Abstract
Ampelopsis grossedentata (Hand.-Mazz.) W. T. Wang is rich in flavonoids and also displays excellent pharmacological activities. The phylogenetic relationship between A. grossedentata and other related Vitaceae family members remains unclear. The chloroplast (cp) genome is a useful model for assessing genome evolution. In this study, we assembled the cp genome of A. grossedentata using the high-throughput Illumina pair-end sequencing data and characterized the genome to providing useful information for future genetic studies. The circular cp genome was 162,147 bp in size, including a large single-copy (LSC) region of 89,244 bp and a small single-copy (SSC) region of 18,439 bp, which were separated by two inverted repeat (IR) regions (27,232 bp each). A total of 135 genes were predicted, including 8 ribosomal RNAs (rRNAs), 37 transfer RNAs (tRNAs), and 90 protein-coding genes (PCGs). Furthermore, phylogenetic analysis revealed that A. grossedentata within Ampelopsis genus and formed a different clade from other three congeneric species. This study provides useful information for future genetic study of A. grossedentata.
Keywords: Ampelopsis grossedentata, Vitaceae, complete chloroplast genome, phylogenetic
Ampelopsis grossedentata (Hand.-Mazz.) W. T. Wang, being classified into Vitaceae family, is widely growing in mountainous areas of southern China. The dried leaves and stems of A. grossedentata, also named vine tea, have extremely high health-promoting benefits due to its strong anti-oxidant (Gao et al. 2017; Ma et al. 2017), anti-inflammatory (Hou et al. 2015), and anti-tumor (Zhou et al. 2014) activities. Although A. grossedentata has received much attention, the complete chloroplast (cp) genome of A. grossedentata has not been reported. In this study, we assembled and determined the cp genome sequence of A. grossedentata as a resource for future genetic, genomic, breeding research.
Young and healthy leaf samples were collected from the A. grossedentata germplasm resource repository of Guizhou Normal University (26°22′51.06″N, 106°38′11.80″E, 1100.5 m above sea level) (Guiyang, Guizhou Province, China). The leaf specimen (accession number: GZNUYZW202001001) was deposited in the herbarium of School of Life Sciences, Guizhou Normal University. The total genomic DNA (No. YZW202001002) was extracted using DNAsecure Plant Kit (TIANGEN, Beijing) and stored at −80 °C in the laboratory (room number: 1403) of School of Life Science, Guizhou Normal University. A total amount of 700 ng DNA per sample was used as input material for the DNA sample preparations. Sequencing libraries were generated using NEB Next® Ultra DNA Library Prep Kit for Illumina® (NEB, USA) following manufacturer’s recommendations. The library preparations were sequenced on an Illumina platform and 150 bp paired-end reads were generated. After removal of adapter sequences, the filtered reads were assembled using the program GetOrganelle (Jin et al. 2019). The assembled cp genome was annotated using the online software GeSeq (https://chlorobox.mpimp-golm.mpg.de/geseq.html) (Tillich et al. 2017). The accurate annotated complete cp genome was submitted to GenBank with accession number MT267294.
The length of the complete cp genome sequence of A. grossedentata is 162,147 bp, consisting of a large single-copy (LSC, 89,244 bp) region, a small single-copy (SSC, 18,439 bp) region and two inverted repeat (IRA and IRB) regions of 27,232 bp each. Totally, 135 genes were predicted, including 90 protein coding genes (PCGs), 8 rRNA genes and 37 tRNA genes. Among of these assembled genes, 4 rRNAs (rrn16, rrn23, rrn4.5 and rrn5), 7 PCGs (rps7, rps19, rpl2, rpl23, ndhB, ycf2 and ycf15), and 7 tRNAs (trnA-UGC, trnI-CAU, trnI-GAU, trnLCAA, trnN-GUU, trnR-ACG and trnV-GAC) with double copies. 1 PCG (rps12) occur in three copies. Intron-exon analysis showed the majority (116 genes, 86%) genes with no introns, whereas 19 (14%) genes contain introns.
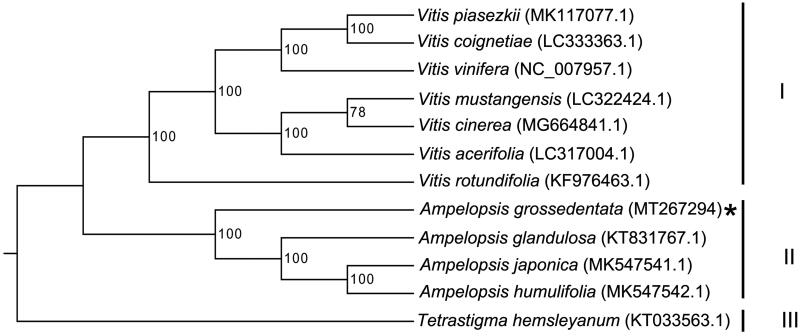
To further understand the chloroplast genome of A. grossedentata, 20 cp genome sequences of Vitaceae family (16 Vitis species, 3 species from Ampelopsis genus, and 1 specie from Tetrastigma genus) were downloaded from GenBank to construct the phylogenetic trees through maximum-likelihood (ML) analysis. The ML tree was performed using RAxML (Version 8.0.19, Model: GTRGAMMA) (Stamatakis 2014) with 1000 bootstrap replicates (confirm the stability of each tree node). The phylogenetic tree indicated that A. grossedentata belongs to Ampelopsis genus and located in a different clade from other three congeneric species (Figure 1). Compared to Tetrastigma hemsleyanum, a member of Tetrastigma genus, A. grossedentata was closely related to Vitis species (Figure 1).
Figure 1.
Maximum likelihood tree based on the complete cp genome sequences of 12 species from the Vitaceae family shows the three distinct clusters. GenBank accession numbers are described in the figure. Shown next to the nodes are bootstrap support values based on 1000 replicates.
Funding Statement
This study was supported by the National Natural Science Foundation of China [U1812401, 31460068], Natural Science Research Project Education Department Guizhou Province [QKHKY [2015]335], Talent Development of the Department of Science and Technology of Guizhou China (Grant No. [2019]5617), Guizhou Provincial Science and Technology Foundation [Grant No. 2020-1Y096], and The Doctoral Foundation of Guizhou Normal University [GZNUD [2018]-6].
Data availability statement
The data (the accurate annotated complete cp genome of A. grossedentata) that support the findings of this study are submitted to public database (NCBI GenBank, https://www.ncbi.nlm.nih.gov/) with GenBank accession number MT267294.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Gao Q, Ma R, Chen L, Shi S, Cai P, Zhang S, Xiang H.. 2017. Antioxidant profiling of vine tea (Ampelopsis grossedentata): Off-line coupling heart-cutting HSCCC with HPLC-DAD-QTOF-MS/MS. Food Chem. 225:55–61. [DOI] [PubMed] [Google Scholar]
- Hou XL, Tong Q, Wang WQ, Shi CY, Xiong W, Chen J, Liu X, Fang JG.. 2015. Suppression of inflammatory responses by dihydromyricetin, a flavonoid from ampelopsis grossedentata, via inhibiting the activation of NF-κB and MAPK signaling pathways. J Nat Prod. 78(7):1689–1696. [DOI] [PubMed] [Google Scholar]
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ.. 2019. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. BioRxiv. 256479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma R, Zhou R, Tong R, Shi S, Chen X.. 2017. At-line hyphenation of high-speed countercurrent chromatography with Sephadex LH-20 column chromatography for bioassay-guided separation of antioxidants from vine tea (Ampelopsis grossedentata). J Chromatogr B Analyt Technol Biomed Life Sci. 1040:112–117. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S.. 2017. GeSeq- versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Shu F, Liang X, Chang H, Shi L, Peng X, Zhu J, Mi M.. 2014. Ampelopsin induces cell growth inhibition and apoptosis in breast cancer cells through ROS generation and endoplasmic reticulum stress pathway. PLOS One. 9(2):e89021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data (the accurate annotated complete cp genome of A. grossedentata) that support the findings of this study are submitted to public database (NCBI GenBank, https://www.ncbi.nlm.nih.gov/) with GenBank accession number MT267294.