Abstract
Apocynum venetum (A. venetum) has high medicinal value that belongs to the family Apocynaceae. Here, we reported the complete chloroplast (cp) genome of A. venetum, which was 150,858 bp in length. The cp genome was characterized by a typical quadripartite structure composed of a large single-copy region (LSC 81,919 bp) and a small single-copy region (SSC 17,257 bp) interspersed by a pair of 25,841 bp inverted repeat regions (IRs), and it contained 86 protein-coding genes, eight rRNAs, and 37 tRNAs. A maximum-likelihood (ML) phylogenetic tree indicated that A. venetum was closely related to Trachelospermum jasminoides.
Keywords: Apocynum venetum, chloroplast genome
Apocynum venetum is a wild plant of subshrub that belongs to the family Apocynaceae. It has a long history as a Chinese traditional medicine with uses to calm the liver, soothe the nerves, dissipate heat, and promote diuresis (Xie et al. 2012). Moreover, research on A. venetum showed that it has antidepressant potential and can reduce anxiety (Grundmann et al. 2007, 2009). To generate the genomic resources of this medicinal plant, we assembled and characterized the complete cp genome of A. venetum. This study will provide valuable resources for species identification, molecular biology, and phylogenetic studies of A. venetum.
Young fresh leaves of A. venetum were collected from Tianjin City (CHN, 38.96°N, 117.06°E). The voucher specimens were deposited in Tianjin State Key Laboratory of Modern Chinese Medicine, Tianjin University of Traditional Chinese Medicine, under the voucher number of LBM20200512. DNA isolation was carried out according to the standard protocol of Extract Genomic DNA Kit. Then, the libraries were sequenced with 2 × 150 bp on the Illumina HiSeq X Ten platform. The generated clean reads were used to assemble the cp genome of A. venetum using NOVOPlasty3.7.2 (Dierckxsens et al. 2017) with kmer length of 39. The cp genome of A. venetum was annotated using GeSeq (Tillich et al. 2017) coupled with manual correction for start and stop codons of protein-coding genes. Finally, the complete cp genome was submitted to GenBank under accession number MT547772.
The cp genome of A. venetum was 150,858 bp in length, including an LSC region of 81,919 bp, an SSC region of 17,257 bp, and a pair of IRs regions of 25,841 bp. Additionally, a total of 131 genes were annotated, including 86 protein-coding genes, eight rRNA genes, and 37 tRNA genes. Overall, the cp genome features of A. venetum were consistent with other angiosperms in terms of genomic structure (Cui et al. 2019; Yan et al. 2019; Yang et al. 2019; Yu et al. 2019).
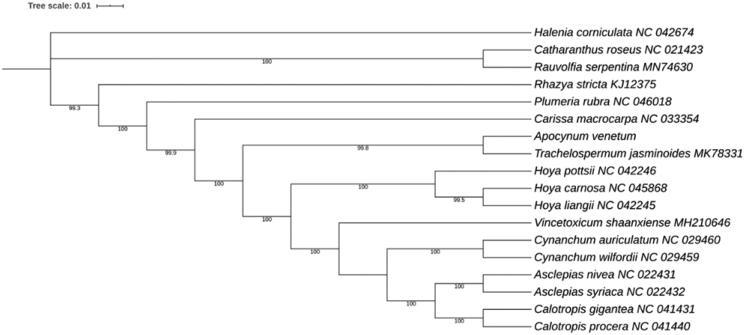
To investigate the phylogenetic position of A. venetum in family Apocynaceae, A. venetum and other 16 Apocynaceae cp genome sequences available in GenBank were used to reconstruct the ML phylogenetic tree. The protein-coding genes were extracted, aligned separately, and recombined to construct a matrix using PhyloSuite_v1.1.15 (Zhang et al. 2020). Then, the matrix was used to conduct ML analyses. The ML tree was constructed using IQ-TREE (Nguyen et al. 2015) under the model automatically selected by IQ-TREE. The result indicated that A. venetum was closely related to Trachelospermum jasminoides (Figure 1).
Figure 1.
ML phylogenetic tree of Apocynaceae inferred from protein-coding genes of cp genomes. The Halenia corniculata was set as the outgroup. The numbers above the lines were bootstrap support values for ML analyses.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study is openly available in Genbank at https://www.ncbi.nlm.nih.gov/genbank/, reference number MT547772.
References
- Cui Y, Nie L, Sun W, Xu Z, Wang Y, Yu J, Song J, Yao H.. 2019. Comparative and phylogenetic analyses of ginger (Zingiber officinale) in the family Zingiberaceae based on the complete chloroplast genome. Plants (Basel). 8(8):283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G.. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundmann O, Nakajima J, Kamata K, Seo S, Butterweck V.. 2009. Kaempferol from the leaves of Apocynum venetum possesses anxiolytic activities in the elevated plus maze test in mice. Phytomedicine. 16(4):295–302. [DOI] [PubMed] [Google Scholar]
- Grundmann O, Nakajima J, Seo S, Butterweck V.. 2007. Anti-anxiety effects of Apocynum venetum L. in the elevated plus maze test. J Ethnopharmacol. 110(3):406–411. [DOI] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S.. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Zhang X, Wang T, Hu J.. 2012. Botany, traditional uses, phytochemistry and pharmacology of Apocynum venetum L. (Luobuma): a review. J Ethnopharmacol. 141(1):1–8. [DOI] [PubMed] [Google Scholar]
- Yan M, Zhao X, Zhou J, Huo Y, Ding Y, Yuan Z.. 2019. The complete chloroplast genomes of Punica granatum and a comparison with other species in Lythraceae. Int J Mol Sci. 20(12):2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Wang G, Ma Q, Ma W, Liang L, Zhao T.. 2019. The complete chloroplast genomes of three Betulaceae species: implications for molecular phylogeny and historical biogeography. PeerJ. 7:e6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Tan W, Zhang H, Gao H, Wang W, Tian X.. 2019. Complete chloroplast genomes of Ampelopsis humulifolia and Ampelopsis japonica: molecular structure, comparative analysis, and phylogenetic analysis. Plants (Basel). 8(10):410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Gao F, Jakovlic I, Zou H, Zhang J, Li WX, Wang GT.. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study is openly available in Genbank at https://www.ncbi.nlm.nih.gov/genbank/, reference number MT547772.