Abstract
Sex chromosomes are classically predicted to stop recombining in the heterogametic sex, thereby enforcing linkage between sex-determining (SD) and sex-antagonistic (SA) genes. With the same rationale, a pre-existing sex asymmetry in recombination is expected to affect the evolution of heterogamety, for example, a low rate of male recombination might favor transitions to XY systems, by generating immediate linkage between SD and SA genes. Furthermore, the accumulation of deleterious mutations on nonrecombining Y chromosomes should favor XY-to-XY transitions (which discard the decayed Y), but disfavor XY-to-ZW transitions (which fix the decayed Y as an autosome). Like many anuran amphibians, Hyla tree frogs have been shown to display drastic heterochiasmy (males only recombine at chromosome tips) and are typically XY, which seems to fit the above expectations. Instead, here we demonstrate that two species, H. sarda and H. savignyi, share a common ZW system since at least 11 Ma. Surprisingly, the typical pattern of restricted male recombination has been maintained since then, despite female heterogamety. Hence, sex chromosomes recombine freely in ZW females, not in ZZ males. This suggests that heterochiasmy does not constrain heterogamety (and vice versa), and that the role of SA genes in the evolution of sex chromosomes might have been overemphasized.
Keywords: linkage mapping, recombination, sex-antagonistic genes, sex-chromosome turnover
Introduction
The evolutionary trajectories of sex chromosomes have attracted much attention from biologists, dating back to Fisher (1931). The several steps that possibly led to their present-day differentiation have been formalized into the so-called canonical model of sex-chromosome evolution (Rice 1984; Charlesworth 1991; Rice 1996; Charlesworth B and Charlesworth D 2000). Accordingly, a sex-determining (SD) mutation first occurs on a chromosome, such that individuals with the mutation develop into one sex (XY males or ZW females), and those without the mutation into the other sex (XX females or ZZ males). Second, this SD mutation attracts sexually antagonistic (SA) mutations: a male-beneficial mutation occurring in the vicinity of a male-determining gene should spread, even if detrimental to females, because genetic linkage makes it more likely to be transmitted to sons than to daughters. This situation in turn selects for an arrest of recombination in the heterogametic sex (XY males or ZW females), as a way to strengthen the link between SD and SA genes. The nonrecombining region then expands along the chromosome pair, possibly via inversions (Kirkpatrick 2010), as new SA mutations occur. However, as a side effect of recombination arrest, the sex-limited chromosome (Y or W) starts to accumulate deleterious mutations, and progressively degenerates.
This model certainly accounts for several features of the highly heteromorphic sex chromosomes found in most mammals and birds, as well as in many insects, such as Drosophila (Charlesworth et al. 2005; Bergero and Charlesworth 2009; Vicoso and Bachtrog 2015). In sharp contrast, however, the sex chromosomes of many fishes, amphibians, and nonavian reptiles appear completely homomorphic (Kikuchi and Hamaguchi 2013; Bachtrog et al. 2014; Miura 2017). One possible reason for this surprising absence of degeneration might be the occurrence of regular sex-chromosome turnovers (Schartl 2004; Volff et al. 2007; Ezaz et al. 2009; Phillips 2013; Gamble et al. 2015), during which a new pair of chromosomes takes over sex determination and replaces established sex chromosomes before they had time to degenerate. Such turnovers can have several causes (van Doorn 2014). First, an SA mutation occurring on an autosome may favor the spread of an SD mutation appearing in its vicinity, in a process symmetrical to the one described above (van Doorn and Kirkpatrick 2007; Roberts et al. 2009; van Doorn and Kirkpatrick 2010). Second, the accumulation of deleterious mutations on nonrecombining Y or W chromosomes may lower their fitness to the point that newly emerged sex chromosomes may take over (Blaser et al. 2013, 2014). Third, sex-ratio biases stemming, for example, from climatic changes, parasites, or any other environmental factor, may favor the spread of a dominant SD mutation supplying the sex in shortage (e.g., Kozielska et al. 2006, 2010; Cordaux et al. 2011; Grossen et al. 2011). Fourth, transitions may occur just by genetic drift (Veller et al. 2017; Saunders et al. 2018).
In an attempt to test among these potential causes, Jeffries et al. (2018) investigated the rate and patterns of sex-chromosome transitions in a radiation of true frogs (family Ranidae). Among 28 investigated taxa, five chromosome pairs (out of 13) were found to determine sex, depending on species and populations. Transitions were not only frequent, but also they were significantly biased toward preserving heterogamety: with the single exception of Glandirana rugosa (which presents both XY and ZW populations, Ogata et al. 2018), all the identified systems were male heterogametic (XY). As argued by Jeffries et al. (2018), this unexpected pattern supports a role for mutation load as a cause of transitions. Indeed, an XY-to-XY transition via the spread and fixation of an epistatically dominant male-determining mutation involves elimination of the established Y (which is favored when this Y is loaded with deleterious mutations); in contrast, an XY-to-ZW transition via the spread and fixation of an epistatically dominant feminizing mutation involves fixation of the Y as an autosome (which should be strongly disfavored if this chromosome is loaded with deleterious mutations). The mutation-load hypothesis thus specifically predicts a systematic bias toward maintenance of heterogamety during transitions.
Jeffries et al. (2018) also pointed out a possible role for sex-specific patterns of recombination in these transitions. Ranidae display a very strong form of heterochiasmy: contrary to females, which show frequent and evenly distributed meiotic crossovers all along their chromosomes, males only show recombination at the tips (Brelsford, Rodrigues, and Perrin 2016; see also fig. S10 in Jeffries et al. 2018). As a result, the vast majority of genes on a chromosome will suddenly and simultaneously stop recombining as soon as this chromosome takes an SD role (i.e., becomes strictly male-limited). This should drastically magnify Hill–Robertson interferences among these genes, and thus precipitate the degeneration of the new Y chromosomes, hence accelerating the turnover process (a boosted version of the “hot-potato” mechanism described by Blaser et al. 2014). Moreover, with such strong heterochiasmy, any male-beneficial gene occurring at any place on a chromosome (except for the tips) will automatically become strictly sex-linked as soon as this chromosome is co-opted for sex determination (Sardell et al. 2018), which might both favor and accelerate XY-to-XY transitions via SA selection.
Male recombination is also much reduced in other frogs (e.g., Hylidae, Brelsford et al. 2016a; Bufonidae, Stöck et al. 2013) as well as in many fishes (e.g., Sutherland et al. 2017; Sardell et al. 2018; Bergero et al. 2019). In fact, the tendency of males to preferentially recombine at chromosome tips seems to be a general feature among vertebrates (Sardell and Kirkpatrick 2020). This striking contrast between sexes might then play a central role in the dynamics of sex chromosomes, for example, potentially contributing to the prevalence of XY systems among amphibians (The Tree of Sex Consortium 2014). Achiasmy (i.e., a complete arrest of recombination in one sex) has been shown to associate with heterogamety (Burt et al. 1991), which was formalized into the so-called Haldane–Huxley rule (Haldane 1922; Huxley 1928). This correlation is classically explained either by the “pleiotropy hypothesis” (Nei 1969), that is, achiasmy as the pleiotropic consequence of selection for a recombination arrest between X and Y (or Z and W), or the “no-recombination hypothesis,” that is, systematic evolution of heterogamety in the sex devoid of recombination (Lenormand 2003). Does the link between male recombination suppression and heterogamety in frogs relate to these hypotheses? Does male recombination arrest favor or constrain transitions toward XY systems (the no-recombination hypothesis), or do XY systems favor or constrain a reduction of male recombination genome-wide (the pleiotropy hypothesis)?
To get insights into the dynamics underlying the evolution of recombination and transitions of SD systems, we focus on Palearctic tree frogs (Hyla), a group of amphibians known for drastically reduced male recombination (Brelsford et al. 2016a) and frequent sex-chromosome turnovers, including one documented change in heterogamety (Dufresnes et al. 2015). Specifically, linkage group 1 (LG1) is sex linked with male heterogamety (XY) in four European species composing the H. arborea group (which shared a common ancestor ∼7 Ma), as well as the African H. meridionalis (diverged from the H. arborea group ∼20 Ma), but it appears autosomal in the Tyrrhenian H. sarda and the Middle-Eastern H. savignyi, which branch at intermediate positions in the phylogeny (∼11 and 9 My, respectively) (Dufresnes et al. 2015; time tree from Dufresnes et al. 2018). Using an RADseq approach, here we investigated sex-linkage and recombination patterns in these two missing pieces of the puzzle. We show that both H. sarda and H. savignyi share a common female-heterogametic system (ZW) since at least 11 My, but retained the same typical pattern of genome-wide heterochiasmy with male recombination restricted to chromosome tips. This opposes the suggestion of a causal link between heterochiasmy and heterogamety in frogs, hence running against both the no-recombination and pleiotropy hypotheses.
Results
Sex-Linked Markers
For H. sarda, no male- or female-specific SNPs were flagged based on heterozygosity patterns (out of 13,622 polymorphic loci). However, five monomorphic tags were specific to females, that is, present in 95% of them but absent in all males, thus pointing to a ZW system. For H. savignyi, the heterozygosity approach did not reveal any sex-linked loci either (out of 16,024 polymorphic loci), whereas three female-specific tags were detected, also indicative of a ZW system.
Linkage Mapping
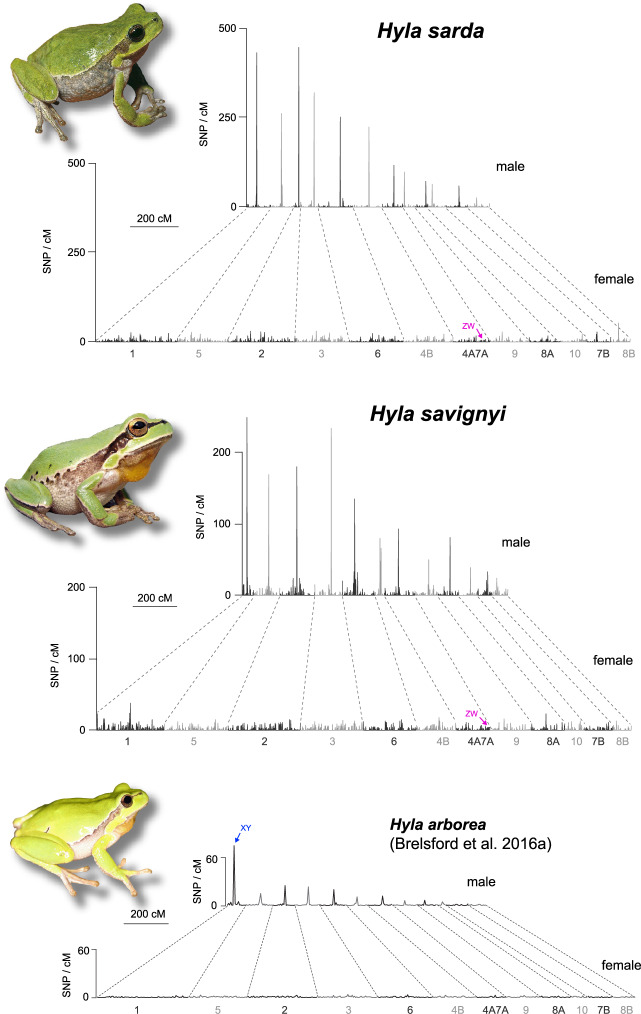
The sex-specific linkage maps of H. sarda and H. savignyi are displayed in figure 1. Twelve major linkage groups were reconstructed for each map, in accordance with the chromosome formula of Palearctic Hyla (2n = 24). For each map, ∼4–5% of markers (directly or via their H. arborea scaffold) were confidently aligned to the Xenopus tropicalis genome, and confirmed the generally conserved synteny (albeit with some rearrangements) documented between Hyla and Xenopus in Brelsford et al. (2016a). Following that study, we labeled linkage groups based on their homologous X. tropicalis chromosomes, and arranged them in the same order for comparison. Table 1 provides the number of mapped markers, and the subsets that could be located on the H. arborea linkage map and the X. tropicalis genome.
Fig. 1.
Sex-specific linkage maps for the ZW species Hyla sarda and H. savignyi, with location of their sex-linked region on LG4A7A. In both species, the male maps are about half shorter than the female maps, and highlight drastically reduced recombination in the central parts of linkage groups (peaks of SNP density), but recombination limited to the edges (with homogenous SNP density). The same patterns of heterochiasmy are observed in the XY species H. arborea, displayed here for comparison (adapted from Brelsford et al. 2016a).
Table 1.
Length and Number of Markers on the Hyla sarda and H. savignyi Sex-specific Maps, and their Numbers Placed on the H. arborea Linkage Map and the Xenopus tropicalis Genome.
|
H. sarda
|
H. savignyi
|
|||
|---|---|---|---|---|
| ♀ | ♂ | ♀ | ♂ | |
| Map length (cM) | 2,209 | 1,004 | 2,412 | 1,199 |
| Informative markers used to build the map | 3,046 | 3,176 | 2,453 | 2,739 |
| Markers located on the H. arborea linkage map | 141 (4.6%) | 115 (3.6%) | 86 (3.5%) | 109 (4.0%) |
| Markers located on the X. tropicalis genome | 138 (4.5%) | 166 (5.2%) | 100 (4.1%) | 132 (4.8%) |
The sexual genotypes (ZZ or ZW, according to the W-specific markers identified above) segregated with linkage group LG4A7A in both species (fig. 1). As suggested by Brelsford et al. (2016a), this linkage group originates from a fusion between parts of Xenopus chromosomes 4 and 7. The SD regions share a very similar position: in H. sarda, it was tightly linked (<3 cM) to markers mapping Xenopus chromosome 4 at positions 88,533,774 and 88,018,899; in H. savignyi, it was fully linked (0 cM) to a marker mapping chromosome 4 at position 90,759,259. Therefore, the nonrecombining sex-linked markers identified independently in H. sarda and H. savignyi fall <3 Mb apart on the X. tropicalis genome.
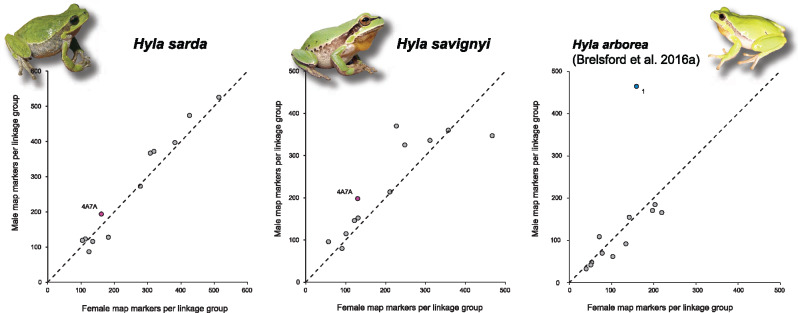
In a similar fashion as H. arborea (Brelsford et al. 2016a), H. sarda and H. savignyi both featured strong patterns of heterochiasmy (fig. 1). In females, SNP density was evenly distributed across the whole length of the linkage groups. Male maps were about twice as short (table 1) and characterized by suppressed recombination across the central parts of linkage groups, with recombining regions restricted to the edges. The ratio of male- to female-markers was similar across all linkage groups in both species, including the sex-linked LG4A7A (fig. 2).
Fig. 2.
Numbers of male versus female markers in the linkage maps of the ZW species Hyla sarda and H. savignyi (left). Unlike in the XY species H. arborea (right, adapted from Brelsford et al. 2016a), all twelve linkage groups, including the sex-linkage group (LG4A7A), show comparable numbers of male- and female-informative markers. Dashed lines show diagonals.
Phylogeny
A SNP-based species tree obtained with SNAPP was identical to the concatenated RAD tag sequences tree previously published by Dufresnes et al. (2018) (supplementary fig. S1, Supplementary Material online). In particular, the phylogeny confirms the independent divergences of H. sarda and H. savignyi from the ancestor of the H. arborea group.
Discussion
Scenarios for Transitions
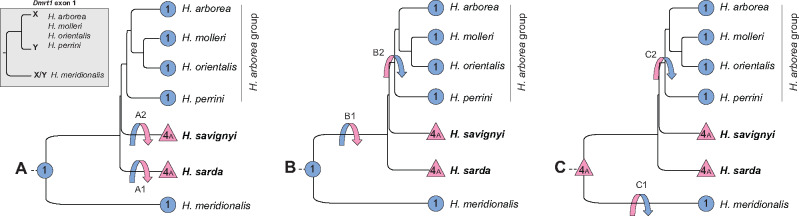
Following up on Dufresnes et al. (2015), our study supports two major SD systems in Western Palearctic tree frogs: an XY system on LG1 (five species) and a ZW system on LG4A7A (two species). The phylogeny has now been fully resolved for all nodes (Dufresnes et al. 2018), as confirmed here by SNP-based analyses. Hence, there are only three possible scenarios of transitions, as follows (fig. 3). (A) The XY system is ancestral: two transitions to ZW occurred independently in H. sarda (A1) and H. savignyi (A2), after their splits from the European ancestor. (B) The XY system is also ancestral: a transition to ZW occurred in the European ancestor, after the split of the branch leading to H. meridionalis but before the split of H. sarda, that is, between 20 and 11 Ma (B1); a back transition then restored the XY system after the split of H. savignyi but before the diversification of the H. arborea group, that is, between 9 and 7 Ma (B2). (C) The ZW system is ancestral: two independent transitions to XY occurred, one in the branch leading to H. meridionalis (C1), and the other in the ancestor of the H. arborea group, between 9 and 7 Ma (C2).
Fig. 3.
Nuclear phylogeny of Western Palearctic Hyla species (based on Dufresnes et al. [2018] and new analyses, supplementary fig. S1, Supplementary Material online) for which sex chromosomes have been identified (blue circles: XY system on LG1—abbreviated “1”; pink triangles: ZW system on LG4A7A—abbreviated “4A”), with possible scenarios A–C of evolutionary transitions (arrows); the focal species of this study are in bold. The phylogeny of X and Y sequences at Dmrt1 exon 1—the presumed sex-determining region of the Hyla arborea group on LG1—is adapted from Brelsford et al. (2016b).
Scenario A seems unlikely because the sex locus shows the same location in both ZW species (at the tip of LG4A7A), which supports homology and shared ancestry (even though convergence cannot be entirely ruled out). Furthermore, the sex locus apparently differs between H. meridionalis and the H. arborea group: alleles at Dmrt1 exon 1 cluster by gametologs (not by species) for all species of the H. arborea group, pointing to a common SD role for this locus; whereas H. meridionalis alleles cluster by species (not by gametologs) (Brelsford et al 2016b; fig. 3). Scenario A would therefore imply one additional (homologous) transition, that is, from one locus to another on LG1. In contrast, scenarios B and C require only two transitions, both changing heterogamety: from XY to ZW and then ZW back to XY (B), or from ZW to XY independently twice (C). With the data in hand, these two scenarios seem equally plausible, and could possibly be distinguished by testing additional species, such as H. carthaginiensis (the sister clade of H. meridionalis, Dufresnes et al. 2019): if this taxon shares the same SD system as H. sarda/savignyi (ZW at LG4A7A), this would increase the likelihood of scenario C. Finally, if, opposing our phylogeny, H. sarda and H. savignyi were to form a sister clade, it would imply a fourth possible scenario also with two turnover events (Dufresnes et al. 2015). Even though this alternative topology is not supported by our data (Dufresnes et al. 2018; supplementary fig. S1, Supplementary Material online), it should be kept in mind that phylogenetic uncertainty has the potential to bias the reconstruction of sex-chromosome evolution (Nielsen et al. 2019).
Evolutionary Causes of Transitions
What possibly drove two heterogametic transitions during the diversification of Western Palearctic Hyla since the Miocene? In an individual-based simulation framework, Saunders et al. (2019) investigated the dynamics of XY-to-ZW transitions, when the nonrecombining Y had accumulated SA genes and deleterious mutations. Their analyses showed that, contrasting with XY-to-XY transitions (which are favored by deleterious mutations, see Introduction), mutation load does not promote the spread of an epistatically dominant W mutation, because the Y would have to be fixed as an autosome during such transitions. Outcomes, however, vary depending on the mechanisms preventing X–Y recombination (Saunders et al. 2019). If recombination is controlled by genetic sex (e.g., due to an inversion on the Y), transitions are much hindered by mutation load, and totally prevented by even small-effect SA genes (because both deleterious and female-detrimental mutations would then be fixed with the Y). If, however, recombination is controlled by phenotypic sex (as appears to be the case in frogs, Rodrigues et al. 2018), X–Y recombination occurring in ZW females that still carry the ancestral X and Y chromosomes (i.e., XY ZW females) can purge the deleterious mutations and female-detrimental alleles from the Y, which can then be fixed as an autosome under a drift-mediated transition. The same reasoning would apply if X–Y recombination would instead occur in sex-reversed XY females (the fountain-of-youth hypothesis; Perrin 2009).
Importantly, as also pointed out by Saunders et al. (2019), a transition from male to female heterogamety can still result from the spread of a weakly masculinizing mutation M (i.e., recessive to the wild-type allele m), allowing an XX mm/XY mm system to evolve toward an XX mM/XX MM system (case 2B in Bull and Charnov 1977). As the mutation-loaded Y is lost in such a transition, this change from male to female heterogamety would actually be favored by mutation-load selection. Thus, the XY-to-ZW transition B1 in Hyla could have been favored by mutation load through the spread of a dosage-dependent masculinizing mutation.
In principle, the arguments outlined above are symmetrical regarding heterogamety, so that the same conclusions should apply to ZW-to-XY transitions. In the present instance, however, the prevalent pattern of heterochiasmy (drastically reduced recombination in males, whatever the pattern of heterogamety) introduces a fundamental asymmetry. The consequences in terms of allelic fixation are obvious when comparing H. sarda and H. savignyi in our figure 2, where LG4A7A lies on the diagonal, with the numbers of SNPs in males and females proportional to the genome average, with a similar plot of the XY species H. arborea (adapted from fig. 1 in Brelsford et al. 2016a), where LG1 is a clear outlier, showing a nearly 3-fold excess of male SNPs due to XY differentiation. These consequences are also exemplified by the striking rarity of W-specific markers in both H. sarda and savignyi. Thus, the high rate of Z–W recombination in females prevents the fixation of W-specific alleles, including deleterious mutations, so the ZW-to-XY transitions B2 and C1-C2 in Hyla cannot result from mutation load.
The expected effects of SA selection are also asymmetrical, along the same rational. SA selection is unlikely to favor an XY-to-ZW transition because female recombination prevents the fixation of female-beneficial mutations on the W. Thus, the XY-to-ZW transition B1 in Hyla cannot have been driven by SA mutations on LG4A7A. Reciprocally, the ZW-to-XY transitions B2 and C1-C2 are compatible with SA selection, because, in an XY system, male recombination arrest would ensure immediate linkage of SA and SD genes on any chromosome.
Of course, and as mentioned above, genetic drift cannot be excluded as the main cause of any of these transitions. Drift-induced transitions are generally 2–4 times more likely to preserve heterogamety than to change it (Saunders et al. 2018), but this likelihood decreases with low effective population sizes. XY-to-ZW transitions can even become more likely than XY-to-XY transitions if the small effective size is due to a low number of breeding males (Saunders et al. 2018). Such demographic situations appear realistic for tree frogs, which are characterized by good-gene mechanisms of sexual selection on males (Jaquiéry et al. 2010), and as ectotherms, are susceptible to strong fluctuations in distribution ranges and thus of effective population sizes, as a response to climate changes during the last million years (e.g., Bisconti et al. 2011 for H. sarda).
To summarize, the XY-to-ZW transition that led to the SD system of H. sarda and H. savignyi could have been mediated by mutation load (but not by SA selection), whereas the reverse ZW-to-XY transition that led to the SD system of the H. arborea group was possibly mediated by SA selection (but not by mutation load); and genetic drift cannot be excluded in either case. As this discussion emphasizes, it remains difficult to disentangle the relative contributions of the diverse and nonexclusive processes potentially involved in sex-chromosome turnovers. More work is certainly needed to formalize proper predictions, and test them using integrative data from multiple organismal groups featuring a variety of labile SD systems.
Heterochiasmy and Heterogamety
From the sex-specific maps presented in figure 1, the ZW Hyla species display the same recombination patterns as their XY relative H. arborea (Brelsford et al. 2016a). That recombination in males is restricted to chromosome tips seems actually a widespread pattern among frogs (Brelsford, Rodrigues, and Perrin 2016; Jeffries et al. 2018). Our present results show that this pattern also extends to ZW systems. Such remarkable similarity between XY and ZW systems strongly suggests that heterochiasmy is independent of heterogamety in frogs. This result argues against the no-recombination hypothesis: the strongly reduced recombination in males does not seem to constrain transitions toward XY systems, so their prevalence among amphibians likely stems from other causes (Jeffries et al. 2018). It also argues against the pleiotropy hypothesis: patterns of recombination in H. sarda and H. savignyi did not adapt to the female heterogamety, despite at least 11 My (i.e., split of H. sarda) since the transition. Note that the absence of male recombination over most of the chromosome length should itself strongly hinder Z–W recombination arrest. An inversion on the W, for instance, would make both W and Z stop recombining (given that Z would not recombine in males either), thus causing both copies of any gene on the nonrecombining segment to degenerate, which is obviously unviable. Therefore, if the absence of male recombination is a constraint stemming from specificities of the male meiosis in frogs, then it should prevent any arrest of Z–W recombination.
What Role for SA Genes in Recombination Arrest?
Our results also run against the classical prediction that sex chromosomes stop recombining in the heterogametic sex as a way to enforce linkage between SD and SA genes. The sex chromosomes of H. sarda and H. savignyi actually recombine in the heterogametic sex (ZW females), but not in the homogametic sex (ZZ males). Despite this, both species display sexual dimorphism comparable to that of XY tree frogs of the same radiation (Dufresnes 2019). This adds to the growing body of evidence that SA genes play little role (if any) in the arrest of recombination and ensuing X–Y (or Z–W) genetic differentiation in amphibians (Ma, Veltsos, Toups, et al. 2018; Ma, Veltsos, Sermier, et al. 2018; Veltsos et al. 2020; reviewed in Perrin 2020). Sexual dimorphism seemingly results more from the differential expression of autosomal genes than from the fixation of sex-limited genes.
It is worth adding that the classical pattern (recombination arrest in the heterogametic sex) might often stem from other causes than SA genes, including genetic drift (Ironside 2010). If, for instance, an inversion occurs on an autosome, recombination will be temporarily stopped in individuals that are heterozygous for the inversion, but it will ultimately resume as soon as the inversion is either fixed or eliminated by drift. If, however, such an inversion occurs and is fixed by drift on the Y or the W, these chromosomes will definitely stop recombining with their homologs. Such neutral models might well account for many cases of recombination arrest of sex chromosomes (e.g., Branco et al. 2017). Neutrality might also explain why sex chromosomes may continue recombining in the heterogametic sex in Hylid frogs.
Whatever its causes, the fact that Z and W did not stop recombining in H. sarda and H. savignyi, despite their putatively old origin, clearly opposes expectations from the canonical model of sex-chromosome evolution, and definitely shows that the arrest of recombination and ensuing degeneration is not the ineluctable destiny of sex-limited chromosomes.
Materials and Methods
Sampling
We analyzed samples of H. sarda collected in Corsica (43.00°N, 9.40°E) and of H. savignyi collected in Cyprus (34.97°N, 32.41°E), all previously included in Dufresnes et al. (2015). For each species, we considered unrelated live-caught adults (n = 15 of each sex, diagnosed by secondary sexual characters) as well as 40 offspring from a single cross, including eight that were raised after metamorphosis and phenotypically sexed by gonad dissection (Haczkiewicz and Ogielska 2013). DNA was obtained from noninvasive buccal swabs (adults, including parents of the crosses), or tissues fixed in 96% ethanol (larvae, muscle pieces from dissected froglets), and extracted with the Qiagen Biosprint Robotic workstation.
Laboratory and Bioinformatic Procedures
A genomic library was prepared following the double-digest Restriction-Associated DNA (ddRAD) protocol of Brelsford et al. (2016a). Briefly, it consisted of enzyme digestion by SbfI and MseI, ligation of barcoded adapters, PCR-amplification of the resulting fragments and size-selection of PCR products between 400 and 500 bp, and was sequenced on a single Illumina HiSeq 2000 lane.
Raw reads were demultiplexed using the process_radtags module of Stacks v. 1.24 (Catchen et al. 2013) and loci were built with the denovo_map.pl pipeline, run separately for each species, with –bound_high 0.05 and other parameters left with default values. The resulting variants were then split into separate files for mapping crosses and unrelated adults. Variants were filtered with VCFtools v0.1.10 (Danecek et al. 2011), retaining genotypes with minimum read depth of 7, and retaining loci genotyped in at least 80% of individuals. We excluded the loci with minor allele frequencies below 0.1 for unrelated adults and below 0.15 for mapping cross parents and offspring. These filters resulted in data sets of 13,622 and 16,024 SNPs for unrelated adults in H. sarda and H. savignyi, respectively, and 12,509 and 11,064 SNPs for mapping crosses in H. sarda and H. savignyi, respectively.
Sex-Linked Markers
Two approaches were followed to find sex-linked markers: 1) sex-differences in heterozygosity, screening for SNPs homozygous in all individuals of one sex but heterozygous in >95% of individuals of the other; 2) sex-linked occurrence, screening for tags absent in all individuals of one sex, but present in >95% of individuals of the other. The rationale and R scripts of these methods are detailed in Brelsford et al. (2017).
Linkage Mapping
Sex-specific linkage maps were generated based on recombination patterns from the cross data (parents + tadpoles/sexed juveniles). To this end, we first produced separate lists of paternal-informative and maternal-informative markers, excluding loci that were heterozygous in both parents or in neither parent, and correcting Mendelian segregation errors (as described in Brelsford et al. 2016a). To locate the SD region on the linkage maps, we predicted the sex of tadpoles based on the presence/absence of the W-specific markers identified in each species (see Results). Linkage groups were then built with MSTmap (Wu et al. 2008), including sex as an additional character, and split at gaps >40 cM.
To determine homology with the linkage map of H. arborea (Brelsford et al. 2016a), tags were aligned to a low-coverage draft genome of that species (doi:10.5061/dryad.n856c) using BlastN. The homology then allowed to assign sets of anonymous markers to their corresponding linkage groups. For each RAD tag, the corresponding H. arborea scaffold was subsequently aligned to the X. tropicalis genome (ftp://ftp.xenbase.org/pub/Genomics/JGI/Xentr7.1/) using BlastN, retaining only the best hits if their e-value was at least five orders of magnitude lower than second-best hits.
SNP-Based Phylogeny
The phylogeny of Western Palearctic tree frog was previously resolved for all nodes based on analyses of concatenated RAD tags (43 kb) in Dufresnes et al. (2018). However, concatenation can sometimes produce incorrect topologies with high posterior probabilities, especially when internal branches are short (Degnan and Rosenberg 2006). To confirm the Hyla species tree, here we performed a phylogenomic reconstruction under a different approach, namely the SNP-based Bayesian framework of SNAPP (Leaché et al. 2014) implemented in the BEAST 2 environment (Bouckaert et al. 2014). For this analysis, an alignment of 2,302 nuclear SNPs was exported from the processed RAD tags of Dufresnes et al. (2018) for 40 samples representative of the ten Western Palearctic taxa plus Dryophytes japonicus as outgroup (3–4 samples per taxa). Model parameters and priors were optimized following the recommendations of Leaché and Bouckaert (2018). The chain was sampled every 1,000 iterations and ran for 5 million iterations, long after large effective sample sizes of parameters (>200) and long-term stationarity were reached (Tracer 1.5, http://beast.community/). The trees obtained were visualized in DensiTree 2 (Bouckaert and Heled 2014), discarding the first 20% as burnin.
Data Availability
The data of this article (individual raw sequence reads) were archived on the NCBI SRA under Bioproject PRJNA542138.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We are grateful to R. Sermier and J. Wassef for help in the field and in the lab. This study was supported by the Swiss National Science Foundation (Grant No. 31003A_166323 to N.P.).
References
- Bachtrog D, Mank JE, Peichel CL, Kirkpatrick M, Otto SP, Ashman TL, Hahn MW, Kitano J, Mayrose I, Ming R, et al. 2014. Sex determination: why so many ways of doing it? PLoS Biol. 12(7):e1001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergero R, Charlesworth D.. 2009. The evolution of restricted recombination in sex chromosomes. Trends Ecol Evol. 24(2):94–102. [DOI] [PubMed] [Google Scholar]
- Bergero R, Gardner J, Bader B, Yong L, Charlesworth D.. 2019. Exaggerated heterochiasmy in a fish with sex-linked male coloration polymorphisms. Proc Natl Acad Sci USA. 116(14):6924–6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisconti R, Canestrelli D, Colangelo P, Nascetti G.. 2011. Multiple lines of evidence for demographic and range expansion of a temperate species (Hyla sarda) during the last glaciation. Mol Ecol. 20(24):5313–5327. [DOI] [PubMed] [Google Scholar]
- Blaser O, Grossen C, Neuenschwander S, Perrin N.. 2013. Sex-chromosome turnovers induced by deleterious mutation load. Evolution 67(3):635–645. [DOI] [PubMed] [Google Scholar]
- Blaser O, Neuenschwander S, Perrin N.. 2014. Sex-chromosome turnovers: the hot-potato model. Am Nat. 183(1):140–146. [DOI] [PubMed] [Google Scholar]
- Bouckaert RR, Heled J.. 2014. DensiTree 2: seeing trees through the forest. BioRxiv 012401; doi: 10.1101/012401.
- Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D, Suchard MA, Rambaut A, Drummond AJ.. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLOS Comput Biol. 10(4):e1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco S, Badouin H, Rodríguez de la Vega RC, Gouzy J, Carpentier F, Aguileta G, Siguenza S, Brandenburg J-T, Coelho MA, Hood ME, et al. 2017. Evolutionary strata on young mating-type chromosomes despite the lack of sexual antagonism. Proc Natl Acad Sci USA. 114(27):7067–7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brelsford A, Dufresnes C, Perrin N.. 2016. a. High-density sex-specific linkage maps of a European tree frog (Hyla arborea) identify the sex chromosome without information on offspring sex. Heredity 116(2):177–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brelsford A, Dufresnes C, Perrin N.. 2016. b. Trans-species variation in Dmrt1 is associated with sex determination in four European tree-frog species. Evolution 70(4):840–847. [DOI] [PubMed] [Google Scholar]
- Brelsford A, Lavanchy G, Sermier R, Rausch A, Perrin N.. 2017. Identifying homomorphic sex chromosomes from wild-caught adults with limited genomic resources. Mol Ecol Resour. 17(4):752–759. [DOI] [PubMed] [Google Scholar]
- Brelsford A, Rodrigues N, Perrin N.. 2016. High-density linkage maps fail to detect any genetic component to sex determination in a Rana temporaria family. J Evol Biol. 29(1):220–22546. [DOI] [PubMed] [Google Scholar]
- Bull JJ, Charnov EL.. 1977. Changes in the heterogametic mechanism of sex determination. Heredity 39(1):1–14. [DOI] [PubMed] [Google Scholar]
- Burt A, Graham B, Harvey PH.. 1991. Sex differences in recombination. J Evol Biol. 4(2):259–277. [Google Scholar]
- Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA.. 2013. Stacks: an analysis tool set for population genomics. Mol Ecol. 22(11):3124–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. 1991. The evolution of sex chromosomes. Science 251(4997):1030–1033. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D.. 2000. The degeneration of Y chromosomes. Phil Trans R Soc Lond B. 355(1403):1563–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B, Marais G.. 2005. Steps in the evolution of heteromorphic sex chromosomes. Heredity 95(2):118–128. [DOI] [PubMed] [Google Scholar]
- Cordaux R, Bouchon D, Grève P.. 2011. The impact of endosymbionts on the evolution of host sex-determination mechanisms. Trends Genet. 27(8):332–341. [DOI] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics 27(15):2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan JH, , Rosenberg NA. 2006. Discordance of species trees with their most likely gene trees. PLoS Genet. 2(5):e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresnes C. 2019. Amphibians of Europe, North Africa and the Middle East. London: Bloomsbury. [Google Scholar]
- Dufresnes C, Beddek M, Skorinov DV, Fumagalli L, Perrin N, Crochet P-A, Litvinchuk SN.. 2019. Diversification and speciation in tree frogs from the Maghreb (Hyla meridionalis sensu lato), with description of a new African endemic. Mol Phylogenet Evol. 134:291–299. [DOI] [PubMed] [Google Scholar]
- Dufresnes C, Borzée A, Horn A, Stöck M, Ostini M, Sermier R, Wassef J, Litvinchuck SN, Kosch TA, Waldman B, et al. 2015. Sex-chromosome homomorphy in Palearctic tree frogs results from both turnovers and X-Y recombination. Mol Biol Evol. 32(9):2328–2337. [DOI] [PubMed] [Google Scholar]
- Dufresnes C, Mazepa G, Rodrigues R, Brelsford A, Litvinchuk SN, Sermier R, Betto-Colliard C, Blaser O, Borzée A, Cavoto E, et al. 2018. Genomic evidence for cryptic speciation in tree frogs from the Apennine Peninsula, with description of Hyla perrini sp. nov. Front Ecol Evol. 6:144. [Google Scholar]
- Ezaz T, Sarre SD, O’Meally D, Graves JAM, Georges A.. 2009. Sex chromosome evolution in lizards: independent origins and rapid transitions. Cytogenet Genome Res. 127(2–4):249–260. [DOI] [PubMed] [Google Scholar]
- Fisher RA. 1931. The evolution of dominance. Biol Rev. 6(4):345–368. [Google Scholar]
- Gamble T, Coryell J, Ezaz T, Lynch J, Scantlebury DP, Zarkower D.. 2015. Restriction site-associated DNA sequencing (RAD-seq) reveals an extraordinary number of transitions among gecko sex-determining systems. Mol Biol Evol. 32(5):1296–1309. [DOI] [PubMed] [Google Scholar]
- Grossen C, Neuenschwander S, Perrin N.. 2011. Temperature-dependent turnovers in sex-determination mechanism: a quantitative model. Evolution 65(1):64–78. [DOI] [PubMed] [Google Scholar]
- Haczkiewicz K, Ogielska M.. 2013. Gonadal sex differentiation in frogs: how testes become shorter than ovaries. Zool Sci. 30(2):125–134. [DOI] [PubMed] [Google Scholar]
- Haldane JBS. 1922. Sex ratio and unisexual sterility in hybrid animals. J Gen. 12(2):101–109. [Google Scholar]
- Huxley JS. 1928. Sexual differences of linkage in Gammarus chevreuxi. J Gen. 20(2):145–156. [Google Scholar]
- Ironside JE. 2010. No amicable divorce? Challenging the notion that sexual antagonism drives sex chromosome evolution. BioEssays 32(8):718–726. [DOI] [PubMed] [Google Scholar]
- Jaquiéry J, Broquet T, Aguilar C, Evanno G, Perrin N.. 2010. Good genes drive female choice for mating partners in the lek-breeding European treefrog. Evolution 64(1):108–115. [DOI] [PubMed] [Google Scholar]
- Jeffries DL, Lavanchy G, Sermier R, Sredl MJ, Miura I, Borzée A, Barrow LN, Canestrelli D, Crochet PA, Dufresnes C, et al. 2018. A rapid rate of sex-chromosome turnover and non-random transitions in true frogs. Nat Comm. 9:4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozielska M, Pen I, Beukeboom LW, Weissing FJ.. 2006. Sex ratio selection and multi-factorial sex determination in the housefly: a dynamic model. J Evol Biol. 19(3):879–888. [DOI] [PubMed] [Google Scholar]
- Kozielska M, Weissing FJ, Beukeboom LW, Pen I.. 2010. Segregation distortion and the evolution of sex-determining mechanisms. Heredity 104(1):100–112. [DOI] [PubMed] [Google Scholar]
- Kikuchi K, Hamaguchi S.. 2013. Novel sex-determining genes in fish and sex chromosome evolution. Dev Dyn. 242(4):339–353. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M. 2010. How and why chromosome inversions evolve. PLoS Biol. 8(9):e1000501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaché AD, Bouckaert RR.. 2018. Species trees and species delimitation with SNAPP: a tutorial and worked example. Available from: http://evomicsorg.wpengine.netdnacdn.com/wpcontent/uploads/2018/01/BFD-tutorial-1.pdf.
- Leaché AD, Fujita MW, Minin VN, Bouckaert RR.. 2014. Species delimitation using genome-wide SNP data. Syst Biol. 63(4):534–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenormand T. 2003. The evolution of sex dimorphism in recombination. Genetics 163(2):811–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W-J, Veltsos P, Sermier R, Parker DJ, Perrin N.. 2018. Evolutionary and developmental dynamics of sex-biased gene expression in common frogs with proto-Y chromosomes. Genome Biol. 19(1):156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W-J, Veltsos P, Toups MA, Rodrigues N, Sermier R, Jeffries DL, Perrin N.. 2018. Tissue specificity and dynamics of sex-biased gene expression in a common frog population with differentiated, yet homomorphic, sex chromosomes. Genes 9(6):294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura I. 2017. Sex determination and sex chromosomes in amphibia. Sex Dev. 11(5-6):298–306. [DOI] [PubMed] [Google Scholar]
- Nei M. 1969. Linkage modification and sex difference in recombination. Genetics 63(3):681–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen SV, Guzmán-Méndez IA, Gamble T, Blumer M, Pinto BJ, Kratochvíl L, Rovatsos M.. 2019. Escaping the evolutionary trap? Sex chromosome turnover in basilisks and related lizards (Corytophanidae: Squamata). Biol Lett. 15(10):20190498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata M, Lambert M, Ezaz T, Miura I.. 2018. Reconstruction of female heterogamety from admixture of XX-XY and ZZ-ZW sex-chromosome systems within a frog species. Mol Ecol. 27(20):4078–4089. [DOI] [PubMed] [Google Scholar]
- Perrin N. 2009. Sex reversal: a fountain of youth for sex chromosomes? Evolution 63(12):3043–3049. [DOI] [PubMed] [Google Scholar]
- Perrin N. 2020. Sex-chromosome evolution in frogs: what role for sex-antagonistic genes? Philos Trans R Soc Lond B Biol Sci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips RB. 2013. Evolution of the sex chromosomes in salmonid fishes. Cytogenet Genome Res. 141(2–3):177–185. [DOI] [PubMed] [Google Scholar]
- Roberts RB, Ser JR, Kocher TD.. 2009. Sexual conflict resolved by invasion of a novel sex determiner in Lake Malawi cichlid fishes. Science 326(5955):998–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice WR. 1984. Sex chromosomes and the evolution of sexual dimorphism. Evolution 38(4):735–742. [DOI] [PubMed] [Google Scholar]
- Rice WR. 1996. Evolution of the Y sex chromosome in animals. BioScience 46(5):331–343. [Google Scholar]
- Rodrigues N, Studer T, Dufresnes C, Perrin N.. 2018. Sex-chromosome recombination in common frogs brings water to the fountain-of-youth. Mol Biol Evol. 35(4):942–948. [DOI] [PubMed] [Google Scholar]
- Sardell JM, Cheng C, Dagilis AJ, Ishikawa A, Kitano J, Peichel CL, Kirkpatrick M.. 2018. Sex differences in recombination in sticklebacks. G3 8(6):1971–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardell JM, Kirkpatrick M.. 2020. Sex differences in the recombination landscape. Am Nat. 195(2):361–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders PA, Neuenschwander S, Perrin N.. 2018. Sex chromosome turnovers and genetic drift: a simulation study. J Evol Biol. 31(9):1413–1419. [DOI] [PubMed] [Google Scholar]
- Saunders PA, Neuenschwander S, Perrin N.. 2019. Impact of deleterious mutations, sexually antagonistic selection, and mode of recombination suppression on transitions between male and female heterogamety. Heredity 123(3):419–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schartl M. 2004. Sex chromosome evolution in non-mammalian vertebrates. Curr Opin Genet Dev. 14(6):634–641. [DOI] [PubMed] [Google Scholar]
- Stöck M, Savary R, Betto-Colliard C, Biollay S, Jourdan-Pineau H, Perrin N.. 2013. Low rates of X-Y recombination, not turnovers, account for homomorphic sex chromosomes in several diploid species of Palearctic green toads (Bufo viridis subgroup). J Evol Biol. 26(3):674–682. [DOI] [PubMed] [Google Scholar]
- Sutherland BJG, Rico C, Audet C, Bernatchez L.. 2017. Sex chromosome evolution, heterochiasmy and physiological QTL in the salmonid brook charr Salvelinus fontinalis. G3 7(8):2749–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Tree of Sex Consortium. 2014. Tree of Sex: a database of sexual systems. Sci Data. 1:140015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn GS. 2014. Patterns and mechanisms of evolutionary transitions between genetic sex-determining systems. Cold Spring Harb Perspect Biol. 6(8):a017681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn GS, Kirkpatrick M.. 2007. Turnover of sex chromosomes induced by sexual conflict. Nature 449(7164):909–912. [DOI] [PubMed] [Google Scholar]
- van Doorn GS, Kirkpatrick M.. 2010. Transitions between male and female heterogamety caused by sex-antagonistic selection. Genetics 186(2):629–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veller C, Muralidhar P, Constable GWA, Nowak MA.. 2017. Drift-induced selection between male and female heterogamety. Genetics 207(2):711–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltsos P, Rodrigues N, Studer T, Ma WJ, Sermier R, Leuenberger J, Perrin N.. 2020. No evidence that Y-chromosome differentiation affects male fitness in a Swiss population of common frogs. J Evol Biol. 33(4):401–409. [DOI] [PubMed] [Google Scholar]
- Vicoso B, Bachtrog D.. 2015. Numerous transitions of sex chromosomes in Diptera. PLoS Biol. 13(4):e1002078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volff JN, Nanda I, Schmid M, Schartl M.. 2007. Governing sex determination in fish: regulatory putsches and ephemeral dictators. Sex Dev. 1(2):85–99. [DOI] [PubMed] [Google Scholar]
- Wu Y, Bhat PR, Close TJ, Lonardi S.. 2008. Efficient and accurate construction of genetic linkage maps from the minimum spanning tree of a graph. PLoS Genet. 4(10):e1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data of this article (individual raw sequence reads) were archived on the NCBI SRA under Bioproject PRJNA542138.