Abstract
Morchella eohespera Beug, Voitk & O’Donnell is a typical black morel species. In this study, using the Nanopore sequencing platform, we characterized its whole mitochondrial (mt) genome sequence. Mt genome of M. eohespera is composed of circular DNA molecules of 243,963 bp, which encoded 102 protein-coding genes (PCGs), two ribosomal RNA genes (rRNA), and 31 transfer RNA (tRNA) genes. The base composition of M. eohespera mitogenome is as follows: A (30.40%), T (29.30%), G (20.8%), and C (19.5%). The phylogenetic analysis suggested that M. eohespera was closely related to the congeneric M. importuna.
Keywords: Morchella eohespera, mitochondrial genome, phylogenetic relationship
Morchella eohespera Beug, Voitk, & O’Donnell (Morchellaceae) is a typical black morel species, which widely distributed in the central Europe, North American, and the central parts of China (Du et al. 2016; Voitk et al. 2016). The modern medical research showed that the morel species possesses various physiologically active properties, such as anti-oxidative, anti-inflammatory, anti-microbial, immunostimulatory, anti-tumor, etc. (Nitha et al. 2007). However, in recent years, the overexploitation and climatic oscillations have resulted in the rapidly decline of the natural resources of morel. Therefore, it is urgent to protect and manage the filed resources of morel species. The genome of species provides the important information to the management and conservation of bio-resources. In the study presented here, we characterized the complete mitochondrial genome of M. eohespera.
The M. eohespera M200 strain was collected from Maco River Forest Farm of Qinghai province, China (N:32.6738; E:101.0773). The tissue materials and voucher specimen (No. MCRFF2019200) of M. eohespera were stored in Northwest University. Total DNA was extracted from the myclia using the fungal DNA kit (Sangon Biotech, Shanghhai, China) according to the manufacturer’s instructions. Four nuclear gene fragments (ITS, EF1-α, RPB1, and RPB2) were amplified to species identification. Mitochondrial genome sequencing was performed using Nanopore platform (GOOLA GENE, Wuhan, China). The long reads were assembled by CANU software (Koren et al. 2017). The mitochondrial ORFs were identified automatically with Mfannot and RNAweasel using genetic code 4 (http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl). Functional assignments were performed based on sequence similarity to characterize fungal mitochondrial proteins using BLASTP searches against Nr and InterPro databases. The tRNAscan-SE version 2.0 (Lowe and Eddy 1997) was used to predict tRNA genes.
The mitogenome of M. eohespera was assembled as circular DNA molecules with 243,963 bp in length, which encoded 102 protein-coding genes (PCGs), 31 tRNA genes, and two ribosomal RNA genes. The overall base composition of M. eohespera is as follows: A (30.40%), T (29.30%), G (20.8%), and C (19.5%). The complete and annotated mitogenome genome of M. eohespera has been submitted to GenBank with the accession number MT635617.
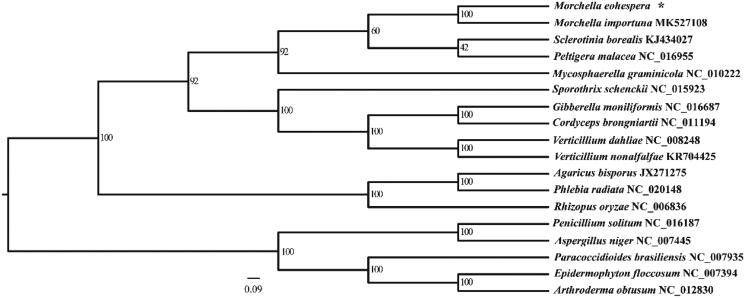
The whole mt genomes of M. importuna and sixteen other fungal species were downloaded from NCBI. The fourteen conserved protein genes (atp6, atp8, atp9, cob, cox1, cox2, cox3, nad1, nad2, nad3, nad4, nad4L, nad5, and nad6) were used for phylogenetic analysis, the multiple sequence alignment was performed by MAFFT version 7.158b software (Katoh et al. 2002). The phylogenetic tree was constructed via RAxML software (Stamatakis 2014) with 1000 bootstrap replicates. The results indicated that M. eohespera was closely related to the congeneric M. importuna with high bootstrap support (Figure 1).
Figure 1.
Phylogenetic tree based on fourteen combined mitochondrial protein-coding genes set. *The newly generated mitochondrial genome of Morchella eohespera.
Funding Statement
This work was co-supported by the Youth program of Natural Science Foundation of Science and Technology Department of Qinghai Province (2018-ZJ-963Q), the Key R & D and transformation projects of science and Technology Department of Qinghai Province (2017-NK-151), the Fourth National Survey of Traditional Chinese Medicine Resources (Grants no. 2017-66, 2018-43 and 2019-39) and the Open Foundation of Key Laboratory of Resource Biology and Biotechnology in Western China (Ministry of Education) (Grants no. ZSK2017007 and ZSK2019008).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the results of this research are openly available in GenBank at https://www.ncbi.nlm.nih.gov, reference number MT635617.
References
- Du XH, Zhao Q, Xu J, Yang ZL.. 2016. High inbreeding, limited recombination and divergent evolutionary patterns between two sympatric morel species in China. Sci Rep. 6:22434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T.. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM.. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27(5):722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR.. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25(5):955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitha B, Meera CR, Janardhanan KK.. 2007. Anti-inflammatory and antitumour activities of cultured mycelium of morel mushroom, Morchella esculenta. Curr Sci India. 92:235–239. [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voitk A, Beug MW, O'Donnell K, Burzynski M.. 2016. Two new species of true morels from Newfoundland and Labrador: cosmopolitan Morchella eohespera and parochial M. laurentiana. Mycologia. 108(1):31–37. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the results of this research are openly available in GenBank at https://www.ncbi.nlm.nih.gov, reference number MT635617.