Abstract
In the present study, the complete mitochondrial genome of a basidiomycetous yeast Cystobasidium sp. was assembled and obtained. The mitochondrial genome of Cystobasidium sp. contains 16 protein-coding genes, 2 ribosomal RNA genes (rRNA), and 24 transfer RNA (tRNA) genes. The complete mitogenome of Cystobasidium sp. has a total length of 24,914 bp, with the base composition as follows: A (30.82), T (32.88%), C (18.37%) and G (17.93%). The Cystobasidium sp. mitogenome exhibited a close relationship with the mitogenome of Microbotryum cf. violaceum, M. lychnidis-dioicae, and Rhodotorula mucilaginosa.
Keywords: Cystobasidium, basidiomycetous yeast, mitochondrial genome, phylogenetic analysis
The genus Cystobasidium belongs to Cystobasidiaceae, Cystobasidiales, which was first emended by Yurkov et al. (2015) to accommodate a group of closely related asexual species. Cystobasidium species is often pink pigmented, with ovoidal to elongate cells and polar budding (Turchetti et al. 2018). Species of this genus were reported to have biotransformation capacities (Vyas and Chhabra 2017; Tanimura et al. 2018). So far, about 16 species have been described in this genus (Chang et al. 2019). The complete mitochondrial genome of Cystobasidium sp. will promote the understanding of the phylogeny and evolution of the basidiomycetous yeast.
The specimen (Cystobasidium sp.) was isolated from a water sample collected in Chengdu, Sichuan, China (103.51E; 30.42N). The specimen was stored in Culture Collection Center of Chengdu Medical College (No. Cys03). The total genomic DNA of Cystobasidium sp. was extracted using Fungal DNA Kit D3390-00 (Omega Bio-Tek, Norcross, GA). Extracted DNA was purified using a Gel Extraction Kit (Omega Bio-Tek, Norcross, GA), and then stored in the Chengdu Medical College (No. DNA_ Cys03). Sequencing libraries were constructed with purified genomic DNA using the NEBNext® Ultra™ II DNA Library Prep Kit (NEB, Beijing, China). Whole genomic sequencing (WGS) was performed by the Illumina HiSeq 2500 Platform (Illumina, San Diego, CA). The complete mitogenome was de novo assembled as implemented by SPAdes 3.9.0 (Bankevich et al. 2012). Gaps among contigs were filled by using MITObim V1.9 (Hahn et al. 2013). The obtained mitogenome was annotated according to the methods described by Li, Chen, et al. (2018), Li, Liao, et al. (2018), Li, Wang, et al. (2018).
The circular mitogenome of Cystobasidium sp. is 24,914 bp in size. The complete mitogenome contains 16 protein-coding genes, 2 ribosomal RNA genes (rns and rnl), and 24 transfer RNA (tRNA) genes. The base composition of the genome is as follows: A (30.82), T (32.88%), C (18.37%) and G (17.93%).
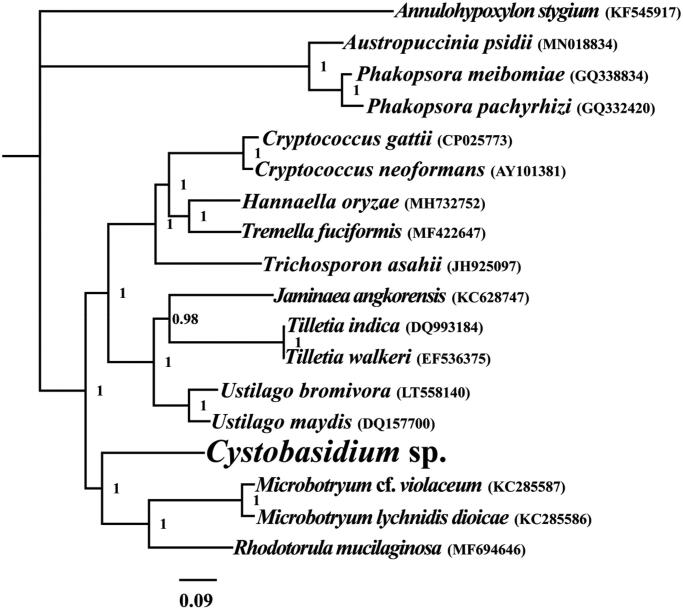
To investigate the phylogenetic status of Cystobasidium sp., we constructed phylogenetic trees of 18 species. Phylogenetic tree was constructed using Bayesian analysis (BI) method based on the combined 14 core protein-coding genes according to Li, Wang, Jin, Chen, Xiong, Li, Liu, et al. (2019), Li, Wang, Jin, Chen, Xiong, Li, Zhao, et al. (2019), Li, Yang, et al. (2020). As shown in the phylogenetic tree (Figure 1), the taxonomic status of the Cystobasidium sp. mitogenome exhibited a close relationship with the mitogenomes of Microbotryum cf. violaceum (KC285587), M. lychnidis-dioicae (KC285586), and Rhodotorula mucilaginosa (Gan et al. 2017).
Figure 1.
Bayesian phylogenetic analysis of 18 species based on the combined 14 core protein-coding genes. Accession numbers of mitochondrial sequences used in the phylogenetic analysis are listed in brackets after species.
Funding Statement
This study was supported by the National Natural Science Foundation of China [81301145], Sichuan Research Center for Applied Psychology [CSXL-192A11], and the Sichuan province College Students’ Innovation and Entrepreneurship [S201913705053 and S201913705064].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
This mitogenome of Cystobasidium sp. was submitted to GenBank under the accession of MT366950 (https://www.ncbi.nlm.nih.gov/nuccore/MT366950).
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. . 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CF, Lee CF, Liu SM.. 2019. Cystobasidium keelungensis sp. nov., a novel mycosporine producing carotenogenic yeast isolated from the sea surface microlayer in Taiwan. Arch Microbiol. 201(1):27–33. [DOI] [PubMed] [Google Scholar]
- Gan HM, Thomas BN, Cavanaugh NT, Morales GH, Mayers AN, Savka MA, Hudson AO.. 2017. Whole genome sequencing of Rhodotorula mucilaginosa isolated from the chewing stick (Distemonanthus benthamianus): insights into Rhodotorula phylogeny, mitogenome dynamics and carotenoid biosynthesis. PeerJ. 5:e4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn C, Bachmann L, Chevreux B.. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads–a baiting and iterative mapping approach. *Nucleic Acids Res. 41(13):e129–e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Chen C, Xiong C, Jin X, Chen Z, Huang W.. 2018. Comparative mitogenomics reveals large-scale gene rearrangements in the mitochondrial genome of two Pleurotus species. Appl Microbiol Biotechnol. 102(14):6143–6153. [DOI] [PubMed] [Google Scholar]
- Li Q, Liao M, Yang M, Xiong C, Jin X, Chen Z, Huang W.. 2018. Characterization of the mitochondrial genomes of three species in the ectomycorrhizal genus Cantharellus and phylogeny of Agaricomycetes. Int J Biol Macromol. 118(Pt A):756–769. [DOI] [PubMed] [Google Scholar]
- Li Q, Wang Q, Chen C, Jin X, Chen Z, Xiong C, Li P, Zhao J, Huang W.. 2018. Characterization and comparative mitogenomic analysis of six newly sequenced mitochondrial genomes from ectomycorrhizal fungi (Russula) and phylogenetic analysis of the Agaricomycetes. Int J Biol Macromol. 119:792–802. [DOI] [PubMed] [Google Scholar]
- Li Q, Wang Q, Jin X, Chen Z, Xiong C, Li P, Liu Q, Huang W.. 2019. Characterization and comparative analysis of six complete mitochondrial genomes from ectomycorrhizal fungi of the Lactarius genus and phylogenetic analysis of the Agaricomycetes. Int J Biol Macromol. 121:249–260. [DOI] [PubMed] [Google Scholar]
- Li Q, Wang Q, Jin X, Chen Z, Xiong C, Li P, Zhao J, Huang W.. 2019. Characterization and comparison of the mitochondrial genomes from two Lyophyllum fungal species and insights into phylogeny of Agaricomycetes. Int J Biol Macromol. 121:364–372. [DOI] [PubMed] [Google Scholar]
- Li Q, Yang L, Xiang D, Wan Y, Wu Q, Huang W, Zhao G.. 2020. The complete mitochondrial genomes of two model ectomycorrhizal fungi (Laccaria): features, intron dynamics and phylogenetic implications. Int J Biol Macromol. 145:974–984. [DOI] [PubMed] [Google Scholar]
- Tanimura A, Sugita T, Endoh R, Ohkuma M, Kishino S, Ogawa J, Shima J, Takashima M.. 2018. Lipid production via simultaneous conversion of glucose and xylose by a novel yeast, Cystobasidium iriomotense. PLoS One. 13(9):e0202164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchetti B, Selbmann L, Gunde-Cimerman N, Buzzini P, Sampaio JP, Zalar P.. 2018. Cystobasidium alpinum sp. nov. and Rhodosporidiobolus oreadorum sp. nov. from European cold environments and Arctic region. Life. 8(2):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas S, Chhabra M.. 2017. Isolation, identification and characterization of Cystobasidium oligophagum JRC1: a cellulase and lipase producing oleaginous yeast. Bioresour Technol. 223:250–258. [DOI] [PubMed] [Google Scholar]
- Yurkov AM, Kachalkin AV, Daniel HM, Groenewald M, Libkind D, de Garcia V, Zalar P, Gouliamova DE, Boekhout T, Begerow D.. 2015. Two yeast species Cystobasidium psychroaquaticum f.a. sp. nov. and Cystobasidium rietchieii f.a. sp. nov. isolated from natural environments, and the transfer of Rhodotorula minuta clade members to the genus Cystobasidium. Antonie Van Leeuwenhoek. 107(1):173–185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This mitogenome of Cystobasidium sp. was submitted to GenBank under the accession of MT366950 (https://www.ncbi.nlm.nih.gov/nuccore/MT366950).