Abstract
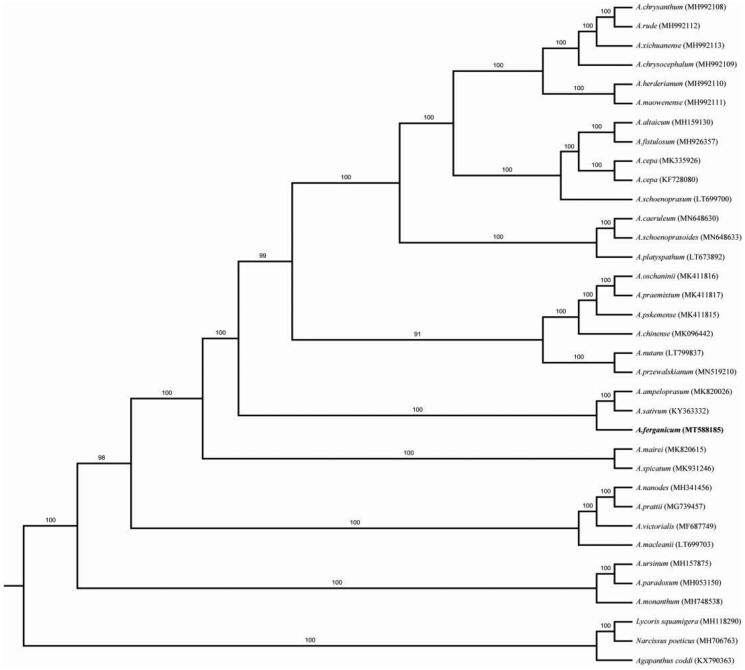
The complete chloroplast (cp) genome of Allium ferganicum was sequenced and annotated. The whole chloroplast genome consists of 153,126 bp with a typical quadripartite structure separated by a pair of 26,556 bp inverted repeat (IR) regions. The structure also includes the large single copy (LSC) − 81,982 bp and the small single copy (SSC) − 18,033 bp. The A. ferganicum chloroplast genome encodes 114 unique genes including 80 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. The phylogenetic trees for 35 plastomes genomes showed that A. ferganicum is closely related to A. sativum (garlic) and A. ampeloprasum (leek).
Keywords: Allium, chloroplast genome, phylogenetic analysis
Species of subgenus Allium (Amaryllidaceae) are economically important plants in the genus Allium L. in the world, such as leek and garlic, which accounts about 40% (375 species) of the total number species of the genus (Khassanov 2018). Allium ferganicum Vved. is an endemic member of subg. Allium of Fergana Valley (Eastern Uzbekistan and partly in Tajikistan and Kyrgyzstan). The A. ferganicum grows from sea level to about 500 − 800 m altitudes in gypsum-bearing strata. (Khassanov 2017). In this study, A. ferganicum cp genome of A.ferganicum was successfully assembled and annotated, and its relationship with closely related species (A. sativum and A. ampeloprasum) were investigated.
The fresh leaves of A. ferganicum were collected from Akchop hills, near the Kairakum reservoir, Sultonobod village, Bobojon Gofurov region, Sugd province, Tajikistan (E69°92′86″, N40°31′78″). Voucher specimen has been deposited at Kunming Institute of Botany (ZD0635). Total DNA was extracted from 100 mg of fresh leaves using the modified CTAB method (Doyle and Doyle 1987), then genomic DNA was fragmented by sonication to a size of 350 bp. These libraries constructed above were sequenced by Illumina HiSeq4000 and 150 bp paired-end reads were generated with insert size around 350 bp at Beijing Novogene Bioinformatics Technology Co., Ltd, Beijing, China. The plastid genome was assembled using as a reference Allium fistulosum (MH926357, Yusupov et al. 2019) by software NovoPlasty version 3.8.3 (Dierckxsens et al. 2017). The cp genome of A. ferganicum was annotated using Geneious v10.2 (Kearse et al. 2012). The annotated chloroplast genome of A. ferganicum has been deposited into the GenBank with the Accession number MT588185. RAxML-HPC BlackBox v8.1.24 software (Stamatakis 2006) was used to conduct for maximum likelihood (ML). The phylogenetic trees for 35 plastomes genomes showed that A. ferganicum is closely related to A. sativum (garlic) and A. ampeloprasum (leek) (Figure 1).
Figure 1.
Phylogenetic analysis of A. fistulosum with 35 related species. Numbers in the nodes are the bootstrap values from 1000 replicates.
The complete cp genome sequence of A. ferganicum is 153,126 bp in length and exhibits a typical quadripartite structure, including a pair of IRs (26,556 bp, GC-42.6% for each) separated by the LSC and SSC regions (LSC: 81,982 bp, GC: 34.5%; SSC: 18,033 bp, GC: 29.1%). The A. ferganicum chloroplast genome encodes 114 unique genes including 80 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. Among these, four genes (rps2, rps16 and ycf15, infA) were pseudogenes. In the plastome of A. ampeloprasum the infA gene is absent, unlike the plastome of A. sativum and A. ferganicum. This plastome sequence of A. ferganicum will provide useful plastid genomic resources for population genetics and can be used for phylogenetic analyses of genus Allium.
Funding Statement
This study was supported by the Fund for Reserve Talents of Young and Middle-aged Academic and Technical Leaders of Yunnan Province [Grant 2014HB027], the Scientific Research Fund Project of Education Department of Yunnan Province [2019J0143] the CAS “Light of West China” Program, the National Natural Science Foundation of China [Grant 31360456, 31060252], and the Fund for Science and Technology Plan of Puer City [Science and technology bureau Puer city, No. 2014-38].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are available in [GenBank] at [https://www.ncbi.nlm.nih.gov/genbank], reference number [MT588185].
References
- Dierckxsens N, Mardulyn P, Smits G.. 2017. NOVOPlasty:de novoassembly of organelle genomes from whole genome data. Nucleic Acids Res,45(4):e18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL.. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15. [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. . 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khassanov FO. 2017. Allium L. In: Sennikov A, editor. Flora of Uzbekistan, Vol. 1; p. 67–68. [Google Scholar]
- Khassanov FO. 2018. Taxonomical and ethnobotanical aspects of Allium species from middle Asia with particular reference to subgenus Allium. In: Shigyo M, Khar A, Abdelrahman M, editors. The Allium genomes. Compendium of plant genomes. Cham: Springer, 11-21. [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690. [DOI] [PubMed] [Google Scholar]
- Yusupov Z, Deng T, Liu C, Lin N, Tojibaev K, Sun H.. 2019. The complete chloroplast genome of Allium fistulosum. Mitochondrial DNA Part B: Resources. 4(1):449–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available in [GenBank] at [https://www.ncbi.nlm.nih.gov/genbank], reference number [MT588185].