ABSTRACT
Background
The majority of existing studies aimed at investigating the incidence and prevalence of multidrug-resistance by bacteria have been performed in healthcare settings. Relatively few studies have been conducted in community settings, but these have consistently shown a high prevalence of multidrug-resistant bacteria in low- and middle-income countries (LMICs).
Objectives
To provide an appraisal of the evidence on the high prevalence of multidrug-, extensive drug-, and pandrug-resistance in commensal Escherichia coli isolates from human sources in community settings in LMICs.
Methods
Using the preferred reporting items for systematic reviews and meta-analyses (PRISMA) guidelines, PubMed, EMBASE, MEDLINE, Web of Science, CINAHL, and Cochrane Library databases were systematically searched with the search string: ‘Enterobacteriaceae’, OR ‘E. coli’, OR ‘Escherichia coli’, AND ‘antibiotic resistance’, OR ‘antimicrobial resistance’, OR ‘drug-resistance’, AND ‘prevalence’, OR ‘incidence’, OR ‘morbidity’, OR ‘odds ratio’, OR ‘risk ratio’, OR ‘confidence interval’, OR ‘p-value’, OR ‘rate’. Data were extracted and proportional meta-analysis was performed using the Freeman–Tukey transformation random effect model.
Results
The prevalence of multidrug-, extensive drug- and pandrug-resistance were extracted from articles that met our inclusion criteria and pooled together after a systematic screening of 9,369 items. The prevalence of multidrug-resistance was 28% of 14,336 total cases of isolates tested, 95% CI: 23–32. Extensive drug-resistance was 24% of 8,686 total cases of isolates tested, 95% CI: 14–36. Lastly, pandrug-resistance was 5% of 5,670 total cases of isolates tested, 95% CI: 3–8.
Conclusion
This paper provides an appraisal of the evidence on the high prevalence of multidrug-, extensive drug- and pandrug-resistance by commensal E. coli in community settings in LMICs. Our results call for greater effort to be placed at the community level in the design of new and improved public health policies to counter the global threat of antibiotic-resistant infections and bacteria.
SPECIAL ISSUE: Antimicrobial Resistance
KEYWORDS: Antibiotic resistance, multidrug resistance, commensal Escherichia coli, community settings, health policy
Background
Antibiotic resistance (ABR) is one of the most significant global health crises of our time[1–4]. Antibiotics are medicines used to treat bacterial infections [5]. Resistance occurs when the antibiotics become ineffective in blocking one or more of the pathways involved in protein, nucleic acid, cell wall, or folate synthesis, that are essential for bacterial survival. In the USA, more than 2.8 million people are infected by resistant bacteria, and at least 35,000 of these patients die each year [1]. In the European Economic Area, 25,000 and 33,000 people died of infections from resistant bacteria in 2007 and 2015, respectively, indicating that the burden caused by antibiotic resistance is increasing [6,7]. In fact, the World Health Organization predicted that by 2050, the annual number of deaths caused by antimicrobial resistance will increase from 700,000 deaths to 10 million deaths per year globally [8]. The worrying fact is that antibiotic resistance is increasing far more rapidly than the development of new antibiotics [9]. Low- and middle-income countries (LMICs) suffer the most from antibiotic resistance because their healthcare systems lack the resources to contain numerous infectious diseases that are becoming very difficult and expensive to treat due to antibiotic resistance. In high-income countries, stringent policies to limit antibiotic prescribing have been extensively employed to contain the spread of resistant bacteria [10]. Unfortunately, this has not been the case in LMICs, especially at the community level, where antibiotics can be purchase from community pharmacists without a doctor’s prescription. Antibiotic use has been associated with the carriage of resistant commensal E. coli in healthy children in community settings worldwide [11].
Extensive antibiotic use leads to an alteration in the gut microbiome [12], and the development of resistance by commensal bacteria to antibiotics through natural selection [13]. One of these commensal bacteria is Escherichia coli, one of the main reservoirs for transmitting antibiotic resistance to pathogenic bacteria through the exchange of mobile genetic elements [14,15]. E. coli is an Enterobacteriaceae, which is on the World Health Organization (WHO) global critical priority list for research, discovery, and development of new antibiotics. Although LMICs bare the highest-burden of ABR, one cannot exclude the fact that a collective approach involving all sectors is required to reverse its course [1,2,16]. It is well recognized that an emphasis has not been placed at the community level to contain the spread of multidrug-resistant bacteria [17], despite some studies consistently showing the existence of a high incidence and prevalence of multidrug-resistant commensal E. coli. If this situation is not addressed, some of the gains made in modern medicine will be reversed due to infections caused by multidrug-resistant bacteria becoming more challenging and expensive to treat. This paper aimed to provide an appraisal on the evidence of a high prevalence of multidrug-, extensive drug-, and pandrug-resistant commensal E. coli isolated from healthy humans in LMICs community settings (i.e. locations outside of a hospital, such as schools and homes). These findings should prove useful to researchers, farmers, community pharmacists, policymakers, and advocacy groups among others, to guide the formulation, amendments, and effective implementation of antibiotic resistance-related policies to prevent and contain the spread of multidrug-resistant bacteria.
Methods
Design
We systematically identified and synthesized studies that met the inclusion criteria using the preferred reporting items for systematic reviews and meta-analyses (PRISMA) guidelines [18].
Type of studies
The studies considered for review were cross-sectional, case-control, cohort, and randomized control trials whose primary outcome was the prevalence of multi-drug resistance in commensal E. coli.
Type of participants (study population)
The participants included persons from the general population of all ages and sex, in community settings in LMICs.
Outcome of interest
Since most of the studies used different definitions for multi-, extensive-, and pandrug resistance, we designed alternative definitions, as described in the primary outcomes sub-section below, to facilitate the inclusion of the studies for meta-analysis.
The primary outcomes were:
• Prevalence of multidrug-resistance, i.e. resistance to one or more agents in two different antibiotic classes (protein, nucleic acid, cell wall, and folate synthesis inhibitors) by commensal E. coli in community settings in LMICs.
• Prevalence of extensive drug-resistance, i.e. resistance to one or more agents in three different antibiotic classes by commensal E. coli in community settings in LMICs.
• Prevalence of pandrug-resistance, i.e. resistance to one or more agents in four different antibiotic classes by commensal E. coli in community settings in LMICs.
The secondary outcomes were odds ratio, risk ratio, rate, 95% confidence interval, and P-value.
Other inclusion and exclusion criteria
We included all articles published in the English language in LMICs. Articles containing experiments conducted in a country that was an LMICs before transformation into a high-income country, according to the World Bank definition, if any, were also included. Although this was an inclusion criterion, no study was performed in a country that had undergone such transformation.
Essential data necessary for inclusion
The studies included were those that utilized the Kirby–Bauer disc diffusion or synergy susceptibility test to study and report at least one of the primary outcomes.
Data sources
The systematic search was performed in PubMed, EMBASE, MEDLINE, Web of Science, CINAHL, and Cochrane Library. Reference lists of selected studies and unpublished data, such as abstracts from conference proceedings, dissertations and theses, were also searched.
Systematic search strategy
On 10 March 2018, we searched PubMed, EMBASE, MEDLINE, Web of Science, CINAHL, and Cochrane Library using MeSH terms for PubMed and the comparable terms for the other databases. The search was updated on 28 June 2020. The search strings were: ‘E. coli’, OR ‘Escherichia coli’, OR ‘Enterobacteriaceae’, AND ‘antibiotic resistance’, OR ‘antimicrobial resistance’, OR ‘drug resistance’, AND ‘prevalence’, OR ‘incidence’, OR ‘morbidity’, OR ‘odds ratio’, OR ‘risk ratio’, OR ‘confidence interval’, OR ‘P-value’, OR ‘rate’. For PubMed, EMBASE, and MEDLINE, studies performed in humans and written in the English language were selected using the language and species filters. While for Web of Science and the Cochrane Library, additional search words were added to select for species (‘human*’ OR ‘infant*’ OR ‘child*’ OR ‘adolescent*’ OR ‘male*’ OR ‘female’ OR ‘age’ OR “adult*) since there was no sorting filter for species. For CINAHL, neither selection for species nor language was performed. Search words were designed from the different categories in the PICO (Population, Intervention, Comparison and Outcome) format.
Study selection
The searched articles were exported to the Rayyan software [19], and screening was performed, firstly by title and abstract, and finally by reading the full articles. Two independent researchers screened every article, and a third researcher was available to resolve any disagreements, thereby avoiding selection mistakes.
Data extraction, statistical analysis and quality assessment
Data were extracted on an Excel spreadsheet. To ensure that no mistakes were made, every article was double-checked by an additional researcher. A third researcher was available to solve any disagreement that arose during the process. Statistical analysis was performed on the Joanna Briggs Institute (JBI) SUMARI [20]. Proportional meta-analysis was performed using a statistical random effect model of Freeman–Tukey. In the analysis, multiple resistance for the different combinations of antibiotics investigated in the studies were included. Quality assessment was performed as previously described [21,22]. Primarily, articles that combined both disc diffusion or synergy tests with polymerase chain reaction (PCR), to confirm both the expression and presence of resistant genes were ranked as ‘high’. Those that utilized only disc diffusion or synergy tests were classified as ‘medium’. Finally, studies with a sample size of less than 15 and those that used single disc diffusion or synergy to determine the expression of resistant genes were classified as ‘low’.
Results
Literature search
We obtained 9363 articles from the database search and an additional 6 were identified through a review of reference lists of the included articles (Figure 1). A total of 7089 articles were screened by title and abstract after duplicates were removed in order to exclude those that did not meet our inclusion criteria. The full text of 33 articles were assessed for eligibility, and 18 met our inclusion criteria. The remaining 15 articles were excluded because the definition of multiple resistance was not clear (n = 3), did not document any multi-, extensive- or pandrug-resistant data (n = 11), and could not be obtained online (n = 1). A meta-analysis was therefore performed from 10 articles.
Figure 1.
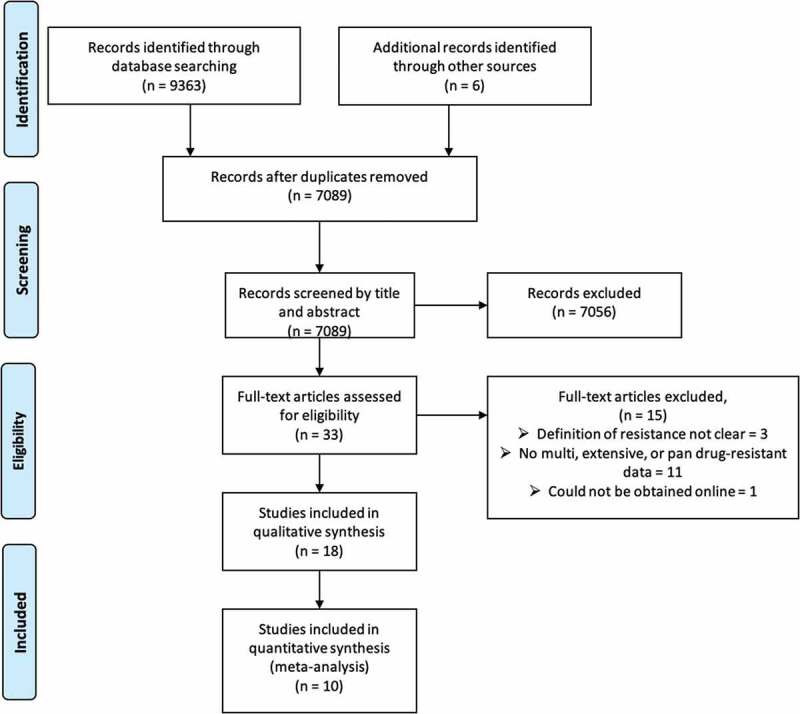
Preferred reporting items for systematic reviews and meta-analyses (PRISMA) flow diagram of the search results and selection of the included studies in the systematic review and meta-analysis. Additional records were identified by screening the reference lists of the included studies
Study location, strength of evidence, and study type
Out of the 10 studies included in the meta-analysis [23–32], six were from Asia and four from South America (Table 1). Since I2 statistics for heterogeneity can be misleading [33,34], we performed an alternative assessment, as described previously [21,22]. From the evaluation, five studies were ranked as high quality, while five were ranked as medium. All of the studies were cross-sectional except for two, which were cohort studies.
Table 1.
Characteristics of studies included for meta-analysis
| Study | Age | Number of Isolates | Number of participants | Gender | Type of study | Country | Strength of evidence | Reference |
|---|---|---|---|---|---|---|---|---|
| Bartoloni et al. 1998 | 6–72 months | 321 | 321 | Male = 165, female = 158 | Cross-sectional | Bolivia | Medium | [23] |
| Bartoloni et al. 2004 | 1–77 years | 108 | 108 | Male = 61, female = 47 | Cross-sectional | Bolivia | High | [24] |
| Bartoloni et al. 2006 | 6–72 months | 1080 | 3174 | Male = 1627, female = 1546 | Cross-sectional | Bolivia | High | [25] |
| Dyar et al., 2012 | 6–60 months | 738 | 818 | Male = 445, female = 373 | Cross-sectional | Vietnam | Medium | [26] |
| Kalter et al. 2010 | 3 months–3 years | 522 | 523 | - | Cross-sectional | Peru | Medium | [27] |
| Mamun et al. 1992 | 1–5 years | 649 | 64 | Male = 33, female = 31 | Cohort | Bangladesh | High | [28] |
| Purohit et al. 2017 | 1–3 years | 127 | 22 | - | Cross-sectional | India | High | [29] |
| Shakya et al. 2013 | 3–14 years | 529 | 529 | Male = 304, female = 225 | Cross-sectional | India | Medium | [30] |
| Singh et al. 2018 | 1–14 years | 550 | 550 | Male = 396, female = 154 | Cohort | India | High | [31] |
| Sahoo et al. 2012 | 3–9 years | 139 | 417 | Male = 230, female = 187 | Cross-sectional | India | Medium | [32] |
Of the 18 studies included for qualitative synthesis (Figure 1), 10 were excluded for the meta-analysis because the definition of multi-, extensive-, and pandrug-resistance was very vague [23–32] (Table 2). However, there was one study in which parts of the data were included in the meta-analysis while others were not [28]. There were three studies conducted in Africa and five in Asia, with four studies classified as high quality and four medium quality.
Table 2.
Prevalence and characteristics of included studies not used for meta-analysis
| Study | Age | Number of Isolates | Number of participants | Gender | Definition of multi-drug resistance | Pooled prevalence (%) | Lower bound 95% CI | Upper bound 95% CI | Type of study | Country | Strength of evidence | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aworh et al. 2019 | Mean age 30.6 ± 9.7 years |
48 | 122 | Male = 121 female = 1 | Resistance to three or more classes of antimicrobials |
79 | 66 | 90 | Cross-sectional | Nigeria | Medium | [35] |
| Al-Dweik and Shehabi, 2009 | - | 108 | 108 | - | Greater than 2 or 3 of the 14 antimicrobials tested | >2 drugs = 51 >3 drugs = 19 |
41 12 |
60 27 |
Cohort | Jordan | High | [36] |
| Hoang et al. 2017 | 20–70 years | 103 | 103 | - | Resistance to more than 3 and 5 classes of antimicrobials | >3 classes = 71 >5 classes = 41 |
62 32 |
80 51 |
Cross-sectional | Vietnam | High | [37] |
| Lamikanra et al. 1996 | 5–6 years | 396 | 450 | - | Resistance to 3, 4, 5, 6, 7, and 8 antibiotics | 3 antibiotics = 11 4 antibiotics = 10 5 antibiotics = 10 6 antibiotics = 10 7 antibiotics = 7 8 antibiotics = 8 |
8 7 7 7 5 5 |
15 14 14 14 10 10 |
Cross-sectional | Nigeria | Medium | [38] |
| Lamikanra et al. 1989 | 19–25 years | 13 | 35 | - | Resistant to at least one other antibiotic | 53 | 22 | 83 | Cross-sectional | Nigeria | Medium | [39] |
| Mamun et al. 1992 | 1–5 years | 649 | 64 | Male = 33, female = 31 | Resistance to 3–5 antibiotics | January to December = 74 | 60 | 86 | Cohort | Bangladesh | High | [28] |
| Phongpaichit et al. 2007 | 12–46 years | 143 (non-pig farmers), 7 (pig farmers) | Non-pig farmers (41), Pig farmers (7) | Non-pig farmers male = 211, female = 20 | Resistance to at least 1 antimicrobial and multidrug-resistance | Non-pig farmers: at least 1 antimicrobial = 67 multidrug-resistance = 74 |
60 66 |
75 81 |
Cross-sectional | Thailand | High | [40] |
| Pathak et al. 2012 | 45 years and above | 241 | 241 | Male = 0, female = 241 | Resistance to at least 1 or 3 different antibiotics | At least 1 = 94, at least 3 = 35 |
92 31 |
96 39 |
Cross-sectional | India | Medium | [43] |
Age, sample size, gender
For all the 18 included studies (Tables 1 and 2), the age of the participants ranged from 3 months to 77 years, with 7633 participants in total, of which approximately 55% were male and 45% female (Tables 1 and 2).
High prevalence of multidrug-resistance (MDR) in community settings in low- and middle-income countries
In this study, multidrug-resistance was defined as the resistance of one or more agents in two out of the four different classes of antibiotics (protein, nucleic acid, cell wall, and folate synthesis). The pooled multidrug-resistance was 27% of 14,336 total cases of isolates tested, with 95% CI: 23–32 (Figure 2).
Figure 2.
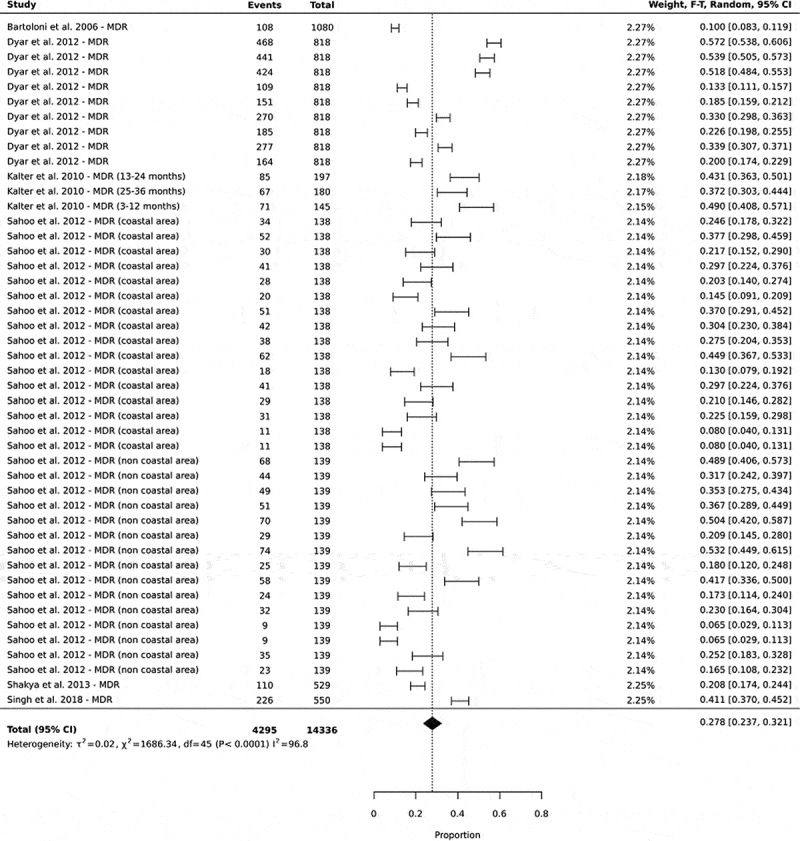
Forest plots showing the prevalence of multidrug-resistance in commensal E. coli isolated from human sources in community settings in low- and middle-income countries. In this analysis, the prevalence of the different antibiotic combinations in a single study were all included
High prevalence of extensive-drug resistance (XDR) in community settings in low- and middle-income countries
In this study, extensive-drug resistance was classified as the resistance of one or more agents in three out of the four different classes of antibiotics (protein, nucleic acid, cell wall, and folate synthesis). The pooled prevalence of extensive drug-resistance was 24% of 8686 total cases of isolates tested, with 95% CI: 14–36 (Figure 3).
Figure 3.
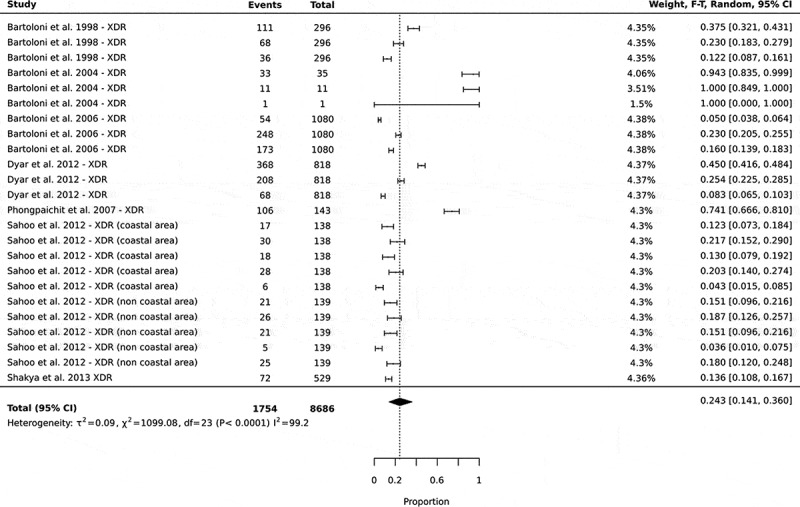
Forest plots showing the prevalence of extensive drug-resistance in commensal E. coli isolated from human sources in community settings in low- and middle-income countries. In this analysis, the prevalence of the different antibiotic combinations in a single study were all included
Emergence of pandrug-resistance (PDR) in community settings in low- and middle-income countries
Pandrug-resistance was defined as the resistance of one or more agents in all four classes of antibiotics (protein, nucleic acid, cell wall, and folate synthesis). Pandrug-resistance had a pooled prevalence of 5% of 5670 total cases of isolates tested, with 95% CI: 3–8 (Figure 4).
Figure 4.
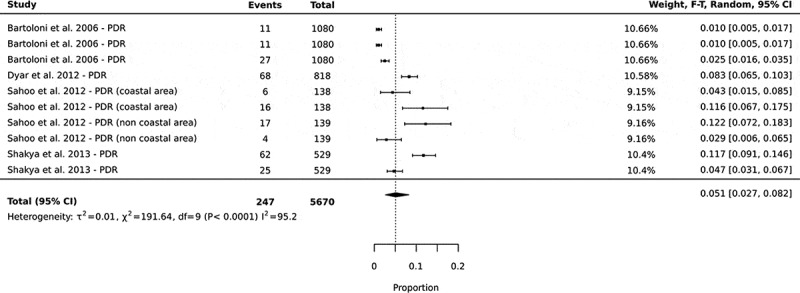
Forest plots showing the prevalence of pandrug-resistance in commensal E. coli isolated from human sources in community settings in low- and middle-income countries. In this analysis, the prevalence of the different antibiotic combinations in a single study were all included
Discussion
To contain the spread of multidrug-resistant bacteria, we must improve our knowledge of how multidrug-resistance is changing over time in LMICs, including the prevalence of multidrug-resistant commensal Escherichia coli in LMIC community settings (i.e. locations outside of, a hospital inpatient, acute care setting or a hospital clinic setting).
While there is an increase in multidrug-resistant pathogenic bacteria in general in community settings [41], we have observed that studies on multidrug-resistance of commensal E. coli isolated from healthy humans have consistently demonstrated very high incidences and prevalences in community settings in LMICs. This review synthesized eighteen studies that reported the prevalence of multidrug-resistance in commensal E. coli isolates from human sources in community settings in LMICs. Our results showed that the prevalence of multidrug- and extensive drug-resistance of E. coli at the community level is high in LMICs, and more concerning is the emergence of pandrug resistance in these settings. This evidence should prove useful to inform researchers, community pharmacists, and public health policymakers, among others, to develop appropriate interventions and to formulate and implement policies aimed at counteracting the global threats of multidrug-resistant infections and bacteria.
Most of the included studies had different definitions of multidrug-, extensive drug-, and pandrug-resistance, as also reported by Magiorakos et al. [42]. For this study, we classified resistance based on the inhibition mechanisms of action (protein, nucleic acid, cell wall, and folate synthesis inhibition). Multidrug-resistance was defined as the nonsusceptibility of at least one agent in two categories of the different mechanisms of action. At least one agent in three groups of the mechanism of action was classified as extensive drug resistance, and at least one agent in all four categories of the mechanism of action was classified as pandrug-resistance. As expected, the heterogeneity between studies was very high which may have been because several factors affect the prevalence of antibiotic resistance, such as previous antibiotic use [11,24,27,43], geographical location [26,32], age [24,26], socioeconomic status [30,32,43], and exposure to animals [27,44]. These factors were different in the included studies (Tables 1 and 2).
There are several drivers of antibiotic resistance whose inter-relationship is very complicated as it cuts across different sectors like health, agriculture, environment, and industry. The primary driver of multidrug-resistance in commensal E. coli in LMICs is the relaxed policies surrounding antibiotic access, prescription, and use [45]. Commensal E. coli are Gram-negative bacteria usually present in the guts of humans, animals, birds, and the environment [46]. When we ingest antibiotics for the treatment of bacterial infections, the commensal E. coli are exposed to these antibacterial agents and can develop resistance through natural selection [13]. Other drivers of multidrug-resistance in commensal E. coli in LMICs have been identified as inappropriate socioecological behaviors, poverty, overcrowding, lack of surveillance systems, food and supply chain safety issues, and highly contaminated waste effluents [45].
This study is in line with WHO recommendations, which call for improved awareness of the multidrug-resistance problem's existence as one of the five strategic objectives to control the problem spelled out in the Global action plan on antimicrobial resistance[47]. Many people in LMICs are aware of the existence of antibiotics, but fewer people are aware of antibiotic resistance [48]. Our findings will raise awareness of the extent of the problem and calls for urgent action to address this global health crisis especially in the current situation of the emergence of pandemics, like the severe acute respiratory coronavirus disease 2019 (COVID 19) that has led to a surge in the use of antimicrobial drugs within the community through self-medication and chemoprophylaxis. For COVID19, there has been an extensive use of not only antibiotics worldwide, but several medicines have also been tested in clinical trials as a potential treatment for COVID19 [49–57]. Such is the case for remdesivir, which has been authorised for the treatment of COVID19 patients even before approval [52]. A combination of both antibiotic and nonantibiotic drugs leads to an alteration of the gut microbiome that could fuel antibiotic resistance by commensal Escherichia coli, which can then be transmitted to other pathogenic enteric bacteria, e.g. Salmonella typhi, Vibrio cholerae, Helicobacter pylori, among others, through the exchange of mobile genetic materials [12,58–64].
Antibiotic resistance (ABR) is a growing problem that requires urgent action to stop its spread. Multiple interventions targeting different drivers of multidrug-resistance have been recommended [47]. Some of these interventions targets community settings and some are tailored to contribute to combating the challenges of multidrug-resistance. Based on the WHO antimicrobial control plan strategic objectives [47], the interventions are classified into the following categories; i) those aimed at improving awareness and understanding of antibiotic resistance, for example, it is recommended that creating participatory videos (participatory documentaries) about antibiotic resistance would be useful in creating awareness in communities [65]; ii) activities strengthening knowledge through active and routine surveillance and research; iii) those focussing on the reduction of infections for example improvement in sanitation and hygiene, to reduce the demand of antibiotics; iv) those that optimize the use of antibiotics, for example, the formulation and implementation of regulations surrounding the use of antibiotics coupled with strict monitoring of antibiotic use as part of the policy, in addition to the development of new screening and diagnostic tools [66]; and v) interventions ensuring that there is a sustainable investment in combating antibiotic resistance. A global and interdisciplinary approach will fast track implementation of some of the interventions that may be universal, for example, the development of new screening and diagnostic tools [66]. The recommended interventions have shown to have an impact on the combating of multidrug-resistance at the community level. However, in order to harness the community’s potential in combating this problem, active community engagement and involvement from the design to the implementation of the selected intervention should be emphasized and carried out in order to allow the implementation of interventions that are well adapted to the local context and easy to scale up at less cost [67].
To conclude, this study provides an appraisal of the evidence on the high prevalence of multidrug-, extensive drug resistance, and the emergence of pandrug-resistant commensal E. coli from healthy human sources in community settings in LMICs. This evidence appraisal calls for urgent action in the fight against multidrug-resistance in community settings to make antibiotic resistance history.
Responsible Editor Stig Wall, Umeå University,Sweden
Author contributions
E.N. designed and supervised the research. E.N. performed the systematic database search and statistical analysis. E.N, N.A.N, J.K., and D.A.K.T. performed the data extraction. E.N. wrote the manuscript with assistance from J.K.
Paper context
Studies conducted to investigate the prevalence of multi-drug resistance in commensal Escherichia coli isolated from humans in community settings in low- and middle-income countries have consistently been high. This study provides an appraisal of the evidence on the high prevalence of multidrug-resistance in commensal E. coli isolated from humans in community settings in low- and middle-income countries. Our findings call for greater effort to be placed at the community level to reverse the course of multidrug-resistance.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Centers for Disease Control and Prevention (U.S.) Antibiotic resistance threats in the USA, 2019 [Internet]. Centers for disease control and prevention (U.S.); 2019. [cited 2020 July11]. Available from: https://stacks.cdc.gov/view/cdc/82532
- [2].Ghebreyesus TA. Making AMR history: a call to action. Glob Health Action. 2019;12:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Rogers BA, Sidjabat HE, Paterson DL. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J Antimicrob Chemother. 2011;66:1–10. [DOI] [PubMed] [Google Scholar]
- [4].Pitout JD, Laupland KB. Extended-spectrum β-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis. 2008;8:159–166. [DOI] [PubMed] [Google Scholar]
- [5].World Health Organization Antibiotic resistance [Internet]. World Health Organization; 2020. [cited 2020 August17]. Available from: https://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance [Google Scholar]
- [6].European Centre for Disease Prevention and Control, editor. The bacterial challenge, time to react: a call to narrow the gap between multidrug-resistant bacteria in the EU and the development of new antibacterial agents Stockholm: ECDC; 2009. [Google Scholar]
- [7].Cassini A, Högberg LD, Plachouras D, et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European economic area in 2015: a population-level modelling analysis. Lancet Infect Dis. 2019;19:56–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].de Kraker MEA, Stewardson AJ, Harbarth S. Will 10 million people die a year due to antimicrobial resistance by 2050? PLOS Med. 2016;13:e1002184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Adam RD. Antimicrobial resistance at a community level. Lancet Planet Health. 2018;2:e473–e474. [DOI] [PubMed] [Google Scholar]
- [10].Sanchez GV, Fleming-Dutra KE, Roberts RM, et al. Core elements of outpatient antibiotic stewardship. MMWR Recomm Rep. 2016;65:1–12. [DOI] [PubMed] [Google Scholar]
- [11].Bryce A, Costelloe C, Hawcroft C, et al. Faecal carriage of antibiotic resistant Escherichia coli in asymptomatic children and associations with primary care antibiotic prescribing: a systematic review and meta-analysis. BMC Infect Dis. 2016;16:359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hanada S, Pirzadeh M, Carver KY, et al. Respiratory viral infection-induced microbiome alterations and secondary bacterial pneumonia. Front Immunol. 2018;9:2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Allocati N, Masulli M, Alexeyev M, et al. Escherichia coli in Europe: an overview. Int J Environ Res Public Health. 2013;10:6235–6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Salyers AA, Gupta A, Wang Y. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 2004;12:412–416. [DOI] [PubMed] [Google Scholar]
- [15].Blake DP, Hillman K, Fenlon DR, et al. Transfer of antibiotic resistance between commensal and pathogenic members of the Enterobacteriaceae under ileal conditions. J Appl Microbiol. 2003;95:428–436. [DOI] [PubMed] [Google Scholar]
- [16].Balkhy HH, Jarlier V, Kluytmans J, et al. Antimicrobial resistance: one world, one fight! Antimicrob resist infect control [Internet]; 2015. [cited 2017 December17]. 4 Available from: https://aricjournal.biomedcentral.com/articles/10.1186/s13756-015-0091-2
- [17].Hooton TM, Levy SB. Antimicrobial resistance: a plan of action for community practice. Am Fam Physician. 2001;63:1087–1098. [PubMed] [Google Scholar]
- [18].Shamseer L, Moher D, Clarke M, et al. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015: elaboration and explanation. BMJ. 2015;350:g7647. [DOI] [PubMed] [Google Scholar]
- [19].Ouzzani M, Hammady H, Fedorowicz Z, et al. Rayyan—a web and mobile app for systematic reviews. Syst Rev. 2016; [cited 2018 April30]; 5 [Internet] Available from http://systematicreviewsjournal.biomedcentral.com/articles/10.1186/s13643-016-0384-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].The System for the Unified Management Assessment and review of information (SUMARI) [Internet]. Joanna briggs institute (JBI); [cited 2020 July13]. Available from: https://auth.jbisumari.org
- [21].Storberg V. ESBL-producing Enterobacteriaceae in Africa – a non-systematic literature review of research published 2008–2012. Infect Ecol Epidemiol. 2014;4:20342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hedin A, Källestål C. Knowl-based public health work part 2 natl inst public health stockh. 2004.
- [23].Bartoloni A, Cutts F, Leoni S, et al. Patterns of antimicrobial use and antimicrobial resistance among healthy children in Bolivia. Trop Med Int Health. 1998;3:116–123. [DOI] [PubMed] [Google Scholar]
- [24].Bartoloni A, Bartalesi F, Mantella A, et al. High prevalence of acquired antimicrobial resistance unrelated to heavy antimicrobial consumption. J Infect Dis. 2004;189:1291–1294. [DOI] [PubMed] [Google Scholar]
- [25].Bartoloni A, Pallecchi L, Benedetti M, et al. Multidrug-resistant commensal Escherichia coli in children, Peru and Bolivia. Emerg Infect Dis. 2006;12:907–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Dyar OJ, Hoa NQ, Trung NV, et al. High prevalence of antibiotic resistance in commensal Escherichia coli among children in rural Vietnam. BMC Infect Dis. 2012;12:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kalter HD, Gilman RH, Moulton LH, et al. Risk factors for antibiotic-resistant Escherichia coli carriage in young children in Peru: community-based cross-sectional prevalence study. Am J Trop Med Hyg. 2010;82:879–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Mamun KZ, Shears P, Hart CA. The prevalence and genetics of resistance to commonly used antimicrobial agents in faecal Enterobacteriaceae from children in Bangladesh. Epidemiol Infect. 1993;110:447–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Purohit M, Chandran S, Shah H, et al. Antibiotic resistance in an Indian rural community: a ‘one-health’ observational study on commensal coliform from humans, animals, and water. Int J Environ Res Public Health. 2017;14:386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Shakya P, Barrett P, Diwan V, et al. Antibiotic resistance among Escherichia coli isolates from stool samples of children aged 3 to 14 years from Ujjain, India. BMC Infect Dis. 2013;13:477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Singh AK, Das S, Singh S, et al. Prevalence of antibiotic resistance in commensal Escherichia coli among the children in rural hill communities of Northeast India. PloS One. 2018;13:e0199179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sahoo KC, Tamhankar AJ, Sahoo S, et al. Geographical variation in antibiotic-resistant Escherichia coli isolates from stool, cow-dung and drinking water. Int J Environ Res Public Health. 2012;9:746–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Iorio A, Spencer FA, Falavigna M, et al. Use of GRADE for assessment of evidence about prognosis: rating confidence in estimates of event rates in broad categories of patients. BMJ. 2015;350:h870. [DOI] [PubMed] [Google Scholar]
- [34].Guyatt GH, Oxman AD, Kunz R, et al. GRADE guidelines: 7. Rating the quality of evidence–inconsistency. J Clin Epidemiol. 2011;64:1294–1302. [DOI] [PubMed] [Google Scholar]
- [35].Aworh MK, Kwaga J, Okolocha E, et al. Prevalence and risk factors for multi-drug resistant Escherichia coli among poultry workers in the federal capital territory, Abuja, Nigeria. PloS One. 2019;14:e0225379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Al-Dweik MR, Shehabi AA. Common antimicrobial resistance phenotypes and genotypes of fecal Escherichia coli isolates from a single family over a 6-month period. Microb Drug Resist Larchmt N. 2009;15:103–107. [DOI] [PubMed] [Google Scholar]
- [37].Hoang PH, Awasthi SP, DO Nguyen P, et al. Antimicrobial resistance profiles and molecular characterization of Escherichia coli strains isolated from healthy adults in Ho Chi Minh City, Vietnam. J Vet Med Sci. 2017;79:479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lamikanra A, Ako-Nai AK, Ogunniyi DA. Transferable antibiotic resistance in Escherichia coli isolated from healthy Nigerian school children. Int J Antimicrob Agents. 1996;7:59–64. [DOI] [PubMed] [Google Scholar]
- [39].Lamikanra A, Fayinka ST, Olusanya OO. Transfer of low level trimethoprim resistance in faecal isolates obtained from apparently healthy Nigerian students. FEMS Microbiol Lett. 1989;59:275–278. [DOI] [PubMed] [Google Scholar]
- [40].Phongpaichit S, Liamthong S, Mathew AG, et al. Prevalence of class 1 integrons in commensal Escherichia coli from pigs and pig farmers in Thailand. J Food Prot. 2007;70:292–299. [DOI] [PubMed] [Google Scholar]
- [41].Roca I, Akova M, Baquero F, et al. The global threat of antimicrobial resistance: science for intervention. New Microbes New Infect. 2015;6:22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Magiorakos A-P, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis. 2012;18:268–281. [DOI] [PubMed] [Google Scholar]
- [43].Pathak A, Mahadik K, Sharma R, et al. Factors associated with carriage of multi-resistant commensal Escherichia coli among postmenopausal women in Ujjain, India. Scand J Infect Dis. 2012;44:973–977. [DOI] [PubMed] [Google Scholar]
- [44].Riccobono E, Pallecchi L, Mantella A, et al. Carriage of antibiotic-resistant Escherichia coli among healthy children and home-raised chickens: a household study in a resource-limited setting. Microb Drug Resist Larchmt N. 2012;18:83–87. [DOI] [PubMed] [Google Scholar]
- [45].Iskandar K, Molinier L, Hallit S et al. Drivers of antibiotic resistance transmissionin low- and middle-income countriesfrom a “one health” perspective – a review. Antibiot Basel Switz. 2020:9:372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Alekshun MN, Levy SB. Commensals upon us. Biochem Pharmacol. 2006;71:893–900. [DOI] [PubMed] [Google Scholar]
- [47].Global Action Plan on Antimicrobial Resistance [Internet] World health organization; 2015. [cited 2020 August17]. Available from: https://apps.who.int/iris/bitstream/handle/10665/193736/9789241509763_eng.pdf?sequence=1
- [48].Haenssgen MJ, Charoenboon N, Zanello G, et al. Antibiotic knowledge, attitudes and practices: new insights from cross-sectional rural health behaviour surveys in low-income and middle-income South-East Asia. BMJ Open. 2019;9:e028224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Okba NMA, Müller MA, Li W, et al. Severe acute respiratory syndrome coronavirus 2−specific antibody responses in coronavirus disease 2019 patients. Emerg Infect Dis. 2020. [cited 2020 May8]; 26 [Internet] Available from: http://wwwnc.cdc.gov/eid/article/26/7/20-0841_article.htm [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Mitjà O, Clotet B. Use of antiviral drugs to reduce COVID-19 transmission. Lancet Glob Health. 2020;8:e639–e640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Poland GA. SARS-CoV-2: a time for clear and immediate action. Lancet Infect Dis. 2020;20:531–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Li Z, Wang X, Cao D, et al. Rapid review for the anti-coronavirus effect of remdesivir. Drug Discov Ther. 2020;14:73–76. [DOI] [PubMed] [Google Scholar]
- [53].Meo SA, Klonoff DC, Akram J. Efficacy of chloroquine and hydroxychloroquine in the treatment of COVID-19. Eur Rev Med Pharmacol Sci. 2020;24:4539–4547. [DOI] [PubMed] [Google Scholar]
- [54].Hashem AM, Alghamdi BS, Algaissi AA, et al. Therapeutic use of chloroquine and hydroxychloroquine in COVID-19 and other viral infections: a narrative review. Travel Med Infect Dis. 2020;35:101735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Retsas S. Clinical trials and the COVID-19 pandemic. Hell J Nucl Med. 2020;23:4–5. [DOI] [PubMed] [Google Scholar]
- [56].Imhann F, Bonder MJ, Vich Vila A, et al. Proton pump inhibitors affect the gut microbiome. Gut. 2016;65:740–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Jackson MA, Goodrich JK, Maxan M-E, et al. Proton pump inhibitors alter the composition of the gut microbiota. Gut. 2016;65:749–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Rogers MAM, Aronoff DM. The influence of non-steroidal anti-inflammatory drugs on the gut microbiome. Clin Microbiol Infect. 2016;22:e1–178.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Bandera A, De Benedetto I, Bozzi G, et al. Altered gut microbiome composition in HIV infection: causes, effects and potential intervention. Curr Opin HIV AIDS. 2018;13:73–80. [DOI] [PubMed] [Google Scholar]
- [60].Monaco CL, Gootenberg DB, Zhao G, et al. Altered virome and bacterial microbiome in human immunodeficiency virus-associated acquired immunodeficiency syndrome. Cell Host Microbe. 2016;19:311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Blaser MJ. Antibiotic use and its consequences for the normal microbiome. Science. 2016;352:544–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Bonten MJM, Prins JM. Antibiotics in pandemic flu. BMJ. 2006;332:248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Rawson TM, Moore LSP, Zhu N, et al. Bacterial and fungal co-infection in individuals with coronavirus: a rapid review to support COVID-19 antimicrobial prescribing. Clin Infect Dis. 2020;ciaa530 DOI: 10.1093/cid/ciaa530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Morens DM, Taubenberger JK, Fauci AS. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J Infect Dis. 2008;198:962–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Cooke P, Shrestha A, Arjyal A, et al. What is ‘antimicrobial resistance’ and why should anyone make films about it? Using ‘participatory video’ to advocate for community-led change in public health. New Cine J Contemp Film. 2020;17:85–107. [Google Scholar]
- [66].Aslam B, Wang W, Arshad MI, et al. Antibiotic resistance: a rundown of a global crisis. Infect Drug Resist. 2018;11:1645–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].King R, Hicks J, Rassi C, et al. Community Dialogue Approach for addressing the drivers of antibiotic resistance in Bangladesh: a mixed methods study on developing a sustainable and scalable approach to community engagement [Internet]. In Review; 2020. [cited 2020 August18]. Available from: https://www.researchsquare.com/article/rs-15638/v1 [DOI] [PMC free article] [PubMed]


