Abstract
Background
The aim of this study was to compare a commercial dosimetry workstation (PLANET® Dose) and the dosimetry approach (GE Dosimetry Toolkit® and OLINDA/EXM® V1.0) currently used in our department for quantification of the absorbed dose (AD) to organs at risk after peptide receptor radionuclide therapy with [177Lu]Lu-DOTA-TATE.
Methods
An evaluation on phantom was performed to determine the SPECT calibration factor variations over time and to compare the Time Integrated Activity Coefficients (TIACs) obtained with the two approaches. Then, dosimetry was carried out with the two tools in 21 patients with neuroendocrine tumours after the first and second injection of 7.2 ± 0.2 GBq of [177Lu]Lu-DOTA-TATE (40 dosimetry analyses with each software). SPECT/CT images were acquired at 4 h, 24 h, 72 h and 192 h post-injection and were reconstructed using the Xeleris software (General Electric). The liver, spleen and kidneys masses and TIACs were determined using Dosimetry Toolkit® (DTK) and PLANET® Dose. The ADs were calculated using OLINDA/EXM® V1.0 and the Local Deposition Method (LDM) or Dose voxel-Kernel convolution (DK) on PLANET® Dose.
Results
With the phantom, the 3D calibration factors showed a slight variation (0.8% and 3.3%) over time, and TIACs of 225.19 h and 217.52 h were obtained with DTK and PLANET® Dose, respectively. In patients, the root mean square deviation value was 8.9% for the organ masses, 8.1% for the TIACs, and 9.1% and 7.8% for the ADs calculated with LDM and DK, respectively. The Lin’s concordance correlation coefficient was 0.99 and the Bland–Altman plot analysis estimated that the AD value difference between methods ranged from − 0.75 to 0.49 Gy, from − 0.20 to 0.64 Gy, and from − 0.43 to 1.03 Gy for 95% of the 40 liver, kidneys and spleen dosimetry analyses. The dosimetry method had a minor influence on AD differences compared with the image registration and organ segmentation steps.
Conclusions
The ADs to organs at risk obtained with the new workstation PLANET® Dose are concordant with those calculated with the currently used software and in agreement with the literature. These results validate the use of PLANET® Dose in clinical routine for patient dosimetry after targeted radiotherapy with [177Lu]Lu-DOTA-TATE.
Keywords: Dosimetry workstation, Peptide receptor radionuclide therapy, [177Lu]Lu-DOTA-TATE, 3D calibration factor, MIRD, Voxel-based dosimetry
Background
Dosimetry applications are expanding and the number of nuclear medicine departments performing patient dosimetry is growing, especially for patients with neuroendocrine tumours receiving peptide receptor radionuclide therapy [1]. This is possible thanks to the multidisciplinary collaboration between nuclear medicine physicians, medical physicists, and nuclear medicine technologists [2].
In the clinic, many medical teams have developed their own methodology using the tools available in their department and according to their own organizational possibilities [3–5]. This has led to the local implementation of in-house developed dosimetry software and programs [6–10]. However, the legislation on Medical Devices restricts their use to clinical trials. For routine clinical use, a software package must have received the CE mark. Therefore, many medical teams are now acquiring commercial packages, because obtaining the CE label is usually beyond the missions/capabilities of academic structures and due to the increasing availability of commercial software tools. In a previous article we evaluated some commercial packages already on the market [11]. An updated table (Table 1) is presented below, but the field is rapidly and constantly evolving. Only CE-marked software tools are presented (therefore, STRATOS from the Philips research station Imalytics is not included), and some features that are still under development may not have been approved yet.
Table 1.
List of commercial packages with CE marking
| Name | Manufacturer | Image format | VOI/voxel dosimetry | Absorbed dose calculation | References |
|---|---|---|---|---|---|
| Dosimetry Toolkit® | GE Healthcare, Waukesha, WI, USA |
SPECT/CT Hybrid Planar |
VOI (volume of interest) | OLINDA/EXM® V1/V2 |
https://www.gehealthcare.com/products/molecular-imaging/nuclear-medicine/xeleris-4-dr Stabin et al. [14] Kupitz et al. [15] |
| PLANET® Dose | DOSIsoft, Cachan, France |
SPECT/CT Hybrid |
Voxel-based absorbed dose rates calculation with integration over the VOI |
Local energy deposition Convolution +/− density correction |
https://www.dosisoft.com/products/planet-dose/ Huizing et al. [16] |
| MIM SurePlan™ MRT | MIM Software Inc., Cleveland, OH, USA |
SPECT/CT Hybrid |
Voxel-based | Convolution |
https://www.mimsoftware.com/mim_sureplan_mrt Maughan et al. [17] |
|
Organ Dosimetry™ Voxel Dosimetry™ |
Hermes Medical Solutions, Stockholm, Sweden |
SPECT/CT Hybrid Planar |
VOI/Voxel-based |
OLINDA/EXM® V2 Monte Carlo method |
https://www.hermesmedical.com/dosimetry/ Hippeläinen et al. [18] |
| QDOSE® | ABX-CRO, Dresden, Germany |
SPECT/CT Hybrid Planar |
VOI |
IDAC-Dose 2.1 Convolution |
https://www.quantitativedose.com Barna et al. [19] |
GE, general electric; VOI, volume of interest; SPECT/CT, single photon emission computed tomography/computed tomography; IDAC, internal dose assessment by computer
In our department, dosimetry is integrated in the clinical routine for patients with neuroendocrine tumours treated by peptide receptor radionuclide therapy with [177Lu]Lu-DOTA-TATE. Up to now, dosimetry analyses of organs at risk (OAR) have been performed using the combination of Dosimetry Toolkit® (GE Healthcare, Milwaukee, USA) and OLINDA/EXM® V1.0 to calculate the Time Integrated Activity Coefficient (TIAC) and organ-level absorbed dose (AD), respectively [12]. Recently, our department acquired PLANET® Dose (DOSIsoft SA, Cachan, France), a new CE-marked commercial dosimetry workstation. The initial motivation for changing was to use a vendor-neutral solution to support multicentric trials and to put in place a central dosimetry system. However, before its clinical routine implementation, we wanted to compare this new dosimetric package with our internal reference. Our validation plan involved comparing the results obtained on phantom (calibration factor, TIAC) and in patients (mean AD to OARs, TIACs and organ volumes), using similar parameters in terms of segmentation, registration, and time activity curve fitting. This allowed assessing the consistency between the dosimetric results obtained using Dosimetry Toolkit® and OLINDA/EXM® V1.0 (our reference) and with PLANET® Dose.
Material and methods
Dosimetry software packages
The characteristics of the two dosimetry software packages used in this study are presented in Table 2.
Table 2.
General characteristics for each step of the dosimetry workflow in the currently used dosimetry “Dosimetry Toolkit + OLINDA” software (the reference in this study), and the new PLANET® Dose package
| Dosimetry Toolkit® + OLINDA/EXM® V1.0 | PLANET® Dose | |
|---|---|---|
| Calibration procedure |
GE recommendations: planar acquisitions Calibration factor: counts.s−1.MBq−1 |
No manufacturer’s recommendations Calibration factor: Bq.Count−1 |
| Clinical imaging expected |
Whole body scans, SPECT/CT or hybrid 5 time points maximum Only from GE devices |
SPECT/CT or hybrid Unlimited time points Import in DICOM format from all devices |
| Reconstruction | « Preparation for Dosimetry Toolkit» application | Non applicable |
| Registration |
« Preparation for Dosimetry Toolkit» application Automatic rigid registration between CT images (Full FOV) |
Rigid or elastic registration Using CT data |
| Segmentation and propagation |
Manual, semi-automatic or automatic segmentation using the first images (NM or CT) Rigid propagation (constant volumes) Segmentation adjusted by translation or rotation on others CT images |
Manual, semi-automatic or automatic segmentation using the first images (NM or CT) Rigid propagation (constant volumes) No adjustment on others CT images |
| TIA fitting | Mono-exponential exclusively | Mono-exponential, Bi-exponential, Xexp, Trapezoidal, Tri-exponential, … |
| Absorbed dose calculation |
Not avalaible on Dosimetry Toolkit® TIAC exported to OLINDA/EXM® V1.0 |
Local Deposition Method (LDM) Dose Kernel (DK) approach with / without density correction |
| Import/export | Not applicable | DICOM format (DICOM-RT-Structure and RT-Dose) |
GE, general electric; FOV, field of view; NM, nuclear medicine; CT, computed tomography; TIA, Time tntegrated activity; TIAC, time integrated activity coefficient; DICOM-RT, digital imaging and communications in medicine-radiotherapy
Dosimetry Toolkit® is an application of the Xeleris® software (GE Healthcare, Milwaukee, USA) [13, 15]. GE Healthcare recommends to use a procedure based on planar acquisition to determine the calibration factor (CF; in counts.s−1.MBq−1). For clinical dosimetry, different scenarios are available: whole-body, hybrid, or multi-SPECT/CT image acquisition. It includes two steps. The first, “Preparation for Dosimetry Toolkit”, is used for the reconstruction of SPECT/CT raw data and registration of the CT or planar whole body images. The second, “Dosimetry Toolkit”, is used to segment the different organs, create the time activity curves fitted by a mono-exponential function, and calculate the TIAC for each of them. These TIACs are then uploaded in OLINDA/EXM® V1.0 [14] to calculate the organ mass-adjusted ADs. At the time of the study, only OLINDA V1 was available. The future version of Dosimetry Toolkit should propose OLINDA V2, but any model-based dosimetry software that includes S values (IDAC-Dose for example) could be used [20].
PLANET® Dose is a treatment planning system from DOSIsoft. The calibration procedure is left to the user’s discretion and a CF (in Bq.count−1) is required. This dosimetry package does not reconstruct SPECT/CT data, but accepts reconstructed data in DICOM format from all devices. It provides multi-time point registration (using rigid and elastic algorithms), organ segmentation (manual and automatic), and TIAC calculation with a wide choice of interpolation methods of the time-activity curve (linear, trapezoidal, mono-, X-, bi-, tri-exponential…). The mean AD can be calculated by assuming the local energy deposition (local deposition model—LDM) or by convolution of AD voxel kernels (dose kernels—DK), with or without media density correction [21–23].
For this study, the segmentation, registration and TIAC steps were carried out on PLANET® Dose using parameters similar to those defined in Dosimetry Toolkit® to allow the comparison.
Dosimetry imaging protocol
All imaging acquisitions were performed with a SPECT/CT Discovery NM/CT 670 (General Electric [GE] Healthcare), including a BrightSpeed 16 CT scanner and a 3/8-inch NaI(Tl) crystal, according to the acquisition protocol previously described [12]. Briefly, nuclear medicine images were acquired using a medium-energy general purpose parallel-hole collimator. A 20% energy window centred on the 208 keV photopeak and a 10% scatter correction window centred on 177 keV were applied. NM acquisitions were performed using a body contour option, rotation of 180° per detector, total of 60 projections and 45 s each. For attenuation correction, CT images were acquired (120 kV, automatic mA regulation with a max at 200 mA, noise index at 6.43, slice thickness of 5 mm, rotation time of 0.8 s, pitch 1.375, 512 × 512 pixels matrix), with standard reconstruction.
The application “Preparation for Dosimetry Toolkit” was used for SPECT/CT image reconstruction for both dosimetry approaches. The Ordered Subset Expectation Maximization iterative reconstruction algorithm was used with 6 iterations and 10 subsets, attenuation, scatter, recovery resolution corrections and a Gaussian post-filter of 0.11 cm [12].
Phantom study
Calibration factor and time integrated activity coefficient
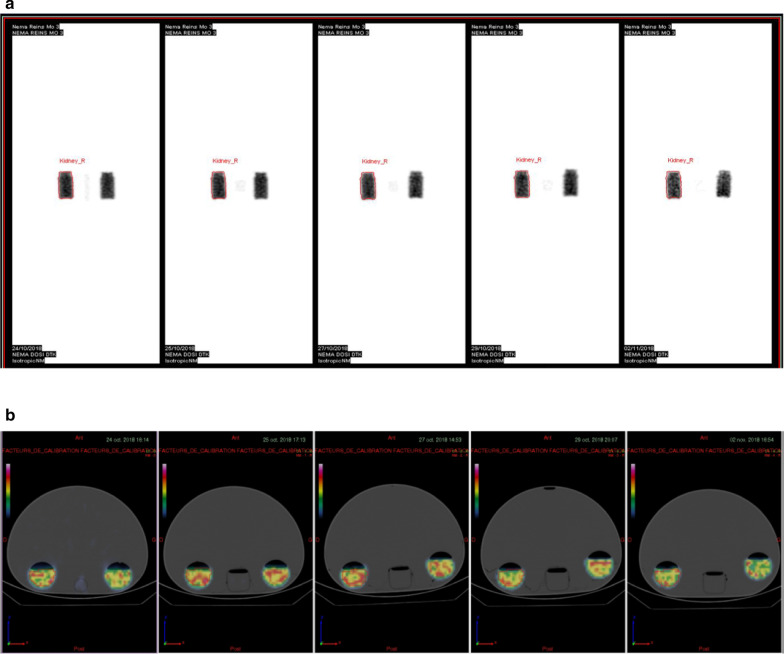
A NEMA IEC body phantom (Body Phantom NU2-2001/2007) that contains two bottles of 250 mL filled with 200 mL of 82.2 ± 4.1 (i.e. maximum activity measurement error of 5%) MBq [177Lu]Lu-DOTA-TATE was chosen with the aim of mimicking the size of kidneys. The background was filled with non-radioactive water. SPECT/CT images were acquired at different time points to evaluate CF variations over time. The CF was estimated using one of the two bottles filled with [177Lu]Lu-DOTA-TATE (Fig. 1).
Fig. 1.
Dosimetry Toolkit® (a) and PLANET® Dose interfaces (b) after imaging of the NEMA IEC body phantom (Body Phantom NU2-2001/2007) that contains two 250 mL bottles filled with 200 mL of [177Lu]Lu-DOTA-TATE at different time points. Only one of the two bottles was segmented
Dosimetry toolkit
A CT rigid registration based on the full phantom was performed with “Preparation for Dosimetry Toolkit”. Then, using the “Dosimetry Toolkit” application, an isocontour representing a volume of 200 mL was automatically segmented on the first nuclear medicine image and was replicated for the images at 24 h, 72 h, 120 h and 216 h. For each time-point, the segmented volume was kept constant. To determine the CF in counts.s−1.MBq−1 at each time point, the number of events in the volume was divided by the acquisition time provided by the DICOM data (2700 s) and by the activity. For radioactivity decay correction, a physical half-life of 6.647 days [24] was applied and the activity at each acquisition time point was corrected for the phantom preparation time. The CF was calculated as the mean of the CF values obtained at the different time points. To obtain the time-activity curve fitted by a mono-exponential function and the TIAC (h), information about the radionuclide and the CF were entered in the appropriate interface.
PLANET® Dose
For the body phantom, CT image rigid registration based on the volume that tightly enclosed the bottle was performed with PLANET® Dose. Similarly, an isocontour that represented a volume of 200 mL was automatically segmented on the first functional image and strictly transferred to the SPECT/CT images acquired at the other time points using a rigid contour propagation algorithm. The segmented volume was maintained over time. To determine the CF in Bq.count−1 at each time point, the number of events in the segmented volume was divided by the activity in Bq, by taking into account the radioactivity decay. The CF was calculated as the mean of the CF values obtained at the different time points.
A mono-exponential fitting function, similar to the Dosimetry Toolkit® approach, was used to calculate the TIA , and the TIAC , as [25]:
where is the initial activity, and is the experimental half-life assessed using a mono-exponential fit.
As in that situation, only physical decay is observed (T = 6.647 days) [24], it is possible to derive the reference TIAC as:
The reference TIAC was compared to the experimental values obtained with the two dosimetry platforms.
Clinical study
Patients and treatment
Twenty-one patients (5 women and 16 men; median age 68 years, range 41–82 years) with a neuroendocrine tumour and treated with [177Lu]Lu-DOTA-TATE, Lutathera® (Advanced Accelerator Applications, Saint Genis Pouilly, France) were evaluated (Additional file 1: Table S1). The treatment consisted in 7.2 ± 0.2 GBq activity (four infusions in total) injected every 8 weeks. Amino acids (lysine + arginine) were administered concomitantly to ensure renal protection by reducing tubular reabsorption of the radiolabelled peptides. All patients were hospitalized in specialized radioprotection rooms for 24 h after injection. Patients were then discharged and had to come back for the other post-infusion imaging sessions. Dosimetry to liver, kidneys and spleen (i.e. the OARs) was performed after the first and second infusion of [177Lu]Lu-DOTA-TATE. Dosimetry data after the first injection were not evaluable in one patient, and another patient died before the second infusion. In total, 40 dosimetry analyses were performed with each dosimetry package.
Dosimetry workflow
The dosimetry workflow for the two packages is presented in Table 3. SPECT/CT images were acquired at 4 h, 24 h, 72 h and 192 h after infusion. For some patients, due to health problems, technical issues or calendar reasons, SPECT/CT images were acquired at only three time points after injection. As dosimetry for the first two injections of [177Lu]Lu-DOTA-TATE is performed routinely in our department, currently with Dosimetry Toolkit + OLINDA, we carried out a retrospective additional analysis of the already available dosimetric data with PLANET® Dose.
Table 3.
Dosimetry workflow in patients using the reference dosimetry approach Dosimetry Toolkit + OLINDA and the new workstation PLANET® Dose
| Dosimetry Toolkit® + OLINDA/EXM® V1.0 | PLANET® Dose | |
|---|---|---|
| Calibration procedure |
SPECT/CT acquisitions Calibration factor (counts.s−1.MBq−1) |
SPECT/CT acquisitions Calibration factor (Bq.Count−1) |
| Clinical imaging | 4 SPECT/CT at 4 h, 24 h, 72 h and 192 h after injection—60 projections of 45 s | |
| Reconstruction | « Preparation for Dosimetry Toolkit» 6 iterations 10 subsets-AC, SC, RR, Gaussian post filter 0.11 cm | |
| Registration |
« Preparation for Dosimetry Toolkit» application Automatic rigid registration using CT images (Full FOV) |
Organ-based rigid registration using CT images |
| Segmentation and propagation |
Manual segmentation using the first CT image Rigid propagation (constant volumes) Segmentation adjusted by translation or rotation for other images |
Manual segmentation using the first CT image Rigid propagation (constant volumes) No adjustment for other images |
| TIA fitting | Mono-exponential | Mono-exponential |
| Absorbed dose calculation |
OLINDA/EXM® V1.0 Patient-adapted organ masses |
Local Deposition Method (LDM) Dose Kernel (DK) approach with/without density correction |
AC, attenuation correction; SC, scatter correction, RR, recovery resolution; FOV, field of view; CT, computed tomography; TIA, time integrated activity
Reference dosimetry method
For both infusions, after the last SPECT/CT image acquisition at 192 h, all SPECT/CT data were loaded on the “Preparation for Dosimetry Toolkit” application. Imaging data were reconstructed and an automatic rigid registration of the CT scans was performed. The results were loaded on the “Dosimetry Toolkit” application. Liver, kidneys and spleen were manually segmented using the CT images collected at 4 h post-injection, and then the segmented contour was rigidly propagated to the 24 h, 72 h, and 192 h images. For each time point, the segmented volume was maintained, but sometimes it was adjusted by translation or rotation. The partial volume effect correction was considered negligible. The administered activity, date and time of administration, radionuclide and CF values (in counts.s−1.MBq−1) were entered. To obtain the TIAC, the time-activity curves were fitted using a mono-exponential function, the only fitting model available in the “Dosimetry Toolkit” application. Then, the TIAC values were exported to OLINDA/EXM® V1.0 to calculate the ADs to liver, kidneys and spleen (i.e. the OARs). The organ masses included in this software were determined from the organ volume defined on the CT images using “Dosimetry Toolkit” and the biological tissue density (1.06 g.cm−3 for liver and spleen; 1.05 g.cm−3 for kidneys).
PLANET® Dose
The transversal slices reconstructed using the “Preparation for Dosimetry Toolkit” application and the corresponding CT images were uploaded on PLANET® Dose. Automatic and rigid registration was performed iteratively for each organ, based on a volume tightly enclosing the organ of interest. Then, the organs of interest on the first CT image were manually segmented, and the volumes were transferred from the reference CT image to the SPECT/CT images acquired at the other time points by rigid propagation. The volume of each OAR remained constant at all time points. As mentioned above, the partial volume effect correction was considered negligible for the OARs under study.
The administered activity, date and time of administration, radionuclide used, and CF value (in Bq.counts−1) were entered. The time-activity curve was fitted using a similar approach as the one used in Dosimetry Toolkit® (i.e. mono-exponential function) to provide the TIAC (h) and the TIA (MBq.s). The mean ADs were calculated using the LDM and DK methods, with correction of density.
Mean absorbed dose calculation
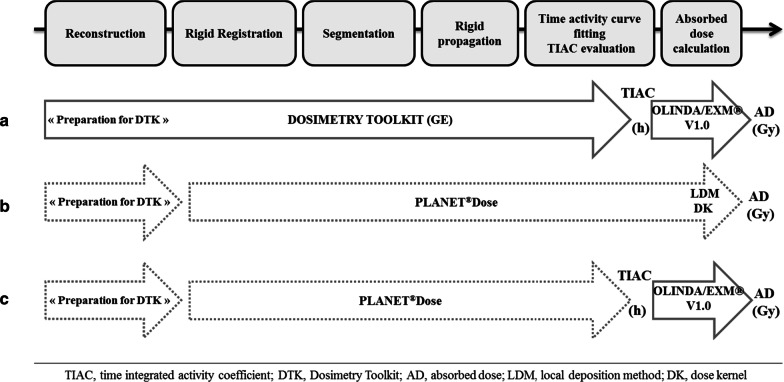
The mean AD was calculated using the first two approaches presented in Fig. 2:
The “Dosimetry Toolkit” approach used TIAC provided by Dosimetry Toolkit®, entered in OLINDA/EXM® V1.0, to derive the mean ADs to liver, kidneys and spleen (Fig. 2a).
The PLANET® Dose approach used reconstructed images (Preparation for Dosimetry Toolkit) and full processing (registration, segmentation, TIAC and AD calculation using the LDM and DK methods, with density correction) on PLANET® Dose (Fig. 2b).
Fig. 2.
Dosimetry workflow using the reference dosimetry approach Dosimetry Toolkit + OLINDA (a) and the PLANET® Dose package with the LDM and DK methods (b). The PLANET® Dose + OLINDA approach (c) was used to evaluate independently the AD calculation methods
A third approach was also used to evaluate independently the previous two AD calculation methods. This approach was similar to the PLANET® Dose method, but after the TIAC step, the ADs were computed with OLINDA/EXM® V1.0 (Fig. 2c) using the same phantom model than in the first approach (adult male or female).
Statistical analyses
Besides the relative difference (in %), the root mean-square deviation (RMSD) of organ masses, TIACs and ADs per cycle obtained with PLANET® Dose and Dosimetry Toolkit + OLINDA (taken as reference) was calculated as follows:
where X(i) was the organ masses, TIACs or ADs obtained for the dosimetry analysis i.
The Lin’s concordance correlation coefficient [26] was used to evaluate the agreement between PLANET® Dose LDM and the reference method (Dosimetry Toolkit + OLINDA). This analysis was performed using the mean values for all patients and organs after the two infusions. Moreover, the absolute differences between the mean AD values obtained with the two approaches were assessed for each organ using the Bland–Altman plot analysis [27]. The 95% limits of agreement, from − 1.96 to + 1.96 SD, were calculated for each organ.
The paired Student’s t-test was used to compare the liver, kidneys and spleen ADs calculated with Dosimetry Toolkit + OLINDA and PLANET® Dose LDM (n = 40). This analysis was performed using the mean values for all patients and organs after the two infusions.
To evaluate independently the AD calculation methods, the mean AD obtained with PLANET® Dose and with “PLANET® Dose + OLINDA/EXM® V1.0” (taken as reference) were compared. A bland–Altman analysis and a correlation evaluation were performed.
Results
Phantom-based study
SPECT/CT CF values of 5.60 ± 0.04 counts.s−1.MBq−1 and 5.53 ± 0.19 counts.s−1.MBq−1 were obtained with Dosimetry Toolkit® and PLANET® Dose, respectively. These values did not vary significantly over time (0.8% and 3.3% of variation, respectively) (Fig. 3). For PLANET® Dose, the CF value was converted to 67 ± 2.2 Bq.counts−1 to be uploaded on the software.
Fig. 3.
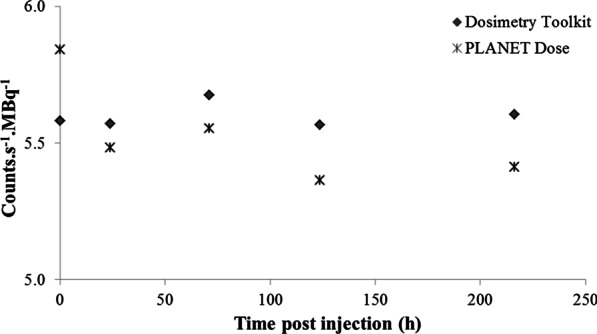
Variation of the SPECT CF values over time after [177Lu]Lu-DOTA-TATE injection obtained with Dosimetry Toolkit® and PLANET® Dose
These CF values were then used in the clinical study to calculate the time activity curves and TIACs for liver, kidneys, and spleen.
TIACs of 217.52 h and 225.19 h were obtained with Dosimetry Toolkit and PLANET® Dose, respectively. These values showed a deviation of − 5.5% and − 2.2% relative to the reference residence time of 230.15 h.
Clinical results
Package comparison
The mean organ masses, TIACs and ADs for liver, kidneys and spleen (i.e. OARs) obtained using each dosimetry package are summarized in Table 4a. The relative differences of the organ mass values and TIACs between Dosimetry Toolkit + OLINDA/EXM® V1.0 (the reference) and PLANET® Dose are presented in Table 4b. These results highlighted RMSD values lower than 10% for organ mass values and TIACs, but for the spleen TIAC (RMSD = 10.4%).
Table 4.
(a) Mean and standard deviation (n = 40 dosimetry analyses) of OAR organ masses, TIACs and ADs calculated using Dosimetry Toolkit + OLINDA/EXM® V1.0 and PLANET® Dose with LDM and DK and density correction. (b) Relative differences (%) of the organ mass values and TIACs between PLANET® Dose and Dosimetry Toolkit + OLINDA, DTK, Dosimetry Toolkit
| (a) | |||||||
|---|---|---|---|---|---|---|---|
| ORGANS | Mass (g) | TIAC (h) | Absorbed dose (Gy) | ||||
| DTK + OLINDA/EXM® V1.0 | PLANET®Dose | DTK + OLINDA/EXM® V1.0 | PLANET® Dose | DTK + OLINDA/EXM® V1.0 | PLANET®Dose | ||
| LDM | DK | ||||||
| Liver | 2141.6 ± 1213.3 | 2191.7 ± 1205 | 14.9 ± 24.1 | 15.1 ± 24.2 | 3.40 ± 3.9 | 3.27 ± 3.7 | 3.21 ± 3.6 |
| Kidneys | 478.3 ± 111.4 | 461.5 ± 108.3 | 2.2 ± 0.7 | 2.4 ± 0.8 | 3.01 ± 0.9 | 3.23 ± 0.9 | 3.16 ± 0.9 |
| Spleen | 290.4 ± 181.3 | 281.7 ± 178.5 | 2.0 ± 1.5 | 2.0 ± 1.4 | 4.15 ± 1 | 4.45 ± 1.2 | 4.36 ± 1.2 |
| (b) | ||||||
|---|---|---|---|---|---|---|
| Relative difference (%)/DTK+OLINDA | ||||||
| Mass | TIAC | |||||
| Liver | Kidneys | Spleen | Liver | Kidneys | Spleen | |
| Mean | 2.9 | −3.4 | −2.4 | 5.7 | 6.4 | 4.9 |
| Min | −4.0 | −18.8 | −23.4 | −6.9 | −9.1 | −18.9 |
| Max | 19.7 | 16.4 | 17.3 | 17.3 | 25.7 | 23.5 |
| RMSD | 5.2 | 7.2 | 8.9 | 8.1 | 9.4 | 10.4 |
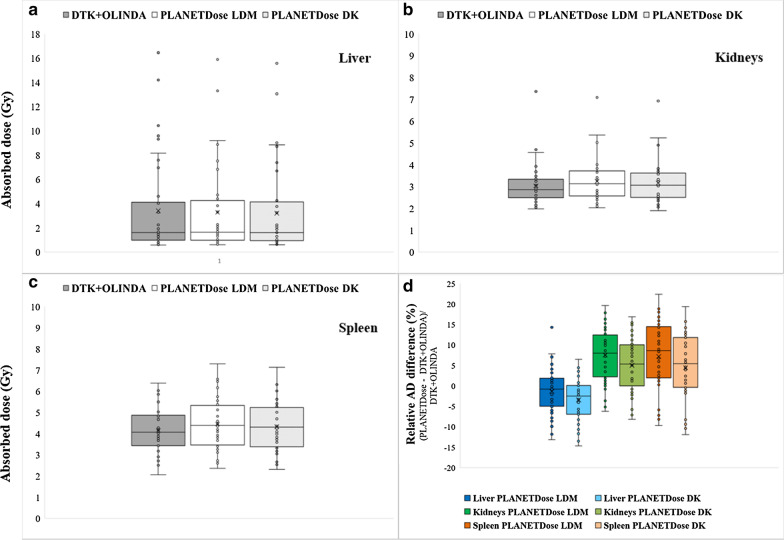
The comparison of the OAR AD values obtained with Dosimetry Toolkit + OLINDA and with PLANET® Dose, LDM and DK, are presented in Table 5a–c and Fig. 4. Again, the mean difference and RMSD values were lower than 10% for all organs, except for spleen (RMSD = 10.9%). For kidneys and spleen, the AD values obtained with PLANET® Dose were slightly, but significantly higher (p < 0.05).
Table 5.
Comparison of the AD values to each OAR: liver (a), kidneys (b) and spleen (c) calculated with Dosimetry Toolkit + OLINDA and PLANET® Dose with density correction. DTK, Dosimetry Toolkit
| (a) | |||||
|---|---|---|---|---|---|
| Dosimetry analyses | Absorbed dose to liver (Gy) | ||||
| DTK + OLINDA/EXM® V1.0 | PLANET®Dose LDM | PLANET®Dose DK | Relative difference (%)/DTK + OLINDA | ||
| PLANET®Dose LDM | PLANET®Dose DK | ||||
| 1 | 1.04 | 1.04 | 1.02 | − 0.1 | − 1.9 |
| 2 | 1.59 | 1.64 | 1.61 | 3.2 | 1.2 |
| 3 | 0.92 | 0.86 | 0.85 | − 6.7 | − 8.3 |
| 4 | 16.44 | 15.89 | 15.56 | − 3.4 | − 5.4 |
| 5 | 14.19 | 13.31 | 13.06 | − 6.2 | − 8.0 |
| 6 | 3.27 | 3.30 | 3.24 | 0.9 | − 1.0 |
| 7 | 4.59 | 4.40 | 4.32 | − 4.1 | − 5.9 |
| 8 | 0.57 | 0.61 | 0.60 | 7.0 | 4.4 |
| 9 | 2.05 | 2.04 | 2.00 | − 0.3 | − 2.2 |
| 10 | 1.26 | 1.09 | 1.07 | − 13.2 | − 14.7 |
| 11 | 1.12 | 1.11 | 1.10 | − 0.9 | − 2.2 |
| 12 | 2.40 | 2.28 | 2.23 | − 5.0 | − 7.0 |
| 13 | 4.02 | 3.83 | 3.76 | − 4.7 | − 6.4 |
| 14 | 0.91 | 0.93 | 0.92 | 2.1 | 0.7 |
| 15 | 0.79 | 0.80 | 0.78 | 1.3 | − 1.0 |
| 16 | 9.58 | 9.00 | 8.83 | − 6.0 | − 7.8 |
| 17 | 9.32 | 8.88 | 8.72 | − 4.7 | − 6.4 |
| 18 | 1.47 | 1.49 | 1.44 | 1.5 | − 1.9 |
| 19 | 1.92 | 1.89 | 1.86 | − 1.8 | − 3.3 |
| 20 | 0.94 | 0.86 | 0.84 | − 8.7 | − 10.7 |
| 21 | 1.13 | 1.18 | 1.16 | 4.1 | 2.4 |
| 22 | 1.65 | 1.68 | 1.65 | 1.8 | 0.1 |
| 23 | 0.88 | 0.93 | 0.91 | 5.7 | 3.5 |
| 24 | 6.95 | 6.89 | 6.75 | − 0.9 | − 2.8 |
| 25 | 2.09 | 2.10 | 2.06 | 0.7 | − 1.1 |
| 26 | 2.34 | 2.29 | 2.25 | − 2.1 | − 3.8 |
| 27 | 7.57 | 6.82 | 6.69 | − 9.9 | − 11.7 |
| 28 | 0.62 | 0.65 | 0.63 | 5.2 | 2.7 |
| 29 | 1.61 | 1.64 | 1.61 | 1.9 | 0.1 |
| 30 | 1.14 | 1.14 | 1.12 | − 0.4 | − 2.3 |
| 31 | 1.39 | 1.31 | 1.29 | − 5.6 | − 7.0 |
| 32 | 2.24 | 2.19 | 2.15 | − 2.1 | − 4.1 |
| 33 | 4.13 | 4.72 | 4.25 | 14.3 | 2.9 |
| 34 | 0.78 | 0.84 | 0.83 | 7.7 | 6.5 |
| 35 | 0.81 | 0.81 | 0.80 | 0.0 | − 1.6 |
| 36 | 10.43 | 9.19 | 9.02 | − 11.9 | − 13.5 |
| 37 | 8.15 | 7.51 | 7.38 | − 7.9 | − 9.5 |
| 38 | 1.42 | 1.36 | 1.34 | − 4.1 | − 5.7 |
| 39 | 1.57 | 1.49 | 1.46 | − 5.1 | − 6.8 |
| 40 | 0.92 | 0.94 | 0.93 | 2.0 | 0.5 |
| Mean | 3.40 | 3.27 | 3.20 | − 1.4 | − 3.5 |
| SD | 3.89 | 3.65 | 3.58 | 5.5 | 4.9 |
| Min | 0.57 | 0.61 | 0.60 | − 13.2 | − 14.7 |
| Max | 16.44 | 15.89 | 15.56 | 14.3 | 6.5 |
| RMSD | 5.6 | 6.0 | |||
| (b) | |||||
|---|---|---|---|---|---|
| Dosimetry analyses | Absorbed dose to kidneys | ||||
| DTK + OLINDA/EXM® V1.0 | PLANET®Dose LDM | PLANET®Dose DK | Relative difference (%)/DTK + OLINDA | ||
| PLANET®Dose LDM | PLANET®Dose DK | ||||
| 1 | 2.78 | 3.14 | 3.07 | 12.8 | 10.3 |
| 2 | 2.79 | 3.10 | 3.03 | 11.1 | 8.5 |
| 3 | 3.26 | 3.41 | 3.33 | 4.6 | 2.1 |
| 4 | 3.68 | 3.45 | 3.38 | − 6.3 | − 8.2 |
| 5 | 2.05 | 2.03 | 1.91 | − 0.7 | − 6.9 |
| 6 | 3.06 | 3.66 | 3.57 | 19.6 | 16.8 |
| 7 | 3.56 | 3.84 | 3.75 | 7.7 | 5.1 |
| 8 | 3.48 | 3.83 | 3.70 | 10.0 | 6.2 |
| 9 | 2.53 | 2.40 | 2.35 | − 5.2 | − 7.2 |
| 10 | 3.07 | 3.29 | 3.22 | 7.2 | 5.0 |
| 11 | 2.10 | 2.40 | 2.34 | 14.3 | 11.5 |
| 12 | 2.51 | 2.61 | 2.56 | 3.9 | 1.8 |
| 13 | 3.24 | 3.35 | 3.28 | 3.4 | 1.1 |
| 14 | 2.44 | 2.72 | 2.66 | 11.5 | 8.8 |
| 15 | 2.20 | 2.50 | 2.45 | 13.6 | 11.1 |
| 16 | 1.98 | 2.10 | 2.06 | 5.8 | 3.8 |
| 17 | 2.96 | 3.49 | 3.41 | 18.0 | 15.4 |
| 18 | 2.90 | 3.13 | 3.05 | 8.1 | 5.4 |
| 19 | 4.69 | 5.37 | 5.24 | 14.5 | 11.7 |
| 20 | 2.48 | 2.46 | 2.40 | − 0.8 | − 3.2 |
| 21 | 3.31 | 3.73 | 3.65 | 12.7 | 10.2 |
| 22 | 2.85 | 3.15 | 3.12 | 10.7 | 9.5 |
| 23 | 3.93 | 4.00 | 3.91 | 1.8 | − 0.5 |
| 24 | 2.60 | 2.60 | 2.54 | 0.0 | − 2.3 |
| 25 | 3.54 | 3.91 | 3.82 | 10.5 | 8.0 |
| 26 | 3.31 | 3.50 | 3.42 | 5.7 | 3.3 |
| 27 | 7.36 | 7.09 | 6.93 | − 3.7 | − 5.8 |
| 28 | 3.35 | 3.86 | 3.76 | 15.2 | 12.3 |
| 29 | 2.51 | 2.55 | 2.49 | 1.6 | − 0.8 |
| 30 | 3.32 | 3.34 | 3.27 | 0.6 | − 1.6 |
| 31 | 2.62 | 2.82 | 2.75 | 7.5 | 5.0 |
| 32 | 2.31 | 2.55 | 2.49 | 10.5 | 8.0 |
| 33 | 3.37 | 3.92 | 3.83 | 16.3 | 13.5 |
| 34 | 2.53 | 2.68 | 2.62 | 5.9 | 3.5 |
| 35 | 2.30 | 2.71 | 2.64 | 17.8 | 14.9 |
| 36 | 2.15 | 2.22 | 2.17 | 3.4 | 1.2 |
| 37 | 2.08 | 2.10 | 2.06 | 1.2 | − 0.8 |
| 38 | 2.87 | 3.11 | 3.04 | 8.5 | 6.0 |
| 39 | 4.57 | 5.02 | 4.90 | 9.8 | 7.2 |
| 40 | 2.52 | 2.80 | 2.73 | 11.1 | 4.6 |
| Mean | 3.03 | 3.25 | 3.17 | 7.5 | 4.9 |
| SD | 0.95 | 0.97 | 0.95 | 6.5 | 6.4 |
| Min | 1.98 | 2.03 | 1.91 | − 6.3 | − 8.2 |
| Max | 7.36 | 7.09 | 6.93 | 19.6 | 16.8 |
| RMSD | 9.9 | 8.0 | |||
| (c) | |||||
|---|---|---|---|---|---|
| Dosimetry analyses | Absorbed dose to spleen | ||||
| DTK + OLINDA/EXM® V1.0 | PLANET®Dose LDM | PLANET®Dose DK | Relative difference (%)/DTK + OLINDA | ||
| PLANET®Dose LDM | PLANET®Dose DK | ||||
| 1 | 3.86 | 4.12 | 4.03 | 6.8 | 4.5 |
| 2 | 4.67 | 5.41 | 5.28 | 15.8 | 13.1 |
| 3 | 3.83 | 4.40 | 4.31 | 15.0 | 12.5 |
| 4 | 4.01 | 3.68 | 3.59 | − 8.2 | − 10.4 |
| 5 | 4.35 | 3.93 | 3.83 | − 9.7 | − 12.0 |
| 6 | 2.98 | 3.30 | 3.23 | 10.9 | 8.3 |
| 7 | 5.03 | 5.37 | 5.28 | 6.7 | 5.0 |
| 8 | 2.07 | 2.36 | 2.31 | 14.3 | 11.7 |
| 9 | 3.67 | 3.40 | 3.32 | − 7.2 | − 9.4 |
| 10 | 4.87 | 4.59 | 4.37 | − 5.7 | − 10.2 |
| 11 | 3.69 | 3.83 | 3.74 | 3.8 | 1.4 |
| 12 | 4.27 | 4.35 | 4.25 | 1.8 | − 0.5 |
| 13 | 4.17 | 4.47 | 4.37 | 7.2 | 4.7 |
| 14 | 3.22 | 3.30 | 3.22 | 2.5 | 0.0 |
| 15 | 2.91 | 3.33 | 3.26 | 14.6 | 12.0 |
| 16 | 2.94 | 3.11 | 3.05 | 5.9 | 3.8 |
| 17 | 2.57 | 2.60 | 2.54 | 1.1 | − 1.2 |
| 18 | 4.75 | 5.19 | 5.07 | 9.3 | 6.8 |
| 19 | 5.50 | 6.17 | 6.02 | 12.2 | 9.5 |
| 20 | 3.77 | 3.94 | 3.86 | 4.6 | 2.4 |
| 21 | 4.83 | 5.64 | 5.51 | 16.8 | 14.2 |
| 22 | 3.88 | 4.38 | 4.35 | 12.9 | 12.1 |
| 23 | 4.91 | 5.55 | 5.42 | 13.0 | 10.4 |
| 24 | 4.05 | 3.81 | 3.71 | − 5.9 | − 8.3 |
| 25 | 6.38 | 7.30 | 7.13 | 14.4 | 11.8 |
| 26 | 4.18 | 4.53 | 4.42 | 8.3 | 5.7 |
| 27 | 5.57 | 5.12 | 5.02 | − 8.1 | − 9.9 |
| 28 | 2.50 | 2.72 | 2.66 | 8.8 | 6.4 |
| 29 | 3.43 | 4.20 | 4.10 | 22.4 | 19.3 |
| 30 | 4.75 | 4.82 | 4.70 | 1.5 | − 1.0 |
| 31 | 5.52 | 6.51 | 5.67 | 18.0 | 2.8 |
| 32 | 4.09 | 4.46 | 4.36 | 9.0 | 6.6 |
| 33 | 5.02 | 5.75 | 5.63 | 14.5 | 12.0 |
| 34 | 2.71 | 2.80 | 2.73 | 3.3 | 0.6 |
| 35 | 3.44 | 4.09 | 3.98 | 18.8 | 15.7 |
| 36 | 3.81 | 4.41 | 4.31 | 15.8 | 13.3 |
| 37 | 3.18 | 3.25 | 3.18 | 2.2 | − 0.1 |
| 38 | 5.86 | 6.46 | 6.32 | 10.2 | 7.8 |
| 39 | 6.03 | 6.57 | 6.40 | 9.0 | 6.1 |
| 40 | 4.84 | 4.85 | 4.75 | 0.2 | − 1.9 |
| Mean | 4.15 | 4.45 | 4.33 | 7.2 | 4.4 |
| SD | 1.03 | 1.19 | 1.14 | 8.3 | 8.0 |
| Min | 2.07 | 2.36 | 2.31 | − 9.7 | − 12.0 |
| Max | 6.38 | 7.30 | 7.13 | 22.4 | 19.3 |
| RMSD | 10.9 | 9.1 | |||
Fig. 4.
Box-and-whisker plots showing the ADs to liver (a), kidneys (b) and spleen (c) calculated using Dosimetry Toolkit + OLINDA and the PLANET® Dose packages, as well as the relative differences of AD values between PLANET® Dose and Dosimetry Toolkit + OLINDA (d). DTK, Dosimetry Toolkit
The mean difference between the AD values calculated with LDM and DK (PLANET® Dose) was 2.2%. The values obtained with the LDM method were always higher than those obtained with the DK method (Fig. 4d). Moreover, whatever the software used, liver presented the highest AD variability, with the highest values reaching 16 Gy (Fig. 4a).
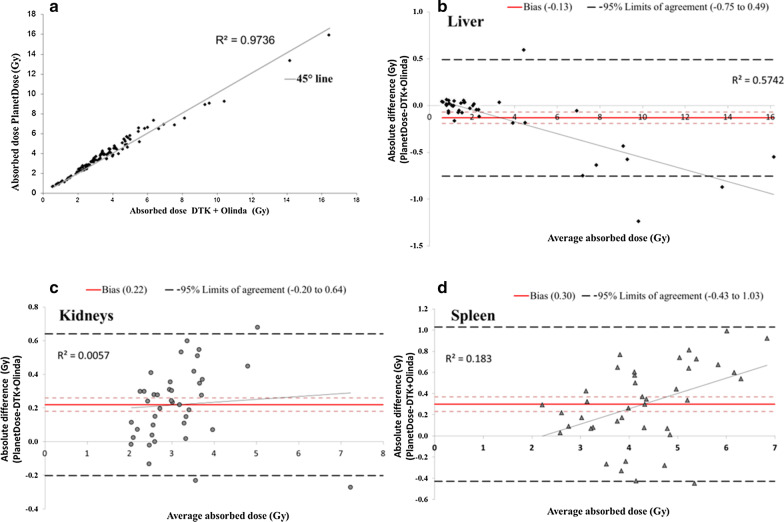
Concordance between packages
For the 40 dosimetry evaluations, the estimated Lin’s concordance correlation coefficient was 0.99 (95% CI 0.99; 0.99; R2 = 0.9736) (Fig. 5a). This result suggests an excellent concordance between our current dosimetry method and PLANET® Dose. According to the Bland–Altman plot method (Fig. 5b–d), the “bias” value (i.e. the mean of the AD differences between PLANET® Dose and Dosimetry Toolkit + OLINDA) was − 0.13 Gy for liver, 0.22 Gy for kidneys, and 0.30 Gy for spleen. Moreover, this approach estimated that the difference of AD values between methods ranged from − 0.75 to 0.49 Gy, from − 0.20 to 0.64 Gy, and from − 0.43 to 1.03 Gy for 95% of the 40 liver, kidneys and spleen dosimetry analyses, respectively. For liver, the Bland–Altman analysis showed that the mean absolute dose differences progressively increased with the dose (R2 = 0.5742).
Fig. 5.
Dispersion around the 45° line of the AD pairs obtained with Dosimetry Toolkit + OLINDA and PLANET® Dose LDM with density correction for all organs combined (a). Bland–Altman plots of the ADs to liver (b), kidneys (c) and spleen (d), calculated with Dosimetry Toolkit + OLINDA and PLANET® Dose LDM with density correction. DTK, Dosimetry Toolkit
Calculation method assessment
The comparison of the mean AD obtained with PLANET® Dose LDM and DK and using the TIACs from PLANET® Dose uploaded on OLINDA/EXM® V1.0 (i.e. the reference in this comparison) (Additional file 2: Fig. S1 and Table 6) showed that the mean relative difference and RMSD values were lower than 5% for all organs, but for spleen (maximum RMSD value of 6.5%).
Table 6.
Mean, SD, range, RMSD value of the AD values to liver, kidneys and spleen calculated with PLANET® Dose + OLINDA and PLANET® Dose with density correction
| Absorbed dose (Gy) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Liver | Kidneys | Spleen | |||||||||||||
| PLANET + OLINDA/EXM® V1.0 | PLANET®Dose LDM | PLANET®Dose DK | Relative deviation (%)/PLANET + OLINDA | PLANET + OLINDA/EXM® V1.0 | PLANET®Dose LDM | PLANET®Dose DK | Relative deviation (%)/PLANET + OLINDA | PLANET + OLINDA/EXM® V1.0 | PLANET®Dose LDM | PLANET®Dose DK | Relative deviation (%)/PLANET + OLINDA | ||||
| PLANET®Dose LDM | PLANET®Dose DK | PLANET®Dose LDM | PLANET®Dose DK | PLANET®Dose LDM | PLANET®Dose DK | ||||||||||
| Mean | 3.43 | 3.27 | 3.20 | − 4.5 | − 6.2 | 3.31 | 3.25 | 3.17 | − 1.9 | − 4.2 | 4.47 | 4.45 | 4.33 | − 1.1 | − 3.4 |
| SD | 3.84 | 3.65 | 3.58 | 3.6 | 3.6 | 3.58 | 3.58 | 3.58 | 3.6 | 3.6 | 3.58 | 3.58 | 3.58 | 3.6 | 3.6 |
| Min | 0.62 | 0.61 | 0.60 | − 6.6 | − 8.5 | 1.97 | 1.94 | 1.91 | − 5.8 | − 7.7 | 2.40 | 2.36 | 2.31 | − 5.5 | − 7.6 |
| Max | 16.40 | 15.89 | 15.56 | 0.9 | − 1.0 | 7.36 | 7.09 | 6.93 | 2.9 | − 0.6 | 7.58 | 7.30 | 7.13 | 6.5 | 4.2 |
| RMSD | 4.9 | 6.5 | 2.5 | 4.4 | 4.6 | 3.3 | 4.6 | ||||||||
The estimated Lin’s concordance correlation coefficient was 1 (R2 = 0.9966) (Additional file 3: Fig. S2a). The “bias” value of the Bland–Altman plot analysis (i.e. the average of the differences between PLANET® Dose LDM and PLANET® Dose + OLINDA) was − 0.16 Gy for liver, − 0.06 Gy for kidneys, and − 0.04 Gy for spleen. The difference of AD values between methods ranged from − 0.57 to 0.24 Gy, from − 0.18 to 0.06 Gy, and from − 0.34 to 0.26 Gy for 95% of the 40 liver, kidneys and spleen dosimetry analyses, respectively (Additional file 3: Fig. S2b–d). For liver, the negative trend (Bland–Altman analysis) showed an R2 value of 0.8358.
Discussion
Due to the need of implementing central dosimetric data processing for multicentre trials, our department acquired a vendor-neutral solution, PLANET® Dose, for dosimetry assessments. However, to take advantage of the experience and data already acquired with Dosimetry Toolkit® and OLINDA/EXM® V1.0, we needed to compare their results. Overall, the two systems gave similar dosimetry results in the phantom study (CF and TIAC) and also when using imaging data from patients who received two injections of [177Lu]Lu-DOTA-TATE (mean organ masses, TIACs and ADs for liver, kidneys and spleen).
In a recent study, Huizing et al. [16] compared dosimetry results obtained with Hybrid Viewer Dosimetry and PLANET® Dose in ten patients using hybrid imaging data obtained from 0.5 h to 72 h post-injection of [177Lu]Lu-DOTA-TATE. Although our study also concerned PLANET® Dose, our reference method was Dosimetry Toolkit®. Furthermore, our study was based on complete 3D imaging data from 4 to 192 h post-injection in 21 patients undergoing [177Lu]Lu-DOTA-TATE treatment, thus representing a total of 40 dosimetry analysis.
In our study, the reconstruction step was performed with the GE application “Preparation for Dosimetry Toolkit” because PLANET® Dose does not include this functionality. The latter accepts reconstructed data supplied by others workstations, unlike the “Dosimetry Toolkit” application.
In a clinical dosimetry study, the preliminary step of calibration is crucial to obtain accurate activity quantification [28, 29]. A detailed calibration protocol is the cornerstone to achieve accurate and reliable image quantification in multicentric studies [30–33]. According to the GE recommendations, CF determined by planar acquisition of a 15 cm diameter Petri dish partially filled with a solution of 177Lu is sufficient for Dosimetry Toolkit®. However as our first aim was to monitor the AD to OARs, we used a large phantom (i.e. a bottle with a volume relatively close to that of kidney). The same methodology based on SPECT/CT imaging was followed for both calibration and clinical imaging. As described by Gustafsson et al. [34], SPECT image segmentation is essential to determine the activity concentration. For our phantom-based study, we selected a fixed volume threshold method based on an automatically drawn isocontour around the bottle with a volume of 200 mL. This volume was segmented on the SPECT images acquired at the first time point and rigidly copied to the others (i.e. rigid propagation). This step is directly affected by the accuracy of the registration of all SPECT/CT images using a rigid algorithm [11, 35, 36]. The obtained CF was similar (within 10%) to the one determined by Peters et al. [31] using a comparable gamma camera model.
Moreover, CFs are expressed in counts.s−1.MBq−1 by Dosimetry Toolkit® and in Bq.count−1 by PLANET® Dose. This implies that the CF must be modified with the acquisition duration for PLANET® Dose. This also highlights a risk of errors because of the lack of a standardized method for introducing CFs in dosimetry software tools. The crucial recommendation at this step is that the conditions used in clinical studies in terms of acquisition and reconstruction parameters must be similar to those used for calibration. We observed a negligible variation of the CF values over time, from T = 0–216 h. This means that the same CF can be used for each time point. This observation is particularly interesting for Dosimetry Toolkit® in which a single CF must be entered, whereas a different CF can be used at each time point with PLANET® Dose.
In the patient study, the liver masses obtained with the two packages were more similar than those obtained for smaller organs, such as kidneys and spleen, and the variability increased with the OAR decreasing size (Table 4b). Actually, as the DICOM-RT-Structure import could not be used in Dosimetry Toolkit®, organ delineation was done manually for each dosimetry package, leading to operator-induced variability. Thus, a small difference between contours could generate a more important relative deviation in smaller than larger organs. Moreover, the results presented in Table 5a–c seem to indicate a volume/mass effect. The RMSD of the AD values increased with the OAR decreasing size. For spleen, this could be explained by our package comparison methodology that used similar parameters in terms of registration and segmentation with constant volumes over time. Indeed, when using Dosimetry Toolkit®, the volume delineated on the first image was maintained, but adjusted by translation or rotation at each successive time point, when necessary. However, this step was not available in PLANET® Dose. Therefore, to use similar approaches with both packages, in PLANET® Dose we chose to perform an organ-based registration followed by rigid propagation (i.e. the contour registered in the first CT image was exactly propagated to the images acquired at the other time points). Thus, for some patients, the initial spleen contour did not fully match the organ contour at the other time points, and included tissues with different density. This implied an important deviation when calculating the AD with density correction. For instance, in the dosimetry analysis n°31, part of the spleen contour was moved to the left lung at other time points, and the AD was overestimated because of the density correction. For kidneys, AD variations between packages could be explained by the fact that in PLANET® Dose, left and right kidney are considered separately, while in OLINDA/EXM, the two kidneys are considered as a single organ. This may affect pharmacokinetic assessments, especially when rigid registration is considered. Therefore, we tested an additional approach that uses the segmentations and TIACs provided by PLANET® Dose and the dosimetric results (ADs) obtained with PLANET® Dose LDM or DK and with OLINDA/EXM® V1.0 (Table 6). The differences between approaches were reduced when the comparison considered only the AD calculation. This is in agreement with the idea that registration and segmentation are major steps, and probably induce more variability then the AD calculation step. Grassi et al. [36] showed that ADs to organs are significantly affected by the registration algorithm used. Indeed, due to respiratory motion during SPECT/CT and organ deformation, registration errors can happen. Although this may not influence the results of our comparison, it is clear that additional studies on registration and time activity curve fitting are needed, by taking advantage of the additional possibilities available in PLANET® Dose. Moreover, for accurate registration, position reproducibility during the four SPECT/CT acquisitions is crucial. Thus, patient set-up and immobilization devices are strongly recommended.
The Bland–Altman plots showed that the biases were quite low for all organs. The limits of agreement were rather tight with a maximum value of approximately 1 Gy for spleen. Moreover, the results for liver highlighted a trend for slightly higher AD values obtained with Dosimetry Toolkit + OLINDA than PLANET® Dose when the AD increased. This trend was more pronounced for patients with liver metastases, i.e. high activity gradients within the liver. One hypothesis is that cross-absorbed doses (from photons) may be different for heterogeneous vs. homogeneous activity distributions. Another point to consider is the density (homogeneous for OLINDA vs. voxel-based and coming from the CT images for PLANET® Dose). This may influence the AD calculation, but it is not clear to which extent. This certainly deserves to be thoroughly investigated, probably using Monte Carlo modelling to take into account also the possible impact of local density corrections [37].
The concordance evaluation (Lin’s coefficient value of 0.99) highlighted an excellent agreement between methods. Moreover, the dosimetry results obtained using PLANET® Dose (AD to liver, kidneys and spleen of 0.45 ± 0.50 Gy/GBq, 0.45 ± 0.13 Gy/GBq and 0.62 ± 0.17 Gy/GBq respectively) are in agreement with those of the literature [38].
As proposed by Gear et al. [39] in a practical guidance paper, the uncertainties at each step of the dosimetry analysis should be determined to express the accuracy of the dosimetry results. Currently, PLANET® Dose allows evaluating the relative proportion of interpolation (between time points) and extrapolation (after the last time point). Regardless of the goodness of fit, this is a good indication of the relevance of time sampling, but it is not sufficient to fully characterize the uncertainty associated with the dosimetric workflow. Such study requires important developments and will be implemented in the future.
From a qualitative point of view, PLANET® Dose is a user-friendly commercial solution that proposes a wide range of tools for segmentation, and several analytic fit functions. The time necessary for a dosimetry analysis is significantly reduced. Therefore, considering the good agreement with our reference dosimetry method, the concordance of the dosimetry results with the literature, the added value of this software (easy contouring, wide choice of time activity curve fitting models, time saving), and the fact that the observed differences were explainable and clinically acceptable, we think that the PLANET® Dose software can replace our current dosimetry package without any correction for dosimetry analysis. We can now start to investigate the different methodological possibilities offered by PLANET® Dose, such as elastic registration and propagation, time integration activity with multiple exponentials, AD rate computation at the voxel level, DICOM-RT import and export of structures. Its potential can now be fully explored, particularly for the determination of the tumour AD and for investigating the AD-response correlations.
Conclusion
In this work we compared the dosimetric results obtained with the software currently used in our department and with a new dosimetry software package available on the market. The ADs to OARs (liver, kidney and spleen) obtained with PLANET® Dose were concordant with those calculated with GE Dosimetry Toolkit® and OLINDA/EXM® V1.0, and in agreement with the literature. These results allow us to use PLANET® Dose in clinical routine for patient dosimetry after targeted radiotherapy with [177Lu]Lu-DOTA-TATE.
Supplementary Information
Additional file 1. Table S1: Patients’ characteristics.
Additional file 2. Fig. S1: Box-and-whisker plots showing the ADs to liver (a), kidneys (b) and spleen (c) calculated using PLANET® Dose and PLANET® Dose+OLINDA, as well as the relative AD differences between PLANET® Dose and PLANET® Dose+OLINDA (d).
Additional file 3. Fig. S2: Dispersion around the 45° line of the AD pairs obtained with PLANET+OLINDA and PLANET® Dose LDM with density correction for all organs combined (a). Bland-Altman plots of the AD to liver (b), kidneys (c) and spleen (d) calculated with PLANET+OLINDA and PLANET® Dose LDM with density correction.
Abbreviations
- AD
Absorbed dose
- CF
Calibration factor
- DICOM-RT
Digital imaging and communications in medicine-radiotherapy
- DK
Dose kernel
- DTK
Dosimetry toolkit®
- FOV
Field of view
- GE
General Electric
- IDAC
Internal dose assessment by computer
- LDM
Local deposition method
- MIRD
Medical internal radiation dose
- NETs
Neuroendocrine tumours
- NM
Nuclear medicine
- OAR
Organ at risk
- OSEM
Ordered subset expectation maximization
- PRRT
Peptide receptor radionuclide therapy
- RMSD
Root mean square deviation
- SD
Standard deviation
- SPECT/CT
Single photon emission computed tomography/computed tomography
- TACs
Time activity curves
- TIA
Time integrated activity
- TIAC
Time integrated activity coefficient
- VOI
Volume of interest
Authors’ contributions
LS, LP, DT, EMR carried out the phantom measurements, image reconstructions and data analysis. MB and ED participated to the design of the study. POK and ED were responsible of the patient acquisitions and treatments. LS was responsible of the dosimetry software implementation and performed patient dosimetry evaluations with LP. LS wrote the manuscript. All authors read and approved the final manuscript.
Funding
No funding was received.
Availability of data and materials
Please contact the author for data requests.
Ethics approval and consent to participate
All procedures performed in this study involving human participants were in accordance with the ethical standards of the COMERE research committee (“commission interne de méthodologie en recherche clinique”, ICMART201801).
Consent for publication
Consent has been obtained from participants to publish this work.
Competing interests
ED was invited in 2018 to attend an international meeting by AAA who sponsored registration fees and travel expenses. Otherwise, there are no potential conflicts of interest to disclose for any of the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Lore Santoro, Email: lore.santoro@icm.unicancer.fr.
L. Pitalot, Email: laurine.pitalot@gmail.com
D. Trauchessec, Email: dorian.trauchessec@icm.unicancer.fr
E. Mora-Ramirez, Email: erick.mora@ucr.ac.cr
P. O. Kotzki, Email: pierre-olivier.kotzki@icm.unicancer.fr
M. Bardiès, Email: manuel.bardies@inserm.fr
E. Deshayes, Email: emmanuel.deshayes@icm.unicancer.fr
Supplementary information
The online version contains supplementary material available at 10.1186/s13550-020-00737-8.
References
- 1.Huizing DMV, de Wit-van der Veen BJ, Verheij M, Stokkel MPM. Dosimetry methods and clinical applications in peptide receptor radionuclide therapy for neuroendocrine tumours: a literature review. EJNMMI Res (Internet) 2018 doi: 10.1186/s13550-018-0443-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flux GD, Sjogreen Gleisner K, Chiesa C, Lassmann M, Chouin N, Gear J, et al. From fixed activities to personalized treatments in radionuclide therapy: lost in translation? Eur J Nucl Med Mol Imaging. 2018;45(1):152–154. doi: 10.1007/s00259-017-3859-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sundlöv A, Gustafsson J, Brolin G, Mortensen N, Hermann R, Bernhardt P, et al. Feasibility of simplifying renal dosimetry in 177Lu peptide receptor radionuclide therapy. EJNMMI Phys (Internet) 2018 doi: 10.1186/s40658-018-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marin G, Vanderlinden B, Karfis I, Guiot T, Wimana Z, Reynaert N, et al. A dosimetry procedure for organs-at-risk in 177Lu peptide receptor radionuclide therapy of patients with neuroendocrine tumours. Phys Med. 2018;56:41–49. doi: 10.1016/j.ejmp.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 5.Del Prete M, Arsenault F, Saighi N, Zhao W, Buteau F-A, Celler A, et al. Accuracy and reproducibility of simplified QSPECT dosimetry for personalized 177Lu-octreotate PRRT. EJNMMI Phys. 2018;5(1):25. doi: 10.1186/s40658-018-0224-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson TK, McClure D, McCourt S. MABDOSE II: validation of a general purpose dose estimation code. Med Phys. 1999;26(7):1396–1403. doi: 10.1118/1.598637. [DOI] [PubMed] [Google Scholar]
- 7.Gardin I, Bouchet LG, Assié K, Caron J, Lisbona A, Ferrer L, et al. Voxeldose: a computer program for 3-D dose calculation in therapeutic nuclear medicine. Cancer Biother Radiopharm. 2003;18(1):109–115. doi: 10.1089/108497803321269386. [DOI] [PubMed] [Google Scholar]
- 8.Grimes J, Uribe C, Celler A. JADA: a graphical user interface for comprehensive internal dose assessment in nuclear medicine: a GUI for comprehensive internal dose assessment. Med Phys. 2013;40(7):072501. doi: 10.1118/1.4810963. [DOI] [PubMed] [Google Scholar]
- 9.Kletting P, Schimmel S, Hänscheid H, Luster M, Fernández M, Nosske D, et al. The NUKDOS software for treatment planning in molecular radiotherapy. Z Für Med Phys. 2015;25(3):264–274. doi: 10.1016/j.zemedi.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 10.Li T, Zhu L, Lu Z, Song N, Lin K-H, Mok GSP. BIGDOSE: software for 3D personalized targeted radionuclide therapy dosimetry. Quant Imaging Med Surg. 2020;10(1):160–170. doi: 10.21037/qims.2019.10.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mora-Ramirez E, Santoro L, Cassol E, Ocampo-Ramos JC, Clayton N, Kayal G, et al. Comparison of commercial dosimetric software platforms in patients treated with 177 Lu-DOTATATE for peptide receptor radionuclide therapy. Med Phys. 2020;1:mp.14375. doi: 10.1002/mp.14375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santoro L, Mora-Ramirez E, Trauchessec D, Chouaf S, Eustache P, Pouget J-P, et al. Implementation of patient dosimetry in the clinical practice after targeted radiotherapy using [177Lu-[DOTA0, Tyr3]-octreotate. EJNMMI Res. 2018;8(1):103. doi: 10.1186/s13550-018-0459-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.GE Healthcare. Organ dose estimates for radio-isotope therapy treatment planning purposes. Dosimetry toolkit package. White paper. 2011.
- 14.Stabin MG, Sparks RB, Crowe E. OLINDA/EXM: the second-generation personal computer software for internal dose assessment in nuclear medicine. J Nucl Med. 2005;46(6):1023–1027. [PubMed] [Google Scholar]
- 15.Kupitz D, Wetz C, Wissel H, Wedel F, Apostolova I, Wallbaum T, et al. Software-assisted dosimetry in peptide receptor radionuclide therapy with 177Lutetium-DOTATATE for various imaging scenarios. Mosley RL, editor. PLoS ONE. 2017;12(11):e0187570. doi: 10.1371/journal.pone.0187570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huizing DMV, Peters SMB, Versleijen MWJ, Martens E, Verheij M, Sinaasappel M, et al. A head-to-head comparison between two commercial software packages for hybrid dosimetry after peptide receptor radionuclide therapy. EJNMMI Phys. 2020;7(1):36. doi: 10.1186/s40658-020-00308-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maughan NM, Garcia-Ramirez J, Arpidone M, Swallen A, Laforest R, Goddu SM, et al. Validation of post-treatment PET-based dosimetry software for hepatic radioembolization of Yttrium-90 microspheres. Med Phys. 2019;46(5):2394–2402. doi: 10.1002/mp.13444. [DOI] [PubMed] [Google Scholar]
- 18.Hippeläinen ET, Tenhunen MJ, Mäenpää HO, Heikkonen JJ, Sohlberg AO. Dosimetry software Hermes Internal Radiation Dosimetry: from quantitative image reconstruction to voxel-level absorbed dose distribution. Nucl Med Commun. 2017;38(5):357–365. doi: 10.1097/MNM.0000000000000662. [DOI] [PubMed] [Google Scholar]
- 19.Barna S, Haug A, Hartenbach M, Rasul S, Grubmüller B, Kramer G, et al. Dose calculations and dose-effect relationships in 177Lu-PSMA I&T radionuclide therapy for metastatic castration-resistant prostate cancer. Clin Nucl Med. 2020;45(9):661–667. doi: 10.1097/RLU.0000000000003157. [DOI] [PubMed] [Google Scholar]
- 20.Andersson M, Johansson L, Eckerman K, Mattsson S. IDAC-Dose 2.1, an internal dosimetry program for diagnostic nuclear medicine based on the ICRP adult reference voxel phantoms. EJNMMI Res. 2017;7(1):88. doi: 10.1186/s13550-017-0339-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pasciak AS, Bourgeois AC, Bradley YC. A comparison of techniques for 90Y PET/CT image-based dosimetry following radioembolization with resin microspheres. Front Oncol (Internet) 2014 doi: 10.3389/fonc.2014.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dieudonne A, Hobbs RF, Lebtahi R, Maurel F, Baechler S, Wahl RL, et al. Study of the impact of tissue density heterogeneities on 3-dimensional abdominal dosimetry: comparison between dose kernel convolution and direct Monte Carlo methods. J Nucl Med. 2013;54(2):236–243. doi: 10.2967/jnumed.112.105825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dieudonne A, Hobbs RF, Bolch WE, Sgouros G, Gardin I. Fine-resolution voxel S values for constructing absorbed dose distributions at variable voxel size. J Nucl Med. 2010;51(10):1600–1607. doi: 10.2967/jnumed.110.077149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kondev FG. Lu-177_comments on evaluation of decay data for beta-decay; 2002.
- 25.Simpkin DJ. Radiation Interactions and Internal Dosimetry in Nuclear Medicine; 1999;19(1):13. [DOI] [PubMed]
- 26.Lin LI-K. A Concordance Correlation Coefficient to Evaluate Reproducibility. Biometrics. 1989;45(1):255. [PubMed]
- 27.Altman DG, Bland JM. Measurement in medicine: the analysis of method comparison studies. Statistician. 1983;32(3):307. [Google Scholar]
- 28.Zhao W, Esquinas PL, Hou X, Uribe CF, Gonzalez M, Beauregard J-M, et al. Determination of gamma camera calibration factors for quantitation of therapeutic radioisotopes. EJNMMI Phys (Internet) 2018 doi: 10.1186/s40658-018-0208-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uribe CF, Esquinas PL, Tanguay J, Gonzalez M, Gaudin E, Beauregard J-M, et al. Accuracy of 177Lu activity quantification in SPECT imaging: a phantom study. EJNMMI Phys (Internet) 2017 doi: 10.1186/s40658-016-0170-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zimmerman BE, Grošev D, Buvat I, Coca Pérez MA, Frey EC, Green A, et al. Multi-centre evaluation of accuracy and reproducibility of planar and SPECT image quantification: an IAEA phantom study. Z Für Med Phys. 2017;27(2):98–112. doi: 10.1016/j.zemedi.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peters SMB, Meyer Viol SL, van der Werf NR, de Jong N, van Velden FHP, Meeuwis A, et al. Variability in lutetium-177 SPECT quantification between different state-of-the-art SPECT/CT systems. EJNMMI Phys. 2020;7(1):9. doi: 10.1186/s40658-020-0278-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lassmann M, Eberlein U. The relevance of dosimetry in precision medicine. J Nucl Med. 2018;2018:1. doi: 10.2967/jnumed.117.206649. [DOI] [PubMed] [Google Scholar]
- 33.Tran-Gia J, Lassmann M. Optimizing image quantification for 177 Lu SPECT/CT based on a 3D printed 2-compartment kidney phantom. J Nucl Med. 2018;59(4):616–624. doi: 10.2967/jnumed.117.200170. [DOI] [PubMed] [Google Scholar]
- 34.Gustafsson J, Sundlöv A, Sjögreen GK. SPECT image segmentation for estimation of tumour volume and activity concentration in 177Lu-DOTATATE radionuclide therapy. EJNMMI Res. 2017;7(1):18. doi: 10.1186/s13550-017-0262-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grassi E, Fioroni F, Ferri V, Mezzenga E, Sarti MA, Paulus T, et al. Quantitative comparison between the commercial software STRATOS® by Philips and a homemade software for voxel-dosimetry in radiopeptide therapy. Phys Med. 2015;31(1):72–79. doi: 10.1016/j.ejmp.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 36.Grassi E, Fioroni F, Berenato S, Patterson N, Ferri V, Braglia L, et al. Effect of image registration on 3D absorbed dose calculations in 177 Lu-DOTATOC peptide receptor radionuclide therapy. Phys Med. 2018;45:177–185. doi: 10.1016/j.ejmp.2017.11.021. [DOI] [PubMed] [Google Scholar]
- 37.Hippeläinen E, Tenhunen M, Sohlberg A. Fast voxel-level dosimetry for 177 Lu labelled peptide treatments. Phys Med Biol. 2015;60(17):6685–6700. doi: 10.1088/0031-9155/60/17/6685. [DOI] [PubMed] [Google Scholar]
- 38.Sandstrom M, Garske-Roman U, Granberg D, Johansson S, Widstrom C, Eriksson B, et al. Individualized dosimetry of kidney and bone marrow in patients undergoing 177Lu-DOTA-octreotate treatment. J Nucl Med. 2013;54(1):33–41. doi: 10.2967/jnumed.112.107524. [DOI] [PubMed] [Google Scholar]
- 39.Gear JI, Cox MG, Gustafsson J, Gleisner KS, Murray I, Glatting G, et al. EANM practical guidance on uncertainty analysis for molecular radiotherapy absorbed dose calculations. Eur J Nucl Med Mol Imaging (Internet) 2018 doi: 10.1007/s00259-018-4136-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Table S1: Patients’ characteristics.
Additional file 2. Fig. S1: Box-and-whisker plots showing the ADs to liver (a), kidneys (b) and spleen (c) calculated using PLANET® Dose and PLANET® Dose+OLINDA, as well as the relative AD differences between PLANET® Dose and PLANET® Dose+OLINDA (d).
Additional file 3. Fig. S2: Dispersion around the 45° line of the AD pairs obtained with PLANET+OLINDA and PLANET® Dose LDM with density correction for all organs combined (a). Bland-Altman plots of the AD to liver (b), kidneys (c) and spleen (d) calculated with PLANET+OLINDA and PLANET® Dose LDM with density correction.
Data Availability Statement
Please contact the author for data requests.