Abstract
The ribosome is an essential cellular machine performing protein biosynthesis. Its structure and composition are highly conserved in all species. However, some bacteria have been reported to have an incomplete set of ribosomal proteins. We have analyzed ribosomal protein composition in 214 small bacterial genomes (<1 Mb) and found that although the ribosome composition is fairly stable, some ribosomal proteins may be absent, especially in bacteria with dramatically reduced genomes. The protein composition of the large subunit is less conserved than that of the small subunit. We have identified the set of frequently lost ribosomal proteins and demonstrated that they tend to be positioned on the ribosome surface and have fewer contacts to other ribosome components. Moreover, some proteins are lost in an evolutionary correlated manner. The reduction of ribosomal RNA is also common, with deletions mostly occurring in free loops. Finally, the loss of the anti-Shine–Dalgarno sequence is associated with the loss of a higher number of ribosomal proteins.
Keywords: ribosome, bacteria, evolution, genome reduction, rRNA, ribosomal protein
Introduction
The ribosome is a universal biosynthesis machine present in all eukaryotes and prokaryotes. A bacterial ribosome comprised the small (30S) and large (50S) subunits which together form the 70S particle (Kurland 1972; Ramakrishnan 2002). The bacterial ribosome consists of multiple proteins (RP, r-proteins) and three ribosomal RNA (rRNA) molecules — 16S in the small subunit, 23S and 5S in the large subunit (Kurland 1972). The main catalytic functions of the ribosome, such as the peptide bond formation, mRNA decoding, and translocation of mRNA and tRNA after the peptide bond formation, are performed by rRNA (Green and Noller 1997; Nissen et al. 2000; Schmeing et al. 2003). Moreover, rRNA molecules also determine the ribosomal spatial organization providing sites for binding of the ribosomal proteins (Khaitovich et al. 1999). The ribosome of Escherichia coli contains 21 proteins in the 30S subunit (bS1–bS21) and 33 proteins in the 50S subunit (uL1–bL36) (Schuwirth et al. 2005; Kaczanowska and Rydén-Aulin 2007; Ban et al. 2014). The role of ribosomal proteins is to stabilize the ribosome and to regulate the ribosomal activity (Aseev and Boni 2011). Although the key role in the protein biosynthesis is played by rRNA, the r-protein composition also tends to be conserved in most bacteria (Roberts et al. 2008). Moreover, 33 r-proteins are conserved among the domains of life (Lecompte et al. 2002; Roberts et al. 2008; Smith et al. 2008).
The protein composition of bacterial ribosomes has been studied intensively. The analysis of single-gene knockout mutants of E. coli has shown that these bacteria are able to grow without proteins bS6, uS15, bS20, bS21, uL1, bL9, uL11, and bL25, but in most cases, the growth rate of these knockout mutants is reduced (Baba et al. 2006). In another study of an E. coli knockout collection, nine r-proteins (uL15, bL21, uL24, bL27, uL29, uL30, bL34, and uS17) have been found to be nonessential for survival in experimental conditions (Shoji et al. 2011). A similar study in Bacillus subtilis has identified 20 r-proteins nonessential for growth in experimental conditions (Akanuma et al. 2012). Based on this result, smaller r-proteins were proposed to have been incorporated into the ribosome relatively recently in evolution, and hence to be less essential. A phylogenetic analysis of 995 completely sequenced bacterial genomes has shown that 44 r-proteins are strictly ubiquitous, proteins bS16, bL9, bL19, bL31, bL34, and bL36 are rarely missing, whereas bS21, S22 (SRA), bThx, bL25, and uL30 are absent in a large fraction of bacteria (Yutin et al. 2012). A comparative analysis of the translation apparatus of Mollicutes has shown that five r-proteins (uS14, S22, bL7, bL25, and bL31) are missing in all studied genomes, uL30 is missing in almost all of them, and bS1 has been lost in seven independent events in different clades (Grosjean et al. 2014). Finally, an analysis of endosymbiotic bacteria with small genomes from a variety of phyla has demonstrated that they lack the largest fraction of r-proteins, as only 17 out of 21 small-subunit and 16 out of 32 large-subunit r-proteins are universally present in these bacteria (McCutcheon and Moran 2011). For example, Candidatus Tremblaya princeps, an endosymbiont of mealybugs (Pseudococcidae family), has only 139 genes, restricting the possibility to carry a complete translation apparatus (McCutcheon and Moran 2011). Although the published lists of essential r-proteins vary to some extent, bL25, uL30, bL31, bS21, S22, and bThx have been consistently reported to be the least essential.
Less is known about whether there exist universal features common to these nonessential proteins, and how the r-protein loss is linked to the genome reduction. Here, we have analyzed the r-protein composition in bacteria with small genomes from a variety of phyla. We have identified a set of frequently lost proteins and found three patterns of evolutionary correlated r-protein loss. A majority of frequently lost proteins have been shown to be located on the ribosome surface and to form fewer contacts with other ribosome components, compared with universally conserved r-proteins. We also show that the loss of the anti-Shine–Dalgarno (SD) sequence is associated with a higher number of lost ribosomal proteins.
Results
Some Ribosomal Proteins Are Missing in Bacteria with Small Genomes
We reannotated all r-proteins in 214 bacterial strains with small genomes (<1 Mb) from 38 genera of the following phyla: Proteobacteria, Bacteroidetes, Spirochaetes, Tenericutes, and Actinobacteria (supplementary table S1, Supplementary Material online and see Materials and Methods). For that, we used 65 Pfam domains: 63 domains for the canonical set of ribosomal proteins, and two domains of the trigger-factor (TF) protein, a ribosome-associated chaperone (supplementary table S2, Supplementary Material online).
Our results (supplementary fig. S1, Supplementary Material online) show that two proteins are absent in almost all considered strains: S22 (all strains) and bThx (present in only Candidatus Walczuchella monophlebidarum).
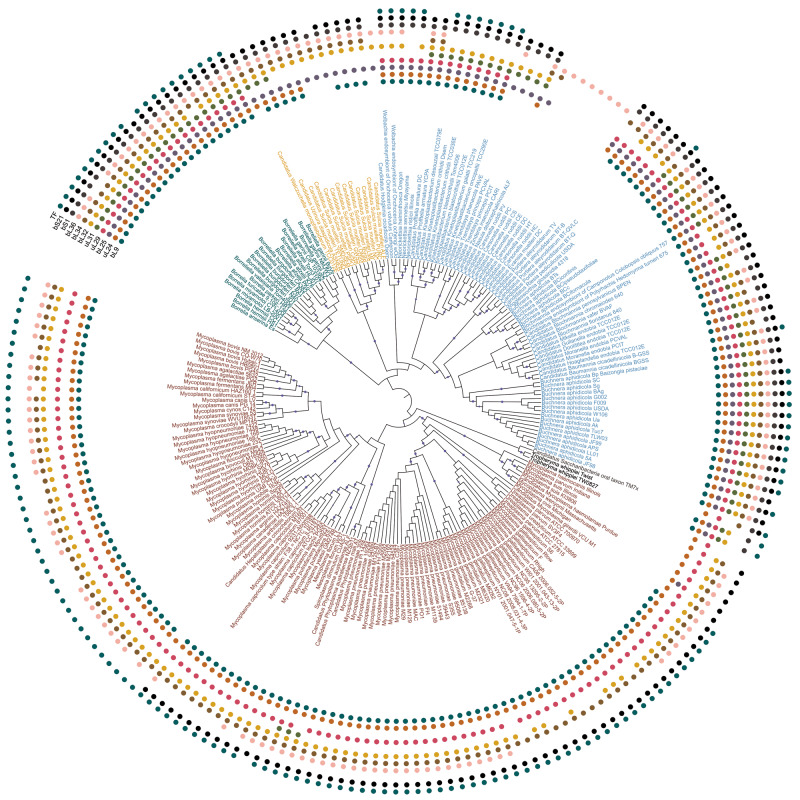
Only nine r-proteins are completely conserved, whereas each of the remaining 48 is lost in at least one strain from our data set. Proteins bL9, uL24, bL25, uL29, uL30, bL32, bL34, bL36, bS1, bS21, and TF are lost frequently, as they each are absent in at least 19 strains from multiple phyla. This set is further referred to as frequently lost proteins. R-proteins of the small subunit are more likely to be retained than the large-subunit r-proteins (supplementary fig. S1, Supplementary Material online). The most frequently lost small-subunit protein bS1 is absent in 116 strains from nine genera, whereas the most frequently lost protein from the large subunit, uL30, is absent in 138 genomes from 18 genera. All frequently lost proteins have been lost independently multiple times in bacteria (fig. 1 and supplementary fig. S2, Supplementary Material online). The largest number of r-protein losses was observed in Candidatus Tremblaya princeps, Candidatus Hodgkinia cicadocola, and Candidatus Carsonella rudii, the three bacteria with the shortest genomes in our data set.
Fig. 1.
Maximum likelihood phylogenetic tree of analyzed bacterial species. The tree was constructed for the concatenated alignment of conserved r-proteins by PhyML with 100 bootstrap replicates. In this representation, the branch lengths are ignored (the tree with branch lengths is provided as supplementary fig. S2, Supplementary Material online). Bootstrap values in the range 0.9–1 are shown by blue circles. The presence of one of 11 frequently lost proteins (bL9, bL21, uL24, bL25, uL29, bL32, bL34, bL36, bS1, bS21, TF) in a strain is marked by a colored circle (inner to outer arcs, respectively). Leaves are colored by the phyla: Actinobacteria, black; Bacteroidetes, yellow; Proteobacteria, blue; Spirochaetes, green; Tenericutes, red; unclassified bacteria, gray.
R-Protein Loss Depends on the Level of Genome Reduction
The comparison of the r-protein composition and the genome size revealed a correlation between the genome size and the number of retained ribosomal proteins (r2 = 0.47, P < 5.4 × 10−13). The slope of this correlation depends on the genome size (supplementary fig. S3a, Supplementary Material online). Indeed, for genomes shorter than 350 kb, the correlation is stronger (r2 = 0.71, P = 6.3 × 10−6). This pattern holds both for the complete ribosome and for the individual subunits (supplementary fig. S3b and c, Supplementary Material online), with a steeper slope for the large subunit.
Patterns of Ribosomal Protein Loss
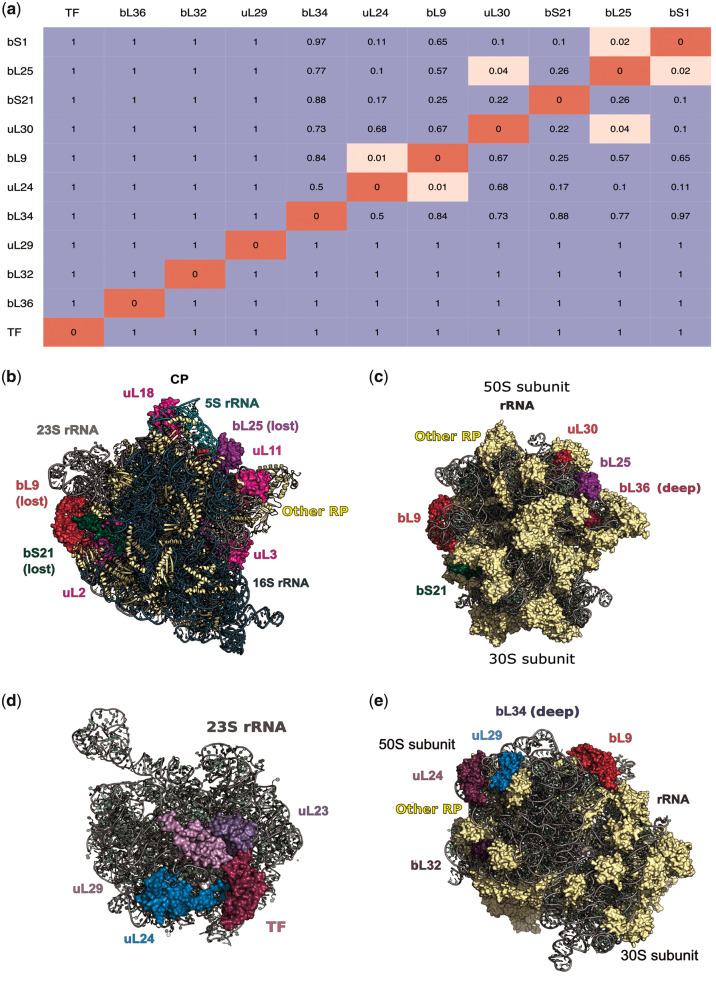
Eleven most frequently lost proteins have been lost independently in parallel in different phyla (fig. 1 and supplementary fig. S2, Supplementary Material online). A general tendency is that frequently lost proteins are usually positioned at the ribosome surface (fig. 2). To analyze possible dependencies between losses of different frequently lost proteins, we estimated the correlation between the vectors of the protein presence/absence in all strains by the Pagel correlation method (Pagel 1994) which allows one to control for the phylogenetic structure of the data set (fig. 2a). We observed three clusters of r-proteins that tend to be lost together: bL25+uL30, bL9+uL24, and bL25+bS1.
Fig. 2.
Patterns of r-protein loss. (a) Pagel’s test of correlated evolution between vectors of ribosomal protein presence/absence in bacterial strains. Insignificant P values are colored blue, the significant ones are colored pink. Only 11 frequently lost proteins are considered. (b) The crown view of the 70S ribosome (the typical crown view position is marked using several ribosomal proteins [magenta], the position of the central protuberance and 5S rRNA [light green], and the position of 16S rRNA [dark blue]) and various perspectives (c and e) of the ribosome showing positions of frequently lost proteins. PDB ID: 5H5U (Escherichia coli) (Ma et al. 2017). (d) 23S rRNA and r-proteins that form contacts with the trigger factor. PDB ID: 2D3O (Deinococcus radiodurans) (Schlünzen et al. 2005). Protein labels are of the same color as the respective proteins in the structure. In (c), (e) only frequently lost ribosomal proteins are labeled.
The first of these clusters is consistent with the location of the r-proteins in the ribosome: bL25 and uL30 are positioned next to each other (fig. 2c), which may explain their correlated loss. The proteins of the other two clusters, however, are spatially separated; in these cases, the correlation is more likely to arise from functional rather than spatial associations. As uL24 binds to the 5′-end of the 23S rRNA, whereas bL9 is positioned close to the 3′-end (Herold and Nierhaus 1987), they do not form a structural cluster (fig. 2e). However, uL24 is reported to be an initiator of the LSU assembly (Spillmann and Nierhaus 1978), whereas bL9 is among the primary 23S rRNA binders. The loss of uL24 may complicate direct binding of bL9 to 23S rRNA, abolishing the need for bL9. The remaining cluster bL25+bS1 is the most challenging to interpret, because these proteins are parts of different subunits and thus spatially and functionally distant. However, both r-proteins have been suggested to be involved in the ribosome recycling (Korepanov et al. 2007; Demo et al. 2017; Loveland and Korostelev 2018).
In the “onion model” of the ribosome structure and evolution (Hsiao et al. 2009; Petrov et al. 2015), the peptidyl transferase center (PTC) is the core and the most ancient part of the ribosome. According to this model, the farther an r-protein is from the PTC, the later it has been added during ribosome evolution. Indeed, some of the frequently lost proteins are positioned in the outer layers of the “onion,” such as bL9, uL24, and uL29 (supplementary table S3, Supplementary Material online). However, there is no systematic difference between conserved and frequently lost r-proteins in their distances from the PTC (P = 0.44).
Frequently Lost R-Proteins Have Fewer Contacts Than Conserved Proteins
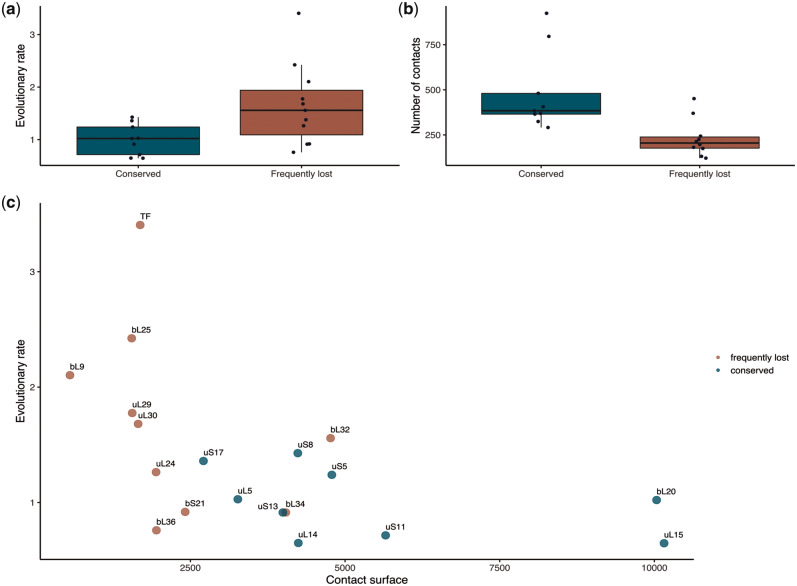
We analyzed the differences in the evolutionary rate (fig. 3a) and the number of contacts (fig. 3b) between conserved and frequently lost proteins (see supplementary table S3, Supplementary Material online). As expected, frequently lost proteins form significantly fewer contacts than conserved proteins (one-sided Wilcoxon test P = 1.5 × 10−3), as the former are mainly positioned on the ribosome surface. Similarly, the contacting surface between each of the studied r-proteins and the rest of the ribosome is significantly smaller for frequently lost r-proteins than for conserved ones (one-sided Wilcoxon test P = 1.4 × 10−3 for the PDB structure 5H5U that does not contain TF, P = 7.3 × 10−4 when the value for TF from the PDB structure 2D3O is added to the data for 5H5U; fig. 3c). However, if the contacting surface values are normalized by the protein’s surface, the difference between frequently lost and conserved r-proteins becomes insignificant; hence, the propensity for loss is associated with the absolute size of the interacting surface rather than the proportion of surface area involved in interactions. Although several PDB structures of the ribosome are available, none of them is complete (i.e., includes the entire ribosome and TF) and for none, all parts of the ribosome are resolved. Therefore, we have considered multiple available PDB structures, and have found that the above results hold for all of them (supplementary fig. S4, Supplementary Material online).
Fig. 3.
Determinants of the r-protein loss. The differences in the evolutionary rate (a) and the number of contacts (b) between the conserved (blue) and frequently lost (red) ribosomal proteins. (c) The dependency between the evolutionary rate and contact surface (Å2) for both conserved and frequently lost ribosomal proteins.
The frequently lost proteins evolve more rapidly than the conserved ones (two-sided Wilcoxon test P = 9.8 × 10−4). This correlation was not driven by differences in protein lengths between conserved and frequently lost r-proteins, because the evolutionary rate was independent of the protein length (r2 = 0.39, P = 0.092) (supplementary fig. S5, Supplementary Material online).
Deletions in rRNAs and the Loss of R-Proteins
To study possible correlations between the loss of r-proteins and the loss of their binding sites in the rRNA, we analyzed 16S and 23S rRNA multiple alignments and identified nine deletion blocks (four in the 23S rRNA and five in the 16S rRNA) that had occurred in more than two genera and were longer than five nucleotides in at least one species. All such deletions affected rRNA free loops not involved in the r-protein binding, indicating that protein loss does not immediately drive the loss of their binding sites in the rRNA (supplementary fig. S6, Supplementary Material online).
Anti-SD Loss Is Frequent among Strains from Different Taxa
The anti-SD sequence in the 16S rRNA is frequently lost in many phyla. Its presence/absence pattern is not strongly linked to the taxonomy, as many phyla have strains both with and without the anti-SD sequence in the 16S rRNA (supplementary fig. S7, Supplementary Material online). The loss is significantly associated with the higher number of lost r-proteins (one-sided Wilcoxon test P = 5.8 × 10−4). However, we have observed no correlation of the anti-SD loss with loss of any specific r-protein.
Discussion
The bacterial genome size is evolutionarily labile, and radical genome reduction has occurred many times independently throughout the bacterial domain. Such multiple independent genome reduction events allow for a systematic study of the patterns of gene loss associated with them.
Although individual r-proteins can be lost in bacteria with genomes of any size, for the tiniest of bacterial genomes further reduction in the genome size is associated with the loss of ribosomal proteins which are generally conserved. The fact that ribosomal proteins are among the last to leave a shrinking genome illustrates that they tend to be less dispensable than most proteins. Two r-proteins absent in the largest number of small-genome bacteria are S22 and bThx. Previous studies indicate that these proteins are largely nonessential. Indeed, the gene encoding S22 is mostly expressed at the stationary growth phase, and its deletion does not affect the viability of E. coli mutants (Izutsu et al. 2001). bThx is found only in thermophilic bacteria and has been shown to stabilize the organization of RNA elements at the top of the 30S subunit head in Thermus thermophilus (Choli et al. 1993; Wimberly et al. 2000), which implies that this protein is only essential for survival at high temperatures.
Among other frequently lost proteins, uL30 is highly conserved in Archaea and Eukarya (Lecompte et al. 2002), where it is thought to be essential for the selenocysteine recognition (Chavatte et al. 2005). The role of this protein in bacteria is not clear. bL25 has been proposed to be essential for interaction with r-protein uL16, the latter being necessary for ribosome stability (Anikaev et al. 2016). Although the loss of bL25 is tolerated in E. coli, mutant bacteria have a reduced growth rate (Baba et al. 2006). bS1 is known to be essential for the translation initiation of mRNAs with structured 5′-UTR (Qu et al. 2012; Duval et al. 2013). bS21 is required for the recognition of native templates, and its function resembles the function of bS1 (Van Duin and Wijnands 1981). R-protein bL9 reduces translation frameshifting (Dunkle et al. 2010; Smith et al. 2019). uL24 is an assembly initiator of the large ribosomal subunit, together with uL3 (Nikolay et al. 2015). The TF is a chaperone associated with the ribosome exit channel (Hoffmann et al. 2010). TF is located at the ribosome surface where it is surrounded by r-proteins uL23, uL24, and uL29 (fig. 2d) directly interacting with uL23 and uL29 (Ferbitz et al. 2004). TF and uL29 are associated with protein folding. Thus, the absence of frequently lost proteins should reduce the ribosome fidelity and overall efficacy of protein translation.
Our list of frequently lost proteins is largely consistent with previous observations (Baba et al. 2006; Shoji et al. 2011; Akanuma et al. 2012; Yutin et al. 2012; Grosjean et al. 2014). For example, among the six nonubiquitous r-proteins reported by Yutin et al. 2012, three (bL25, uL30, bS21) are also among the r-proteins identified as frequently lost in our data set, in addition to two proteins (S22 and bThx) that only occur in some bacterial lineages (see above). However, our focus on bacteria with tiny genomes allowed us to study less dispensable r-proteins. Proteins bL34 and bL36 identified previously as present in almost all genomes (Yutin et al. 2012) are frequently lost in our data set. Although these proteins are generally considered essential, they may be dispensable under certain circumstances. The absence of bL34 affects cell growth, but the cell function can be restored by increasing the magnesium ion flow (Shoji et al. 2011; Akanuma et al. 2014). The absence of bL36 is only essential for cell growth at high temperatures (Ikegami et al. 2005). Moreover, bL36 is missing in Bacteroidetes (Yutin et al. 2012). R-proteins uL24 and uL29 identified here as frequently lost have been lost in at least two independent events, but only in bacteria with drastically reduced genomes. The knockouts of both these proteins are viable (Shoji et al. 2011). Interestingly, although the r-proteins bL31 and uS14 have been identified as lost in all Mollicutes (Grosjean et al. 2014), we find that these proteins are in fact mostly retained, and only a few strains of Mollicutes lack them.
Variation in the ribosome composition is common not only in endosymbiotic bacteria but also in cell organelles, mitochondria, and plastids. Although the patterns of loss seem superficially similar, plastids, mitochondria, and bacteria with reduced genomes cannot be compared directly (supplementary note 1, Supplementary Material online).
What factors affect the propensity of a protein to be lost? Previously, proteins frequently lost in evolution have been shown to evolve rapidly, have fewer interactions with other proteins, and lower expression levels (Krylov et al. 2003). Although there are some data on ribosomal heterogeneity in bacteria (Byrgazov et al. 2013), the complete ribosome requires all r-proteins, and r-proteins are organized in operons with a variety of regulatory feedback loops providing tight coregulation (Lemke et al. 2011), which allowed us not to consider the expression level. Taking into account other factors, we observe that the number of contacts is significantly smaller, and the evolutionary rate, significantly higher for frequently lost r-proteins compared with the conserved ones. The number of contacts is associated with exposure of an r-protein on the ribosome surface. Indeed, r-proteins that are frequently lost tend to be located on the ribosome surface, the exceptions being bL34 and bL36 (fig. 2c and e).
The order in which r-proteins are assembled and their distance from PTC (the “onion” model) provide indirect evidence of the order of r-proteins incorporation in the ribosome during evolution. We observe that frequently lost r-proteins tend to appear late in ribosome evolution (supplementary note 2, Supplementary Material online).
Another interesting question is whether r-proteins are lost at random or there are certain patterns of ribosome simplification, where the loss of one protein facilitates the loss of others. As our data set contained many genomes with incomplete sets of r-proteins, we could study the patterns of common protein loss, and we observed several such correlated groups of proteins.
Surprisingly, all deletions in 23S and 16S rRNAs happened in free loops, meaning that the loss of ribosomal proteins was not accompanied by the reduction of their respective binding sites on the rRNAs. However, the shortening of rRNA is a tendency shared also by mitochondria—the SSU mt-rRNA is 40% shorter than the bacterial 16S rRNA, whereas the LSU mt-rRNA represents a half of the bacterial 23S rRNA (Anderson et al. 1981). Interestingly, deletions in 23S rRNAs from our data set are located in helices H10, H63, H79, and H98, which have been reported to be lost frequently in mt-rRNA of various species (Petrov et al. 2019). Degeneration and losses of a particular element of 16S rRNA, the anti-SD sequence, have been reported in various species including symbionts with small genomes (Lim et al. 2012). The anti-SD loss is recurrent, as the sequence may be present or absent in representatives of all considered phyla, consistent with previous observations (Amin et al. 2018). Although the loss of the anti-SD sequence is a common event among intracellular symbionts, some symbionts with reduced genomes, such as Buchnera aphidicola, retain anti-SD (Lim et al. 2012). Nevertheless, we have observed that the loss of anti-SD is significantly associated with the loss of ribosomal proteins.
In conclusion, the consideration of radically reduced bacterial genomes expands the list of r-proteins with propensity for loss, and allows one to study the forces underlying these losses. We find that frequently lost proteins evolve at a higher rate, have fewer contacts, are located on the ribosomal surface, and have been incorporated in the ribosome late in evolution. This suggests that these losses are neutral or only slightly deleterious, and do not affect essential ribosomal functions, especially in symbiotic bacteria that live in stable host environments.
Materials and Methods
Data Set of Small Bacterial Genomes
The list of all bacterial species with complete genomes not exceeding 1 Mb was compiled from the IMG/M database (Chen et al. 2017). The genomic data for all strains of these species (214 genomes in total, supplementary table S1, Supplementary Material online) were downloaded from the NCBI FTP; files with protein sequences in February 2017 (ftp://ftp.ncbi.nlm.nih.gov/genomes/refseq/bacteria/), files with RNA sequences in September 2017 (ftp://ftp.ncbi.nlm.nih.gov/genomes/genbank/bacteria/).
The genomes of some strains in the selected species exceeded 1 Mb, but were still retained in the data set to check whether the gene loss tendency is consistent throughout the species.
Annotation of Protein Domains
Ribosomal proteins and TF were reannotated in the downloaded genomes (the complete list of proteins is given in supplementary table S2, Supplementary Material online) using HMM matrices from the Pfam-A database (Finn et al. 2016). The HMM-profiles for the selected domains were scanned against each genome with the HMMER software (Mistry et al. 2013). A protein was considered to be present in a given genome if the respective domain had a hit in the HMMER search with E-value <0.001; only the best such hit was retained for further analysis. If the HMMER domain bias had the same order of magnitude as the similarity score, additional filtering was performed as described below.
For frequently lost and conserved r-proteins, we created multiple alignments of proteins with confident HMMer predictions (available at https://github.com/darianick/ribo-simpler). Protein alignments were built using Muscle (Edgar 2004) and subsequently manually curated. All truncated sequences shorter than 50% of the full-length neighbor sequences and all nonribosomal hits were removed.
Two-domain proteins (uL2, uL5, bL12, bL9, uL11, uS4, uS5, TF) were considered to be present if both domains were found and encoded in the same genome locus.
Identification of Independent R-Protein Losses
To map the phyletic patterns of r-protein losses, we constructed the maximum likelihood tree of concatenated multiple alignments of all conserved r-proteins (see below) using PhyML v. 3.1 (Guindon et al. 2010) with tree topology, root, and branch lengths optimized (“-o tlr” parameters) and 100 bootstrap replicates (available at https://github.com/darianick/ribo-simpler). Prior to the tree construction, all columns containing gaps were removed from the concatenated alignment. This tree was used to calculate the number of independent losses of each protein.
Estimation of the Evolutionary Rate
The evolutionary rate of a protein was defined as the average branch length in the respective phylogenetic tree. The maximum likelihood phylogenetic tree was built for each ribosomal protein using PhyML with 100 bootstrap replicates. The LG+G model was used, with tree topology and branch lengths optimized (“-o tl” parameters) and with gamma parameter (supplementary table S4, Supplementary Material online) identified for each protein alignment using ProtTest (version 3.4.2) (Darriba et al. 2011). The evolutionary rate for a protein was compared with the average rates for the set of conserved proteins (uL5, uL14, uL15, bL20, uS5, uS8, uS11, uS13, and uS17). For each studied protein, the corresponding rates for every conserved protein from the set were calculated in the subtree with the same set of species as in the tree of the studied protein. To compare the results for proteins with different phyletic profiles, we calculated the relative evolutionary score, that is, the protein’s evolutionary rate divided by the corresponding average rate for the set of conserved r-proteins.
Detection of Patterns of Ribosomal Protein Loss
The P values for pairwise correlations between losses of r-proteins were estimated using Pagel’s test for correlated evolution (Pagel 1994) with R-package phytools v0.6-99 (Revell 2012) based on vectors of ribosomal protein presence/absence in all studied bacterial strains and the phylogenetic tree of conserved r-proteins (see above). The resulting P values were organized in a heatmap with significant hits grouped together and representing patterns of evolutionary correlated r-protein loss.
Calculation of the Number of Contacts
The number of contacts was estimated by measuring pairwise distances between atoms in ribosome PDB structures (PDB ID: 5H5U [Ma et al. 2017] and 2D3O [Schlünzen et al. 2005]). Atoms were defined as contacting if the distance between them was at most 5 Å. All protein self-contacts were ignored. When multiple atoms of a studied protein were connected with the same atom in the ribosome structure, only one such contact was retained. The pairwise distances were calculated with the PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC, and script pairwise_dist.py. For each protein, the total number of contacts was calculated.
The contacting surface between each studied r-protein and the remaining ribosome was calculated using the “get_area” function in PyMOL by the following formula:
where A is the r-protein surface, R is the surface of a complete ribosome, and L is the surface of the complete ribosome lacking r-protein A.
The distance between r-proteins and PTC was measured using the “distance” function in PyMOL with the parameter “mode = 4” to obtain the distance between the centroids of r-proteins and the PTC. The PTC was defined as residues 2050 2076, 2244 2279, and 2396 2649 of 23S rRNA.
rRNA Analysis
16S and 23S rRNA alignments (available at https://github.com/darianick/ribo-simpler) were built using SINA Alignment Service (Pruesse et al. 2012), and then all common gaps were removed. We considered rRNA deletions if they occurred in more than two genera and were longer than five nucleotides in at least one species. Two deletion blocks where deletions were present in the same set of strains and separated by not more than five nucleotides in the reference sequence were considered as a single deletion block. As a reference, we selected 23S and 16S rRNA sequences from PDB ID: 5H5U.
A strain was defined as having the anti-SD sequence if there was an anti-SD motif CCUCCU at the 3′-end of the strain’s 16S rRNA.
Statistical Analysis and Data Visualization
All statistical analyses were performed using R. The Pearson correlation test was performed in R with the cor.test() function. The Wilcoxon(–Mann–Whitney) unpaired rank sum test was performed using the “wilcox.exact()” function from “exactRankTests” R package. The dependency between the total number of r-proteins and the genome length (supplementary fig. S3, Supplementary Material online) was built in R using the ggplot2 package. Curve smoothing was performed using the “loess” method. Phylogenetic trees were visualized with the iTOL server (Letunic and Bork 2011).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
The project was initiated with Sofya Sherstneva and Anna Ergemlidze at the Summer School of Molecular and Theoretical Biology for high school students, supported by the Zimin Foundation. We thank Arthur Zalevsky (FBB MSU) and Georgii Bazykin (IITP, Skoltech) for useful discussions and suggestions. This work was supported by the Russian Science Foundation (Grant No. 18-14-00358).
References
- Akanuma G, Kobayashi A, Suzuki S, Kawamura F, Shiwa Y, Watanabe S, Yoshikawa H, Hanai R, Ishizuka M.. 2014. Defect in the formation of 70S ribosomes caused by lack of ribosomal protein L34 can be suppressed by magnesium. J Bacteriol. 196(22):3820–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akanuma G, Nanamiya H, Natori Y, Yano K, Suzuki S, Omata S, Ishizuka M, Sekine Y, Kawamura F.. 2012. Inactivation of ribosomal protein genes in Bacillus subtilis reveals importance of each ribosomal protein for cell proliferation and cell differentiation. J Bacteriol. 194(22):6282–6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin MR, Yurovsky A, Chen Y, Skiena S, Futcher B.. 2018. Re-annotation of 12,495 prokaryotic 16S rRNA 3′ ends and analysis of Shine-Dalgarno and anti-Shine-Dalgarno sequences. PLoS One 13(8):e0202767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. 1981. Sequence and organization of the human mitochondrial genome. Nature 290(5806):457–465. [DOI] [PubMed] [Google Scholar]
- Anikaev AY, Isaev AB, Korobeinikova AV, Garber MB, Gongadze GM.. 2016. Role of protein L25 and its contact with protein L16 in maintaining the active state of Escherichia coli ribosomes in vivo. Biochemistry (Mosc) 81(1):19–27. [DOI] [PubMed] [Google Scholar]
- Aseev LV, Boni IV.. 2011. Extraribosomal functions of bacterial ribosomal proteins. Mol Biol. 45(5):739–750. [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H.. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban N, Beckmann R, Cate JH, Dinman JD, Dragon F, Ellis SR, Lafontaine DL, Lindahl L, Liljas A, Lipton JM, et al. 2014. A new system for naming ribosomal proteins. Curr Opin Struct Biol. 24:165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrgazov K, Vesper O, Moll I.. 2013. Ribosome heterogeneity: another level of complexity in bacterial translation regulation. Curr Opin Microbiol. 16(2):133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavatte L, Brown BA, Driscoll DM.. 2005. Ribosomal protein L30 is a component of the UGA-selenocysteine recoding machinery in eukaryotes. Nat Struct Mol Biol. 12(5):408–416. [DOI] [PubMed] [Google Scholar]
- Chen IA, Markowitz VM, Chu K, Palaniappan K, Szeto E, Pillay M, Ratner A, Huang J, Andersen E, Huntemann M, et al. 2017. IMG/M: integrated genome and metagenome comparative data analysis system. Nucleic Acids Res. 45(D1):D507–D516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choli T, Franceschi F, Yonath A, Wittmann-Liebold B.. 1993. Isolation and characterization of a new ribosomal protein from the thermophilic eubacteria, Thermus thermophilus, T. aquaticus and T. flavus. Biol Chem Hoppe-Seyler. 374(1–6):377–384. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27(8):1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demo G, Rasouly A, Vasilyev N, Svetlov V, Loveland AB, Diaz-Avalos R, Grigorieff N, Nudler E, Korostelev AA.. 2017. Structure of RNA polymerase bound to ribosomal 30S subunit. Elife 6:e28560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkle JA, Xiong L, Mankin AS, Cate J.. 2010. Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc Natl Acad Sci U S A. 107(40):17152–17157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval M, Korepanov A, Fuchsbauer O, Fechter P, Haller A, Fabbretti A, Choulier L, Micura R, Klaholz BP, Romby P, et al. 2013. Escherichia coli ribosomal protein S1 unfolds structured mRNAs onto the ribosome for active translation initiation. PLoS Biol. 11(12):e1001731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferbitz L, Maier T, Patzelt H, Bukau B, Deuerling E, Ban N.. 2004. Trigger factor in complex with the ribosome forms a molecular cradle for nascent proteins. Nature 431(7008):590–596. [DOI] [PubMed] [Google Scholar]
- Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, et al. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44(D1):D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R, Noller HF.. 1997. Ribosomes and translation. Annu Rev Biochem. 66(1):679–716. [DOI] [PubMed] [Google Scholar]
- Grosjean H, Breton M, Sirand-Pugnet P, Tardy F, Thiaucourt F, Citti C, Barré A, Yoshizawa S, Fourmy D, de Crécy-Lagard V, et al. 2014. Predicting the minimal translation apparatus: lessons from the reductive evolution of Mollicutes. PLoS Genet. 10(5):e1004363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O.. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321. [DOI] [PubMed] [Google Scholar]
- Herold M, Nierhaus KH.. 1987. Incorporation of six additional proteins to complete the assembly map of the 50 S subunit from Escherichia coli ribosomes. J Biol Chem. 262(18):8826–8833. [PubMed] [Google Scholar]
- Hoffmann A, Bukau B, Kramer G.. 2010. Structure and function of the molecular chaperone trigger factor. Biochim Biophys Acta. 1803(6):650–661. [DOI] [PubMed] [Google Scholar]
- Hsiao C, Mohan S, Kalahar BK, Williams LD.. 2009. Peeling the onion: ribosomes are ancient molecular fossils. Mol Biol Evol. 26(11):2415–2425. [DOI] [PubMed] [Google Scholar]
- Ikegami A, Nishiyama K, Matsuyama S, Tokuda H.. 2005. Disruption of rpmJ encoding ribosomal protein L36 decreases the expression of secY upstream of the spc operon and inhibits protein translocation in Escherichia coli. Biosci Biotechnol Biochem. 69(8):1595–1602. [DOI] [PubMed] [Google Scholar]
- Izutsu K, Wada C, Komine Y, Sako T, Ueguchi C, Nakura S, Wada A.. 2001. Escherichia coli ribosome-associated protein SRA, whose copy number increases during stationary phase. J Bacteriol. 183(9):2765–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczanowska M, Rydén-Aulin M.. 2007. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol Mol Biol Rev. 71(3):477–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P, Mankin AS, Green R, Lancaster L, Noller HF.. 1999. Characterization of functionally active subribosomal particles from Thermus aquaticus. Proc Natl Acad Sci U S A. 96(1):85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korepanov AP, Gongadze GM, Garber MB, Court DL, Bubunenko MG.. 2007. Importance of the 5 S rRNA-binding ribosomal proteins for cell viability and translation in Escherichia coli. J Mol Biol. 366(4):1199–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krylov DM, Wolf YI, Rogozin IB, Koonin EV.. 2003. Gene loss, protein sequence divergence, gene dispensability, expression level, and interactivity are correlated in eukaryotic evolution. Genome Res. 13(10):2229–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland CG. 1972. Structure and function of the bacterial ribosome. Annu Rev Biochem. 41(1):377–408. [DOI] [PubMed] [Google Scholar]
- Lecompte O, Ripp R, Thierry J-C, Moras D, Poch O.. 2002. Comparative analysis of ribosomal proteins in complete genomes: an example of reductive evolution at the domain scale. Nucleic Acids Res. 30(24):5382–5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke JJ, Sanchez-Vazquez P, Burgos HL, Hedberg G, Ross W, Gourse RL.. 2011. Direct regulation of Escherichia coli ribosomal protein promoters by the transcription factors ppGpp and DksA. Proc Natl Acad Sci U S A. 108(14):5712–5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P.. 2011. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 39(Suppl):W475–W478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K, Furuta Y, Kobayashi I.. 2012. Large variations in bacterial ribosomal RNA genes. Mol Biol Evol. 29(10):2937–2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loveland AB, Korostelev AA.. 2018. Structural dynamics of protein S1 on the 70S ribosome visualized by ensemble cryo-EM. Methods 137:55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Kurita D, Li N, Chen Y, Himeno H, Gao N.. 2017. Mechanistic insights into the alternative translation termination by ArfA and RF2. Nature 541(7638):550–553. [DOI] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA.. 2011. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 10(1):13–26. [DOI] [PubMed] [Google Scholar]
- Mistry J, Finn RD, Eddy SR, Bateman A, Punta M.. 2013. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 41(12):e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolay R, Bruck D, Achenbach J, Nierhaus K.. 2015. Ribosomal proteins: role in ribosomal functions. eLS, Chichester, UK: John Wiley & Sons, Ltd. [Google Scholar]
- Nissen P, Hansen J, Ban N, Moore PB, Steitz TA.. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289(5481):920–930. [DOI] [PubMed] [Google Scholar]
- Pagel M. 1994. Detecting correlated evolution on phylogenies: a general method for the comparative analysis of discrete characters. Proc R Soc Lond B. 255(1342):37–45. [Google Scholar]
- Petrov AS, Gulen B, Norris AM, Kovacs NA, Bernier CR, Lanier KA, Fox GE, Harvey SC, Wartell RM, Hud NV, et al. 2015. History of the ribosome and the origin of translation. Proc Natl Acad Sci U S A. 112(50):15396–15401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov AS, Wood EC, Bernier CR, Norris AM, Brown A, Amunts A.. 2019. Structural patching fosters divergence of mitochondrial ribosomes. Mol Biol Evol. 36(2):207–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Peplies J, Glöckner FO.. 2012. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28(14):1823–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Lancaster L, Noller HF, Bustamante C, Tinoco I Jr.. 2012. Ribosomal protein S1 unwinds double-stranded RNA in multiple steps. Proc Natl Acad Sci U S A. 109(36):14458–14463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan V. 2002. Ribosome structure and the mechanism of translation. Cell 108(4):557–572. [DOI] [PubMed] [Google Scholar]
- Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol. 3(2):217–223. [Google Scholar]
- Roberts E, Sethi A, Montoya J, Woese CR, Luthey-Schulten Z.. 2008. Molecular signatures of ribosomal evolution. Proc Natl Acad Sci U S A. 105(37):13953–13958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlünzen F, Wilson DN, Tian P, Harms JM, McInnes SJ, Hansen HA, Albrecht R, Buerger J, Wilbanks SM, Fucini P.. 2005. The binding mode of the trigger factor on the ribosome: implications for protein folding and SRP interaction. Structure 13(11):1685–1694. [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Moore PB, Steitz TA.. 2003. Structures of deacylated tRNA mimics bound to the E site of the large ribosomal subunit. RNA 9(11):1345–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH.. 2005. Structures of the bacterial ribosome at 3.5 A resolution. Science 310(5749):827–834. [DOI] [PubMed] [Google Scholar]
- Shoji S, Dambacher CM, Shajani Z, Williamson JR, Schultz PG.. 2011. Systematic chromosomal deletion of bacterial ribosomal protein genes. J Mol Biol. 413(4):751–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Costello MS, Kettring AH, Wingo RJ, Moore SD.. 2019. Ribosome collisions alter frameshifting at translational reprogramming motifs in bacterial mRNAs. Proc Natl Acad Sci U S A. 116(43):21769–21779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TF, Lee JC, Gutell RR, Hartman H.. 2008. The origin and evolution of the ribosome. Biol Direct. 3(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillmann S, Nierhaus KH.. 1978. The ribosomal protein L24 of Escherichia coli is an assembly protein. J Biol Chem. 253(19):7047–7050. [PubMed] [Google Scholar]
- Van Duin J, Wijnands R.. 1981. The function of ribosomal protein S21 in protein synthesis. Eur J Biochem. 118(3):615–619. [DOI] [PubMed] [Google Scholar]
- Wimberly BT, Brodersen DE, Clemons WM Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V.. 2000. Structure of the 30S ribosomal subunit. Nature 407(6802):327–339. [DOI] [PubMed] [Google Scholar]
- Yutin N, Puigbò P, Koonin EV, Wolf YI.. 2012. Phylogenomics of prokaryotic ribosomal proteins. PLoS One 7(5):e36972. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.