Abstract
Morchella conica Pers. is a highly-prized mushroom for its edible and medical values. In this study, we determined the complete mitochondrial genome of M. conica combining both PacBio and Illumina sequencing technologies. The complete mitochondrial genome is 280,763 bp in length with a GC content of 39.88%. We identified a total of 14 core conserved protein-coding genes, 127 non-conserved open reading frames (ncORFs) and 30 tRNA genes in the M. conica mitogenome. However, no large or small rRNA subunits (rnl or rns) were identified in this mitogenome. In addition, we detected two mitochondrial RNase P (rnpB) genes and one ribosomal protein genes (rps3). Phylogenetic analysis was performed among M. conica and 18 other representative fungi from Ascomycota, Basidiomycota and Mucoromycota. Our results showed that M. conica was most closely related to M. importuna. The availability of the M. conica mitochondrial genome will form the basis of genetic breeding program and enhance our understanding of the evolution of this species.
Keywords: Morchella conica, mushroom, mitochondrial genome, PacBio, evolution
Wild mushrooms have increasingly become an important part of human diet for centuries owing to its pleasant taste and nutritional value (Kalač 2009), among which morels were the most highly-prized for their immense economic and scientific values, widely consumed as food and medicine (Elmastas et al. 2007; Gençcelep et al. 2009; Wong and Chye 2009). Morchella conica Pers. is an edible mushroom belonging to the Ascomycota phylum (Masaphy and Zabari 2013). However, large-scale cultivation of M. conica has not been successful, and excessive harvesting without replenishment has led to a reduced effective population size, resulting in a global shortage and soaring market prices (Turkoglu et al. 2006). In this study, we assembled and annotated the complete mitochondrial genome of M. conica to provide genetic information for the future breeding programs.
The M. conica sample was collected from Shizong (103.99E, 24.83 N), Yunnan Province, China. Fresh fruit bodies were harvested and immediately frozen in liquid nitrogen after collection. Genomic DNA was extracted using a modified CTAB method (Porebski et al. 1997). One paired-end library was constructed following the Illumina’s instructions and sequenced on Illumina Hiseq2000 platform. For PacBio sequencing, a 40-kb SMRTbell DNA library was prepared and sequenced on Sequel II platform. The PacBio long reads were then assembled using CANU v1.6 (Koren et al. 2017). The assembled contigs were further polished using the paired-end Illumina reads with Pilon v1.22 (Walker et al. 2014). The mitochondrial genome of M. importuna (NC_045397) (Liu et al. 2020) was used as the reference to extract the mitogenome contigs using blast searches. One contig representing the complete mitochondrial genome was identified based on the sequence similarity. The mitochondrial genome of M. conica was annotated using MFannot webserver (http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl). The whole mitogenome sequence data reported here has been deposited in the Genome Warehouse in National Genomics Data Center (BIG Data Center Members 2020), Beijing Institute of Genomics (China National Center for Bioinformation), the Chinese Academy of Sciences, under accession number GWHANVR01000000 that is publicly accessible at https://bigd.big.ac.cn/gwh.
The mitochondrial genome of M. conica was 280,763 bp in length with a GC content of 39.88%. Fourteen conserved protein-coding genes encoded 3 ATP synthases (atp6, apt8 and apt9), 3 cytochrome oxidases (cox1, cox2 and cox3), apocytochrome b (cob) and 7 subunits of NAD dehydrogenase (nad1, nad2, nad3, nad4, nad5, nad6 and nad4L). Two mitochondrial RNase P (rnpB) genes and one ribosomal protein genes (rps3) were identified in the mitochondrial genome. Besides, 127 non-conserved open reading frames (ncORFs) were identified to be uniformly dispersed in the M. conica mitogenome. We also detected 30 tRNA genes, but no rRNA genes were found in the genome.
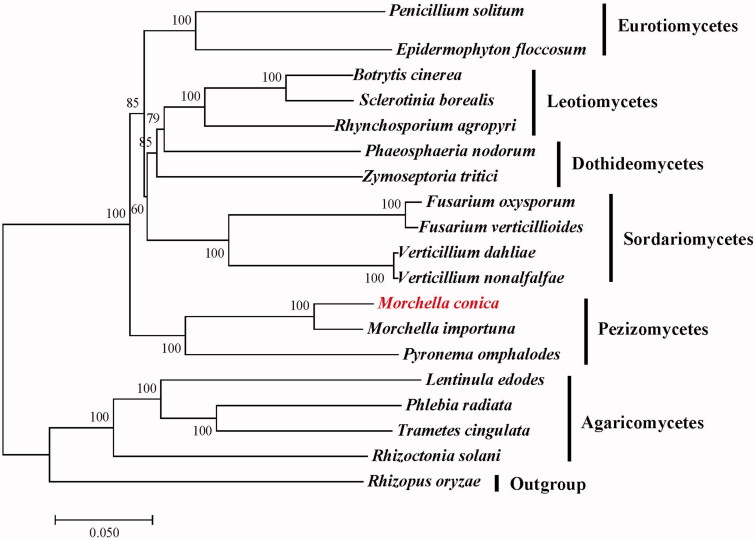
To investigate the phylogenetic position of M. conica we aligned 14 conserved protein-coding genes using MAFFT v7.305 (Katoh and Standley 2013). A maximum likelihood (ML) tree was constructed using Mega 6.0 (Tamura et al. 2013) with 1,000 bootstrap replicates. Our results showed that M. conica is closely related to M. importuna in the genus Morchella (Figure 1).
Figure 1.
Maximum-likelihood (ML) phylogenetic tree of M. conica and related fungal species. The ML-tree is based on 14 conserved core mitochondrial proteins. Bootstraps values (1,000 replicates) are shown at the nodes. All the sequences are currently available in the GenBank database: Botrytis cinerea (KC832409), Epidermophyton floccosum (NC_007394), Fusarium oxysporum (NC_017930), F. verticillioides (NC_016687), Lentinula edodes (NC_018365), M. importuna (NC_045397), Zymoseptoria tritici (NC_010222), Penicillium solitum (NC_016187), Phaeosphaeria nodorum (NC_009746), Phlebia radiata (NC_020148), Pyronema omphalodes (NC_029745), Rhizoctonia solani (NC_021436), Rhynchosporium agropyri (NC_023125), Sclerotinia borealis (NC_025200), Trametes cingulata (NC_013933), Verticillium dahliae (NC_008248) and V. nonalfalfae (NC_029238). Rhizopus oryzae (NC_006836) was served as an outgroup.
Funding Statement
This work was supported by Yunnan Innovation Team Project [2015FA030].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in the Genome Warehouse Database at https://bigd.big.ac.cn/gwh, accession number GWHANVR01000000.
References
- BIG Data Center Members. 2020. Database resources of the national genomics data center in 2020. Nucleic Acids Res. 48(D1):D24–D33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmastas M, Isildak O, Turkekul I, Temur N.. 2007. Determination of antioxidant activity and antioxidant compounds in wild edible mushrooms. J Food Compost Anal. 20(3–4):337–345. [Google Scholar]
- Gençcelep H, Uzun Y, Tunçtürk Y, Demirel K.. 2009. Determination of mineral contents of wild-grown edible mushrooms. Food Chem. 113(4):1033–1036. [Google Scholar]
- Kalač P. 2009. Chemical composition and nutritional value of European species of wild growing mushrooms: a review. Food Chem. 113(1):9–16. [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM.. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27(5):722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Cai Y, Zhang Q, Chen L, Shu F, Ma X, Bian Y.. 2020. The mitochondrial genome of Morchella importuna (272.2 kb) is the largest among fungi and contains numerous introns, mitochondrial non-conserved open reading frames and repetitive sequences. Int J Biol Macromol. 143:373–381. [DOI] [PubMed] [Google Scholar]
- Masaphy S, Zabari L.. 2013. Observations on post-fire black morel ascocarp development in an Israeli burnt forest site and their preferred micro-sites. Fungal Ecol. 6(4):316–318. [Google Scholar]
- Porebski S, Bailey LG, Baum BR.. 1997. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep. 15(1):8–15. [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S.. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkoglu A, Kivrak I, Mercan N, Duru ME, Gezer K, Turkoglu H.. 2006. Antioxidant and antimicrobial activities of Morchella conica Pers. Afr J Biotechnol. 5(11):1146–1150. [Google Scholar]
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 9(11):e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JY, Chye FY.. 2009. Antioxidant properties of selected tropical wild edible mushrooms. J Food Compos Anal. 22(4):269–277. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are openly available in the Genome Warehouse Database at https://bigd.big.ac.cn/gwh, accession number GWHANVR01000000.