Abstract
Correspondence between evolution and development has been discussed for more than two centuries. Recent work reveals that phylogeny−ontogeny correlations are indeed present in developmental transcriptomes of eukaryotic clades with complex multicellularity. Nevertheless, it has been largely ignored that the pervasive presence of phylogeny−ontogeny correlations is a hallmark of development in eukaryotes. This perspective opens a possibility to look for similar parallelisms in biological settings where developmental logic and multicellular complexity are more obscure. For instance, it has been increasingly recognized that multicellular behavior underlies biofilm formation in bacteria. However, it remains unclear whether bacterial biofilm growth shares some basic principles with development in complex eukaryotes. Here we show that the ontogeny of growing Bacillus subtilis biofilms recapitulates phylogeny at the expression level. Using time-resolved transcriptome and proteome profiles, we found that biofilm ontogeny correlates with the evolutionary measures, in a way that evolutionary younger and more diverged genes were increasingly expressed toward later timepoints of biofilm growth. Molecular and morphological signatures also revealed that biofilm growth is highly regulated and organized into discrete ontogenetic stages, analogous to those of eukaryotic embryos. Together, this suggests that biofilm formation in Bacillus is a bona fide developmental process comparable to organismal development in animals, plants, and fungi. Given that most cells on Earth reside in the form of biofilms and that biofilms represent the oldest known fossils, we anticipate that the widely adopted vision of the first life as a single-cell and free-living organism needs rethinking.
Keywords: biofilms, Bacillus, evo-devo, phylogeny−ontogeny correlations, transcriptome, proteome
Introduction
Multicellular behavior is wide-spread in bacteria and it was proposed that they should be considered multicellular organisms (Shapiro 1998). However, this idea has not been generally adopted likely due to the widespread laboratory use of domesticated bacterial models selected against multicellular behaviors, the long tradition of viewing early diverging groups as simple, and the lack of evidence for system-level commonalities between bacteria and multicellular eukaryotes (Claessen et al. 2014; van Gestel et al. 2015a; Lyons and Kolter 2015; O’Malley et al. 2016). Recently developed phylo-transcriptomic tools for tracking evolutionary signatures in animal development (Domazet-Lošo and Tautz 2010a; Kalinka et al. 2010; Irie and Kuratani 2011) were also successfully applied in the analysis of developmental processes in plants and fungi (Quint et al. 2012; Cheng et al. 2015; Drost et al. 2017). Although development evolved independently in these three major branches of eukaryotic diversity (Rensing 2016), their ontogenies showed similar phylogeny−ontogeny correlations indicating that possibly all eukaryotic developmental programs have an evolutionary imprint. Transferability of the phylo-transcriptomic tools across clades and likely universal patterns of phylogeny−ontogeny correlations in eukaryotic multicellularity prompted us to test this approach on bacterial biofilms—a multicellular, and the most common, form of bacterial existence in nature (Flemming and Wuertz 2019).
Bacillus subtilis NCIB3610, a frequently used model organism in bacterial biofilm research (Vlamakis et al. 2013), shows many properties readily found in multicellular eukaryotes including cell differentiation, division of labor, cell signaling, morphogenesis, programmed cell death, and self-recognition (Vlamakis et al. 2013; van Gestel et al. 2015a; Kalamara et al. 2018). These properties of B. subtilis biofilms are often seen as collective behaviors of independent cells. Alternatively, one can also take a top-down view and look at these characteristics as organizing features of a multicellular individual. We took the latter perspective and approached the biofilm as an individual organism by applying phylo-transcriptomics as it would be used for studying ontogeny of an embryo-forming eukaryote. In this respect, we looked for a suitable genome-wise expression data set that covers the full ontogeny of a biofilm growth with relatively dense temporal sampling of entire biofilms. Surprisingly, there is general scarcity of such longitudinal biofilm studies in any bacterial species, and those that exist (e.g., Pisithkul et al. 2019) cover only the initial phase of biofilm growth. We therefore generated a new expression data set by sampling B. subtilis NCIB3610 biofilms grown on a solid−air interface.
Results
Biofilm Growth is a Stage-Organized Process
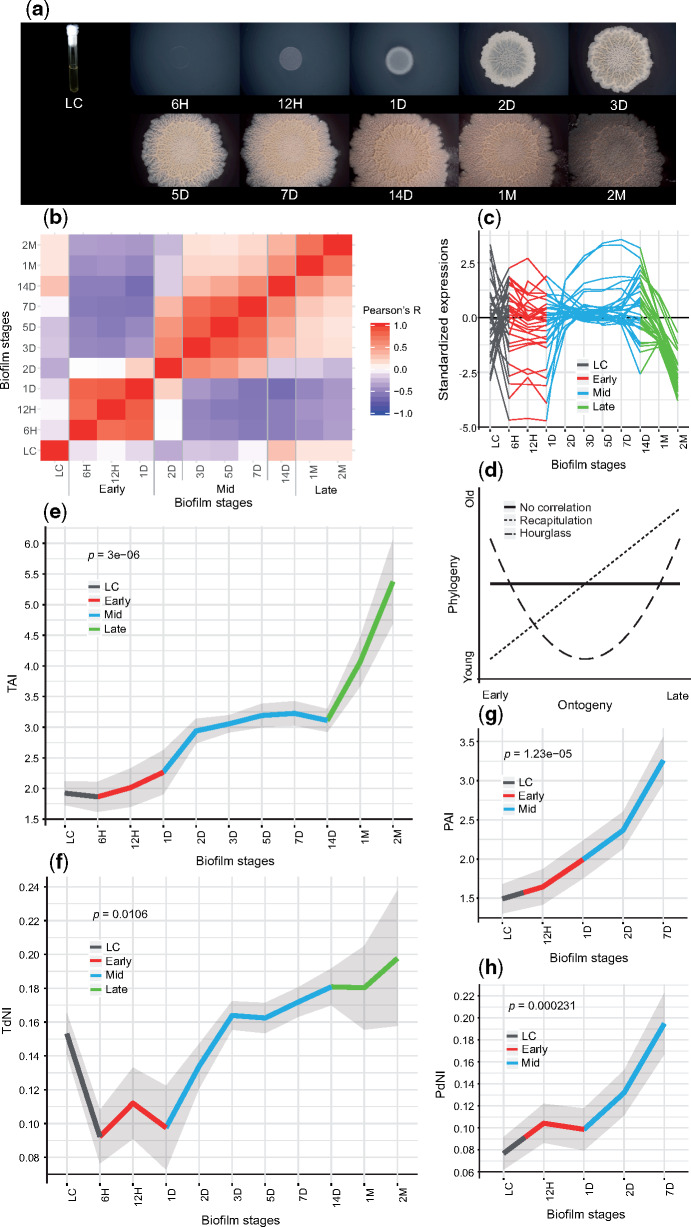
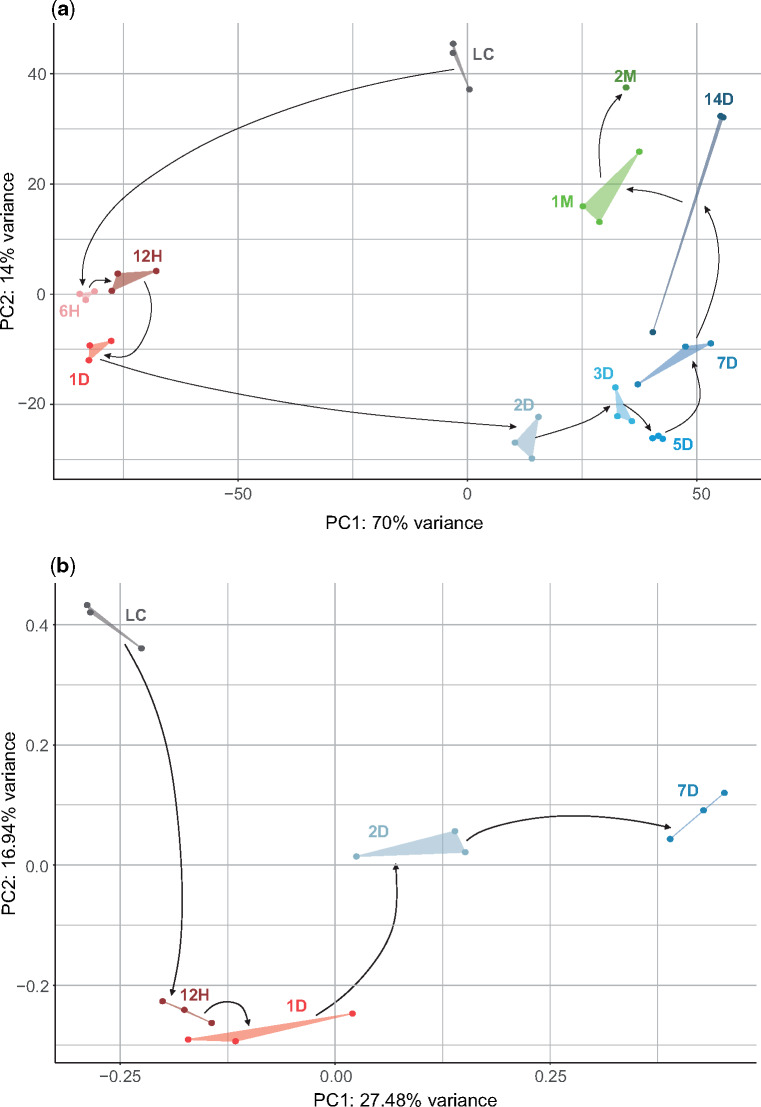
To measure transcriptome expression levels during B. subtilis biofilm formation, we sampled 11 timepoints covering a full span of biofilm ontogeny from its inoculation, until 2 months of age (fig. 1a). We recovered transcript expression values for 4,316 (96%) B. subtilis genes by RNAseq, which revealed three distinct periods of biofilm ontogeny: early (6H−1D), mid (3D−7D), and late period (1M−2M), linked by 2 transition stages at 2D and 14D (figs. 1b and c and 2; supplementary file S1, Supplementary Material online). Biofilm transcriptomes showed a time-resolved principal component analysis (PCA) profile (fig. 2) and poor correlation to the liquid culture (LC) used for inoculation of biofilms (fig. 1b), indicating that biofilm makes a distinct part of the B. subtilis life cycle. When we considered all ontogeny timepoints, 4,263 (99%) genes were differentially expressed (supplementary file S2, Supplementary Material online). This number stayed similar (4,190 genes, 97%) when we looked only at biofilm growth sensu stricto (6H−14D, supplementary files S2 and S3, Supplementary Material online) by excluding the starting LC and late-period timepoints (1M−2M) that show biofilm growth arrest. When we retained only genes with 2-fold or higher expression change, the numbers still remained high: 2,546 genes (59%) in biofilm growth sensu lato and 2,798 genes (65%) in biofilm growth sensu stricto. These values reflect highly dynamic regulation of transcription in biofilm ontogeny, comparable to those seen in animal embryos (Domazet-Lošo and Tautz 2010a; Yanai 2018). Pairwise comparisons between successive ontogeny timepoints uncovered that most genes (around 70%) change their transcription at biofilm inoculation (LC-6H), indicating that transition from a LC to solid agar plates represents a dramatic shift in B. subtilis lifestyle (supplementary file S4, Supplementary Material online). The two most dynamic steps during biofilm growth are transitions at 1D−2D and 7D−14D where, respectively, around 30% and 25% B. subtilis genes change transcription (supplementary file S4, Supplementary Material online). Together with correlation (fig. 1b), PCA (fig. 2) and clustering analyses (fig. 1c;supplementary file S5, Supplementary Material online), this shows that expression during biofilm growth is not a continuous process (Monds and O’Toole 2009). Instead, like development in animals (Levin et al. 2012; Yanai 2018), it is punctuated with bursts of transcriptional change that define discrete ontogeny phases (the early, mid, and late period).
Fig. 1.
Bacillus subtilis biofilm growth is a highly regulated and punctuated process exhibiting a phylogeny−ontogeny recapitulation pattern. (a) Gross morphology of B. subtilis biofilms on solid agar plates at 6 h (6H), 12 h (12H), 1 day (1D), 2 days (2D), 3 days (3D), 5 days (5D), 7 days (7D), 14 days (14D), 1 month (1M), and 2 months (2M) after inoculation with LC (see supplementary file S3, Supplementary Material online). (b) Pearson’s correlation coefficients between timepoints of biofilm ontogeny in all-against-all comparison. Early (6H−1D), mid (3D−7D), and late (1M−2M) periods, together with transition stages at 2D and 14D, are marked. (c) Average transcript expression profiles of the 31 most populated gene clusters (for all 64 clusters see supplementary file S5, Supplementary Material online). (d) Hypothetical profiles of phylogeny−ontogeny correlations. Solid line displays no correlation, dotted line the recapitulation model and dashed line the hourglass model. (e) Transcriptome age index (TAI) and (g) proteome age index (PAI) profiles of B. subtilis biofilm ontogeny show recapitulation pattern. (f) Transcriptome nonsynonymous divergence index (TdNI) and (h) proteome nonsynonymous divergence index (PdNI) profiles show that genes conserved at nonsynonymous sites are used early in the biofilm ontogeny, whereas more divergent ones later during biofilm ontogeny. Nonsynonymous divergence rates were estimated in B. subtilis–B. licheniformis comparison. Depicted P values are obtained by the flat line test and gray shaded areas represent ±one standard deviation estimated by permutation analysis (see Materials and Methods, e−h). Early (red), mid (blue), and late (green) periods of biofilm growth are color coded (c, e−h).
Fig. 2.
Principal component analysis (PCA) of transcriptomes and proteomes shows a punctuated organization of biofilm growth. PCA of (a) transcriptome and (b) proteome data (see Materials and Methods). Biofilm growth timepoints (LC, 6H, 12H, 1D, 2D, 3D, 5D, 7D, 14D, 1M, and 2M) are shown in different colors, where gray represents the liquid culture (LC), different shades of red early (6H−1D), blue mid (2D−14D), and green late (1M−2M) biofilm period. Replicates are in the same color and connected with lines. Black arrows correspond to the experimental timeline of biofilm growth that starts with LC and ends at 2M.
Evolutionary Expression Measures Show a Recapitulation Pattern
To assess whether biofilm growth has some evolutionary directionality, or if this process is macroevolutionary naive, we linked transcriptome profiles to evolutionary gene age estimates to obtain the transcriptome age index (TAI); a cumulative measure that gives overall evolutionary age of an expressed mRNA pool (Domazet-Lošo and Tautz 2010a; Quint et al. 2012; Cheng et al. 2015). Evolutionary gene age estimates are obtained by phylostratigraphic procedure (Domazet-Lošo et al. 2007) using consensus phylogeny (supplementary files S6−S8, Supplementary Material online). If one assumes that expression patterns across biofilm ontogeny are independent of evolutionary age of genes, then the TAI profile should show a trend close to a flat line; that is, TAI and ontogeny should not correlate (fig. 1d). Alternatively, there are many possible phylogeny−ontogeny correlation scenarios that would reflect a macroevolutionary imprint, but hourglass and recapitulation pattern are the two mostly considered (fig. 1d). It is important to note that the recapitulation term has historical burden linked to Ernst Haeckel who applied it in the morphological context to express the idea that the ontogeny of extant species advances through the ancestral adult forms (Olsson et al. 2017). This rigid view has been abandoned very early, but the recapitulation term has remained useful in discussing development (Abzhanov 2013, Olsson et al. 2017), especially at the molecular level to describe situations where, in statistical terms, the transcriptional activation of genes along ontogeny recapitulates the macroevolutionary sequence of gene emergence (Domazet-Lošo and Tautz 2010a). Although the recapitulation pattern historically was the first one to be proposed (Abzhanov 2013), recent studies of eukaryotic development mainly support the hourglass model (Domazet-Lošo and Tautz 2010a; Kalinka et al. 2010; Irie and Kuratani 2011; Kalinka and Tomancak 2012; Quint et al. 2012; Cheng et al. 2015; Drost et al. 2017; Hu et al. 2017). Surprisingly, in B. subtilis we found a recapitulation pattern where early timepoints of biofilm growth express evolutionary older transcriptomes compared with mid and late timepoints that exhibit increasingly younger transcriptomes (fig. 1e). This correlation between biofilm timepoints (ontogeny) and TAI (phylogeny) indicates that, like in complex eukaryotes, a macroevolutionary logic plays a role in B. subtilis biofilm formation. We also examined how the TAI profile relates to the evolutionary age of genes (phylostrata—ps) and found that recapitulation pattern is significant already from the origin of Firmicutes (ps4; supplementary file S9, Supplementary Material online), reflecting its rather deep roots in the bacterial phylogeny (supplementary file S6, Supplementary Material online).
Phylostratigraphic procedure used in determining gene ages is based on detecting the emergence of founder genes (Domazet-Lošo et al. 2007; Domazet-Lošo and Tautz 2010a) (supplementary files S6 and S8, Supplementary Material online). However, one could also analyze the data set by looking at more recent evolutionary history via estimating evolutionary divergence rates of coding sequences (Quint et al. 2012). It is usually assumed that nonsynonymous substitution rates (dN) reflect selective pressures, in contrast to synonymous substitution rates (dS) that provide an estimate of neutral evolution in coding sequences. However, due to the strong codon usage bias, selection also acts on synonymous sites in B. subtilis, and therefore its dS rates cannot be considered neutral (Sharp et al. 2005). To account for this, we looked at substitution rates separately by devising transcriptome nonsynonymous (TdNI) and synonymous (TdSI) divergence indices (see Materials and Methods). In B. subtilis−B. licheniformis comparison, from 1D onwards TdNI showed a recapitulation pattern where genes conserved at nonsynonymous sites tend to be used early, whereas more divergent ones are used later during the biofilm ontogeny (fig. 1f;supplementary file S10, Supplementary Material online). Comparably, TdSI displays more complex correlation which clearly resembles the pattern of the transcriptome codon bias index (TCBI) indicating dependence of synonymous substitution rates and codon usage bias (supplementary file S11a and c, Supplementary Material online). Nevertheless, TdSI recapitulation profile is evident in mid-period biofilms (1D−14D), where genes with more divergent synonymous sites gradually increase in transcription from 1D to 14D (supplementary file S11a, Supplementary Material online). Together, these divergence-ontogeny parallelisms in B. subtilis biofilms further corroborate the recapitulative evolutionary imprint and show that it is actively maintained by relatively recent evolutionary forces in mid-period biofilms.
Regulated mRNA transcription is the essential step in gene expression. However, a full molecular phenotype visible to selection is reached only after protein translation. In nonsteady state processes like ontogeny, a plethora of opposing factors influence mRNA and protein levels, resulting in their relatively low correlations (Liu et al. 2016). To test if the recapitulation pattern also exists at the proteome level, we quantified proteomes of representative stages (LC, 12H, 1D, 2D, 7D), which cover the most dynamic part of macroscopic morphology change (supplementary file S3, Supplementary Material online). We obtained protein expression values for 2,907 (67%) predicted proteins (supplementary file S1, Supplementary Material online) and used them to calculate the proteome age index (PAI); a cumulative measure analogous to TAI (see Materials and Methods) that gives an overall evolutionary age of a protein pool. Regardless of the relatively poor correspondence between transcriptome and proteome levels within timepoints (supplementary file S12a−e, Supplementary Material online), and across ontogeny (supplementary file S12f and g, Supplementary Material online), the PAI profile also showed a significant recapitulation pattern where evolutionary older proteins have higher expression early and younger ones later during biofilm ontogeny (fig. 1g). Similar to TdNI and TdSI for transcriptome, proteome nonsynonymous (PdNI; fig. 1h) and synonymous (PdSI; supplementary file S11b, Supplementary Material online) divergence indices in B. subtilis–B. licheniformis comparison revealed that recapitulation pattern also holds at shallower evolutionary levels (see Materials and Methods). Jointly, this demonstrates that phylogeny−ontogeny dependence, beside transcriptomes, is also visible in biofilm proteomes.
Multicellularity Important Genes Dominate in Mid-Period Biofilms
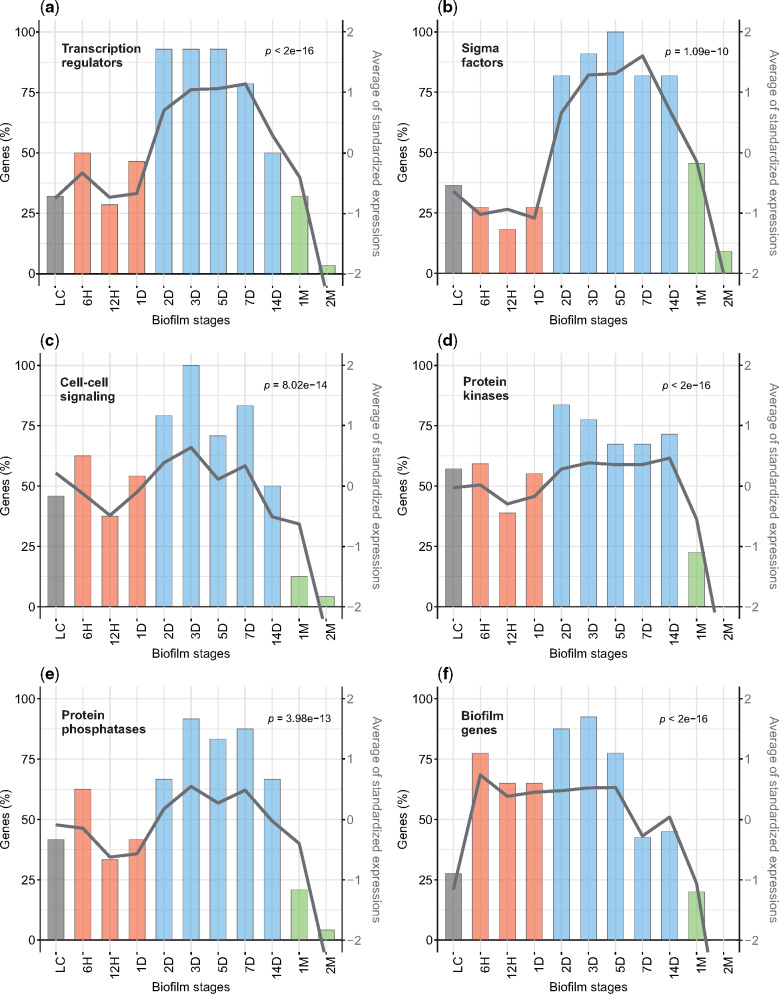
To find further parallels between biofilm growth and multicellular development, we looked for expression patterns of transcription factor and cell-cell signaling genes, which are defining features of development in complex eukaryotes (de Mendoza et al. 2013). We found that B. subtilis transcription regulators cumulatively have the highest transcription in mid-period biofilms (2D to 7D), and that during this period almost all of them are transcribed above the median of their overall expression profiles (fig. 3a). This holds even if we narrow down the analysis to sigma factors only (fig. 3b), and mimics developing transcriptomes in animals where embryos show increased expression of transcription factors (de Mendoza et al. 2013). Similarly, quorum sensing genes peak in transcription at 3D (fig. 3c), suggesting the most elaborate cell-cell communication at the timepoint when the biofilm gets the typical wrinkled morphology (fig. 1a;supplementary file S3, Supplementary Material online). Protein phosphorylation is another important mechanism involved in cell signaling and differentiation both in eukaryotes and bacteria, and it plays a critical role in B. subtilis biofilm formation (Vlamakis et al. 2013; van Gestel et al. 2015a; Kalamara et al. 2018). Again, we found that protein phosphorylation genes (kinases and phosphatases) cumulatively have the highest transcription in mid-period biofilms (fig. 3d and e), likely reflecting various types of cell differentiation in this growth phase (van Gestel et al. 2015a; Kalamara et al. 2018). Another feature of animal development is the enrichment of temporally pleiotropic genes in the mid-embryonic period (Hu et al. 2017). Following described methodology in vertebrates (Hu et al. 2017), we selected temporally pleiotropic genes in B. subtilis at different cutoffs and looked at their cumulative use during biofilm ontogeny (supplementary file S25a−c). We found that temporally pleiotropic genes tend to be expressed in the mid-period biofilms, additionally confirming the central role of mid-development in the growth of B. subtilis biofilms.
Fig. 3.
Multicellularity-important genes show cumulatively the strongest transcription in the mid-biofilm period. Left y-axis shows percentage of genes that are transcribed above the median of their overall transcription profile (histogram). Right y-axis shows the average standardized transcription values for all considered genes (line). Significance of the average expression profile is tested by repeated measures ANOVA and respective P values are shown. (a) Transcription regulators that regulate ≥10 operons (see Materials and Methods; n = 28, F(10, 270) = 17.33); (b) Sigma factors (n = 11, F(10, 100) = 9.257); (c) Cell to cell signaling genes (n = 24, F(10, 230) = 9.947); (d) Protein kinases (n = 49, F(10, 480) = 41.71); (e) Protein phosphatases (n = 24, F(10, 230) = 9.452); (f) Key biofilm genes (n = 40, F(10, 390) = 30.74). Coloring of bars in histograms follows biofilm growth periods: LC (gray), early (red), mid (blue), and late (green). See supplementary files S13 and S22, Supplementary Material online, for full profiles of all considered genes.
Functional annotation of B. subtilis genes, including the gene regulatory networks underlying biofilm formation (Vlamakis et al. 2013; van Gestel et al. 2015a; Kalamara et al. 2018), is comparably advanced in terms of quality and completeness (Zhu and Stülke 2018). This allowed us to follow the expression of key biofilm genes and analyze specific functional patterns in biofilm ontogeny (figs. 3f and 4). Collectively, the key biofilm genes are increasingly transcribed from the onset of biofilm formation (6H), maintain high values over early and mid-period, and progressively decline in late biofilms (fig. 3f). Their individual profiles, however, reflect their specific roles. For instance, extracellular matrix genes show highest transcription in early biofilms (6H−1D), sporulation and cannibalism genes have highest transcription in the mid-period (2D−14D), surfactin has bimodal distribution with peaks at 6H and 14D and protease production increases from 2D to 14D (supplementary files S13−S15, Supplementary Material online).
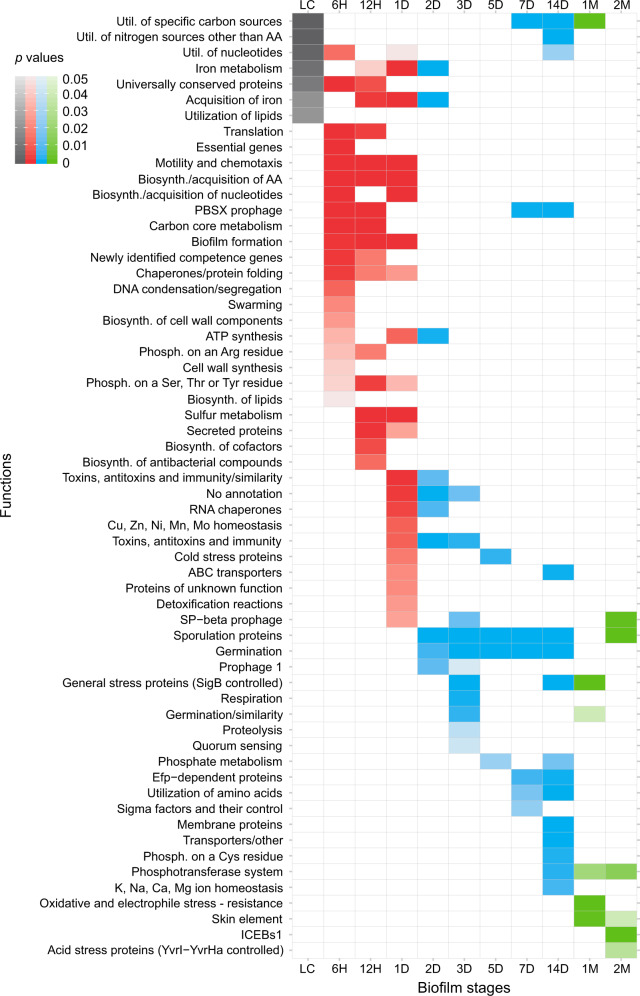
Fig. 4.
Biofilm ontogeny is a punctuated process organized in functionally discreate stages. Enrichment analysis of SubtiWiki functional categories (ontology depth 3) in a respective biofilm growth timepoint for genes that are transcribed 0.5 times (log2 scale) above the median of their overall transcription profile. Similar results are obtained for other transcription level cutoffs and SubtiWiki functional annotation ontology depths (see supplementary file S18, Supplementary Material online). Coloring follows biofilm growth periods: LC (gray), early (red), mid (blue), and late (green). Functional enrichment is tested by one-tailed hypergeometric test and P values are adjusted for multiple testing (see Materials and Methods).
These expression profiles largely follow community-accumulated knowledge on the biochemical networks governing biofilm development (Vlamakis et al. 2013; van Gestel et al. 2015a; Kalamara et al. 2018), but our data set also offers new insights, especially related to the later stages of biofilm growth. Surge of surfactin expression at 14D could be linked to sliding motility observable at the edge of the biofilm around this time point (van Gestel et al. 2015b) (supplementary file S3, Supplementary Material online). Another example is iron homeostasis that is tightly linked to the biofilm formation (Pi and Helmann 2017; Rizzi et al. 2018; Pisithkul et al. 2019). We recovered the full transcription dynamics of this process (supplementary file S16, Supplementary Material online) and found that the expression of siderophores-related pathways has bimodal profile with peaks of expression at the early-to-mid biofilm transition (12H−2D) and at the transition to late biofilms (14D). This second peak at 14D is preceded by the highest expression of iron detoxification genes (Guan et al. 2015) (supplementary file S16, Supplementary Material online). All of this shows that the later stages of biofilm growth, similar to metazoan ontogeny (Domazet-Lošo and Tautz 2010a), display highly specific expression dynamics that have been largely underexplored.
The DNA methylation status of enhancers has important role in the control of gene expression during animal development (Bogdanović et al. 2016). In contrast, bacterial DNA methylation mainly relates to the restriction−modification systems, and only recently it was discovered that YeeA methyltransferase participates in the regulation of gene expression in B. subtilis (Nye et al. 2020). We found that yeeA has the highest transcription at the onset of biofilm growth (supplementary file S25d, Supplementary Material online), suggesting that methylation in the context of gene regulation plays some role in the initiation of B. subtilis development.
Biofilm Growth Has a Stepwise Functional Architecture
The functional category enrichment analysis of biofilm timepoints reveals a tight control where every timepoint expresses a specific battery of functions (fig. 4; supplementary files S17 and S18, Supplementary Material online). Some illustrative examples of enriched functions include PBSX prophage (eDNA production), antibacterial compounds and swarming in early biofilms (6H−12H), zinc metabolism at 1D, iron uptake by siderophores and functionally unannotated genes at transition stage 2D, quorum sensing at 3D, sporulation and toxins/antitoxins in the mid-period (2D−14D), general stress at transition stage 14D, and mobile genetic elements in late biofilms (1M−2M). The enrichment of genes that lack functional annotation at 2D probably reflects the incomplete knowledge on the molecular mechanisms that govern early-to-mid biofilm transition. Statistical analysis of these genes on the phylostratigraphic map (supplementary file S19, Supplementary Material online) revealed that they preferentially originate from the ancestors of B. subtilis strains (ps10-ps12). This parallels development in animals where phylogenetically restricted genes (orphans) are involved in the embryonic transitions and the generation of morphological diversity (Khalturin et al. 2009; Domazet-Lošo and Tautz 2010a; Tautz and Domazet-Lošo 2011). When observed in total, functional enrichment patterns show that biofilm growth at the functional level has discrete hierarchical organization with even finer temporal grading compared with the pure transcription profiles (figs. 1b and 4; supplementary files S17 and S18, Supplementary Material online). This modular nature of biofilm growth is analogous to the noncontinuous and stage-organized architecture of development in animals (Monds and O’Toole 2009; Levin et al. 2012; Yanai 2018).
Discussion
Methodological Considerations
In this study we used phylostratigraphy-based tools (TAI, PAI) that uncovered recapitulation pattern in biofilm ontogeny. In metazoans, these tools were successfully applied in detecting phylogeny−ontogeny correlations and several unrelated approaches independently revealed the similar patterns (Domazet-Lošo and Tautz 2010a; Kalinka et al. 2010; Irie and Kuratani 2011; Levin et al. 2016), thus phylostratigraphy-based tools are considered generally reliable. However, there is an ongoing debate on the sensitivity of the BLAST algorithm in searching for deep homologs within the phylostratigraphy framework (Moyers and Zhang 2015; Domazet-Lošo et al. 2017; Moyers and Zhang 2018). We repeatedly argued that search for deep homologs is not essential in phylostratigraphic approach if a study aims to detect significant shifts in the protein sequence space and statistically correlate them to biological patterns (Domazet-Loso and Tautz 2003; Domazet-Lošo et al. 2007; Domazet-Lošo and Tautz 2010b; Tautz and Domazet-Lošo 2011; Domazet-Lošo et al. 2017). The very first phylostratigraphic study introduced this concept within the framework of punctuated evolution of protein families and recovered divergent evolutionary trajectories between Drosophila germ layers (Domazet-Lošo et al. 2007). Later re-evaluations of this finding confirmed its robustness (Moyers and Zhang 2016; Domazet-Lošo et al. 2017). Moreover, recent work shows that the standard usage of BLAST in phylostratigraphy is adequate in most situations (Shi et al. 2020; Vakirlis et al. 2020). Particularly important is the B. subtilis study where phylostratigraphy, with BLAST e-value cutoff set at 10−3, is used to predict novel sporulation genes, which phenotype is then experimentally confirmed (Shi et al. 2020). This clearly shows that the standard phylostratigraphic approach correctly recovers evolutionary patterns and has predictive power that could guide experiments in bacteria.
Regardless of this argumentation line, to overcome any uncertainty related to the BLAST error rates, we tested the stability of the obtained TAI recapitulation pattern in B. subtilis by shifting e-value cutoff values in the broad range from 10 to 10−30 (supplementary files S8 and S20, Supplementary Material online). Although e-values around 10−3 were repeatedly found to be optimal (Domazet-Loso and Tautz 2003; Domazet-Lošo et al. 2017; Moyers and Zhang 2018; Vakirlis et al. 2020), this test allowed us to evaluate the robustness of the recapitulation pattern by intentionally inflating false positive (e-values closer to 10) and false negative rates (e-values closer to 10−30). Expectedly, high e-value cutoffs pushed gene ages toward older phylostrata (ps1), whereas low e-value cutoffs pulled them toward younger phylostrata (ps12) (supplementary file S8, Supplementary Material online). However, regardless of these substantial underlying shifts in the distribution of the gene ages, the TAI recapitulation pattern remained stable and significant (supplementary file S20, Supplementary Material online), demonstrating a strong macroevolutionary imprint in the biofilm ontogeny that is resilient to the changes in e-value thresholds. Although BLAST is considered an algorithm of choice for obtaining initial phylostratigraphic information (Domazet-Lošo et al. 2017, Moyers and Zhang 2018, Vakirlis et al. 2020), we further tested the stability of the recapitulation pattern in B. subtilis biofilm ontogeny using MMseqs2—a next generation sequence similarity search tool (Steinegger and Söding 2017). In contrast to BLAST, we used a clustering strategy of MMseqs2 to find significant matches. Again, we found that MMseqs2 returns stable and significant recapitulation pattern in a broad parameter space (supplementary files S8 and S21, Supplementary Material online). Finally, we emphasize that our results obtained by phylostratigraphy-based tools (TAI, PAI) are in congruence with the results obtained by divergence-based tools (TdNI, TdSI, PdNI, and PdSI) that are methodologically completely independent.
Future Directions
In this study, we notice that the temporal quantitative dynamics of gene expressions in growing B. subtilis biofilms mimics the succession of early developmental stages in animals. However, in animals the gene expressions of many developmentally relevant genes are strictly spatially localized (Tomancak et al. 2007), so the question arises whether similar spatial localization of gene expressions also exists in developing B. subtilis biofilms. Animal development is extensively studied by RNA whole mount in situ hybridizations that are a very handy high-throughput tool in revealing spatial organization of expression patterns (Tautz and Pfeifle 1989). Similar protocols have yet to be established in biofilms, but previous work that relies on transgenic lines with fluorescent reporter genes and florescence microscopy in biofilms clearly indicates that the expression of some important biofilm genes is spatially localized (Vlamakis et al. 2008; van Gestel et al. 2015a, Srinivasan et al. 2018). An especially interesting finding is that the genes involved in motility, matrix, and sporulation phenotypes of B. subtilis display distinct expression waves that create spatiotemporally localized expression patterns (Srinivasan et al. 2018). The future work should explore expression patterns of promising candidate genes uncovered in this study, especially those with an unknown function and expression peaks in the mid biofilm period. This effort will help in constructing the coherent spatiotemporal picture of gene regulation in biofilms.
In our experimental setup environmental parameters of biofilm growth are tightly controlled, thus it is important to discuss the possibility of applying our approach to biofilms that exist in natural settings. The essential step in our pipeline is the recovery of quality biofilm material for RNA extraction along progressing stages of biofilm growth. We grew our B. subtilis on a solid−air interface of agar plates, but biofilms could be studied by various cultivation techniques and some naturally occurring biofilms could be to some extent replicated in such controlled settings (Franklin et al. 2015). An example is laboratory culturing of B. subtilis biofilms on plant roots; an environment where B. subtilis biofilms persist in nature (Vlamakis et al. 2013; Dragoš et al. 2018). However, we could also imagine that sampling of biofilm development for RNA extractions could be achieved directly in the environments where biofilms naturally occur. For instance, platforms that support biofilm growth could be set in a natural aquatic environment and then samples could be taken in situ at intervals that cover critical phases of biofilm development.
Obviously, the presence and precise shape of phylogeny−ontogeny correlations in more natural settings has yet to be discovered. However, universal strategy of biofilm formation, that is not dependent on the species and environments, was recognized long ago and formalized in the form of the general model of biofilm growth by Costerton (Stoodley et al. 2002, Hall-Stoodley et al. 2004, Lappin-Scott et al. 2014). A broad applicability of this model, irrespective of the species composition and environmental conditions of biofilm formation (Monds and O’Toole 2009, Vlamakis et al. 2013, Hall-Stoodley et al. 2004, Stoodley et al. 2002), encourages us to believe that the ontogeny−phylogeny correlations we discovered in biofilm development of B. subtilis will also be found in other in situ and ex situ biofilms. However, it is also likely that symbiotic interactions within multispecies biofilms as well as specific temporal ecological fluctuations, will bring some idiosyncrasies on the top of the general trend.
For example, patients with chronic respiratory diseases often suffer from recalcitrant multi-species biofilms. These are composed of several bacterial species engaging in cooperative and competitive interactions within the microbial community (Bomberger and Welp 2020). Interestingly, the development of the respiratory tract microbiota that will ultimately lead to persistent biofilms is critically influenced by events at early stages of infancy (Bosch et al. 2016). Therefore, our methodology could be useful in deciphering the early decision points in such complex biofilms by detecting possible fluctuations in the ontogeny−phylogeny correlations. On top of this, by stratifying genes with respect to their evolutionary origin and importance for various stages of biofilm development, our study also provides a basis for identifying new genes that are crucial for this process. The predictive power of phylostratigraphy has already been demonstrated in discovering new sporulation genes in B. subtilis (Shi et al. 2020).
Macroevolutionary Implications
Bonner proposed that polarity and heritable pattern formation, including cell differentiation with division of labor, are fundamental properties of any development (Bonner 2000). Bacillus subtilis biofilms indeed show polarity along bottom-top (Vlamakis et al. 2013; van Gestel et al. 2015a) and central-distal axes (Srinivasan et al. 2018), along with remarkably complex cell differentiation with division of labor (Vlamakis et al. 2013; van Gestel et al. 2015a; Dragoš et al. 2018). Long range electrical signaling (Humphries et al. 2017), control of cheater cells (Kalamara et al. 2018), and recent discovery of cancer-like processes in aging biofilms (Hashuel and Ben-Yehuda 2019) are additional features that go far beyond these minimal conditions that define multicellular development. In this study, we showed that phylogeny−ontogeny correlations and stage-organized gene expression architecture should be added to the list of properties that qualify B. subtilis biofilm growth as a true multicellular developmental process, analogous to developmental processes in complex eukaryotes. It is somewhat surprising that the recapitulation pattern originally proposed to be present in animals by 19th century zoologists (Abzhanov 2013) is now actually found in bacteria. However, this proves that the cross-talk between zoology and microbiology could bring new and exciting insights (McFall-Ngai et al. 2013). For example, in the light of multicellular individuality of B. subtilis biofilms, a host-bacterial symbiosis that involves biofilms (Vlamakis et al. 2013; McFall-Ngai 2014) could be viewed as an interaction of the two multicellular individuals.
Multicellularity is not a rare evolutionary transition as it has independently evolved many times in various lineages (Claessen et al. 2014; Rensing 2016). However, at every independent occurrence, it seems to be governed by the similar basic principles that include a macroevolutionary imprint. Future work should establish generality of these findings across bacterial and archaeal diversity as well as ecological conditions including microbial community biofilms. Yet, the results of our study, pervasiveness of bacterial (Flemming and Wuertz 2019) and archaeal multicellular behavior (van Wolferen et al. 2018), and the fact that the first fossils were bacterial biofilms (Javaux 2019), encourage us to call for the re-evaluation of the widely adopted idea that the first life on the Earth was unicellular. It is undisputable that the cell is the basic unit of life; however, that does not readily imply that the first life was strictly unicellular. At least some models envisage that protocells were organized in biofilm-like structures (Koonin and Martin 2005), and that unicellular part of the life cycle could evolve in parallel as an efficient dispersion mechanism in early oceans (Stoodley et al. 2002, McDougald et al. 2012; Tocheva et al. 2016).
Materials and Methods
Biofilm Growth
Bacillus subtilis subsp. subtilis str. NCIB3610 (B. subtilis) was obtained from the Bacillus Genetic Stock Center (BGSC, Ohio State University, Columbus, OH, USA) and stored in 25% glycerol stocks at −80 °C. Bacteria from the stock were plated on a LB agar plate (1% Bacto tryptone, 0.5% Bacto yeast extract, 1% NaCl, 1 mM NaOH solidified with 1.5% agar) and incubated for 24 h at 37 °C. Liquid LB medium (10 ml) was inoculated with a single colony and incubated with shaking for 24 h at 37 °C and 250 rpm. Petri dishes (90 mm) containing MSgg agar (5 mM potassium phosphate pH 7, 100 mM MOPS pH 7, 2 mM MgCl2, 700 μM CaCl2, 50 μM MnCl2, 50 μM FeCl3, 1 μM ZnCl2, 2 μM thiamine, 0.5% glycerol, 0.5% glutamate, 50 μg/ml tryptophan, 50 μg/ml phenylalanine solidified with 1.5% agar) were inoculated with four drops (5 µl) of LB culture. The drops on each plate were approximately equidistantly distributed. The plates were incubated at 30 °C and the biofilms were harvested for RNA extraction at 6 and 12 h, and at 1, 2, 3, 5, 7, 14, 30, and 60 days post-inoculation time (transcriptome samples 6H, 12H, 1D, 2D, 3D, 5D, 7D, 14D, 1M, and 2M, respectively). For protein extraction biofilms were harvested at 12 h and at 1, 2, and 7 days post-inoculation time (proteome samples 12H, 1D, 2D, and 7D, respectively).
RNA Extraction
To reach a satisfactory amount of biomass for RNA extraction, 102 (6H), 34 (12H), and 4 (1D, 2D, 3D, 5D, 7D, 14D, 1M, and 2M) biofilms were pooled per sample. The starting liquid LB culture (LC) used for biofilm inoculation was pelleted by centrifugation. All samples, excluding 2M, were taken in three biological replicates per timepoint. We succeeded to get only one replicate for 2M due to technically demanding RNA extraction from aged biofilms. To prevent changes in RNA composition due to biofilm harvesting procedure, 1 ml of stabilization mix (RNAprotect Bacteria Reagent—Qiagen diluted with PBS in a 2:1 volume ratio) was applied on plates. Soaked biofilms were gently removed from the agar surface using a sterile Drigalski spatula and a pipette tip and together with the unabsorbed stabilization mix transferred into a 2-ml tube (Eppendorf). An additional 1 ml of stabilization buffer was added to the tubes and the content was homogenized with a sterile pestle. The total RNA was extracted by applying a modified version of the RNeasy Protect Mini Kit (Qiagen) protocol. The homogenized samples were vortexed for 10 s, resuspended by pipetting and incubated for 5 min at RT. Three hundred microliters of the homogenate was transferred into a new 1.5 ml tube (Eppendorf), centrifuged for 10 min at 5,000×g at RT and 220 µl of the mix containing 200 µl of TE buffer, 3 mg of lysozyme and 20 µl of Proteinase K were added. The tubes were incubated in a shaker for 30 min at 25 °C and 550 rpm. Seven hundred microliters of RLT buffer was added into tubes, vortexed for 10 s, and the suspension was transferred into a 2-ml tube containing 50 mg of 425–600 µm acid-washed glass beads (Sigma-Aldrich). The bacterial cells were disrupted using a Fast prep FP120 homogenizer (Thermo Savant Bio101) at 6.5 m/s shaking speed for 5 min. After homogenization, the tubes were centrifuged for 15 s at 13,000×g and 760 µl of the supernatant was transferred into a new 2-ml tube. Three hundred microliters of chloroform was added, followed by a vigorous shaking by hand for 15 s. After incubation for 10 min at RT, the tubes were centrifuged for 15 min at 13,000×g and 4 °C. The upper phase was gently removed into new 1.5-ml tubes, and 590 µl of 80% ethanol was added. The suspension was gently mixed by pipetting and 700 µl of it was transferred into a mini spin column and centrifuged for 15 s at 13,000×g. This step was repeated until the total volume of suspension was filtered through the mini spin column. The columns were washed in three steps using 700 µl of RW1 buffer (Qiagen) and two times 500 µl or RPE buffer (Qiagen) with centrifugation for 15 s at 13,000×g. After the last washing step, the columns were centrifuged for 2 min at 13,000×g. The columns were transferred into new 1.5 ml tubes. Fifty microliters of RNase-Free water (Qiagen) was applied directly on the column filter and incubated for 1 min at RT. The columns were centrifuged for 1 min at 13,000×g and discarded afterwards. Ten microliters of the RDD buffer (Qiagen), 2.5 µl of DNase I (Qiagen), and 37.5 µl of RNase-Free water were added to the filtrate. After 10 min of incubation at RT, 50 µl of 7.5M LiCl solution was added. The tubes were incubated for 1 h at −20 °C. After incubation, the tubes were centrifuged for 15 min at 13,000×g and 4 °C, the supernatant was discarded and 150 µl of 80% ethanol was added. After another centrifugation step for 15 min at 13,000×g and 4 °C, the supernatant was discarded, and samples were resuspended in 30 µl of RNase-Free water. The RNA quantification was performed using a NanoDrop 2000 spectrophotometer (ThermoFisher Scientific). The RNA was stored at −80 °C until sequencing.
RNA Sequencing
Ribosomal RNA was removed from the total RNA samples by the Ribo-Zero rRNA Removal Kit (Illumina). RNA-seq libraries were prepared using the Illumina TruSeq RNA Sample Preparation v2 Kit (Illumina). The RNA sequencing was performed bi-directionally on the Illumina NextSeq 500 platform at the EMBL Genomics Core Facility (Heidelberg, Germany), generating ∼450 million reads per run. Before mapping, the sequence quality and read coverage were checked using FastQC V0.11.7 (Andrews 2010) with satisfactory outcome for each of the samples. In total 1,448,793,058 paired-end sequences (75 bp) were mapped onto the B. subtilis reference genome (NCBI Assembly accession: ASM205596v1; GCA_002055965.1) using BBMap V37.66 (Bushnel 2014) with an average of 93.46% mapped reads per sample (supplementary file S1, Supplementary Material online). We mapped in average 49 million reads per replicate with rather low variation between the samples (supplementary file S1, Supplementary Material online). The mapping was performed using the standard settings with the option of trimming the read names after the first whitespace enabled. The SAMtools package V1.6 (Li et al. 2009) was used to generate, sort and index BAM files for downstream data analysis. Subsequent RNAseq data processing was performed in R V3.4.2 (R Development Core Team 2008) using custom-made scripts. Briefly, mapped reads were quantified per each B. subtilis open reading frame using the R rsamtools package V1.30.0 (Morgan et al. 2017) and raw counts for 4,515 open reading frames were retrieved using the GenomicAlignments R package V1.14.2 (Lawrence et al. 2013). Expression similarity across timepoints and replicates was assessed using PCA implemented in the R package DESeq2 V1.18.1 (Love et al. 2014) and visualized using custom-made scripts based on the R package ggplot2 V3.1.0 (Wickham 2016) (fig. 2a).
Protein Digestion
To reach a satisfactory amount of biomass for protein digestion, four biofilms were pooled per sample (12H, 1D, 2D, and 7D). The LC sample was obtained by pelleting starting liquid LB culture. The samples were taken in three replicates at each timepoint. After corresponding incubation periods, 1 ml of cell lysis buffer (4% w/v SDS, 100 mM TEAB pH 8.6, 5 mM β-glycerophosphate, 5 mM NaF, 5 mM Na3VO4, 10 mM EDTA, and 1/10 tablet of Mini EDTA-free Protease Inhibitor Cocktail, Sigma-Aldrich) was used to harvest biofilms from the plates. Soaked biofilms were gently removed from the agar surface using a sterile Drigalski spatula or a pipette tip and together with the unabsorbed lysis buffer were transferred into a 2-ml tube. The bacterial biomass was resuspended in the cell lysis buffer and boiled for 10 min at 90 °C. After boiling, the samples were sonicated on ice for 30 min at 40% amplitude using the homogenizer Ultrasonic processor (Cole Parmer), and finally centrifuged for 30 min at 13,000×g and 4 °C. The supernatant was discarded and the pellet was cleaned by chloroform/methanol precipitation (4 volumes of 99.99% methanol, 1 volume of chloroform, and 3 volumes of milliQ water). The lysate was centrifuged for 10 min at 5,000×g and 4 °C. The upper, aqueous phase was discarded without disturbing the interphase. Four volumes of methanol were added to the tube, vortexed, and centrifuged for 10 min at 5,000×g and 4 °C. The supernatant was discarded and the pellet was air-dried for 1 min. The air-dried pellet was dissolved in a denaturation buffer (8 M Urea, 2 M Thiourea in 10 mM Tris−HCl, pH 8.0). Samples were separated on a NuPage Bis-Tris 4–12% gradient gel (Invitrogen) based on the manufacturer’s instructions; 100 µg of total protein at each timepoint was loaded on the gel and run for a short duration. The gel was stained with Coomassie blue and subsequently cut into three slices (fractions). Resulting gel slices were destained by washing thrice with 5 mM ammonium bicarbonate (ABC) and 50% acetonitrile (ACN). Gel pieces were next dehydrated in 100% ACN. Proteins were then reduced with 10 mM Dithiothreitol in 20 mM ABC for 45 min at 56 °C and alkylated with 55 mM iodoacetamide in 20 mM ABC for 30 min at RT in dark. This was followed by two more washes with 5 mM ABC and 50% ACN and once with 100% ACN. Proteins were digested with Trypsin protease (PierceTM) at 37 °C overnight. Resulting peptides were extracted from the gel in three successive steps using the following solutions—Step 1: 3% trifluoroacetic acid in 30% ACN; Step 2: 0.5% acetic acid in 80% ACN; Step 3: 100% ACN. Extracted peptides were next concentrated in a vacuum centrifuge and desalted using C18 stage-tips (Ishihama et al. 2006). Briefly, C18 discs (Empore) were activated with 100% methanol and equilibrated with 2% ACN, 1% TFA. Samples were loaded onto the membrane and washed with 0.5% acetic acid. Peptides were eluted in 80% ACN, 0.5% acetic acid, concentrated in a vacuum centrifuge, and analyzed on the mass spectrometer.
Mass Spectrometry
The MS analyses were performed at The Proteomics Core Facility at the Sahlgrenska Academy (University of Gothenburg, Sweden). Samples were analyzed on an QExactive HF mass spectrometer interfaced with Easy-nLC1200 liquid chromatography system (Thermo Fisher Scientific). Peptides were trapped on an Acclaim Pepmap 100 C18 trap column (100 μm x 2 cm, particle size 5 μm, Thermo Fischer Scientific) and separated using an in-house constructed C18 analytical column (300 x 0.075 mm I.D., 3 μm, Reprosil-Pur C18, Dr Maisch) using the gradient from 6% to 38% acetonitrile in 0.2% formic acid over 45 min followed by an increase to 80% acetonitrile in 0.2% formic acid for 5 min at a flow of 300 nl/min. The instrument was operated in data-dependent mode where the precursor ion mass spectra were acquired at a resolution of 60,000, the 10 most intense ions were isolated in a 1.2 Da isolation window and fragmented using collision energy HCD settings at 28. MS2 spectra were recorded at a resolution of 15,000 (for the 12H and 1D timepoints) or 30,000 (for the 2D, 7D, and LC timepoints), charge states 2 − 4 were selected for fragmentation and dynamic exclusion was set to 20 s and 10 ppm. Triplicate injections (technical replicates) were carried out for each of the sample fractions for label free quantitation. Acquired MS spectra were processed with the MaxQuant software suite V1.5.1.0 (Cox and Mann 2008; Tyanova et al. 2016) integrated with an Andromeda (Cox et al. 2011) search engine. Database search was performed against a target-decoy database of B. subtilis (NCBI Assembly accession: ASM205596v1; GCA_002055965.1) containing 4,333 protein entries, and including additional 245 commonly observed laboratory contaminant proteins. Endoprotease Trypsin/P was set as the protease with a maximum missed cleavage of two. Carbamidomethylation (Cys) was set as a fixed modification. Label free quantification was enabled with a minimum ratio count of two. A false discovery rate of 10% was applied at the peptide and protein level individually for filtering identifications. Initial mass tolerance was set to 20 ppm. Intensity-based absolute quantitation (iBAQ) option was enabled, but with log fit turned off. All other parameters were maintained as in the default settings. Finally, we obtained iBAQ values for 2,915 proteins at 10% false discovery rate.
Transcriptome Data Analyses
Out of 4,333 protein coding genes with mapped reads we analyzed 4,316 which passed the phylostratigraphic procedure (see below). First, we normalized raw counts of these 4,316 protein coding genes by calculating the fraction of transcripts (τ) (Li et al. 2010). This measure of relative expression, if multiplied with 106, gives the widely used transcripts per million quantity (Li et al. 2010). As multiplication with the constant 106 only serves to make values more human intuitive and does not influence further analysis in any way, we omitted the multiplication step and worked directly with τ. Given that this normalization allows cross-sample comparison, by controlling for fluctuations in sequencing depth, and retains native expression variability (Conesa et al. 2016), a property crucial for correct estimation of partial concentrations in calculating phylo-transcriptomic measures like TAI (Domazet-Lošo and Tautz 2010a), we chose this approach as the most suitable for downstream calculations of evolutionary measures. After this normalization step, replicates were resolved by calculating their median, whereby we omitted replicates that had zero raw values. These normalized transcript expression values were than used in all analyses except for assessing differential expression (see below). While preparing the transcriptome data set for RNA expression profile correlations, visualization, and clustering, we discarded the genes which had zero expression values in more than one stage, reducing thus the data set to 4,296 genes. If a gene had only one stage with a zero-expression value, we imputed the zero-value by interpolating the mean of the two neighboring stages (two genes in total). In the case a zero-expression value was present in the first or the last stage of the biofilm ontogeny, we directly assigned the value of the only neighbor to it (134 genes in total). We decided on this procedure primarily with the aim to avoid erratic patterns in the visualization and clustering of mRNA expression profiles. To bring the gene expression profiles to the same scale, for every gene we performed the normalization to median and log2 transformed the obtained values. This procedure yielded per-gene normalized expression values for 4,296 genes (standardized expressions) which were visualized using the R ggplot2 package V3.1.0 (supplementary file S14, Supplementary Material online) and clustered with DP_GP_cluster (McDowell et al. 2018) with the maximum Gibbs sampling iterations set to 500 (supplementary files S5 and S24, Supplementary Material online). For transcription regulator and sigma factor expression profiles (fig. 3a and b; supplementary files S22a, b and S24, Supplementary Material online) we selected genes which are regulating ≥10 operons based on the DBTBS database (Sierro et al. 2008). Biofilm-important genes (fig. 3f;supplementary files S13 and S24, Supplementary Material online), cell-to-cell signaling genes (fig. 3c;supplementary files S22c and S24, Supplementary Material online), and methyltransferase genes (supplementary file S25d, Supplementary Material online) used for profile visualization were selected following relevant papers (Vlamakis et al. 2013; van Gestel et al. 2015a; Kalamara et al. 2018; Nye et al. 2020). Protein phosphatase and protein kinase genes (fig. 3d and e; supplementary files S22d, e and S24, Supplementary Material online) were selected for profile visualization following the SubtiWiki database annotations (Zhu and Stülke 2018). Temporally pleiotropic genes (Hu et al. 2017) were chosen as genes with normalized transcript expression values greater than a specific cutoff value (10−6, 10−5, 10−4) in six or more biofilm stages (supplementary file S25, Supplementary Material online). The statistical significance of difference between average standardized expressions was assessed by repeated measures ANOVA (fig. 3; supplementary file S25a−c, Supplementary Material online). To determine the similarity of transcriptomes across stages of biofilm ontogeny, we used Pearson’s correlation coefficients (R) calculated in an all-against-all manner and visualized in a heat map (fig. 1b). Pairwise differential gene expression between individual stages of biofilm ontogeny (supplementary file S4, Supplementary Material online) was estimated using a procedure implemented in the DESeq2 V1.18.1 package. Using the likelihood ratio test implemented in the same package, we also tested the overall differential expression for every gene across all stages of the biofilm ontogeny (supplementary file S2, Supplementary Material online).
Proteome Data Analyses
Out of 2,915 quantified proteins we analyzed 2,907 which passed the phylostratigraphic procedure (see below). First, we calculated partial concentrations by dividing every iBAQ value by sum of all iBAQ values in the sample. After this normalization step replicates were resolved by calculating their median whereby we omitted replicates that had zero iBAQ values. This yielded normalized protein expression values that were used for calculating evolutionary indices. In preparing the proteome data set for protein expression profile correlation, visualization, and clustering, we further discarded genes which had zero-expression values in more than one stage, reducing thus the data set to 2,543 proteins. If a protein had only one stage with a zero-expression value, we interpolated it by taking the mean of the 2 neighboring stages (134 genes in total). In the case a zero-expression value was present in the first or the last stage of the biofilm ontogeny, we directly assigned it the value of the only neighbor (355 genes in total). To assess correlations between normalized transcriptome and proteome expression values, we calculated the Pearson’s correlation coefficient (R) on the matching 2,543 genes/proteins (supplementary files S12f, g and S24, Supplementary Material online). To bring the protein expression profiles to the same scale, for every gene we performed the normalization to median and log2 transformed the obtained values. This procedure yielded per-gene normalized expression values for 2,543 proteins (standardized expressions). Expression similarity across time points and replicates for 2,910 proteins was assessed using PCA in R V3.4.2 (R Development Core Team 2008) stats package. The PCA plot was visualized using the R package ggplot2 V3.1.0 (Wickham 2016), with log2 transformed iBAQ values previously increased by 1 (fig. 2b).
Enrichment Analysis
Due to the poor gene annotations of our focal B. subtilis strain, we transferred annotations from B. subtilis subsp. subtilis str. 168 (NCBI Assembly accession: ASM904v1; GCA_000009045.1) by establishing orthologs between the two strains (supplementary file S23, Supplementary Material online). This was performed by calculating their reciprocal best hit using the blastp algorithm V2.7.1+ (Altschul et al. 1990) with 10−5 e-value cutoff. Functional annotations for the B. subtilis 168 strain were retrieved from the SubtiWiki database (Zhu and Stülke 2018) (October 23, 2019). Functional enrichment of transcriptome and proteome clusters, and individual biofilm timepoints was estimated using the one-way hypergeometric test. For each timepoint genes that had standardized expression in that timepoint at least 0.5 times (log2 scale) above the median of their overall expression profile were tested for functional enrichment. P values were adjusted for multiple comparisons using the Yekutieli and Benjamini procedure (Yekutieli and Benjamini 1999).
Evolutionary Measures
The phylostratigraphic procedure was performed as previously described (Domazet-Lošo et al. 2007; Domazet-Lošo and Tautz 2010a). Following the recent phylogenetic literature (Xu and Côté 2003; Wu et al. 2009; Kubo et al. 2011; Williams and Embley 2014; Raymann et al. 2015; Wang and Ash 2015; Zhang and Lu 2015; Hug et al. 2016; Fan et al. 2017; Mukherjee et al. 2017; Zaremba-Niedzwiedzka et al. 2017; Hernández-González et al. 2018; Lazarte et al. 2018; Parks et al. 2018), we constructed a consensus phylogeny that covers divergence from the last common ancestor of cellular organisms to the B. subtilis as a focal organism (supplementary files S6 and S7, Supplementary Material online). We chose the nodes based on their phylogenetic support in the above listed literature, their importance of evolutionary transitions and availability of reference genomes for terminal taxa. The full set of protein sequences for 926 terminal taxa were retrieved from ENSEMBL (Zerbino et al. 2018) (922) and NCBI (4) databases. After checking consistencies of the files, leaving only the longest splicing variant per gene for eukaryotic organisms and adding taxon tags to the sequence headers of all sequences, we prepared a protein sequence database for sequence similarity searches (supplementary file S8, Supplementary Material online). To construct the phylostratigraphic map (Domazet-Lošo et al. 2007; Domazet-Lošo and Tautz 2010a) of B. subtilis, we compared 4,333 B. subtilis proteins with the protein sequence database by blastp algorithm V2.7.1+ with the 10−3 e-value threshold. After discarding all protein sequences which did not return their own sequence as a match, 4,317 protein sequences left in the sample. These 4,317 protein sequences were than mapped on the 12 internodes (phylostrata) of the consensus phylogeny using a custom-made pipeline (supplementary file S8, Supplementary Material online). We assigned a protein to the oldest phylostratum on the phylogeny where a protein still had a match (Dollo’s parsimony) (Domazet-Lošo et al. 2007; Domazet-Lošo and Tautz 2010a). Using expression values for 4,316 protein coding genes we calculated the TAI for each ontogenetic stage (fig. 1e;supplementary file S10, Supplementary Material online):
where psi is an integer which represents the phylostratum of the protein i, ei is the normalized transcript expression value of the gene i and n is the total number of genes analyzed. TAI is the weighted mean of phylogenetic ranks (phylostrata) that statistical properties and the biological interpretation are previously described (Domazet-Lošo and Tautz 2010a). To test for the robustness of the phylostratigraphic procedure and the TAI profile, we also constructed several phylostratigraphic maps with different e-value cutoffs (10, 1, 10−1, 10−2, 10−5, 10−10, 10−15, 10−20, and 10−30) using the blastp algorithm V2.7.1+ (supplementary file S8, Supplementary Material online). The stability of the recapitulation pattern was further tested using the MMseqs2 tool in the clustering mode (Steinegger and Söding 2017). We clustered protein sequences of 926 terminal taxa and retained clusters that contained at least one B. subtilis gene. For each of these clusters, we determined a gene that comes from the species embedded in the oldest phylostratum. The phylogenetic age of that gene was then assigned to the B. subtilis genes in that cluster. The clustering was performed with 10 different minimum coverage parameters (-c), ranging from 0 to 0.9 (supplementary files S8 and S21, Supplementary Material online). In all clustering iterations, the mode of clustering was set to transitive connection clustering, the maximum e-value threshold was set to 0.001, whereas all other parameters were set to their default values. For each of the clustering iterations we calculated TAI profiles (supplementary files S8 and S21, Supplementary Material online). To test the resilience of the TAI profiles to the sequencing depth related stochastic fluctuations at the lower end of expression levels, we removed from the data set genes with low expression values and recalculated TAI. The obtained TAI profiles remained essentially unchanged at the range of cutoffs (supplementary file S26, Supplementary Material online). To analyze the proteome data in similar fashion, we introduced the PAI (fig. 1g;supplementary file S10, Supplementary Material online), that is, the weighted mean of phylogenetic ranks (phylostrata):
where psi is an integer which represents the phylostratum of the protein i, qi is normalized protein expression value of the protein i and n is the total number of proteins analyzed. In total, we used 2,907 proteins for PAI calculation. To test the resilience of the PAI profiles to the iBAQ-related stochastic fluctuations at the lower end of expression levels, we removed from the data set proteins with low expression values and recalculated PAI. The obtained PAI profiles remained essentially unchanged at the range of cutoffs (supplementary file S27, Supplementary Material online). To estimate evolutionary divergence rates of B. subtilis proteins, we found 3,094 orthologs in Bacillus licheniformis str. DSM 13 (NCBI Assembly accession: ASM1164v1; GCA_000011645.1) by reciprocal best hits using blastp with 10−5 e-value threshold (supplementary file S10, Supplementary Material online). Of the available Bacillus species, B. licheniformis provided the best balance between the number of detected orthologues and the evolutionary distance of species pair for calculating evolutionary rates. We globally aligned B. subtilis–B. licheniformis orthologous pairs using the Needleman–Wunsch algorithm and then constructed codon alignments in pal2nal (Suyama et al. 2006). The nonsynonymous substitution rate (dN) and the synonymous substitution rate (dS) were calculated using Comeron’s method (Comeron 1995). The whole procedure of obtaining dN and dS was performed in the R package orthologr V0.3.0.9000 (Drost et al. 2015). Using dN values of 3,091 genes, we calculated the transcriptome nonsynonymous divergence index (TdNI; fig. 1f;supplementary file S10, Supplementary Material online), that is, the weighted mean of nonsynonymous gene divergence:
where dNi is a real number which represents the nonsynonymous divergence of gene i, ei is the normalized transcript expression value of the gene i and n is the total number of genes analyzed. Using dS values of 2,212 genes, we calculated transcriptome synonymous divergence index (TdSI; supplementary files S10 and S11a, Supplementary Material online), that is, the weighted mean of synonymous gene divergence:
where dSi is a real number which represents the synonymous divergence of gene i, ei is the normalized transcript expression value of the gene i and n is the total number of genes analyzed. To analyze proteome data in similar fashion, we introduced the PdNI and PdSI. Using dN of 2,329 proteins, we calculated the proteome nonsynonymous divergence index (PdNI; fig. 1h), that is, the weighted mean of nonsynonymous divergence:
where dNi is a real number which represents the nonsynonymous divergence of protein i, qi is the normalized protein expression value of the protein i and n is the total number of proteins analyzed. Using expression values of 1,755 proteins, we calculated proteome synonymous divergence index (PdSI; supplementary files S10 and S11b, Supplementary Material online), that is, the weighted mean of synonymous divergence:
where dSi represents the synonymous divergence value of protein i, qi is the normalized protein expression value of protein i that acts as weight factor and n is the total number of proteins analyzed. Using 4,316 transcript expression values and “measure independent of length and composition” (MILC) (Supek and Vlahoviček 2005) values we calculated TCBI (supplementary files S10 and S11c, Supplementary Material online), that is, the weighted arithmetic mean of codon usage bias:
where MILC is a real number which represents the codon usage bias of gene i, ei is the normalized transcript expression value of the gene i that acts as weight factor and n is the total number of genes analyzed. Using 2,907 protein expression and MILC values we calculated proteome codon usage bias index (PCBI; supplementary files S10 and S11d, Supplementary Material online), that is, the weighted arithmetic mean of codon usage bias:
where MILC is a real number which represents the codon usage bias of protein i, qi is the normalized protein expression value of protein i that acts as weight factor and n is the total number of proteins analyzed. MILC values were obtained from R package coRdon V1.3.0 (Elek et al. 2019), with respect to codon usage bias of ribosomal genes. The whole procedure of obtaining TdNI, TdSI, PdNI, and PdSI was made in R package orthologr V0.3.0.9000. The statistical analysis for TAI, PAI, TdNI, TdSI, PdNI, PdSI, TCBI, and PCBI was calculated using the R package myTAI V0.9.0 (Drost et al. 2018).
Imaging
Images of biofilms at sampling timepoints (fig. 1a) along with the time-lapse video (supplementary file S3, Supplementary Material online) were taken using the Sony Alpha a7 II mirrorless camera attached to a Zeiss Stemi 2000-C stereo-microscope with a NEX/T2 adapter (Novoflex) at 30 °C and average relative humidity of 74.5% using the automatic camera settings. The time-lapse video was produced from 1,345 shots taken during 14 days in 15 min intervals using the Adobe After Effects CC 2017 software at 24 fps.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Ž. Pezer-Sakač, P. Štancl, B. Pavletić and I. Šutevski for assistance with protein isolation, T.C.G. Bosch, A. Klimovich, and G. Klobučar for discussions. This work was supported by the Novo Nordisk Foundation (NNF10CC1016517), the Independent Research Fund Denmark (9040-00075A) to I.M.; the City of Zagreb, the Croatian Science Foundation under the project IP-2016-06-5924, the Adris Foundation, and the European Regional Development Fund (KK01.1.1.01.0008 CERRM and KK.01.1.1.01.0009 DATACROSS) to T.D.-L.
Author Contributions
T.D-L. initiated and conceptualized the study; T.D-L. and I.M. supervised the study; L.O. and N.Č. optimized protocols for culturing bacteria, RNA and proteome isolation; L.O. and N.Č. isolated transcriptomes and proteomes; M.F., K.V., and S.K. processed raw transcriptome data, mapped reads, and established expression levels; I.M., V.R., and A.T. performed LC-MS experiments and quantified proteomes; M.F., K.V., S.K., T.Š., and T.D-L. analyzed the data and prepared the figures; M.D-L. and T.Š. developed the phylostratigraphic pipeline; N.Č., M.D-L., D.K., and T.D-L. constructed consensus phylogeny and built the database; M.F., S.K., N.Č., M.L., and M.D-L. made phylostratigraphic maps; T.D.-L. and M.F. wrote the manuscript with contributions of all authors. All authors read and approved the manuscript.
Data Availability
All transcriptome data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE141305. All mass spectrometry proteomics data have been deposited in the PRIDE database and are accessible through the data set identifier PXD016656. The supplementary file S3, Supplementary Material online, contains a video available at https://youtu.be/em5PvOKEj-E. All custom-made scripts used in this study are available at https://github.com/bacillus-biofilms/biofilm-data-analysis.
References
- Abzhanov A. 2013. von Baer’s law for the ages: lost and found principles of developmental evolution. Trends Genet. 29(12):712–722. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. Available from: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed September 7, 2020.
- Bogdanović O, Smits AH, de la Calle Mustienes E, Tena JJ, Ford E, Williams R, Senanayake U, Schultz MD, Hontelez S, van Kruijsbergen I, et al. 2016. Active DNA demethylation at enhancers during the vertebrate phylotypic period. Nat Genet. 48(4):417–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomberger JM, Welp AL.. 2020. Bacterial community interactions during chronic respiratory disease. Front Cell Infect Microbiol. 10:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner JT. 2000. First signals: the evolution of multicellular development. Princeton: Princeton University Press. [Google Scholar]
- Bosch AATM, Levin E, van Houten MA, Hasrat R, Kalkman G, Biesbroek G, de Steenhuijsen Piters WAA, de Groot P-K, Pernet P, Keijser BJF, et al. 2016. Development of upper respiratory tract microbiota in infancy is affected by mode of delivery. EBioMedicine 9:336–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnel B. 2014. BBMap: a fast, accurate, splice-aware aligner. Available from: https://sourceforge.net/projects/bbmap/. Accessed September 7, 2020.
- Cheng X, Hui JHL, Lee YY, Wan Law PT, Kwan HS.. 2015. A “Developmental Hourglass” in Fungi. Mol Biol Evol. 32(6):1556–1566. [DOI] [PubMed] [Google Scholar]
- Claessen D, Rozen DE, Kuipers OP, Søgaard-Andersen L, van Wezel GP.. 2014. Bacterial solutions to multicellularity: a tale of biofilms, filaments and fruiting bodies. Nat Rev Microbiol. 12(2):115–124. [DOI] [PubMed] [Google Scholar]
- Comeron JM. 1995. A method for estimating the numbers of synonymous and nonsynonymous substitutions per site. J Mol Evol. 41(6):1152–1159. [DOI] [PubMed] [Google Scholar]
- Conesa A, Madrigal P, Tarazona S, Gomez-Cabrero D, Cervera A, McPherson A, Szcześniak MW, Gaffney DJ, Elo LL, Zhang X, et al. 2016. A survey of best practices for RNA-seq data analysis. Genome Biol. 17(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M.. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 26(12):1367–1372. [DOI] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M.. 2011. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 10(4):1794–1805. [DOI] [PubMed] [Google Scholar]
- Domazet-Loso T, Tautz D.. 2003. An evolutionary analysis of orphan genes in Drosophila. Genome Res. 13(10):2213–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domazet-Lošo T, Brajković J, Tautz D.. 2007. A phylostratigraphy approach to uncover the genomic history of major adaptations in metazoan lineages. Trends Genet. 23(11):533–539. [DOI] [PubMed] [Google Scholar]
- Domazet-Lošo T, Carvunis A-R, Albà MM, Šestak MS, Bakarić R, Neme R, Tautz D.. 2017. No evidence for phylostratigraphic bias impacting inferences on patterns of gene emergence and evolution. Mol Biol Evol. 34(4):843–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domazet-Lošo T, Tautz D.. 2010. a. A phylogenetically based transcriptome age index mirrors ontogenetic divergence patterns. Nature 468(7325):815–818. [DOI] [PubMed] [Google Scholar]
- Domazet-Lošo T, Tautz D.. 2010. b. Phylostratigraphic tracking of cancer genes suggests a link to the emergence of multicellularity in metazoa. BMC Biol. 8(1):66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragoš A, Kiesewalter H, Martin M, Hsu C-Y, Hartmann R, Wechsler T, Eriksen C, Brix S, Drescher K, Stanley-Wall N, et al. 2018. Division of labor during biofilm matrix production. Curr Biol. 28(12):1903–1913.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost H-G, Gabel A, Grosse I, Quint M.. 2015. Evidence for active maintenance of phylotranscriptomic hourglass patterns in animal and plant embryogenesis. Mol Biol Evol. 32(5):1221–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost H-G, Gabel A, Liu J, Quint M, Grosse I.. 2018. myTAI: evolutionary transcriptomics with R. Bioinformatics 34(9):1589–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost H-G, Janitza P, Grosse I, Quint M.. 2017. Cross-kingdom comparison of the developmental hourglass. Curr Opin Genet Dev. 45:69–75. [DOI] [PubMed] [Google Scholar]
- Elek A, Kuzman M, Vlahoviček K.. 2019. coRdon: codon usage analysis and prediction of gene expressivity. Available from: https://github.com/BioinfoHR/coRdon. Accessed September 7, 2020.
- Fan B, Blom J, Klenk H-P, Borriss R.. 2017. Bacillus amyloliquefaciens, Bacillus velezensis, and Bacillus siamensis Form an “Operational Group B. amyloliquefaciens” within the B. subtilis Species Complex. Front Microbiol. 8:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming H-C, Wuertz S.. 2019. Bacteria and archaea on Earth and their abundance in biofilms. Nat Rev Microbiol. 17(4):247–260. [DOI] [PubMed] [Google Scholar]
- Franklin MJ, Chang C, Akiyama T, Bothner B.. 2015. New technologies for studying biofilms. Microbiol Spectr. 3(4):1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gestel J, Vlamakis H, Kolter R.. 2015. a. Division of labor in biofilms: the ecology of cell differentiation. Microbiol Spectr. 3(2):MB-0002-2014. [DOI] [PubMed] [Google Scholar]
- van Gestel J, Vlamakis H, Kolter R.. 2015. b. From cell differentiation to cell collectives: Bacillus subtilis uses division of labor to migrate. PLOS Biol. 13(4):e1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan G, Pinochet-Barros A, Gaballa A, Patel SJ, Argüello JM, Helmann JD.. 2015. PfeT, a P1B4 -type ATPase, effluxes ferrous iron and protects Bacillus subtilis against iron intoxication. Mol Microbiol. 98(4):787–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Stoodley L, Costerton JW, Stoodley P.. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2(2):95–108. [DOI] [PubMed] [Google Scholar]
- Hashuel R, Ben-Yehuda S.. 2019. Aging of a bacterial colony enforces the evolvement of nondifferentiating mutants. mBio 10(5):e01414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-González IL, Moreno-Hagelsieb G, Olmedo-Álvarez G.. 2018. Environmentally-driven gene content convergence and the Bacillus phylogeny. BMC Evol Biol. 18(1):148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Uesaka M, Guo S, Shimai K, Lu T-M, Li F, Fujimoto S, Ishikawa M, Liu S, Sasagawa Y, EXPANDE Consortium, et al. 2017. Constrained vertebrate evolution by pleiotropic genes. Nat Ecol Evol. 1(11):1722–1730. [DOI] [PubMed] [Google Scholar]
- Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, Castelle CJ, Butterfield CN, Hernsdorf AW, Amano Y, Ise K, et al. 2016. A new view of the tree of life. Nat Microbiol. 1:16048. [DOI] [PubMed] [Google Scholar]
- Humphries J, Xiong L, Liu J, Prindle A, Yuan F, Arjes HA, Tsimring L, Süel GM.. 2017. Species-independent attraction to biofilms through electrical signaling. Cell 168(1 − 2):200–209.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie N, Kuratani S.. 2011. Comparative transcriptome analysis reveals vertebrate phylotypic period during organogenesis. Nat Commun. 2:248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama Y, Rappsilber J, Mann M.. 2006. Modular stop and go extraction tips with stacked disks for parallel and multidimensional peptide fractionation in proteomics. J Proteome Res. 5(4):988–994. [DOI] [PubMed] [Google Scholar]
- Javaux EJ. 2019. Challenges in evidencing the earliest traces of life. Nature 572(7770):451–460. [DOI] [PubMed] [Google Scholar]
- Kalamara M, Spacapan M, Mandic-Mulec I, Stanley-Wall NR.. 2018. Social behaviours by Bacillus subtilis: quorum sensing, kin discrimination and beyond. Mol Microbiol. 110(6):863–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinka AT, Tomancak P.. 2012. The evolution of early animal embryos: conservation or divergence? Trends Ecol Evol (Amst). 27(7):385–393. [DOI] [PubMed] [Google Scholar]
- Kalinka AT, Varga KM, Gerrard DT, Preibisch S, Corcoran DL, Jarrells J, Ohler U, Bergman CM, Tomancak P.. 2010. Gene expression divergence recapitulates the developmental hourglass model. Nature 468(7325):811–814. [DOI] [PubMed] [Google Scholar]
- Khalturin K, Hemmrich G, Fraune S, Augustin R, Bosch TCG.. 2009. More than just orphans: are taxonomically-restricted genes important in evolution? Trends Genet. 25(9):404–413. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Martin W.. 2005. On the origin of genomes and cells within inorganic compartments. Trends Genet. 21(12):647–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo Y, Rooney AP, Tsukakoshi Y, Nakagawa R, Hasegawa H, Kimura K.. 2011. Phylogenetic Analysis of Bacillus subtilis strains applicable to Natto (Fermented Soybean) production. Appl Environ Microbiol. 77(18):6463–6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappin-Scott H, Burton S, Stoodley P.. 2014. Revealing a world of biofilms - the pioneering research of Bill Costerton. Nat Rev Microbiol. 12(11):781–787. [DOI] [PubMed] [Google Scholar]
- Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ.. 2013. Software for computing and annotating genomic ranges. PLoS Comput Biol. 9(8):e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarte JN, Lopez RP, Ghiringhelli PD, Berón CM.. 2018. Bacillus wiedmannii biovar thuringiensis: a specialized mosquitocidal pathogen with plasmids from diverse origins. Genome Biol Evol. 10(10):2823–2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin M, Anavy L, Cole AG, Winter E, Mostov N, Khair S, Senderovich N, Kovalev E, Silver DH, Feder M, et al. 2016. The mid-developmental transition and the evolution of animal body plans. Nature. 531(7596):637–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin M, Hashimshony T, Wagner F, Yanai I.. 2012. Developmental milestones punctuate gene expression in the Caenorhabditis embryo. Dev Cell. 22(5):1101–1108. [DOI] [PubMed] [Google Scholar]
- Li B, Ruotti V, Stewart RM, Thomson JA, Dewey CN.. 2010. RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics 26(4):493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Beyer A, Aebersold R.. 2016. On the dependency of cellular protein levels on mRNA abundance. Cell 165(3):535–550. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons NA, Kolter R.. 2015. On the evolution of bacterial multicellularity. Curr Opin Microbiol. 24:21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDougald D, Rice SA, Barraud N, Steinberg PD, Kjelleberg S.. 2012. Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat Rev Microbiol. 10(1):39–50. [DOI] [PubMed] [Google Scholar]
- McDowell IC, Manandhar D, Vockley CM, Schmid AK, Reddy TE, Engelhardt BE.. 2018. Clustering gene expression time series data using an infinite Gaussian process mixture model. PLOS Comput Biol. 14(1):e1005896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, Domazet-Lošo T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, et al. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci U S A. 110(9):3229–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai MJ. 2014. The importance of microbes in animal development: lessons from the Squid-Vibrio Symbiosis. Annu Rev Microbiol. 68(1):177–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Mendoza A, Sebé-Pedrós A, Šestak MS, Matejčić M, Torruella G, Domazet-Lošo T, Ruiz-Trillo I.. 2013. Transcription factor evolution in eukaryotes and the assembly of the regulatory toolkit in multicellular lineages. Proc Natl Acad Sci. 110(50):E4858–E4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monds RD, O’Toole GA.. 2009. The developmental model of microbial biofilms: ten years of a paradigm up for review. Trends Microbiol. 17(2):73–87. [DOI] [PubMed] [Google Scholar]
- Morgan M, Hervé P, Valerie O, Nathaniel H.. 2017. Rsamtools: binary alignment (BAM), FASTA, variant call (BCF), and tabix file import. Available from: http://bioconductor.org/packages/release/bioc/html/Rsamtools.html. Accessed September 7, 2020.
- Moyers BA, Zhang J.. 2015. Phylostratigraphic bias creates spurious patterns of genome evolution. Mol Biol Evol. 32(1):258–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyers BA, Zhang J.. 2016. Erratum phylostratigraphic bias creates spurious patterns of genome evolution. Mol Biol Evol. 33(11):3031–3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyers BA, Zhang J.. 2018. Toward reducing phylostratigraphic errors and biases. Genome Biol Evol. 10(8):2037–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Seshadri R, Varghese NJ, Eloe-Fadrosh EA, Meier-Kolthoff JP, Göker M, Coates RC, Hadjithomas M, Pavlopoulos GA, Paez-Espino D, et al. 2017. 1,003 reference genomes of bacterial and archaeal isolates expand coverage of the tree of life. Nat Biotechnol. 35(7):676–683. [DOI] [PubMed] [Google Scholar]
- Nye TM, van Gijtenbeek LA, Stevens AG, Schroeder JW, Randall JR, Matthews LA, Simmons LA.. 2020. Methyltransferase DnmA is responsible for genome-wide N6-methyladenosine modifications at non-palindromic recognition sites in Bacillus subtilis. Nucleic Acids Res. 48(10):5332–5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson L, Levit GS, Hoßfeld U.. 2017. The “Biogenetic Law” in zoology: from Ernst Haeckel’s formulation to current approaches. Theory Biosci. 136(1 − 2):19–29. [DOI] [PubMed] [Google Scholar]
- O’Malley MA, Wideman JG, Ruiz-Trillo I.. 2016. Losing complexity: the role of simplification in macroevolution. Trends Ecol Evol. 31(8):608–621. [DOI] [PubMed] [Google Scholar]
- Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil P-A, Hugenholtz P.. 2018. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol. 36(10):996–1004. [DOI] [PubMed] [Google Scholar]
- Pi H, Helmann JD.. 2017. Sequential induction of Fur-regulated genes in response to iron limitation in Bacillus subtilis. Proc Natl Acad Sci U S A. 114(48):12785–12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisithkul T, Schroeder JW, Trujillo EA, Yeesin P, Stevenson DM, Chaiamarit T, Coon JJ, Wang JD, Amador-Noguez D.. 2019. Metabolic remodeling during biofilm development of Bacillus subtilis. mBio 10(3):e00623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint M, Drost H-G, Gabel A, Ullrich KK, Bönn M, Grosse I.. 2012. A transcriptomic hourglass in plant embryogenesis. Nature 490(7418):98–101. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. 2008. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing Available from: http://www.R-project.org. Accessed September 7, 2020.
- Raymann K, Brochier-Armanet C, Gribaldo S.. 2015. The two-domain tree of life is linked to a new root for the Archaea. Proc Natl Acad Sci U S A. 112(21):6670–6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing SA. 2016. ( Why) does evolution favour embryogenesis? Trends Plant Sci. 21(7):562–573. [DOI] [PubMed] [Google Scholar]
- Rizzi A, Roy S, Bellenger J-P, Beauregard PB.. 2018. Iron homeostasis in Bacillus subtilis requires siderophore production and biofilm formation. Appl Environ Microbiol. 85(3):e02439 − 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro JA. 1998. Thinking about bacterial populations as multicellular organisms. Annu Rev Microbiol. 52(1):81–104. [DOI] [PubMed] [Google Scholar]
- Sharp PM, Bailes E, Grocock RJ, Peden JF, Sockett RE.. 2005. Variation in the strength of selected codon usage bias among bacteria. Nucleic Acids Res. 33(4):1141–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Derouiche A, Pandit S, Rahimi S, Kalantari A, Futo M, Ravikumar V, Jers C, Mokkapati VRSS, Vlahoviček K, et al. 2020. Evolutionary analysis of the Bacillus subtilis genome reveals new genes involved in sporulation. Mol Biol Evol. 37(6):1667–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierro N, Makita Y, de Hoon M, Nakai K.. 2008. DBTBS: a database of transcriptional regulation in Bacillus subtilis containing upstream intergenic conservation information. Nucleic Acids Res. 36(Suppl 1):D93–D96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S, Vladescu ID, Koehler SA, Wang X, Mani M, Rubinstein SM.. 2018. Matrix production and sporulation in Bacillus subtilis biofilms localize to propagating wave fronts. Biophys J. 114(6):1490–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinegger M, Söding J.. 2017. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat Biotechnol. 35(11):1026–1028. [DOI] [PubMed] [Google Scholar]
- Stoodley P, Sauer K, Davies DG, Costerton JW.. 2002. Biofilms as complex differentiated communities. Annu Rev Microbiol. 56(1):187–209. [DOI] [PubMed] [Google Scholar]
- Supek F, Vlahoviček K.. 2005. Comparison of codon usage measures and their applicability in prediction of microbial gene expressivity. BMC Bioinformatics. 6(1):182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P.. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34(Web Server):W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D, Domazet-Lošo T.. 2011. The evolutionary origin of orphan genes. Nat Rev Genet. 12(10):692–702. [DOI] [PubMed] [Google Scholar]
- Tautz D, Pfeifle C.. 1989. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma 98(2):81–85. [DOI] [PubMed] [Google Scholar]
- Tocheva EI, Ortega DR, Jensen GJ.. 2016. Sporulation, bacterial cell envelopes and the origin of life. Nat Rev Microbiol. 14(8):535–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomancak P, Berman BP, Beaton A, Weiszmann R, Kwan E, Hartenstein V, Celniker SE, Rubin GM.. 2007. Global analysis of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 8(7):R145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Cox J.. 2016. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc. 11(12):2301–2319. [DOI] [PubMed] [Google Scholar]
- Vakirlis N, Carvunis A-R, McLysaght A.. 2020. Synteny-based analyses indicate that sequence divergence is not the main source of orphan genes. eLife 9:e53500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlamakis H, Aguilar C, Losick R, Kolter R.. 2008. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev. 22(7):945–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlamakis H, Chai Y, Beauregard P, Losick R, Kolter R.. 2013. Sticking together: building a biofilm the Bacillus subtilis way. Nat Rev Microbiol. 11(3):157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Ash GJ.. 2015. Whole genome phylogeny of Bacillus by feature frequency profiles (FFP). Sci Rep. 5:13644. [Google Scholar]
- Wickham H. 2016. ggplot2: elegant graphics for data analysis. New York: Springer-Verlag. Available from: http://ggplot2.org. Accessed September 7, 2020. [Google Scholar]
- Williams TA, Embley TM.. 2014. Archaeal “Dark Matter” and the origin of eukaryotes. Genome Biol Evol. 6(3):474–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wolferen M, Orell A, Albers S-V.. 2018. Archaeal biofilm formation. Nat Rev Microbiol. 16(11):699–713. [DOI] [PubMed] [Google Scholar]
- Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova NN, Kunin V, Goodwin L, Wu M, Tindall BJ, et al. 2009. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature 462(7276):1056–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Côté JC.. 2003. Phylogenetic relationships between Bacillus species and related genera inferred from comparison of 3’ end 16S rDNA and 5’ end 16S-23S ITS nucleotide sequences. Int J Syst Evol Microbiol. 53(3):695–704. [DOI] [PubMed] [Google Scholar]
- Yanai I. 2018. Development and evolution through the lens of global gene regulation. Trends Genet. 34(1):11–20. [DOI] [PubMed] [Google Scholar]
- Yekutieli D, Benjamini Y.. 1999. Resampling-based false discovery rate controlling multiple test procedures for correlated test statistics. J Stat Plan Inference 82(1 − 2):171–196. [Google Scholar]
- Zaremba-Niedzwiedzka K, Caceres EF, Saw JH, Bäckström D, Juzokaite L, Vancaester E, Seitz KW, Anantharaman K, Starnawski P, Kjeldsen KU, et al. 2017. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 541(7637):353–358. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Girón CG, et al. 2018. Ensembl 2018. Nucleic Acids Res. 46(D1):D754–D761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Lu Z.. 2015. Phylogenomic evaluation of members above the species level within the phylum Firmicutes based on conserved proteins. Environ Microbiol Rep. 7(2):273–281. [DOI] [PubMed] [Google Scholar]
- Zhu B, Stülke J.. 2018. SubtiWiki in 2018: from genes and proteins to functional network annotation of the model organism Bacillus subtilis. Nucleic Acids Res. 46(D1):D743–D748. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All transcriptome data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE141305. All mass spectrometry proteomics data have been deposited in the PRIDE database and are accessible through the data set identifier PXD016656. The supplementary file S3, Supplementary Material online, contains a video available at https://youtu.be/em5PvOKEj-E. All custom-made scripts used in this study are available at https://github.com/bacillus-biofilms/biofilm-data-analysis.