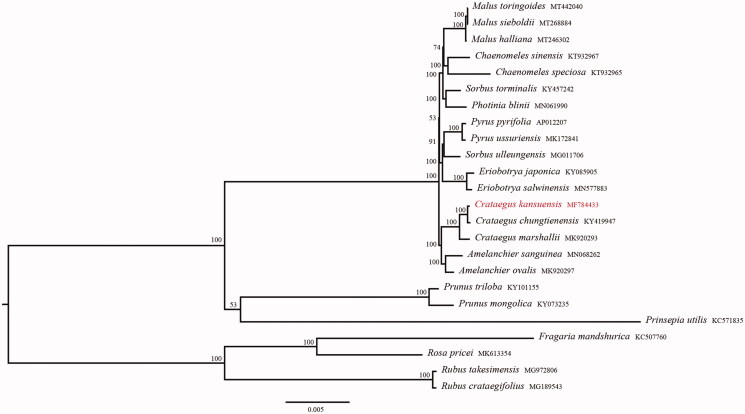
A phylogenetic tree was constructed to confirm the location of C. kansuensis (Figure 1). We used 77 genes within the family Rosaceae to conduct a maximum-likelihood (ML) analysis using IQ-tree with 1000 bootstrap replicates (Nguyen et al. 2015; Zhang et al. 2020). The phylogenetic analysis revealed that tribe Maleae containing genera Malus, Chaenomeles, Sorbus, Photinia, Pyrus, Eriobotrya, Crataegus, and Amelanchier was strongly supported as monophyletic, and the genus Crataegus L. was sister to the genus Amelanchier Medik.. The cpDNA of C. kansuensis is closely related to that of C. chungtienensis and C. marshallii. The complete chloroplast genome reported in this study will be a valuable resource for future studies on genetic diversity, taxonomy, and phylogeny of family Rosaceae.
Figure 1.
Phylogenetic tree reconstruction of 24 taxa using maximum-likelihood (ML) methods based on 77 genes in the chloroplast genome sequences. ML bootstrap support value presented at each node.
Funding Statement
This work was supported by the Fundamental Research Funds for the Central Public Welfare Research Institutes [ZZXT201802].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/nuccore/MF784433.1/, reference number MF784433.
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong W, Xu C, Li W, Xie X, Lu Y, Liu Y, Jin X, Suo Z.. 2017. Phylogenetic resolution in juglans based on complete chloroplast genomes and nuclear DNA sequences. Front Plant Sci. 8:1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JE, Brown PN, Talent N, Dickinson TA, Shipley PR.. 2012. A review of the chemistry of the genus Crataegus. Phytochemistry. 79:5–26. [DOI] [PubMed] [Google Scholar]
- Li JL, Wang S, Jing Y, Wang L, Zhou SL.. 2013. A modified CTAB protocol for plant DNA extraction. Chin Bull Bot. 48(1):72–78. [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL.. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255. [DOI] [PubMed] [Google Scholar]
- Zhang D, Gao F, Jakovlic I, Zou H, Zhang J, Li WX, Wang GT.. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/nuccore/MF784433.1/, reference number MF784433.