Abstract
We developed an orally administered, engineered, bacterium-based, RNA interference-mediated therapeutic method to significantly reduce the symptoms in the most frequently used animal model of inflammatory bowel disease. This bacterium-mediated RNA interference strategy was based on the genomically stable, non-pathogenic E. coli MDS42 strain, which was engineered to constitutively produce invasin and the listeriolysin O cytolysin. These proteins enabled the bacteria first to invade the colon epithelium and then degrade in the phagosome. This allowed the delivery of a plasmid encoding small hairpin RNA (shRNA) targeting tumor necrosis factor (TNF) into the cytoplasm of the target cells. The expression levels of TNF and other cytokines significantly decreased upon this treatment in dextran sulfate sodium (DSS)-induced colitis, and the degree of inflammation was significantly reduced. With further safety modifications this method could serve as a safe and side effect-free alternative to biologicals targeting TNF or other inflammatory mediators.
Keywords: RNA interference; engineered E. coli, treatment of ulcerative colitis; bacterial therapeutic delivery
Graphical Abstract
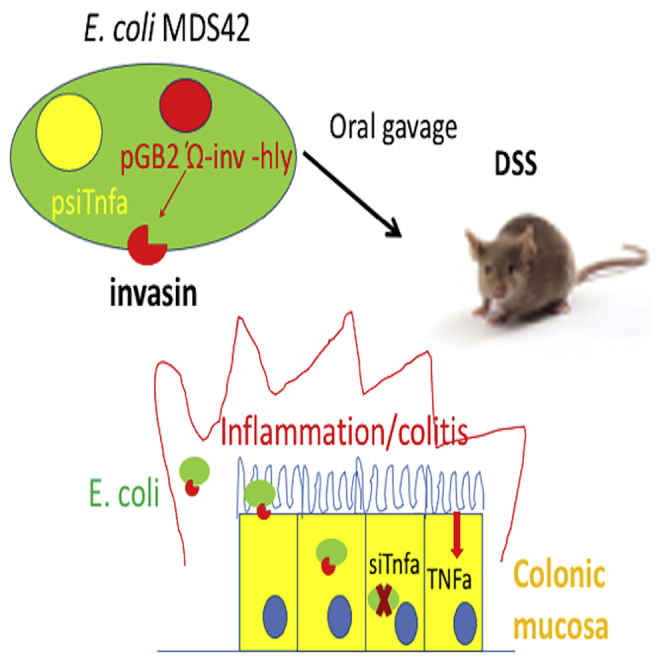
We engineered an orally administered, genomically stable, non-pathogenic E. coli MDS42 strain to constitutively produce invasin and listeriolysin O in order to deliver shRNA to downregulate TNF in colon cells. This bacterium-mediated, RNA interference-based therapeutic method significantly reduced the symptoms of inflammatory bowel disease in animal models.
Introduction
Inflammatory bowel disease (IBD) is an often severe, chronic disease of the colon and small intestine. It is characterized by recurrent inflammation of the gastrointestinal tract and has serious, potentially lethal consequences. Its exact pathomechanism is not known, but it is well established that cytokines play a central role in the development of the disease.1 Several biologics and small-molecule inhibitors were developed to inhibit the pathological activity of the various cytokine pathways.2 Of those, monoclonal antibodies targeting tumor necrosis factor (TNF) have become a cornerstone of IBD therapy.2,3 This treatment option has several drawbacks that warrant the development of alternative treatments, such as high cost, severe potentially lethal side effects, and the fact that up to 50% of patients do not respond to, or lose sensitivity to, this treatment over time.3 While some of the patients resistant to anti-TNF antibody therapy respond to other biologics, such as vedolizumab,4 significant portions of the resistant cases are not responsive to these alternative therapies or become resistant to those with time.5
An alternative approach to the antibody-mediated inhibition of cytokine action is the use of genetically engineered bacteria that could deliver inhibitory molecules, such as small hairpin RNA (shRNA), directly into the target cells specifically against inflammatory mediators. This approach was successfully applied for downregulating COX2 in experimental animals,6 and bacteria were also used to produce elafin inside the gut in order to reduce inflammation.7 In order to directly target one of the cytokines centrally involved in the pathomechanism of IBD, we developed a method to downregulate TNF using a non-pathogenic Escherichia coli MDS42 strain with significantly reduced evolvability.8,9 This strain was modified to constitutively express invasin that allows bacteria to invade mammalian cells by a β integrin-dependent manner and listeriolysin O that destroys phagosomal membranes.6,10,11 These proteins enabled the bacteria first to invade the colon epithelium and then degrade in the phagosome and thus deliver the plasmid encoding shRNA targeting TNF into the cytoplasm of the target cells. The shRNA silencing of TNF significantly reduced intestinal inflammation in a widely used preclinical model of IBD.
Results
Dextran sulfate sodium (DSS)-induced colitis is one of the most commonly used models of ulcerative colitis (UC).12 It induces inflammatory infiltration, a reactive shortening of the colon, and an increase in the expression of inflammatory mediators. In our experiments, we used two different concentrations, 2.5% and 3.5% DSS, to induce colon inflammation of two different levels of severity. (For treatment protocols, see Figure S1A.) As expected, both concentrations of DSS induced a significant increase in the Nancy histological index of UC,13 especially in the distal colon (Figures 1, 2A, and 2B). (For inflammation scores and ulceration scores separately and changes in the distal colon, see Figure S2.) DSS-induced inflammation is accompanied by a reactive shortening of the colon. Both DSS concentrations induced a significant, shortening of the colon (Figures 2C and 2D). Finally, we isolated RNA from the whole colon and measured by quantitative real-time PCR the expression level of several proinflammatory cytokines known to be elevated in IBD.1 3.5% DSS induced a 2-fold induction of TNF, a 100-fold induction of interleukin (IL)-1a, an approximately 150-fold increase of IL-1b, and a 1.4-fold increase of IL-6 (Figure 3). We also investigated the spatial distribution of increased TNF production in the colon by immunohistochemistry (IHC). The diffuse pattern of increased expression suggested that TNF was present both in epithelial cells as well as the immune cells, such as macrophages, infiltrating the lamina propria (Figure 4).
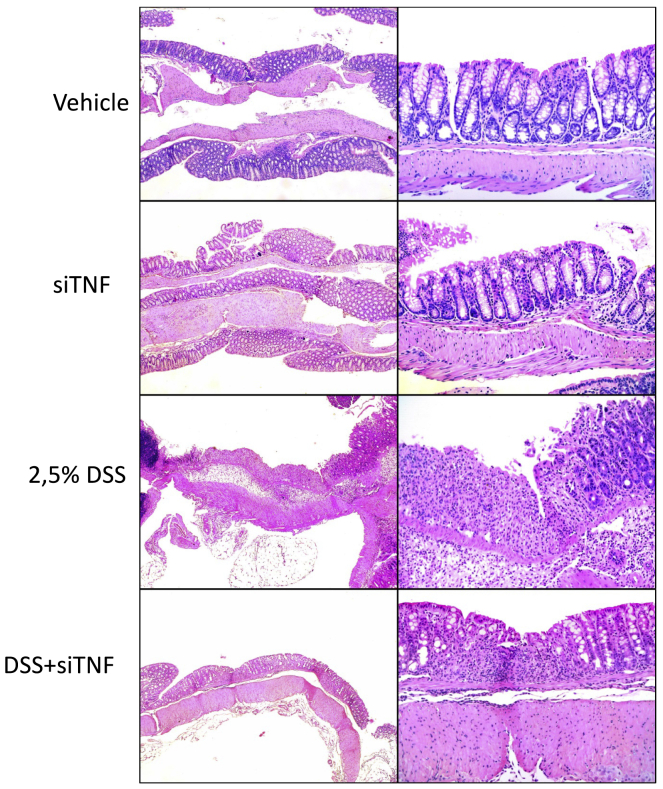
Figure 1.
Histological analysis of the colon obtained from the various experimental groups
Representative histological fields at two magnifications (×50 and ×200) of the various treatment groups as indicated. Vehicle, tap water-administered animal group; siTNF, animals treated with modified E. coli MDS42 strains containing psiTNF and invasive plasmids; DSS, colon specimen of the 2.5% DSS-treated animals; DSS-siTNF, animals administered 2.5% DSS and treated by modified E. coli MDS42 strains containing psiTNF and invasive plasmids.
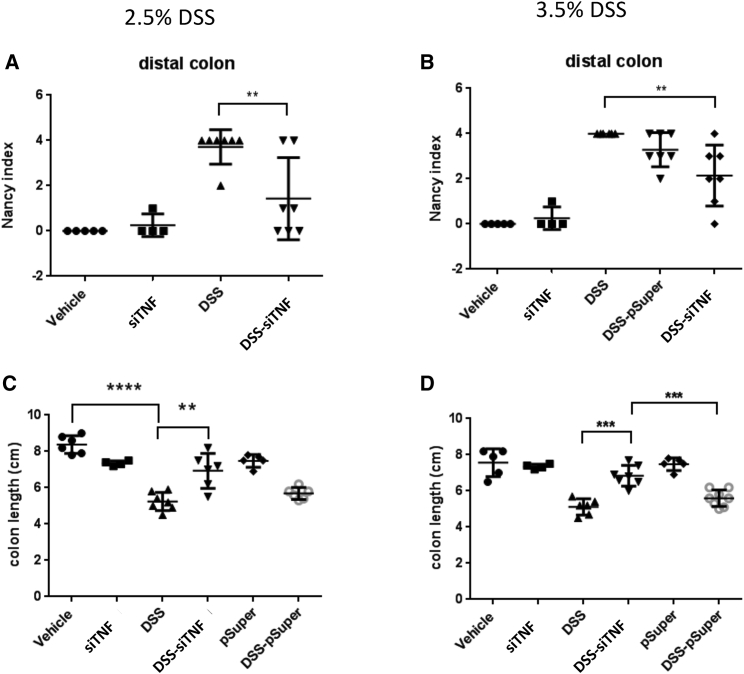
Figure 2.
The Nancy histological index and colon length of the 2.5% and 3.5% DSS-administered animals
Histological analysis was carried out on formalin-fixed, paraffin-embedded (FFPE) colon samples from experimental groups, and the levels of inflammation and ulceration were scored as described in Materials and Methods. (A) The Nancy histological index (UC) of the distal colon for the various treatment groups with 2.5% DSS. (B) The Nancy histological index (UC) of the distal colon for the various treatment groups with 3.5% DSS. (C) Effect of the modified E. coli MDS42 strains containing psiTNF and invasive plasmids on the colon length in the treatment group with 2.5% DSS. (D) Effect of the modified E. coli MDS42 strains containing psiTNF and invasive plasmids on the colon length in the treatment group with 3.5% DSS. Vehicle: tap water-administered animal group; siTNF, animals treated by modified E. coli MDS42 strains containing psiTNF and invasive plasmids; DSS, colon specimen of the DSS-treated animals; DSS-siTNF, animals administered DSS and treated by modified E. coli MDS42 strains containing psiTNF and invasive plasmids; pSuper, animals treated by modified E. coli MDS42 strains containing pSuper (empty vector) and invasive plasmids; DSS-pSuper, animals administered 2.5% DSS and treated by modified E. coli MDS42 strains containing pSuper (empty vector) and invasive plasmids. Data represent the mean (SD). ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
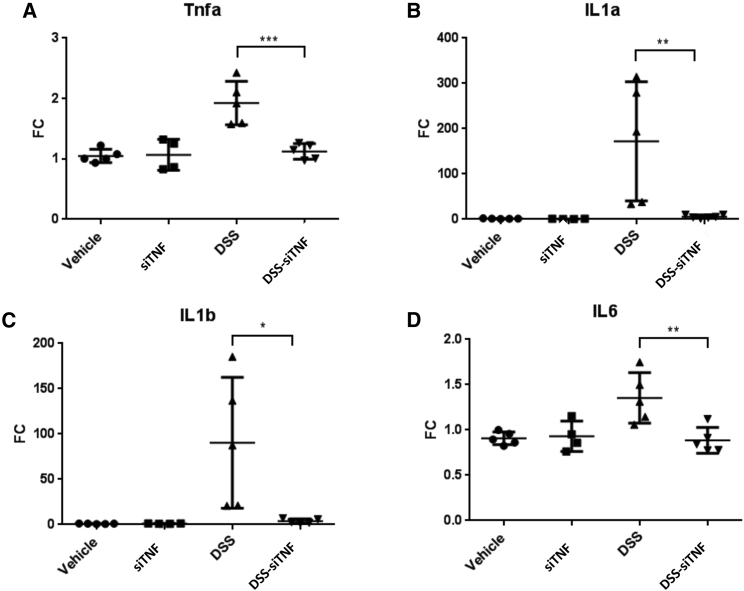
Figure 3.
mRNA expression levels of proinflammatory cytokines in vehicle treated controls and 3,5% DSS induced colitis with and without treatment with the modified E. coli MDS42 strains containing psiTNF
(A) TNF mRNA expression measured on distal colon samples. (B) IL-1a mRNA expression measured on distal colon samples. (C) IL-1b mRNA expression measured on distal colon samples. (D) IL-6 mRNA expression measured on distal colon samples. Vehicle, tap water-administered animal group; DSS, colon specimen of the 3.5% DSS-treated animals; DSS-siTNF, animals administered 3.5% DSS and treated by modified E. coli MDS42 strains containing psiTNF and invasive plasmids. Data represent the means (SD). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
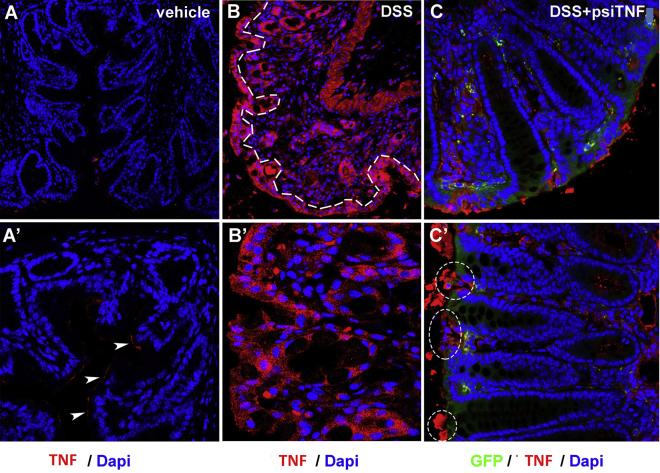
Figure 4.
Epithelial expression of TNF in DSS- and psiTNF-treated mice
(A and A′) Mice treated with vehicle show minimal expression of TNF-α in the distal colon, limited to the apical surface of mucosal epithelial cells (A′, arrowheads). (B and B′) After 7 days of DSS treatment, TNF production increases significantly, where every cell in the epithelial lining of the intestinal mucosa (B, dashed line) shows strong expression of TNF. (C and C′) Animals treated simultaneously with psiTNF plasmid (GFP) and DSS exhibit moderate levels of TNF expression. In contrast to animals not treated with the plasmid, distal colon of psiTNF-treated mice display only non-continuous patches of epithelial TNF expression (C′, encircled areas).
Next, we tested whether the engineered bacteria were able to deliver to the colon the pSuper plasmid and its derivative psiTNF vector, specifically designed to silence TNF. In order to do this, the plasmid was engineered to incorporate a mouse pgk promoter-driven gfp reporter whose expression was detected using both specific PCR primers (Figure S3) and IHC (Figure 5). In particular, we assessed the capacity of the engineered bacteria to deliver their cargo to macrophages, a key inflammatory population that sustains colitis. Double staining with anti-GFP and intestinal macrophage marker anti-Iba1 antibodies shows the presence of GFP-expressing psiTNF plasmid in Iba1+ macrophages in lymphoid aggregates and the lamina propria. GFP was also expressed in mucosal epithelial cells (Figure 5). In order to test the efficacy of the bacterium-delivered gene silencing system, the animals with the DSS-induced IBD were then administered the E. coli MDS42 bacteria specifically engineered to silence TNF. This treatment had a dramatic effect on the DSS-induced colitis. The increase of inflammatory mediators was significantly reduced. For each of the inflammatory mediators measured, the expression returned to baseline levels (Figure 3). Consequently, the Nancy index of UC was also significantly reduced (Figures 2A and 2B). In the case of 2.5% DSS treatment the inflammation score returned to baseline or near baseline levels in the distal colon (Figure S2A). The ulceration score increase was also reduced by 64% (Figure S2C). For the higher DSS concentration (3.5%), both the inflammation and ulceration score were reduced by 50% in the distal colon (Figures S2B and S2D). Finally, the colon length shortening was also significantly reduced to around the level seen in the control animals (Figures 2C and 2D).
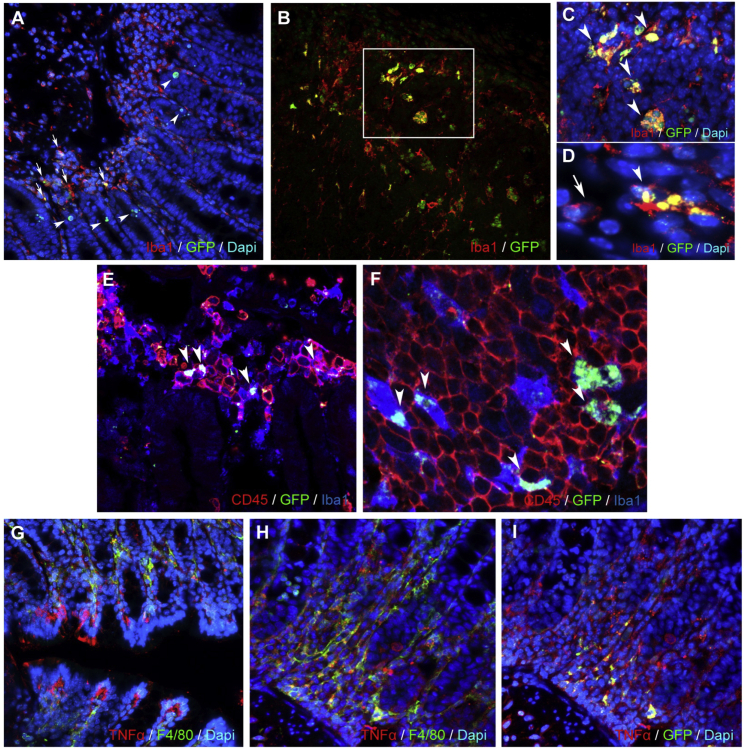
Figure 5.
Immunohistochemical analysis of the distal colon of DSS-administered (8 days), psiTNF-treated (4 days) animals
(A) Distal colon of DSS-administered (8 days), psiTNF-treated (4 days) animals (mucosa, submucosa): psiTNF plasmid (GFP), Iba1+ mucosal macrophages (arrows), and epithelial cells (arrowheads). (B–D) Distal colon of DSS-administered (8 days), psiTNF-treated (4 days) animals (lymphatic aggregate in mucosa): psiTNF plasmid (GFP); GFP/IBA double-positive macrophages (arrowheads). (C and D) High-magnification images: Iba1+GFP+ (arrowheads) and Iba1+GFP− macrophages (arrow in D). (E and F) Distal colon of DSS-administered (8 days), psiTNF-treated (4 days) animals (lymphatic aggregate in mucosa): psiTNF plasmid (GFP); GFP is expressed in CD45+Iba1+ mucosal macrophages (arrowheads in E); only pycnotic and ramified CD45/Iba1 double-positive macrophages express GFP, whereas CD45+Iba1− lymphocytes of round morphology show no plasmid incorporation (F). (G–I) Distal colon of DSS-administered (8 days), psiTNF-treated (4 days) animals (mucosa); psiTNF plasmid (GFP); F4/80+ mucosal macrophages and epithelial cells show strong TNF production (G). (H and I) Expression of TNF and GFP of F4/80+ mucosal macrophages in consecutive sections.
As a control, we engineered the same bacteria carrying an empty vector, without the shRNA for TNF. This treatment did not reduce the various measures of inflammation. Similarly, the colon length of the animals treated with only pSuper and psiTNF plasmids (with invasion plasmid) showed no significant alteration in comparison with the control (water only) (Figures 2C and 2D). The mucosa thickness did not show any significant change in the proximal or distal colon regions for any of the above listed treatments (data not shown).
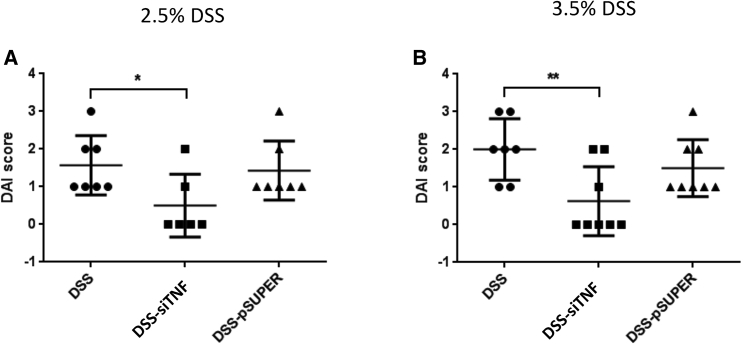
We also determined the DAI (disease activity index) value indicating the severity of the inflammatory processes. Again, this was significantly reduced by the engineered bacteria delivering the TNF-specific shRNA responsible for TNF silencing (Figure 6).
Figure 6.
Effects of DSS-induced colitis on the disease activity index (DAI) of colitis
(A) Effect of the modified E. coli MDS42 strains containing psiTNF and invasive plasmids on the DAI. The animals were administered 2.5% DSS. (B) Biological effect of modified E. coli MDS42 strains containing psiTNF and invasive plasmids were evaluated considering the DAI of 3.5% DSS-induced colitis. DSS, colon specimen of the DSS-treated animals; DSS-siTNF, animals administered DSS and treated by modified E. coli MDS42 strains containing psiTNF and invasive plasmids; DSS-pSuper, animals administered DSS and treated by modified E. coli MDS42 strains containing pSuper (empty vector) and invasive plasmids. Data represent the mean (SD). ∗p < 0.05, ∗∗p < 0.01.
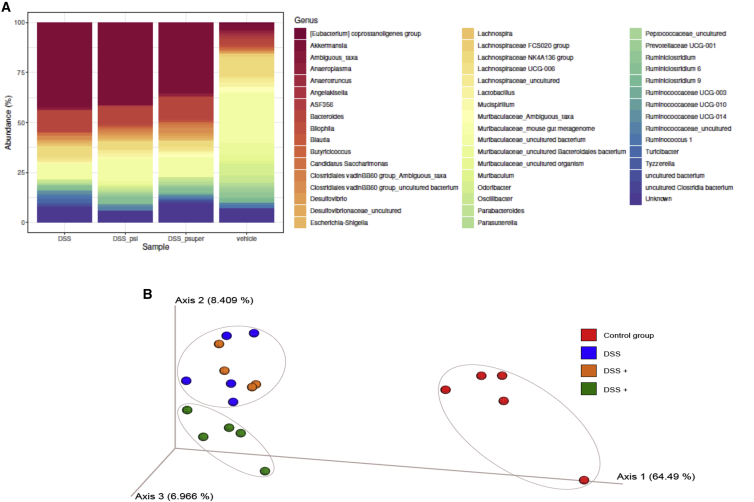
We investigated changes induced by the various experimental conditions in the gut microbiome by sequencing the 16S RNA V3 and V4 hypervariable regions. DSS-induced colitis significantly altered the microbiome relative to control (Figures 7A and 7B). The microbiome composition of the DSS-treated and the DSS + pSuper-treated animals were similar. The DSS-induced animals treated with the bacteria carrying the shRNA for TNF, however, showed differences relative to the disease model, as was demonstrated by a principal component analysis (Figure 7B). These changes were driven by a reduction in the abundance of the predominantly harmful, colitogenic Clostridia, Desulfovibrio, and Parabacteroides (Figure 7A).
Figure 7.
Sequencing of the 16S RNA V3 and V4 hypervariable regions
Characterizing microbiome changes by sequencing the bacterial 16S RNA V3 and V4 hypervariable regions in the vehicle treated control, DSS induced colitis, and the DSS induced colitis that were treated with the modified E. coli MDS42 strain that either carried an empty plasmid or the psiTNF. (A) the relative abundance of the various bacterial genera; (B) Differences in the microbioime composition as expressed by the weighted principal component analysis (wPcoA).
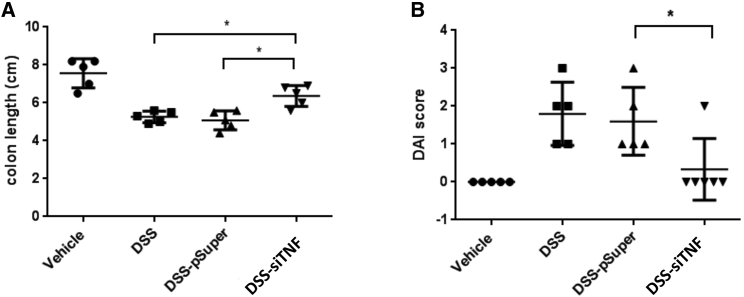
Encouraged by these results we also wanted to test whether the engineered bacteria have a similar therapeutic effect when they are applied after the disease had already developed. Animals were pretreated for 4 days with 2.5% DSS alone, and then the same treatment was continued in the presence or absence of the therapeutic administration of an engineered E. coli strain for 10 days (Figure S1B). The 35%–40% colon length reduction induced by the DSS treatment was significantly reduced to ∼15% in this treatment regimen as well (Figure 8A). DAI measurements showed similar results. The DSS treatment alone induced a DAI of 1.8, which was reduced to 0.3 by the therapeutic bacterial treatment (Figure 8B). Bacteria carrying the empty expression vector had no effect on the DSS-induced disease (Figures 8A and 8B).
Figure 8.
Effect of the 4 + 10 days of 2.5% DSS-induced colitis on colon length and the DAI of colitis
(A) Effect of the modified E. coli MDS42 strains containing psiTNF and invasive plasmids on colon length. The animals were administered 2.5% DSS. (B) Biological effect of modified E. coli MDS42 strains containing psiTNF and invasive plasmids were evaluated considering the DAI of 2.5% DSS-induced colitis. DSS, colon specimen of the DSS-treated animals; DSS-siTNF, animals administered DSS and treated by modified E. coli MDS42 strains containing psiTNF and invasive plasmids; DSS-pSuper, animals administered DSS and treated by modified E. coli MDS42 strains containing pSuper (empty vector) and invasive plasmids. Data represent the mean (SD). ∗p < 0.05, ∗∗p < 0.01.
The therapeutic effect of the engineered E. coli was also reflected by the reduced production of inflammatory mediators and increased production of some of the anti-inflammatory cytokines (Figure S4). Consequently, the Nancy index, the inflammation score, and the ulceration score were significantly reduced (Figure S5).
Discussion
During the past decade, engineered bacteria have emerged as a potential “living” vehicle delivering both diagnostic and therapeutic molecules to the gastrointestinal tract.14 There are several approaches to the therapeutic exploitation of such a biological system in terms of how directly the bacteria are interacting with the host biological system. For example, bacteria were engineered to express phenylalanine-metabolizing enzymes that, upon administration to the digestive tract, would reduce the symptoms of phenylketonuria.15 In this case, the bacteria exert their therapeutic effect inside the colon lumen by breaking down the phenylalanine introduced as part of the diet. Similarly, lactic acid bacteria exerted their therapeutic effect by producing elafin inside the gut, thereby reducing the inflammation in the same preclinical model that we used in this work.7 In these cases, both the bacteria and the therapeutic effectors produced by them remained in the intestinal lumen and did not invade the colon wall. These approaches, therefore, did not change directly the intracellular levels of disease mediators. Considering the often aggressive and therapy resistant nature of IBD,16 we hypothesized that suppressing the expression of TNF inside the target cells by delivering shRNA may exert a more dramatic therapeutic effect. Efficient intracellular silencing of genes by engineered bacteria was first demonstrated by Xiang et al.17 using E. coli-expressing invasin, listeriolysin O cytolysin, and specific shRNA that could silence the β-1 catenin gene in the colon of experimental animals. Spisni et al.6 used a strategy to silence cyclooxygenase-2 to treat experimental models of IBD by introducing the therapeutic bacteria in enemas. In the present study, we further adjusted this approach to target one of the key mediators of IBD, TNF, using an orally applicable delivery method. Our results suggest that this minimally invasive direct delivery of therapeutic modulators can cause a significant reduction of inflammation.
An appealing feature of this technology is its flexibility to target other genes. There are rare forms of IBD, such as the very early onset IBD (VEOIBD), in children.18 These patients often fail to respond to standard therapy, but the disease can be linked to well-defined mutations or gene dysfunctions, which could form the basis of alternative therapeutic approaches. Since the bacterial delivery technology presented here can either suppress or overexpress specific genes, our delivery method can be engineered to correct specific gene malfunctions.
There are serious safety concerns about the use of live bacteria in the therapeutic setting. Bacteria may exchange genetic material with other bacteria in the colon, which may lead to uncontrollable or difficult to control consequences.19 Therefore, in contrast to earlier efforts, we used the genetically stable E. coli MDS 42 strain. More than 20% of the genome of this strain, including transposons, was deleted, and the recombination and conjugation abilities of the bacteria were eliminated. Thus, the risk of horizontal gene transfers was minimized. This risk could be further reduced by strategies that would introduce mechanisms into the bacterial vectors leading to their self-destruction within a few days in the colon but still able to deliver the therapeutic effect.
Materials and methods
Animals
Adult (8–10 weeks old) male FVB/Ant mice were obtained from the local colony bred at the Medical Gene Technology Unit (specific pathogen-free [SPF] level) at the Institute of Experimental Medicine (Budapest, Hungary). Animals were housed at the minimal disease (MD) level, three to five mice per cage, under controlled environmental conditions: temperature, 21°C ± 1°C; humidity, 65%; 12-h light/12-h dark cycle; lights on at 7:00 am. Mice had free access to rodent food and water. All procedures were conducted in accordance with the guidelines set by the European Communities Council (86/609/EEC/2 and 2010/63 Directives of European Community), and the protocol was approved by the Institutional Animal Care and Use Committee of the Institute of Experimental Medicine (Budapest, Hungary) (permit no. PEI/001/29-4/2013).
UC model generation and animal treatment
In animal models, UC can be induced by DSS administration. The experimental animals were 10-week-old FVB/Ant male mice. Mice were treated with drinking water mixed with 2.5% and 3.5% DSS, which was administered for 8 days or 14 days. The engineered bacterial cells were grown in Luria-Bertani (LB) liquid medium at 37°C. The overnight bacterial culture was harvested by centrifugation and washed twice with sterile tap water to eliminate the antibiotic contamination (ampicillin and spectinomycin). Finally, the cell pellet was suspended in sterile tap water and administered to the animals. The engineered bacterial cells (2–5 × 108 colony-forming units [CFU]) and vehicle (sterile tap water) were administered daily by oral gavages. During treatment, DAI values were observed. At the end of the experiment, the animals were anesthetized and decapitated. The length of the colon samples was measured by a digital caliper. Colon samples were frozen and fixed for histological processing. Intestinal contents were frozen.
Cloning procedures
The nucleotide sequences encoding shRNAS for efficiently silencing the TNF were designed using Serial Cloner 2.6.1 software.
The forward (5′-GATCCCCGACAACCAACTAGTGGTGCTTTTCAAGAGAAAGCACCACTAGGTGGTTGTCTTTTTG-3′) and reverse (5′-AGCTCAAAAAGACAACCAACTAGTGGTGCTTTCTCTTGAAAAGCACCACTAGTTGGTTGTCGGG-3′) synthetic DNA oligonucleotides (Microsynth Seqlab, Germany) were heated in annealing buffer (100 mM NaCl, 50 mM HEPES [pH 7.4]) at 90°C for 5 min and allowed to cool to room temperature. The annealed double-stranded synthetic DNA oligonucleotides were ligated into the pSUPER.retro.neo GFP vector system (Oligoengine, USA) after linearization with BglII and HindIII enzymes. During the cloning steps, the ligation was heated to 90°C for 5 min and allowed to cool to room temperature. Subsequently, conventional heat shock transformation was introduced into the prepared E. coli MDS42 strain. Recombinant bacteria were selected on LB medium containing ampicillin. Plasmid constructs encoding the resulting psiTNF-α (shRNA) precursors were verified by nucleotide sequence assay and restriction enzyme digestion. Using the high-speed plasmid mini kit (Geneaid), a plasmid was isolated from recombinant colonies and digested with EcoRI and HindIII enzymes. The digested plasmids were checked by agarose gel electrophoresis (1% agarose [Sigma], 1× Tris-boric acid EDTA buffer [pH 8.5]). The empty vector carries a 1,000-bp-long insert, while the plasmids containing the ligated synthetic oligonucleotides are 281-bp-long inserts that result after double digestion. Plasmids from positive colonies were analyzed by Sanger’s nucleotide assay using a specific sequencing primer (5′-GGAAGCCTTGGCTTTTG-3′). The invasin gene of the pGB2-Ω plasmid was verified by PCR amplification and restriction enzymatic digestion PstI. PstI removes the fragment from the plasmid that contains the invasin and listeriolysin.
Quantitative real-time PCR
Frozen tissue samples were homogenized in TRI reagent solution (Ambion, USA), and total RNA was isolated with a QIAGEN RNeasy mini kit (QIAGEN, Valencia, CA, USA) according the manufacturers’ instructions. To eliminate genomic DNA contamination, DNase I treatment was used and 100 μL of RNase-free DNase I (1 U of DNase) (Thermo Scientific, USA) solution was added. Sample quality control and the quantitative analysis were carried out by NanoDrop 2000 (Thermo Scientific, USA). Amplification was not detected in the RT-minus controls. The cDNA synthesis was performed with a high-capacity cDNA reverse transcription kit (Applied Biosystems, USA). Primers for the comparative Ct experiments were designed by the Primer Express 3.0 program and Primer-BLAST software. The primers (Microsynth, Balgach, Switzerland) were used in the real-time PCR reaction with Fast EvaGreen qPCR master mix (Biotium, USA) on an ABI StepOnePlus instrument (Applied Biosystems, USA) and are listed in Table S1. The gene expression was analyzed by the ABI StepOne 2.3 program. The amplicon was tested by melt curve analysis on an ABI StepOnePlus instrument. Experiments were normalized to gapdh expression.
Library construction for Illumina MiSeq sequencing of the 16S rRNA amplicons
Standard library preparation was performed according to the Illumina (San Diego, CA, USA) 16S metagenomic sequencing library preparation protocol (15044223 rev. B). The V3 and V4 hypervariable regions of the bacterial 16S rRNA gene were sequenced with Illumina MiSeq benchtop sequencer generating amplicons of ∼460 by using the universal primer set as follows: 341F (5′-CCTACGGGNGGCWGCAG-3′) and 785R (5′-GACTACHVGGGTATCTAATCC-3′) primers flanked by Illumina overhang adaptor sequences (forward overhang, 5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG-3′; reverse overhang, 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG-3′). After completion of the amplicon PCR with 2× KAPA HiFi HotStart ReadyMix, dual indexing of the 96 with adaptor sequences (i7-N7xx, 12 items; i5-S5xx, 8 items) was performed using the Illumina Nextera XT index kit (FC-131-1001/2). PCR cleanups and amplicon size selections were carried out with KAPA Pure Beads (KAPA Biosystems) based on the technical data sheet (KR1245, v3.16) of the manufacturer, resulting in final ∼580- to 630-bp libraries. Every time, verifications were done with PCR Agilent D1000 screen tapes (5067-5582) and D1000 reagents (5067-5583). The 16S amplicon libraries for each sample were quantified with qPCR, normalized with respect to amplicon sizes, and pooled into a single library in equal molar quantities. Finally, 5 μL of the pooled 4 nM DNA library pool was prepared for sequencing on the Illumina MiSeq platform. The library pool was denatured with 0.2 M NaOH and diluted to 8 pM final concentration. Sequencing was carried out with MiSeq reagent kit v3 (618 cycle; MS-102-3003) following the manufacturer’s protocols (Illumina, San Diego, CA, USA). All data are publicly available and can be accessed through the PRJNA687617 from the NCBI Sequence Read Archive (SRA).
Sequencing read preparation for downstream analysis
Paired-end reads were demultiplexed by the integrated software of the Illumina MiSeq sequencing machine. The FastQ files were imported into the QIIME 2 pipeline (https://qiime2.org/) according to the “Atacama soil microbiome” tutorial. Residual adaptor sequences (5′-CTGTCTCTTATACACATCT-3′) were trimmed from the 3′ end of the reads with Cutadapt software integrated in the QIIME 2 pipeline. Quality trimming was performed by using DADA2 software,20 and the denoising parameters were set as follows: forward read length was set to 299 bases; for the reverse reads, the length was set to 249 bases. No trimming from the start of the reads was applied.
Operational taxonomic unit (OTU) generation
Multiple sequence alignment was performed with MAFFT software,21 and reads were taxonomically classified using a naive Bayesian classifier trained with the Greengenes reference database (v13_8) by selecting mapping points according to the forward-reverse primer set that was used for amplifying the 16S V3–V4 regions of the bacterial community (341F, 806R). Aligned sequences were clustered into OTUs with a threshold level of 99% sequence identity. Singletons were discarded in order to reduce the likelihood of sequence artifacts interfering with farther downstream analysis. A phylogenetic tree was constructed with a FastTree plugin.22
Biodiversity analysis
For sample normalization, 8,300 sequencing (read) depths were set. Beta diversity analysis involved measuring weighted/unweighted UNIFRAC distances.23 For visualization of beta diversity matrices, principal coordinate analysis (PCoA) plots were generated using the EMPeror24 plugin.
Data visualization
For data mining (preparation), the QIIME 2 pipeline was used. QIIME artifact files were exported from the pipeline resulting BIOM files that were converted to TSV files, which were used with external visualization packages. Pie charts were constructed with R programming language (https://www.R-project.org)25 using the ggplot2 package (http://ggplot2.org).26
GFP tracing in the colon
Total RNA was isolated and cDNA synthesis was performed from the frozen colon samples. The gfp expression in the colon tissue was demonstrated by PCR using specific oligonucleotide primers (forward, 5′-GGA CGA CGG CAA CTA CAA GA-3′; reverse, 5′-AAG TCG ATG CCC TTC AGC TC-3′). The PCR product was separated and stained on agarose gel.
Pathology (histology)
Formalin-fixed colon samples were processed and embedded in paraffin using the standard protocol. Sections of 4 μm were stained with hematoxylin and eosin. Slides were analyzed by an expert pathologist in a blinded manner using the Nancy histological index for UC.13
Histological procedures
For immunofluorescence (IF), distal colon samples of adult (8- to 10-week-old) FVB/Ant mice, treated with 2.5% DSS and psiTNF-α plasmid, were fixed in 4% paraformaldehyde (PFA) in PBS for 24 h. After extensive washing in PBS, the samples were infiltrated with 15% sucrose/PBS overnight at 4°C. The medium was changed to 7.5% gelatin containing 15% sucrose at 37°C for 2 h, and the tissues were rapidly frozen at −60°C in isopentane (Sigma). Frozen sections were cut at 10 μm with a Shandon cryostat for confocal laser-scanning microscopy, collected on poly-l-lysine-coated slides (Sigma).
Immunofluorescence and image analysis
IF was performed as previously described.27 Briefly, primary antibodies anti-GFP (R&D Systems), anti-TNF-α (Abcam), anti-Iba1 (Invitrogen), and anti- F4/80 (Invitrogen) were diluted 1:200 in 1% PBS-BSA. For IF, frozen sections were incubated with primary antibodies overnight at 4°C, followed by relevant secondary antibodies (Alexa Fluor 647-, 546-, and 488-conjugated anti-rabbit immunoglobulin G [IgG] and Alexa Fluor 488-conjugated anti-goat IgG, from Invitrogen) used for 1 h at room temperature (RT). Cell nuclei were visualized by DAPI (4′,6-diamidino-2-phenylindole; Vector Laboratories). Sections were covered with aqueous Poly/Mount (Polysciences, Warrington, PA, USA) and examined with a Zeiss LSM 780 laser-scanning confocal microscope (Zeiss). The ZEN software package (black edition, Zeiss) was used for the scanning of 4-μm and 1.8-μm-thick optical slices of 10 megapixel (MP) resolution for low- and high-magnification images, respectively. Images were compiled with ImageJ and Adobe Photoshop CS6 software packages.
Statistical analysis
Data are expressed as means ± SD. The data were first subjected to a Kolmogorov-Smirnov normality test. Data passing this test were analyzed by one-way ANOVA followed by the Tukey’s post hoc test. When the data showed non-Gaussian distribution, the Kruskal-Wallis test was used. Statistical analysis was performed using GraphPad Prism version 6 software (GraphPad, USA). p ≤0.05 was considered significant.
Acknowledgments
This work was supported by the National Research Development and Innovation Office Hungary (grants KFI_16-1-2017-0245, 109622 and 124424 to K. J. K., and 109744 to S. F.).
Author contributions
Conception and design: S.F., Z.S., and K.J.K. Development of methodology: S.F., D.K., Z.W., and D.D. Acquisition of data: S.F., N. Szeőcs, Z.G., D.K., B.J., T.P., F.S., and D.D. Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): S.F., N. Solymosi, Z.S., I.H., Z.G., A.S., M.P., D.D., N.N, K.J.K., and F.S. Writing, review, revision of the manuscript: S.F., N. Solymosi, I.Z., Z.S., D.D., and K.J.K.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtm.2020.11.010.
Supplemental information
References
- 1.Neurath M.F. Cytokines in inflammatory bowel disease. Nat. Rev. Immunol. 2014;14:329–342. doi: 10.1038/nri3661. [DOI] [PubMed] [Google Scholar]
- 2.Lamb C.A., Kennedy N.A., Raine T., Hendy P.A., Smith P.J., Limdi J.K., Hayee B., Lomer M.C.E., Parkes G.C., Selinger C., IBD guidelines eDelphi consensus group British Society of Gastroenterology consensus guidelines on the management of inflammatory bowel disease in adults. Gut. 2019;68(Suppl 3):s1–s106. doi: 10.1136/gutjnl-2019-318484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Papamichael K., Lin S., Moore M., Papaioannou G., Sattler L., Cheifetz A.S. Infliximab in inflammatory bowel disease. Ther. Adv. Chronic Dis. 2019;10 doi: 10.1177/2040622319838443. 2040622319838443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pagnini C., Pizarro T.T., Cominelli F. Novel pharmacological therapy in inflammatory bowel diseases: beyond anti-tumor necrosis factor. Front. Pharmacol. 2019;10:671. doi: 10.3389/fphar.2019.00671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feagan B.G., Rutgeerts P., Sands B.E., Hanauer S., Colombel J.-F., Sandborn W.J., Van Assche G., Axler J., Kim H.-J., Danese S., GEMINI 1 Study Group Vedolizumab as induction and maintenance therapy for ulcerative colitis. N. Engl. J. Med. 2013;369:699–710. doi: 10.1056/NEJMoa1215734. [DOI] [PubMed] [Google Scholar]
- 6.Spisni E., Valerii M.C., De Fazio L., Cavazza E., Borsetti F., Sgromo A., Candela M., Centanni M., Rizello F., Strillacci A. Cyclooxygenase-2 silencing for the treatment of colitis: a combined in vivo strategy based on RNA interference and engineered Escherichia coli. Mol. Ther. 2015;23:278–289. doi: 10.1038/mt.2014.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Motta J.-P., Bermúdez-Humarán L.G., Deraison C., Martin L., Rolland C., Rousset P., Boue J., Dietrich G., Chapman K., Kharrat P. Food-grade bacteria expressing elafin protect against inflammation and restore colon homeostasis. Sci. Transl. Med. 2012;4:158ra144. doi: 10.1126/scitranslmed.3004212. [DOI] [PubMed] [Google Scholar]
- 8.Umenhoffer K., Fehér T., Balikó G., Ayaydin F., Pósfai J., Blattner F.R., Pósfai G. Reduced evolvability of Escherichia coli MDS42, an IS-less cellular chassis for molecular and synthetic biology applications. Microb. Cell Fact. 2010;9:38. doi: 10.1186/1475-2859-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pósfai G., Plunkett G., 3rd, Fehér T., Frisch D., Keil G.M., Umenhoffer K., Kolisnychenko V., Stahl B., Sharma S.S., de Arruda M. Emergent properties of reduced-genome Escherichia coli. Science. 2006;312:1044–1046. doi: 10.1126/science.1126439. [DOI] [PubMed] [Google Scholar]
- 10.Young V.B., Falkow S., Schoolnik G.K. The invasin protein of Yersinia enterocolitica: internalization of invasin-bearing bacteria by eukaryotic cells is associated with reorganization of the cytoskeleton. J. Cell Biol. 1992;116:197–207. doi: 10.1083/jcb.116.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grillot-Courvalin C., Goussard S., Huetz F., Ojcius D.M., Courvalin P. Functional gene transfer from intracellular bacteria to mammalian cells. Nat. Biotechnol. 1998;16:862–866. doi: 10.1038/nbt0998-862. [DOI] [PubMed] [Google Scholar]
- 12.Perše M., Cerar A. Dextran sodium sulphate colitis mouse model: traps and tricks. J. Biomed. Biotechnol. 2012;2012:718617. doi: 10.1155/2012/718617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marchal-Bressenot A., Salleron J., Boulagnon-Rombi C., Bastien C., Cahn V., Cadiot G., Diebold M.-D., Danese S., Reinisch W., Schreiber S. Development and validation of the Nancy histological index for UC. Gut. 2017;66:43–49. doi: 10.1136/gutjnl-2015-310187. [DOI] [PubMed] [Google Scholar]
- 14.Riglar D.T., Silver P.A. Engineering bacteria for diagnostic and therapeutic applications. Nat. Rev. Microbiol. 2018;16:214–225. doi: 10.1038/nrmicro.2017.172. [DOI] [PubMed] [Google Scholar]
- 15.Isabella V.M., Ha B.N., Castillo M.J., Lubkowicz D.J., Rowe S.E., Millet Y.A., Anderson C.L., Li N., Fisher A.B., West K.A. Development of a synthetic live bacterial therapeutic for the human metabolic disease phenylketonuria. Nat. Biotechnol. 2018;36:857–864. doi: 10.1038/nbt.4222. [DOI] [PubMed] [Google Scholar]
- 16.Nielsen O.H., Ainsworth M.A. Tumor necrosis factor inhibitors for inflammatory bowel disease. N. Engl. J. Med. 2013;369:754–762. doi: 10.1056/NEJMct1209614. [DOI] [PubMed] [Google Scholar]
- 17.Xiang S., Fruehauf J., Li C.J. Short hairpin RNA-expressing bacteria elicit RNA interference in mammals. Nat. Biotechnol. 2006;24:697–702. doi: 10.1038/nbt1211. [DOI] [PubMed] [Google Scholar]
- 18.Moran C.J., Klein C., Muise A.M., Snapper S.B. Very early-onset inflammatory bowel disease: gaining insight through focused discovery. Inflamm. Bowel Dis. 2015;21:1166–1175. doi: 10.1097/MIB.0000000000000329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wegmann U., Carvalho A.L., Stocks M., Carding S.R. Use of genetically modified bacteria for drug delivery in humans: revisiting the safety aspect. Sci. Rep. 2017;7:2294. doi: 10.1038/s41598-017-02591-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price M.N., Dehal P.S., Arkin A.P. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lozupone C., Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vázquez-Baeza Y., Pirrung M., Gonzalez A., Knight R. EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience. 2013;2:16. doi: 10.1186/2047-217X-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.R Core Team . R Foundation for Statistical Computing; 2014. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 26.Wickham H. Springer; 2016. ggplot2: Elegant Graphics for Data Analysis, Second Edition. [Google Scholar]
- 27.Dora D., Arciero E., Hotta R., Barad C., Bhave S., Kovacs T., Balic A., Goldstein A.M., Nagy N. Intraganglionic macrophages: a new population of cells in the enteric ganglia. J. Anat. 2018;233:401–410. doi: 10.1111/joa.12863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.