Abstract
The complete chloroplast genome of Corydalis inopinata Prain ex Fedde was assembled and characterized in this study. The chloroplast genome was 181,335 bp in length, consisting of a large single-copy (LSC) region of 91,727 bp, a small single-copy (SSC) region of 1502 bp, and a pair of inverted repeat (IR) regions of 44,053 bp. It encoded 110 unique genes, including 68 protein-coding genes, 29 tRNA genes, 4 rRNA genes, and 9 pseudogenes. Phylogenetic analyses by maximum likelihood (ML) and Bayesian inference (BI) revealed that C. inopinata was closely related to C. conspersa Maxim. with full support in the present sampling. The complete plastid genome provided in this work would be useful for elucidating the taxonomy, phylogeny, and evolution of C. inopinata and other related species.
Keywords: Corydalis inopinata, chloroplast genome, phylogeny
Corydalis DC., one of the largest genus in Chinese flora, stands out for its complex, variable, interconnected and intergrading morphological characters (Wu et al. 1996). To date, relatively few molecular researches have been done on this taxon, leaving us with many unresolved questions about its taxonomy, phylogeny, and evolution. In the present study, we characterized and reported the complete plastid genome of C. inopinata Prain ex Fedde, a cushion-like alpine perennial herb, and investigated its phylogeny within Ranunculales.
The fresh leaves and voucher specimens of C. inopinata were collected from its wild population (Jiangzi, Xizang, China, 28°54′N, 90°9′E, D. Wang et al. 140599), and deposited in the herbarium of Central China Normal University (CCNU). Total genomic DNA was isolated using CTAB method (Doyle and Doyle 1987). Library with an insert size of about 350 bp was constructed, and sequenced using Illumina NovaSeq 6000 (Illumina, San Diego, CA) at Novogene (Tianjin, China). Approximately 7.63 Gb raw data were generated, and then the raw reads were filtered using fastp v0.20.1 (Chen et al. 2018). The cleaned reads were assembled de novo using GetOrganelle v1.6.2 (Jin et al. 2018) with chloroplast genome of Lamprocapnos spectabilis (L.) Fukuhara (NC_039756) as reference. The assembled chloroplast genome was annotated using PGA (Qu et al. 2019) with the chloroplast genome of L. spectabilis (NC_039756) as reference, checked manually, and submitted to National Center for Biotechnology Information (NCBI, accession number: MT755641). To investigate the phylogeny of C. inopinata, 99 shared genes among C. inopinata and another 16 representative Ranunculales species from NCBI (accession numbers were provided in Figure 1) were aligned using MAFFT v.7 (Katoh and Standley 2013). The best-fit nucleotide substitution model was determined by jModeltest v2.1.10 (Darriba et al. 2012) using the Akaike information criterion (AIC). Maximum likelihood (ML) and Bayesian inference (BI) phylogenetic analyses were conducted using RAxML v8.2.12 (Stamatakis 2014, -m GTRGAMMA -N 1000 -f a) and MrBayes v3.2.7 (Ronquist et al. 2012, GTR + I + G model, ngen = 1,000,000, samplefreq = 100, nchains = 4, burnin = 2500), respectively.
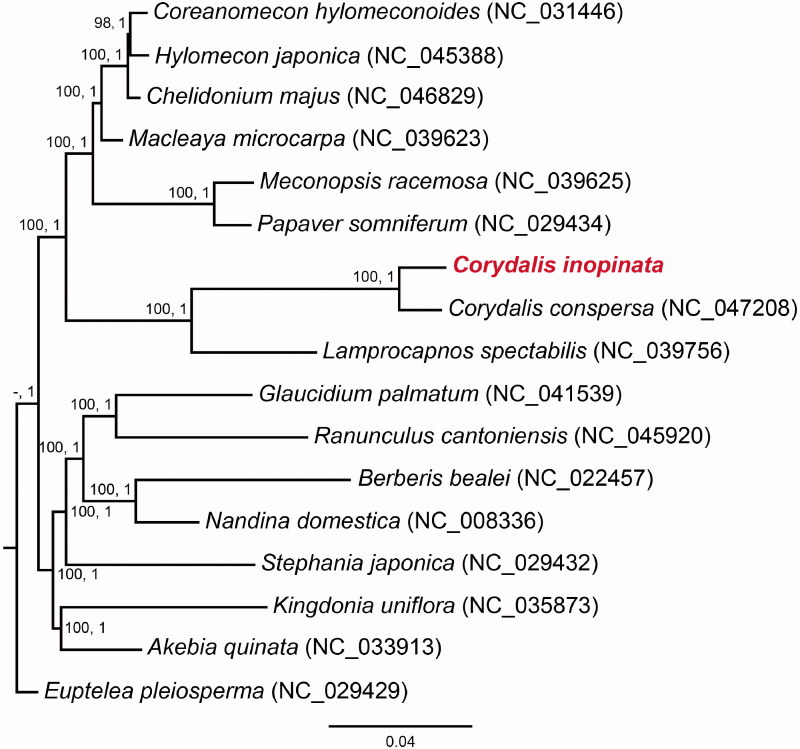
Figure 1.
ML and BI phylogenetic tree based on C. inopinata and another 16 Ranunculales species, with ML bootstrap values (left) and BI posterior probability (right) indicated near the nodes.
The complete chloroplast genome of C. inopinata was 1,81,335 bp in length, exhibiting typical angiosperm quadripartite structure. The large single-copy (LSC), inverted repeat (IR), and small single-copy (SSC) were 91,727 bp, 44,053 bp, and 1,502 bp, respectively. Inversion and translocation of about 5 kb (rbcL, atpB, atpE, and trnM-CAU) was observed in the LSC. The IRs expanded markedly into the SSC, resulting in the IRs 15 kb longer than the average. In total, 110 genes were predicted, including 68 protein-coding genes, 29 tRNA genes, four rRNA genes, and nine pseudogenes (eight ndh gene and trnV-UAC). Three genes (accD, ndhJ, and ndhI) were absent in this chloroplast genome. Twenty-four genes were duplicated in the IR, including eight protein-coding genes, four rRNA genes, eight tRNA genes, and four pseudogenized ndh genes. The GC content of the complete chloroplast genome, LSC, IR, and SSC regions were 40.93%, 39.85%, 42.17%, and 34.38%, respectively. The ML and BI analyses both revealed that C. inopinata belonged to Fumarioideae (Papaveraceae) clade, and C. inopinata was closely related to C. conspersa Maxim. with full support in the present sampling (Figure 1).
The complete plastid genome provided in this work would be useful for elucidating the taxonomy, phylogeny, and evolution of C. inopinata, and providing molecular data for future research in Corydalis as well.
Funding Statement
The work was supported by Grants from the National Natural Science Foundation of China [31170310], Science and Technology Basic Work [2013FY112100, 2013FY112300], the Special Foundation for the Specimen Platform of China, Teaching Specimen Sub-Platform, Web, http://mnh.scu.edu.cn/ [2005DKA21403-JK], and Fundamental Research Funds for the Central Universities [Excellent Doctoral Dissertation Development Program of CCNU, 202050185085].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in National Center for Biotechnology Information (NCBI) at https://www.ncbi.nlm.nih.gov, reference number MT755641.
References
- Chen SF, Zhou YQ, Chen YR, Gu J.. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34(17):i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL.. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15. [Google Scholar]
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Dz L.. 2018. GetOrganelle: a simple and fast pipeline for de novo assemble of a complete circular chloroplast genome using genome skimming data. bioRxiv. 256479. [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu XJ, Moore MJ, Li DZ, Yi TS.. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Teslenk M, Van der Mark P, Ayres D, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP.. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu ZY, Zhuang X, Su ZY.. 1996. The systematic evolution of Corydalis in relation to florogenesis and floristic regionalization in the world. Acta Bot Yunnan. 18:241–267. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are openly available in National Center for Biotechnology Information (NCBI) at https://www.ncbi.nlm.nih.gov, reference number MT755641.