Abstract
The genus Acropora comprises the most diverse and abundant scleractinian corals (Anthozoa, Cnidaria) in coral reefs, the most diverse marine ecosystems on Earth. However, the genetic basis for the success and wide distribution of Acropora are unknown. Here, we sequenced complete genomes of 15 Acropora species and 3 other acroporid taxa belonging to the genera Montipora and Astreopora to examine genomic novelties that explain their evolutionary success. We successfully obtained reasonable draft genomes of all 18 species. Molecular dating indicates that the Acropora ancestor survived warm periods without sea ice from the mid or late Cretaceous to the Early Eocene and that diversification of Acropora may have been enhanced by subsequent cooling periods. In general, the scleractinian gene repertoire is highly conserved; however, coral- or cnidarian-specific possible stress response genes are tandemly duplicated in Acropora. Enzymes that cleave dimethlysulfonioproprionate into dimethyl sulfide, which promotes cloud formation and combats greenhouse gasses, are the most duplicated genes in the Acropora ancestor. These may have been acquired by horizontal gene transfer from algal symbionts belonging to the family Symbiodiniaceae, or from coccolithophores, suggesting that although functions of this enzyme in Acropora are unclear, Acropora may have survived warmer marine environments in the past by enhancing cloud formation. In addition, possible antimicrobial peptides and symbiosis-related genes are under positive selection in Acropora, perhaps enabling adaptation to diverse environments. Our results suggest unique Acropora adaptations to ancient, warm marine environments and provide insights into its capacity to adjust to rising seawater temperatures.
Keywords: genome sequencing, gene duplicatoin, scleractinian corals, environment
Introduction
Coral reefs support the most diverse marine ecosystems on Earth (Wilkinson 2008). Coral reef structure depends upon calcium carbonate deposition by anthozoan cnidarians known as scleractinian corals. Corals form obligate endosymbioses with photosynthetic dinoflagellates of the family Symbiodiniaceae, which supply the vast majority of their photosynthetic products to the host corals (Yellowlees et al. 2008). However, corals face a range of anthropogenic challenges, including ocean acidification and increasing seawater temperatures (Hoegh-Guldberg et al. 2007). Tropical storms, predation by crown-of-thorns starfish, and coral bleaching, a breakdown of the mutualism between corals and their symbiotic dinoflagellates caused by high ocean temperatures, are major causes of coral reef decline (De'ath et al. 2012). Bleaching has been observed around the world with increasing frequency (Hughes et al. 2017; Nakamura 2017). Loss of coral reefs also destroys the habitats of diverse marine species, making extensive loss of reef habitats one of the most pressing environmental issues of our time.
The genus Acropora (Family Acroporidae) is a keystone reef taxon globally, distributed from the Red Sea through the Indo-Pacific Ocean to the Caribbean. It is also the most diverse and abundant taxon, with more than 100 described species (Wallace 1999). The high growth rate of Acropora corals contributes significantly to reef growth, island formation, coastal protection, and support for fisheries (Shinn 1966; Bruckner 2002). The complex, 3D structures of Acropora corals provide habitat and refuge for more than a million species of marine organisms (Hinrichsen 1997; Knowlton et al. 2010). Acropora species are highly susceptible to coral bleaching induced by increasing seawater temperatures (Marshall and Baird 2000; Loya et al. 2001; Hughes et al. 2018); hence, they are expected to decline in the near future (Alvarez-Filip et al. 2013). Due to their bleaching susceptibility, more than 70% of acroporid species are listed as near threatened or threatened in the International Union for Conservation of Nature Red List (Carpenter et al. 2008).
The evolutionary history of Acropora is complex, with gaps in molecular data and fossil records. Although a molecular phylogenetic analysis using mitochondrial genes suggested that modern diversification of Acropora from a single Pliocene ancestor probably occurred after the Miocene, around 2 Ma (Fukami et al. 2000), recent molecular phylogenetic analysis using nuclear and mitochondrial genes suggested that divergence of Acropora started around 34 Ma (Richards et al. 2013). Despite its bleaching susceptibility, the first appearances of Acropora in the fossil record are from Somalia (Carbone et al. 1993) and Austria (Baron-Szabo 2006) during the Paleocene (66 Ma), a warmer period than the present, in which there was no sea ice. An Acropora-dominated fossil assemblage is first seen in the Oligocene of Greece (Schuster 2000). It has been reported that 12 extant species were already present in the Indo-Pacific in the Early Miocene, suggesting that speciation and diversification of Acropora occurred throughout the Cenozoic, in different world regions, including the Indo-Pacific (Santodomingo et al. 2015). In addition to Acropora fossil records from the Paleogene period (Paleocene, Eocene, and Oligocene), the presence of Acropora corals in seasonal warm water environments (the Southern Red Sea, Persian Gulf) as well as locations with large daily thermal fluctuations (reef pools in Ofu, Samoa) also suggest that they have the potential to cope with elevated ocean temperatures (Barshis et al. 2013; Coles and Riegl 2013). The Intergovernmental Panel on Climate Change (IPCC) intermediate RCP 6.0 scenarios predicts that the global mean temperature will rise by average of 2.2 °C by AD 2100 (IPCC 2013). How did Acropora corals survive under past warm ocean conditions and how will they cope with climate changes occurring today?
Because of the ecological significance of Acropora, the complete genome of Acropora digitifera was the first coral genome sequenced (Shinzato et al. 2011), and additional coral genomic data are becoming available (Prada et al. 2016; Voolstra et al. 2017; Cunning et al. 2018; Ying et al. 2018, 2019; Helmkampf et al. 2019; Shumaker et al. 2019). In order to identify genomic novelties that enabled Acropora to disperse widely and thrive, and to adapt to warmer environments, we sequenced genomes of 15 Acropora species (A. acuminata, A. awi, A. cytherea, A. digitifera, A. echinata, A. florida, A. gemmifera, A. hyacinthus, A. intermedia, A. microphthalma, A. muricata, A. nasta, A. selago, A. tenuis, and A. yongei) (fig. 1). We further sequenced genomes of confamilial taxa, Montipora cactus, M. efflorescens, and Astreopora myriophthalma. Montipora is another speciose genus (fig. 1) (Veron 2000), and Astreopora represents the basal clade of the Acroporidae, based on molecular data (Fukami et al. 2000). Together with available coral and anthozoan cnidarian genomic data, we examine genomic novelties that could shed light on the evolutionary success of Acropora. Understanding such genetic mechanisms may facilitate predictions about whether and how can they survive current global warming.
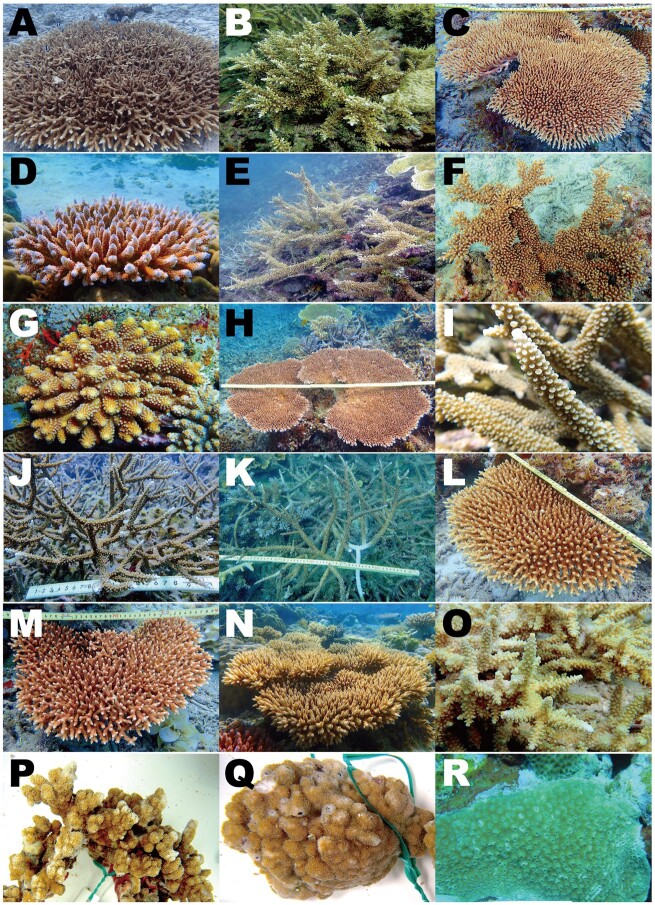
Fig. 1.
Fifteen Acropora, two Montipora, and Astreopora species for which we sequenced complete genomes in this study. (A) Acropora acuminata, (B) A. awi, (C) A. cytherea, (D) A. digitifera, (E) A. echinata, (F) A. florida, (G) A. gemmifera, (H) A. hyacinthus, (I) A. intermedia, (J) A. microphthalma, (K) A. muricata, (L) A. nasta, (M) A. selago, (N) A. tenuis, (O) A. yongei, (P) Montipora cactus, (Q) M. efflorescens, (R) Astreopora myriophthalma.
Results and Discussion
Whole-Genome Assembly and Gene Predictions for Acroporid Corals
For the 15 Acropora species, we obtained draft genome assemblies of 384–447 Mb with N50 sizes from 575 kb to 3 Mb (table 1). These represent significant improvements over the first version of the A. digitifera genome assembly (N50, 484 kb) (Shinzato et al. 2011), and are of comparable or better quality than other coral genomes reported in the NCBI Reference Sequence (RefSeq) database, in terms of N50 sizes and numbers of scaffold sequences (table 1). In contrast to the ∼30,000 gene models in previous Acropora genome assemblies, without performing error correction or removing haplotype sequences (Mao et al. 2018), we predicted ∼22,000 genes from each Acropora species (table 1).
Table 1.
Genome Assembly and Gene Prediction Statistics for Acropora, Montipora, and Astreopora Species (family Acroporidae) Used in This Study and Comparisons with Publicly Available Scleractinian Coral Genomes (NCBI RefSeq).
| Coral Species | Total Assembly Size (Mb) | Gap Rate (%) | No. Scaffolds | Scaffold N50 (kb) | No. Predicted Genes | BUSCO Completeness % (upper: genome assembly, lower: gene model) | Accession Numbers or Reference | |
|---|---|---|---|---|---|---|---|---|
| This study | Acropora acuminata | 395 | 5.3 | 3,293 | 1,005 | 21,904 |
C:91.5 [S:89.8, D:1.7], F:1.4, M:7.1 C:93.8 [S:92.2, D:1.6], F:2.0, M:4.2 |
BLEZ01000001-BLEZ01003293 |
| Acropora awi | 429 | 13.3 | 2,821 | 1,089 | 22,491 |
C:89.2 [S:88.2, D:1.0], F:1.8, M:9.0 C:92.7 [S:91.8, D:0.9], F:3.5, M:3.8 |
BLFA01000001-BLFA01002821 | |
| Acropora cytherea | 426 | 6.1 | 4,046 | 1,084 | 22,584 |
C:90.4 [S:86.5, D:3.9], F:1.9, M:7.7 C:91.9 [S:90.2, D:1.7], F:3.4, M:4.7 |
BLFB01000001-BLFB01004046 | |
| Acropora digitifera | 416 | 0.4 | 955 | 1,856 | 22,221 |
C:90.2 [S:88.7, D:1.5], F:1.6, M:8.2 C:92.4 [S:90.6, D:1.8], F:3.7, M:3.9 |
BLFC01000001-BLFC01000955 | |
| Acropora echinata | 401 | 14.7 | 2,002 | 1,917 | 21,554 |
C:87.7 [S:86.4, D:1.3], F:2.5, M:9.8 C:91.4 [S:89.9, D:1.5], F:3.7, M:4.9 |
BLFD01000001-BLFD01002002 | |
| Acropora florida | 442 | 9.1 | 6,979 | 751 | 23,237 |
C:88.6 [S:86.2, D:2.4], F:2.8, M:8.6 C:92.6 [S:90.8, D:1.8], F:3.5, M:3.9 |
BLFE01000001-BLFE01006979 | |
| Acropora gemmifera | 401 | 9.5 | 2,274 | 1,141 | 21,983 |
C:88.6 [S:87.3, D:1.3], F:3.0, M:8.4 C:91.1 [S:90.0, D:1.1], F:4.3, M:4.6 |
BLFF01000001-BLFF01002274 | |
| Acropora hyacinthus | 447 | 7.8 | 2,758 | 1,584 | 22,464 |
C:90.7 [S:87.8, D:2.9], F:1.5, M:7.8 C:93.4 [S:92.1, D:1.3], F:1.7, M:4.9 |
BLFG01000001-BLFG01002758 | |
| Acropora intermedia | 417 | 5.3 | 6,224 | 577 | 22,835 |
C:90.6 [S:87.9, D:2.7], F:1.4, M:8.0 C:93.0 [S:91.6, D:1.4], F:2.9, M:4.1 |
BLFH01000001-BLFH01006224 | |
| Acropora microphthalma | 384 | 9.3 | 4,878 | 1,061 | 22,016 |
C:88.6 [S:86.9, D:1.7], F:2.6, M:8.8 C:91.1 [S:89.9, D:1.2], F:3.6, M:5.3 |
BLFI01000001-BLFI01004878 | |
| Acropora muricata | 421 | 6.8 | 6,861 | 575 | 23,103 |
C:88.3 [S:85.8, D:2.5], F:3.2, M:8.5 C:92.0 [S:90.3, D:1.7], F:3.4, M:4.6 |
BLFJ01000001-BLFJ01006861 | |
| Acropora nasta | 416 | 7.2 | 4,717 | 1,051 | 22,545 |
C:90.5 [S:86.4, D:4.1], F:1.7, M:7.8 C:92.4 [S:91.4, D:1.0], F:2.7, M:4.9 |
BLFL01000001-BLFL01004717 | |
| Acropora selago | 393 | 6.2 | 5,816 | 657 | 22,616 |
C:87.6 [S:85.5, D:2.1], F:3.5, M:8.9 C:90.1 [S:89.0, D:1.1], F:4.9, M:5.0 |
BLFM01000001-BLFM01005816 | |
| Acropora tenuis | 403 | 7.4 | 1,538 | 1,166 | 22,802 |
C:90.5 [S:89.4, D:1.1], F:1.8, M:7.7 C:94.6 [S:93.9, D:0.7], F:2.8, M:2.6 |
BLAZ01000001-BLAZ01001538 | |
| Acropora yongei | 438 | 6.7 | 1,010 | 3,033 | 23,044 |
C:88.6 [S:86.2, D:2.4], F:2.8, M:8.6 C:93.6 [S:92.0, D:1.6], F:3.3, M:3.1 |
BLFN01000001-BLFN01001010 | |
| Astreopora myriophthalma | 373 | 5.4 | 1,149 | 1,634 | 28,712 |
C:89.3 [S:87.7, D:1.6], F:2.6, M:8.1 C:83.2 [S:82.0, D:1.2], F:6.4, M:10.4 |
BLFK01000001-BLFK01001149 | |
| Montipora cactus | 653 | 7.9 | 4,925 | 899 | 21,983 |
C:88.5 [S:86.7, D:1.8], F:2.7, M:8.8 C:86.0 [S:85.4, D:0.6], F:5.2, M:8.8 |
BLFO01000001-BLFO01004925 | |
| Montipora efflorescens | 643 | 9.0 | 5,162 | 1,132 | 21,370 |
C:86.4 [S:84.8, D:1.6], F:2.7, M:10.9 C:84.3 [S:84.0, D:0.3], F:5.2, M:10.5 |
BLFP01000001-BLFP01005162 | |
| NCBI RefSeq | Acropora digitifera | 419 | 15.2 | 2,420 | 484 | 26,060 |
C:75.9 [S:71.0, D:4.9], F:8.5, M:15.6 C:80.7 [S:74.2, D:6.5], F:9.2, M:10.1 |
Shinzato et al. (2011) |
| Acropora millepora | 387 | 9.7 | 3,869 | 494 | 23,710 |
C:91.3 [S:89.5, D:1.8], F:1.5, M:7.2 C:96.0 [S:94.5, D:1.5], F:1.5, M:2.5 |
Ying et al. (2019) | |
| Montipora capitata | 614 | 6.9 | 27,865 | 185 | NA |
C:82.1 [S:80.9, D:1.2], F:6.6, M:11.3 NA |
Helmkampf et al. (2019) | |
| Stylophora postillata | 400 | 10.5 | 5,687 | 457 | 24,833 |
C:88.3 [S:86.8, D:1.5], F:3.3, M:8.4 C:96.7 [S:81.3, D:15.4], F:1.5, M:1.8 |
Voolstra et al. (2017) | |
| Orbicella faveolata | 486 | 26.7 | 1,932 | 1,162 | 25,916 |
C:85.8 [S:83.2, D:2.6], F:4.5, M:9.7 C:90.0 [S:87.1, D:2.9], F:5.0, M:5.0 |
Prada et al. (2016) | |
| Pocillopora damicornis | 234 | 3.7 | 4,392 | 326 | 19,935 |
C:89.2 [S:88.7, D:0.5], F:2.4, M:8.4 C:94.0 [S:93.5, D:0.5], F:2.5, M:3.5 |
Cunning et al. (2018) |
Note.—C, complete BUSCOs; S, complete and single-copy BUSCOs; D, complete and duplicated BUSCOs; F, fragmented BUSCOs; M, missing BUSCOs; NA, not available in NCBI RefSeq100.
Benchmarking Universal Single-Copy Orthologs (BUSCO) analyses (Simao et al. 2015; Waterhouse et al. 2018), which assess whether universal single-copy orthologous genes observed in more than 90% of metazoan species from the OrthoDB database of orthologs (www.orthodb.org, version 9) are recovered in a genome/transcriptome assembly, yielded completeness scores of genome assemblies and gene models of around 89% and 92% (average of Complete BUSCO %), respectively, in all of these Acropora species (table 1). The Montipora and Astreopora genome assemblies were of comparable quality (table 1). BUSCO completeness scores of both genome assemblies and gene models of the acroporid genomes were also comparable to those of other coral genomes available in NCBI RefSeq (table 1), indicating that these draft genome assemblies and gene predictions are of reasonable quality.
Genome Organization of Acroporid Genomes
Not surprisingly, proportions of various repetitive elements and repeat landscapes were similar among the acroporid genomes (supplementary fig. S1, Supplementary Material online). In Acropora, about 40–45% of the genomes consist of interspersed repeats (supplementary fig. S1, Supplementary Material online). In Montipora, 50% of the genomes comprise repeats, possibly reflecting larger assembled sizes than those of Acropora (supplementary fig. S1, Supplementary Material online, table 1). The most abundant repeat types were long interspersed nuclear element (LINE) and short interspersed nuclear element (SINE), among the annotated elements, but the majority of the repeats, comprising 28–30% of the genomes, seem to be novel and possibly acroporid- or anthozoan-specific (supplementary fig. S1, Supplementary Material online). In order to compare genome organization of Acropora and other anthozoan genomes, A. digitifera scaffolds containing at least 100 orthologous groups (OGs, see below) shared with other Acropora species, resulting in 38 scaffolds (125.7 Mb, 30% of the genome), were used to evaluate synteny. Genome alignments to individual scleractinian genomes revealed high conservation of genome organization within Acropora (supplementary fig. S2, Supplementary Material online). Commensurate with phylogenetic distances to Acropora, conservation of genome organization among acroporids (Montipora and Astreopora), another scleractinian (Orbicella), and a sea anemone (Nematostella) diminished progressively (supplementary fig. S2, Supplementary Material online).
To determine whether large-scale (whole genome or chromosomal level) genome duplication occurred in the anthozoan lineage, we performed phylogenetic analyses of anthozoan genes (24 proteomes) using 300 randomly selected protein sequences from A. digitifera. We show 22 examples of phylogenetic analyses that were based on an alignment of ≥150 AAs in more than 80 gene sequences or ≥200 AAs in more than 40 gene sequences from 24 anthozoan proteomes (supplementary fig. S3, Supplementary Material online). Almost all nodes supported by high bootstrap values (>80%) contained one sequence from each anthozoan species (supplementary fig. S3, Supplementary Material online). In addition, there was no clear signature of large-scale duplications in genome alignment dotplots (supplementary fig. S2, Supplementary Material online). Consequently, in contrast to a suggested whole-genome duplication event in the common ancestor of Acropora (Mao and Satoh 2019), we detected no incontrovertible evidence of whole-genome or large-scale duplication events in any anthozoan lineage, including scleractinians, the Acroporidae, or the genus Acropora, in this study. Thus, we did not take genome duplication events into account in subsequent analyses.
Common Characteristics of Gene Repertoires of Scleractinians and Acroporids
In addition to the acroporid genomes, we used publicly available gene models of two anemones, Nematostella vectensis (Putnam et al. 2007) and Exaiptasia palida (Baumgarten et al. 2015), two corallimorpharians, Amplexidiscus fenestrafer and Discosoma spp. (Wang et al. 2017), and two scleractinians, Stylophora pistillata (Voolstra et al. 2017) and Orbicella faveolata (Prada et al. 2016) for OG clustering. Examination of OGs in anthozoan genomes allowed us to identify 21,697 OGs in all taxa, 20,765 in scleractinians, 19,737 in acroporids, and 18,692 in Acropora. Approximately 98% of Acropora genes and 93–97% of those of other acroporids (Montipora and Astreopora) belonged to OGs identified in other scleractinian species (fig. 2A, supplementary tables S1 and S2, Supplementary Material online), indicating high conservation of gene repertoires among scleractinians. Of the 21,697 OGs, 27 were exclusive to all scleractinians, but not observed in other groups, 48 to the Acroporidae, and 90 to Acropora (supplementary tables S3–S5, Supplementary Material online). Among the 27 OGs exclusive to scleractinians, a known coral calcification gene, skeletal aspartic acid-rich protein 2 (Ramos-Silva et al. 2013), was included (OG0002460, supplementary table S3, Supplementary Material online), suggesting that this gene might be essential for coral skeleton formation. In addition, we also identified OGs that were not found in any corals (supplementary tables S6–S8, Supplementary Material online), suggesting that these OGs were either lost in the coral clade, or that they arose after its divergence. In a previous study, we reported that cystathionine ß-synthase, an essential enzyme for cysteine biosynthesis, was possibly lost from the A. digitifera genome (Shinzato et al. 2011). As Acropora species are sensitive to bleaching (Loya et al. 2001), it is likely that Acropora depends upon symbiotic dinoflagellates to produce cysteine. In this study, we were unable to detect this gene in any acroporid genome, but we did identify it in other coral, corallimorpharian, and sea anemone genomes (OG0014971, supplementary table S7, Supplementary Material online), supporting the notion that this enzyme was lost in the common ancestor of the Acroporidae and that differences in dependency on symbiotic algae could partially explain the high sensitivity of Acropora to bleaching.
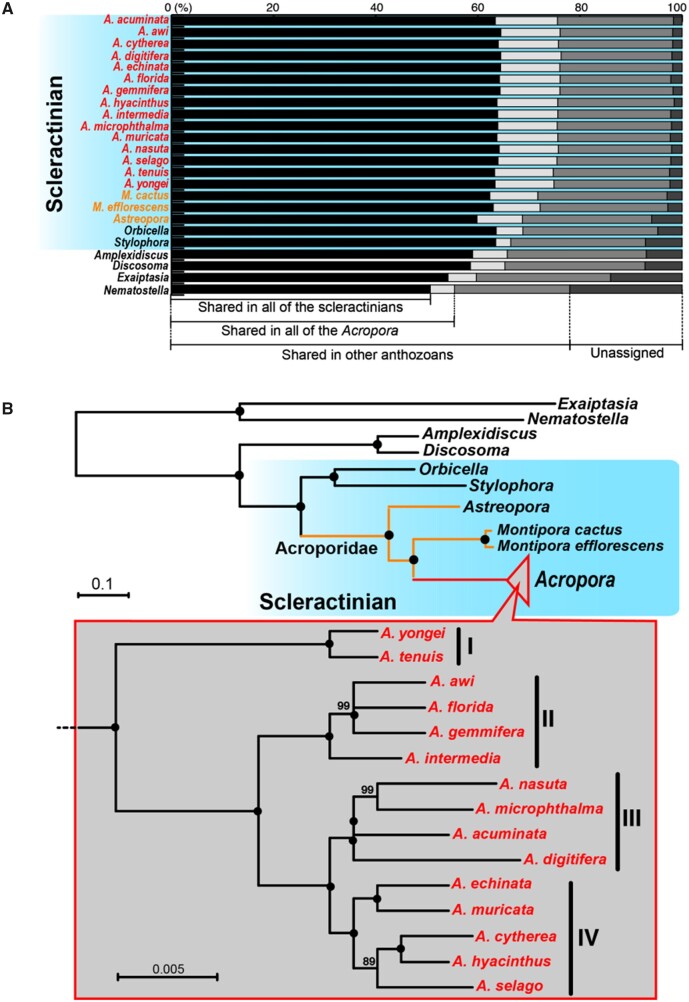
Fig. 2.
Comparisons of orthologous groups and phylogenetic relationships of anthozoan genomes. (A) Proportions of shared orthologous group genes among anthozoans. Scleractinians are shaded in blue. Astreopora and Montipora species are in yellow, and Acropora species are in red. (B) Molecular phylogeny of anthozoans using 818 single-copy orthologous genes (176,160 amino acids). Nodes with 100% bootstrap support are shown with black circles.
Phylogenomic Analysis Revealed That the Common Ancestor of Acropora Survived Warm Periods without Sea Ice from the Mid or Late Cretaceous to the Early Eocene
Phylogenomic analysis of these anthozoan genomes using 818 single-copy OGs yielded robust phylogenetic relationships, with the major anthozoan cnidarian clades being supported by 100% bootstrap values (fig. 2B). Almost all nodes among the 15 Acropora species in four distinct clades were also supported by 100% bootstrap values (fig. 2B), indicating that these molecular phylogenetic relationships are well supported and are likely to reflect evolutionary relationships of the 15 Acropora species. Acropora corals exhibit diverse morphologies (arborescent, hispidose, corymbose, table, etc.) (Wallace 1999), and each clade contains species with different morphologies. For instance, hispidose branching corals, A. awi and A. echinata belong to Clades II and IV, respectively (figs. 1 and 2), indicating that the diverse colony forms of Acropora are the result of convergent evolution in each clade.
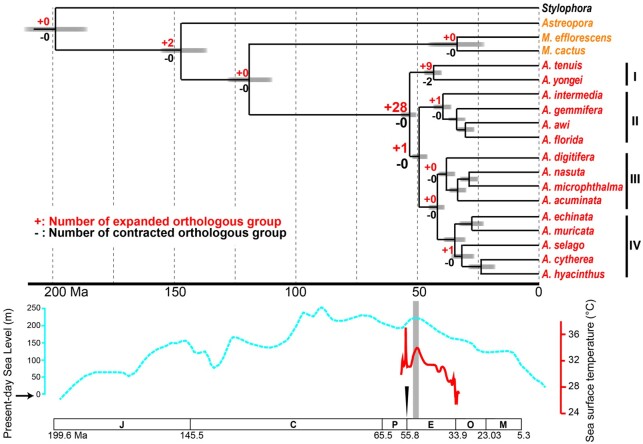
Molecular dating analysis using 2,126 single-copy OGs indicates that common ancestors of the Family Acroporidae emerged 199–147 Ma, whereas those of the genus Acropora appeared later, between 119 and 52 Ma (fig. 3). In contrast to the suggested divergence timing of Acropora in a previous study using five genomes (<15 Ma) without using scleractinian fossil records for dating calibration (Mao et al. 2018), diversification of Acropora was thought to have occurred during the Eocene and Oligocene (around 25–50 Ma), possibly accounting for the high species diversity of Acropora fossils known from the Miocene (5.3–23 Ma) and the existence of 12 extant species in the Early Miocene (fig. 3) (Santodomingo et al. 2015). Although these molecular dating estimates could shift as additional fossils are discovered, our data suggest that the Acropora common ancestor originated and survived in warm environments during the mid–late Cretaceous and the Paleocene–Eocene Thermal Maximum (55.8 Ma), when global temperatures rose 5–8 °C in 20,000 years (McInerney and Wing 2011), until the Early Eocene Climatic Optimum (EECO, 51–53 Ma) when they reached a long-term maximum (Zachos et al. 2001, 2008). Then a 17-Ma cooling trend occurred until the beginning of the Oligocene (33.9 Ma), which may have facilitated diversification of Acropora.
Fig. 3.
Divergence time estimates for acroporid corals using 2,126 single-copy orthologous genes (621,659 amino acid length) and evolution of gene family size changes in scleractinians. Numbers of significantly (P < 0.01) expanded or contracted orthologous groups with more or less than three genes are shown at each node. Expected sea level changes based on Olde et al. (2015) are shown with a blue dotted line, and tropical sea surface temperature of the Eocene (Cramwinckel et al. 2018) is shown with a red line. The Paleocene–Eocene Thermal Maximum is indicated with an arrowhead, and the EECO is highlighted in light gray. An approximate geological time scale is shown at the bottom. Abbreviations of geologic periods are as follows; J, Jurassic Period; C, Cretaceous Period; P, Paleocene; E, Eocene; O, Oligocene; M, Miocene.
Gene Expansions Unique to Acropora Include Possible Coral Stress Response Genes
We detected 48 and 90 OGs that are restricted to the Acroporidae and Acropora, but not observed in the two other scleractinians, two corallimorpharians, or two anemones, respectively (supplementary tables S4 and S5, Supplementary Material online). Of the 90 OGs observed exclusively in Acropora, four genes are involved in coral calcification (galaxins, aspartic and glutamic acid-rich proteins, uncharacterized skeletal organic matrix protein 6) (Ramos-Silva et al. 2013; Takeuchi et al. 2016), implying independent evolutionary mechanisms of calcification in Acropora, in addition to shared mechanisms in scleractinians and possible involvement of these genes in the great diversity of morphologies and high levels of calcification rates in Acropora.
Gene duplication is a major driving force of genome evolution and facilitates acquisition of novel gene functions (Ohno 1970). In contrast to the lack of expanded or contracted genes in the common ancestor of scleractinians (fig. 3), 28 OGs were predicted to have expanded in the common ancestor of Acropora, which is the largest number of expanded OGs in the entire scleractinian lineage (fig. 3, supplementary table S9, Supplementary Material online). These genes may contribute to adaptations to past warm environments, wide distributions, and ecological success of Acropora corals. These include genes possibly restricted to corals or cnidarians, small cysteine-rich peptides (SCRiPs) (OG0000795, only observed in Acropora and Montipora) (Sunagawa et al. 2009) and a novel coral caspase type, Caspase-X (OG0000692, supplementary table S3, Supplementary Material online) (Moya et al. 2016). Phylogenetic analyses of SCRiPs and Caspase-X genes demonstrated that expansions originated by tandem duplication in Acropora genomes (supplementary fig. S4, Supplementary Material online). Gene expression analysis showed that most of these genes were more highly expressed in adults than in embryos (supplementary fig. S4, Supplementary Material online), suggesting that they function in adult corals. Phylogenetic analysis showed that the tandemly duplicated SCRiPs did not cluster together with reported SCRiPs in UniProt, suggesting that these belong to a novel class (supplementary fig. S4A, Supplementary Material online). Some SCRiP genes were downregulated and are highly responsive to thermal stress (Sunagawa et al. 2009) and are thought to be potent neurotoxins (Jouiaei et al. 2015). It has been proposed that suppression of a caspase-mediated apoptotic cascade in host corals, induced by endogenous production of reactive oxygen species from symbiotic algae, is important in thermal stress responses (Kvitt et al. 2011; Tchernov et al. 2011). Caspase-X genes possess both inactive and active caspase domains, probably interacting and controlling caspase activity (Moya et al. 2016), as in cooperative and hierarchical binding of c-FLIP and caspase-8, in promoting or inhibiting apoptotic cell death (Hughes et al. 2016), and may function in thermal stress responses of Acropora. Although detailed functions of Acropora-specific expanded SCRiPs and Caspase-X genes under thermal stress remain to be revealed, these genes may enable Acropora corals to cope with thermal stress and to disperse widely and thrive globally.
Dimethlysulfonioproprionate Lyases, Which Promote Cloud Formation and Which May Have Been Acquired by Horizontal Gene Transfer from Algal Symbionts, Are the Most Duplicated Genes in the Acropora Ancestor
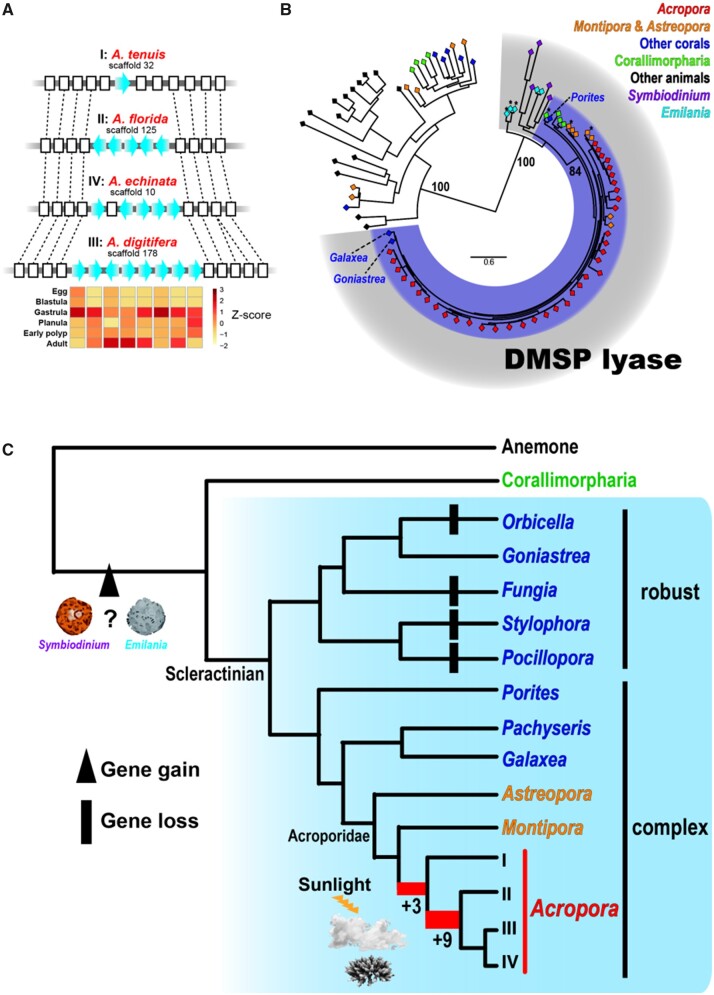
Among the 28 expanded OGs in the Acropora ancestor, the most diversified (OG0000129) is similar to dimethlysulfonioproprionate (DMSP) lyase of a coccolithophore, Emiliania huxleyi (Alcolombri et al. 2015) (figs. 3 and 4, supplementary tables S9 and S10, Supplementary Material online). This enzyme mediates cleavage of DMSP into dimethyl sulfide (DMS) and acrylate. DMS is the principal form of sulfur that is released from oceans into the atmosphere; thus, it is a key component of the ocean sulfur cycle (Quinn and Bates 2011). DMS may be crucial for cloud formation, and may serve to reduce light levels and water temperatures in marine environments (Vallina and Simo 2007). Thus, DMSP lyases in marine organisms participate in atmosphere-ocean feedback and may influence local climate regulation. Interestingly, concentrations of DMSP and DMS in corals are the highest reported among marine organisms, suggesting that corals are important sources of these two sulfur compounds (Broadbent et al. 2002; Broadbent and Jones 2004).
Our analysis shows that DMSP lyase gene expansions occurred first in the common ancestor of Acropora and again after divergence of the basal clade (supplementary tables S9 and S10, Supplementary Material online). Synteny analysis revealed that the second expansion occurred by tandem duplication (fig. 4A). Relative expression levels of tandemly located genes in A. digitifera showed higher expression levels in gastrula and/or adult stages (fig. 4A), corresponding to higher expression of a DMSP biosynthesis gene in larval stages of an Acropora coral (Raina et al. 2013). Extant scleractinians comprise two major clades, “complex” and “robust,” based on molecular analyses (Romano and Palumbi 1996; Kitahara et al. 2010). Although DMSP lyase could not be detected in genomes of “robust” corals, except for Goniastrea aspera, we did detect it in genomes of “complex” corals, including Astreopora and Montipora, and the two corallimorpharians (Amplexidiscus and Discosoma) (fig. 4B and C). We detected a single DMSP lyase locus in each scleractinian genome except for acroporids. Interestingly, ORTHOSCOPE analysis, for detecting orthologs, showed that among metazoans, DMSP lyases occur only in the Scleractinia and Corallimorpharia (fig. 4B). Molecular phylogeny showed that DMSP lyases from Emiliania, Symbiodiniaceae, Scleractinia, and Corallimorpharia cluster together (fig. 4B). Moreover, genes similar to Acropora DMSP lyase are only found in Acropora, Emiliania, and the Symbiodiniaceae, among eukaryotes in the NCBI NR database to date (BlastP, 1e−5). These results suggest that anthozoan DMSP lyases may have been acquired by the common ancestor of scleractinians and corallimorpharians via horizontal gene transfer from symbiotic Symbiodiniaceae or Emiliania (fig. 4C) and were later possibly lost in variety species of the “robust” clade (fig. 4C). Then, expansions occurred in the common ancestor of Acropora. It has been suggested that DMSP participates in a wide range of coral stress responses, including those to heat, sunlight, air exposure, and hyposalinity (Sunda et al. 2002; Raina et al. 2010; Deschaseaux et al. 2014; Aguilar et al. 2017). Higher DMSP concentrations were observed in Acropora than in other corals (Broadbent et al. 2002). These species-specific phenomena may be supported by Acropora-specific gene expansions. Although the functions of expanded DMSP lyases in Acropora remain to be determined, Acropora-specific expansions suggest that warmer and shallower environments from the Cretaceous to the EECO may have enhanced gene duplication in the Acropora ancestor. Diversified functions of Acropora DMSP lyase may enable adaptation to stresses, such as intense heat, light, and salinity, probably by forming clouds to minimize ocean heating due to insolation.
Fig. 4.
Expansions of DMSP lyase specific to the genus Acropora. (A) Examples of tandem duplication of DMSP lyase in Acropora genomes. Genomic sequences of Acropora species from four clades in which neighboring genes of DMSP lyases (both 5′ and 3′) were correctly assembled are shown. DMSP lyase genes are shown with blue arrows and other genes are shown as white boxes. Genes belonging to the same orthologous groups are connected by dot lines. Relative gene expression levels of A. digitifera tandemly located genes in embryonic and adult stages are shown in Z-scores. (B) Maximum likelihood analysis of DMSP lyase using homologous eukaryote genes identified with ORTHOSCOPE (Inoue and Satoh 2019). Asterisks indicate query sequences in ORTHOSCOPE analysis (BlastP, 1e−4), including Montiopra, Astreopora, Discosoma, and Emilania DMSP lyase genes. Species are colored as shown at the top right. Bootstrap support for representative nodes is shown. The eukaryote DMSP lyase clade is highlighted in gray and the cnidarian clade is in purple. Scleractinian DMSP lyase sequences identified from Goniastrea, Galaxea, and Porites were also included in the analysis. (C) Proposed evolutionary history of DMSP lyase in the Anthozoa. Two gene expansion events are shown in red in the phylogenetic tree, and numbers represent the number of genes expanded at each node. Phylogenetic relationships of scleractinian corals are derived from Kitahara et al. (2016).
Possible Antimicrobial Peptides and Symbiosis Genes Are under Positive Selection in Acropora
In order to explore the genetic bases of Acropora diversification, we compared gene repertoires of Acropora corals. We identified 17 OGs for which amino acid sequences are identical among Acropora species (supplementary table S11, Supplementary Material online), indicating that these serve fundamental functions in Acropora. Conserved genes included Homeobox, Forkhead, and Ras-related genes.
In contrast, fast-evolving genes (Ka/Ks > 1) may be essential for Acropora adaptation to diverse or changing environments. Despite the highly conserved protein sequences of Acropora single-copy genes (supplementary fig. S5, Supplementary Material online), manifesting Ka/Ks ratios below 1 (supplementary fig. S6, Supplementary Material online), we identified 35 rapidly evolving candidate OGs (supplementary table S12, Supplementary Material online). Among 14 OGs in which Ka/Ks ratios >1 were detected in more than three species combinations, seven OGs were exclusive to scleractinians (supplementary table S3, Supplementary Material online) or Acropora (supplementary table S5, Supplementary Material online). Interestingly, two candidate genes possibly involved in coral-alga symbiosis, prosaposin (OG0014095, Acropora restricted) and NHL domain-containing gene (OG0010986, scleractinian restricted), were included (supplementary table S12, Supplementary Material online). These genes were exclusively upregulated when planula larvae of Acropora tenuis were infected with native algal symbionts (Y. Yoshioka et al., under review) and Ka/Ks ratios >1 were detected from all species combinations between Clade I and Clade II in prosaposin (supplementary table S12, Supplementary Material online), suggesting possible diverse symbiotic mechanisms within the Acropora clade. Seven fast-evolving OGs with no similarity to proteins in the Swissprot/Pfam databases and shorter than 200 AA were predicted to be antimicrobial peptides (probability >0.95, iAMPpred), facilitating Acropora responses to diverse pathogens.
Conclusion
These comparative genomic analyses reveal genomic novelties that could have allowed Acropora ancestors to survive dynamic environmental changes during geological periods much warmer than the present. Acropora-specific gene duplications in probable stress responsive genes (Caspase-X and SCRiPs), and DMSP lyases may also enable Acropora to cope with elevated ocean temperatures and to disperse and thrive around the world. Although further investigation will be needed, we identified candidate genes involved in Acropora diversification. Genetic mechanisms that enabled Acropora corals to survive past global warming periods may permit them to cope with current global warming. However, the speed of modern climate change may exceed their capacity to adapt, particularly when also confronting local anthropogenic stressors, such as coastal pollution and overexploitation. The present genomic resources, together with further molecular studies, will provide a powerful resource to understand how Acropora diversity originated and has been maintained.
Materials and Methods
Sampling and Genomic DNA Isolation from Acroporid Corals
Specimens of 14 Acropora species (A. acuminata, A. awi, A. cytherea, A. echinata, A. florida, A. gemmifera, A. hyacinthus, A. intermedia, A. microphthalma, A. muricata, A. nasta, A. selago, A. tenuis, and A. yongei) were collected in Sekisei Lagoon, Okinawa, Japan in May 2015, and were maintained in aquaria at the Research Center for Subtropical Fisheries, Seikai National Fisheries Research Institute, until spawning. As reported in Shinzato et al. (2011), each coral colony was separated into different buckets in order to avoid mixing gametes from different individuals, and buoyant egg-sperm bundles were collected to isolate sperm, which were used for isolation of high-quality genomic DNA for sequencing. DNA of A. digitifera was isolated from sperm from a colony collected at Onna Village, Okinawa, in June 2015. We collected eggs, blastulae, gastrulae, planula larvae, early polyps, and adult branches from A. tenuis and A. digitifera for RNA extraction. We also collected three acroporid corals (Astreopora myriophthalma, M. cactus, and M. efflorescens) in Sekisei Lagoon in June 2012 and May 2015. Gametes were collected during spawning and genomic DNA was isolated from sperm. Permits for coral collection were kindly provided by the Okinawa Prefectural Government for research use (Permits #23-47, 25-49, 25-67, 26-68, 27-73, and 27-12).
Genome Assembly and Repetitive Element Analysis
DNA from each species was isolated using the phenol–chloroform method and was fragmented into ∼600 bp lengths. Two hundred nanograms of DNA was used for PCR-free shotgun library preparation. For mate-pair libraries, different sizes of DNA (∼3, 7, 10, and 15 kb) were separated using SageELF (Sage Science). Nextera Mate Pair Library Prep Kits (Illumina) were used for library preparation, following manufacturer instructions. Each 250-bp paired-end library was sequenced using a HiSeq 2500 in Rapid mode (Illumina). Illumina adaptor sequences in raw sequencing data were removed using Trimmomatic (Bolger et al. 2014), and cleaned sequencing data were assembled using Platanus genome assembler ver. 1.2.440 (Kajitani et al. 2014). For A. digitifera, we used a PacBio for genome sequencing and data were assembled with FALCON_unzip (Chin et al. 2016). Sequencing errors in assembled sequences were corrected with Arrows (SMRT Link ver. 4.0.0) using PacBio raw data. All raw genome sequencing data are available under BioProject Accession PRJDB8519. Assembled genomes were further improved by merging possible haploid sequences with HaploMerger2 (Huang et al. 2017). Then, possible errors in all genome assemblies were corrected with Pilon version 1.22 (Walker et al. 2014) using Illumina shotgun and 3-kb mate-pair data. In the end we identified scaffold sequences with high or low coverage or those that may have originated from one of the two allelic copies of heterozygous regions, using Purge Haplotigs (Roach et al. 2018), and excluded these from subsequent analyses. We assessed completeness of genome assembly with BUSCO ver. 3.0.2 (Simao et al. 2015; Waterhouse et al. 2018) and the Metazoan set (978 genes). Repetitive elements in the draft genomes of Acroporidae (Acropora, Montipora, and Astreopora) were identified de novo with RepeatScout v.1.0.5 (Price et al. 2005) and annotated with BlastN and BlastX searches against RepeatMasker.lib and RepeatPeps.lib bundled with RepeatMasker v.4.0.6 (Smit et al. 1996–2010), as reported in Luo et al. (2015, 2018). For nonannotated or putative novel repeats, one additional class “Novel” was introduced. The expansion history of repetitive elements were calculated and visualized using perl scripts from RepeatMasker package (calcDivergenceFromAlign.pl and createRepeatLandscape.pl) as reported in Khalturin et al. (2019).
Gene Prediction and Annotation
We isolated RNA from eggs, blastulae, gastrulae, planula larvae, early polyps, and adult branches of A. tenuis and A. digitifera using RNeasy Mini Kits (Qiagen) and performed RNA-Seq using a HiSeq 2000 platform (Illumina). For gene prediction of Acropora genome assemblies, Augustus version 3.2.3 (Stanke et al. 2006) was first trained using 2,000 high-quality A. digitifera-assembled transcriptome sequences (Shinzato et al. 2011) selected by PASA (Haas et al. 2003). Then, the trained Augustus was used for gene prediction from repeat-masked genome assemblies produced by RepeatMasker (Smit et al. 1996–2010) together with the A. tenuis and A. digitifera RNA-Seq data as gene structure hints. For Montipora and Astreopora, we used predicted protein sequences from A. digitifera and A. tenuis as hints. In order to remove gene models that sometimes originate from different haplotypes, each proteome was clustered using CDHIT (98% sequence identity) (Li and Godzik 2006), and proteins shorter than 30 amino acids were excluded from subsequent analyses. We also assessed completeness of repertoires of predicted genes (mRNA) using BUSCO with the “transcriptome” setting. All proteomes were BLASTed against the Uniprot/Swissprot (UniProt Consortium 2018) database and were analyzed with InterProScan 5 (Jones et al. 2014). Genome browsers for the 18 acroporid genomes are available from the Marine Genomics Unit web site (https://marinegenomics.oist.jp/gallery).
Clustering of Orthologous Anthozoan Genes
In addition to the acroporid genomes, we used publicly available gene models of two anemones, N. vectensis (Putnam et al. 2007) and E. palida (Baumgarten et al. 2015), two corallimorpharians, Amplexidiscus fenestrafer and Discosoma spp. (Wang et al. 2017), and two scleractinians, S. pistillata (Voolstra et al. 2017) and O. faveolata (Prada et al. 2016). For the S. pistillata, O. faveolata, and E. palida genomes, we downloaded data from the NCBI RefSeq database. For Acropora, Montipora, and Astreopora gene models, we selected the longest transcript variant from each gene and used it for subsequent analyses. Then, using OrthoFinder version 2.1.2 (Emms and Kelly 2015), we performed clustering of OGs, the genes descended from a single gene in the last common ancestor of a group of species. CAFE (Computational Analysis of gene Family Evolution, version 4.1) (Han et al. 2013) was used to analyze changes in OG family size in order to account for phylogenetic history. In addition, we prepared a special updated version of ORTHOSCOPE (Inoue and Satoh 2019) for gene tree and orthogroup estimation in publicly available metazoan genomes by incorporating acroporid genomic data (https://www.orthoscope.jp), from which nucleotide and translated amino acid sequences of acroporid gene models used in this study are available. For DMSP lyase analysis, we used Montiopra, Astreopora, Discosoma, and Emilania DMSP genes for BlastP (1e−4) homologous gene search of eukaryote genomes in ORTHOSCOPE, and numbers of expanded genes in each node were predicted using NOTUNG (Chen et al. 2000). We also searched DMSP lyase (TBLASTN, 1e−5) in genomes of Fungia spp., Goniastrea aspera, Pocillopora damicornis, Porites lutea, Galaxea fascicularis, and Pachyseris speciosa deposited in Reefgenomics website (http://reefgenomics.org/) (Voolstra et al. 2015; Liew et al. 2016). For molecular phylogenetic analysis of DMSP lyase, we retrieved possible DMSP lyase sequences from available G. aspera and G. fascicularis gene models. Although no probable DMSP lyase locus was identified in the P. lutea genome and gene models in Reefgenomics, we included a probable DMSP lyase (accession number: FX437344.1) identified from the Porites australiensis transcriptome assembly (Shinzato et al. 2014) for the molecular phylogenetic analysis. For Acropora sequences, we used single species from each of the four clades as follows; I: A. tenuis, II; A. intermedia, III: A. digitifera, IV: A. selago.
Comparison of Anthozoan Genome Organization and Assessment of Genome Duplication
In order to select highly conserved Acropora genome sequences, we selected scaffolds from the A. digitifera genome assembly that share more than 100 OGs in at least one scaffold of the other 14 Acropora genomes, resulting in 38 scaffolds (total 125.7 Mb, 30% of the total assembly size). Conserved 38 A. digitifera scaffold sequences were aligned against each acroporid (A. tenuis, A. intermedia, A. microphthalma, A. selago, Montipora cactus, and Astreopora myriophthalma), scleractinian (S. pistillata), and sea anemone (E. palida) genome using LAST (version 956) (Kielbasa et al. 2011). For each genome alignment, we selected genome sequences showing the three highest alignment scores for each query sequence, and alignments with error probabilities >10−5 were discarded. Dot plots of the alignments sorted by alignment order and orientation of the query were drawn using last-dotplot (Kielbasa et al. 2011). To analyze whether genome- or chromosome-level genome duplication events occurred in anthozoans, we used 300 randomly selected protein sequences of A. digitifera as queries in subsequent BLAST searches. Each selected A. digitifera protein was searched against each anthozoan proteome using BlastP with an e-value cutoff of e−5. Then the top five hits were retrieved. Retrieved protein sequences were aligned using MAFFT (ver. 7.310. with –auto option) (Katoh and Standley 2013), and gaps in the aligned sequences were trimmed using TrimAL (Capella-Gutierrez et al. 2009) with the –gappyout option. After that, poorly aligned sequences were removed (-resoverlap 0.75 -seqoverlap 80). Finally all gaps in alignments were removed using the –nogaps option. In order to restrict protein sequences to those with high-quality alignments and to increase the number of genes for subsequent phylogenetic analyses, we selected alignments containing >200 AAs and more than 40 sequences or >150 AA with more than 80 sequences. Then, we performed molecular phylogenetic analysis of the selected alignments using RAxML (maximum likelihood method) with 100 bootstrap replicates (Stamatakis 2014).
Molecular Phylogeny and Divergence Time Estimation
For molecular phylogenetic analysis of anthozoans, we used 818 OGs that were assigned by OrthoFinder as single-copy genes in all of the above anthozoan genomes. All amino acid sequences belonging to same OG were aligned with MAFFT (Katoh and Standley 2013) and all gaps in the alignment were removed with TrimAL (Capella-Gutierrez et al. 2009). Then all sequences from the same species were concatenated, and finally, a maximum likelihood analysis was performed using concatenated sequences (176,160 amino acids in length) from RAxML with 100 bootstraps. To estimate acroporid divergence times, we performed molecular phylogenetic analysis using 2,126 single-copy OGs among S. pistillata and acroporid corals, and concatenated sequences (621,659 amino acids in length) were analyzed using PhyloBayes v.1.6j (Lartillot et al. 2013) with the -cat -gtr model. We used the oldest fossil records of Astreopora, Montipora, Acropora, and the Suborder Astrocoeniina, the parent taxon of the Acroporidae and Pocilloporidae, found in the Fossilworks database (Behrensmeyer and Turner 2013). For the Stylopora (Pocilloporidae) and Astreopora (Acroporidae) divergence calibration, we applied the oldest fossil record (225.1 Ma) of suborder Astrocoeniina, parent taxon of both the Acroporidae and Pocilloporidae as the upper limit and considered the oldest fossil record of the Acroporidae (164.7 Ma) as the lower limit. For the Astreopora and Montipora divergence calibration, we applied the oldest fossil record of the Acroporidae for the upper limit and used the oldest fossil record of Astreopora (136.4 Ma) for the lower limit. To calibrate the divergence of Acropora and Montipora, we applied the oldest fossil record of Astreopora for the upper limit, because Astreopora is a more basal clade than Acropora and Montipora, and applied the oldest fossil record of Montipora (70.6 Ma) for the lower limit. For calibrations between Acropora species, we employed the oldest fossil record of Acropora (55.8 Ma) as the upper limit and applied the oldest fossil record of each species as the lower limit. Fossil calibrations used are shown in supplementary table S13, Supplementary Material online.
Identification of Highly Conserved and Fast-Evolving Acropora Genes
In order to avoid comparisons of paralogs, we restricted the following calculations to OGs assigned in this study as single-copy genes in Acropora species (4,548 OGs). In addition, to avoid comparing different transcript variants between species, nucleotide and translated amino acid sequences of the longest transcript variant from each species were retrieved for each single-copy OG. Translated amino acid sequences of each group were aligned with MAFFT (with –auto option). Then, aligned nucleotide codon sequences without alignment gaps were retrieved using the PAL2NAL script (Suyama et al. 2006). In order to analyze single-copy OGs with reliable alignment among Acropora, we selected OGs as follows: 1) Alignment nucleotide sequence lengths had to be longer than 90 bp, 2) species for which aligned nucleotide residues were shorter than 90% of alignment were removed using TrimAL “-seqoverlap 90” option, and 3) alignment had to include at least 14 of the 15 species. Average percent identities of aligned nucleotides were calculated using TrimAL. In order to identify possible fast-evolving genes, we calculated nonsynonymous (Ka) and synonymous (Ks) substitution rates of Acropora single-copy OGs by pairwise species comparisons of the 15 Acropora species (105 species combinations in total) using KaKs_Calculator 2.0 (Wang et al. 2010), which incorporates 17 methods for calculation of Ka and Ks substitution rates with the –MA option. To exclude paralogous gene comparisons, we ignored OGs showing synonymous substitution rates >0.1 (Bustamante et al. 2005), and poorly aligned genes (PAL2NAL codon alignment length was shorter than 95% of the average length of the two sequences) were removed from the calculation. Finally, OGs showing Ka/Ks >1 with P < 0.05 (Fisher’s exact test, KaKs_Calculator 2.0) were identified for each pairwise combination. For genes without homology to any sequence in the Uniprot/Swissprot database, we predicted transmembrane helices in translated amino acid sequences using the TMHMM Server v. 2.0 (Krogh et al. 2001) and antimicrobial peptides using iAMPpred (Meher et al. 2017).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Dr Takeshi Takeuchi, Dr Asuka Arimoto, Dr Eiichi Shoguchi, and Dr Yi-Jyun Luo for fruitful discussions. We thank Kanako Hisata for creating the genome browsers. We thank Keiichi Nomura and Go Shimada for helping with species identification of Montipora and Astreopora, respectively. This study was partially supported by JSPS KAKENHI grants (17KT0027, 17K07949, 17H03861 and 20H03235 for C.S., 17K15179 for Y.Z., 26291094 and 18H02270 for H.Y., 15H04538 for G.S.).
Author Contributions
C.S. and G.S. conceptualized, and C.S. and N.S. designed the project. Y.Z., H.Y., and G.S. performed animal collection, acroporid species identification, coral culturing, and sample collection. Mi.K. and Ma.K. prepared sequencing libraries and produced sequencing data. C.S. assembled genomes and performed gene predictions. C.S., K.K., J.I., and Y.Y. performed data analyses. C.S. and N.S. wrote the manuscript. All authors reviewed and approved the final version of the manuscript.
Data Availability
The data underlying this article have been registered at GenBank under the BioProject accession PRJDB8519. Genome assemblies have been deposited at the DNA DataBank of Japan/European Nucleotide Archive/GenBank under accession numbers BLEZ01000000 (Acropora acuminata), BLFA01000000 (Acropora awi), BLFB01000000 (Acropora cytherea), BLFC01000000 (Acropora digitifera), BLFD01000000 (Acropora echinata), BLFE01000000 (Acropora florida), BLFF01000000 (Acropora gemmifera), BLFG01000000 (Acropora hyacinthus), BLFH01000000 (Acropora intermedia), BLFI01000000 (Acropora microphthalma), BLFJ01000000 (Acropora muricata), BLFL01000000 (Acropora nasta), BLFM01000000 (Acropora selago), BLAZ01000000 (Acropora tenuis), BLFN01000000 (Acropora yongei), BLFK01000000 (Astreopora myriophthalma), BLFO01000000 (Montipora cactus), and BLFP01000000 (Montipora efflorescens). Genome browsers for the 18 acroporid genomes are available at the Marine Genomics Unit web site (https://marinegenomics.oist.jp/gallery), and nucleotide and translated amino acid sequences of the acroporid gene models are available at ORTHOSCOPE (https://www.orthoscope.jp).
References
- Aguilar C, Raina JB, Motti CA, Foret S, Hayward DC, Lapeyre B, Bourne DG, Miller DJ.. 2017. Transcriptomic analysis of the response of Acropora millepora to hypo-osmotic stress provides insights into DMSP biosynthesis by corals. BMC Genomics 18(1):612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcolombri U, Ben-Dor S, Feldmesser E, Levin Y, Tawfik DS, Vardi A.. 2015. MARINE SULFUR CYCLE. Identification of the algal dimethyl sulfide-releasing enzyme: a missing link in the marine sulfur cycle. Science 348(6242):1466–1469. [DOI] [PubMed] [Google Scholar]
- Alvarez-Filip L, Carricart-Ganivet JP, Horta-Puga G, Iglesias-Prieto R.. 2013. Shifts in coral-assemblage composition do not ensure persistence of reef functionality. Sci Rep. 3(1):3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron-Szabo RC. 2006. Corals of the K/T-boundary: scleractinian corals of the suborders Astrocoeniina, Faviina, Rhipidogyrina and Amphiastraeina. J Syst Palaeontol. 4:1–108. [Google Scholar]
- Barshis DJ, Ladner JT, Oliver TA, Seneca FO, Traylor-Knowles N, Palumbi SR.. 2013. Genomic basis for coral resilience to climate change. Proc Natl Acad Sci U S A . 110(4):1387–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarten S, Simakov O, Esherick LY, Liew YJ, Lehnert EM, Michell CT, Li Y, Hambleton EA, Guse A, Oates ME, et al. 2015. The genome of Aiptasia, a sea anemone model for coral symbiosis. Proc Natl Acad Sci U S A. 112(38):11893–11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrensmeyer AK, Turner A.. 2013. Taxonomic occurrences of Suidae recorded in the Paleobiology Database. Fossilworks [Internet]. 2013. Available from: http://fossilworks.org.
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadbent AD, Jones GB.. 2004. DMS and DMSP in mucus ropes, coral mucus, surface films and sediment pore waters from coral reefs in the Great Barrier Reef. Mar Freshwater Res. 55(8):849–855. [Google Scholar]
- Broadbent AD, Jones GB, Jones RJ.. 2002. DMSP in corals and benthic algae from the Great Barrier Reef. Estuar Coast Shelf Sci. 55(4):547–555. [Google Scholar]
- Bruckner AW. 2002. Proceedings of the Caribbean Acropora Workshop: Potential Application of the U.S. Endangered Species Act as a Conservation Strategy. NOAA Technical Memorandum NMFS-OPR-24; 2002 April 16–18; Silver Spring (MD): U.S. Department of Commerce.
- Bustamante CD, Fledel-Alon A, Williamson S, Nielsen R, Hubisz MT, Glanowski S, Tanenbaum DM, White TJ, Sninsky JJ, Hernandez RD, et al. 2005. Natural selection on protein-coding genes in the human genome. Nature 437(7062):1153–1157. [DOI] [PubMed] [Google Scholar]
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T.. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25(15):1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone F, Matteucci R, Pignatti JS, Russo A.. 1993. Facies analysis and biostratigraphy of the Auradu Limestone Formation in the Berbera-Sheikh area, northwestern Somalia. Geol Romana. 29:213–235. [Google Scholar]
- Carpenter KE, Abrar M, Aeby G, Aronson RB, Banks S, Bruckner A, Chiriboga A, Cortes J, Delbeek JC, Devantier L, et al. 2008. One-third of reef-building corals face elevated extinction risk from climate change and local impacts. Science 321(5888):560–563. [DOI] [PubMed] [Google Scholar]
- Chen K, Durand D, Farach-Colton M.. 2000. NOTUNG: a program for dating gene duplications and optimizing gene family trees. J Comput Biol. 7(3–4):429–447. [DOI] [PubMed] [Google Scholar]
- Chin CS, Peluso P, Sedlazeck FJ, Nattestad M, Concepcion GT, Clum A, Dunn C, O'Malley R, Figueroa-Balderas R, Morales-Cruz A, et al. 2016. Phased diploid genome assembly with single-molecule real-time sequencing. Nat Methods. 13(12):1050–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles SL, Riegl BM.. 2013. Thermal tolerances of reef corals in the Gulf: a review of the potential for increasing coral survival and adaptation to climate change through assisted translocation. Mar Pollut Bull. 72(2):323–332. [DOI] [PubMed] [Google Scholar]
- Cramwinckel MJ, Huber M, Kocken IJ, Agnini C, Bijl PK, Bohaty SM, Frieling J, Goldner A, Hilgen FJ, Kip EL, et al. 2018. Synchronous tropical and polar temperature evolution in the Eocene. Nature 559(7714):382–386. [DOI] [PubMed] [Google Scholar]
- Cunning R, Bay RA, Gillette P, Baker AC, Traylor-Knowles N.. 2018. Comparative analysis of the Pocillopora damicornis genome highlights role of immune system in coral evolution. Sci Rep. 8(1):16134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De'ath G, Fabricius KE, Sweatman H, Puotinen M.. 2012. The 27-year decline of coral cover on the Great Barrier Reef and its causes. Proc Natl Acad Sci U S A. 109(44):17995–17999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschaseaux ESM, Jones GB, Deseo MA, Shepherd KM, Kiene RP, Swan HB, Harrison PL, Eyre BD.. 2014. Effects of environmental factors on dimethylated sulfur compounds and their potential role in the antioxidant system of the coral holobiont. Limnol Oceanogr. 59(3):758–768. [Google Scholar]
- Emms DM, Kelly S.. 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16(1):157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukami H, Omori M, Hatta M.. 2000. Phylogenetic relationships in the coral family acroporidae, reassessed by inference from mitochondrial genes. Zool Sci. 17(5):689–696. [DOI] [PubMed] [Google Scholar]
- Haas BJ, Delcher AL, Mount SM, Wortman JR, Smith RK Jr, Hannick LI, Maiti R, Ronning CM, Rusch DB, Town CD, et al. 2003. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31(19):5654–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MV, Thomas GW, Lugo-Martinez J, Hahn MW.. 2013. Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Mol Biol Evol. 30(8):1987–1997. [DOI] [PubMed] [Google Scholar]
- Helmkampf M, Bellinger MR, Geib SM, Sim SB, Takabayashi M.. 2019. Draft genome of the rice coral Montipora capitata obtained from linked-read sequencing. Genome Biol Evol. 11(7):2045–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichsen D. 1997. Coral reefs in crisis: an overview of these vanishing ecosystems, the problems that plague them, and the means for saving them. BioScience 47(9):554–558. [Google Scholar]
- Hoegh-Guldberg O, Mumby PJ, Hooten AJ, Steneck RS, Greenfield P, Gomez E, Harvell CD, Sale PF, Edwards AJ, Caldeira K, et al. 2007. Coral reefs under rapid climate change and ocean acidification. Science 318(5857):1737–1742. [DOI] [PubMed] [Google Scholar]
- Huang S, Kang M, Xu A.. 2017. HaploMerger2: rebuilding both haploid sub-assemblies from high-heterozygosity diploid genome assembly. Bioinformatics 33(16):2577–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes MA, Powley IR, Jukes-Jones R, Horn S, Feoktistova M, Fairall L, Schwabe JW, Leverkus M, Cain K, MacFarlane M.. 2016. Co-operative and hierarchical binding of c-FLIP and caspase-8: a unified model defines how c-FLIP isoforms differentially control cell fate. Mol Cell. 61(6):834–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TP, Kerry JT, Alvarez-Noriega M, Alvarez-Romero JG, Anderson KD, Baird AH, Babcock RC, Beger M, Bellwood DR, Berkelmans R, et al. 2017. Global warming and recurrent mass bleaching of corals. Nature 543(7645):373–377. [DOI] [PubMed] [Google Scholar]
- Hughes TP, Kerry JT, Baird AH, Connolly SR, Dietzel A, Eakin CM, Heron SF, Hoey AS, Hoogenboom MO, Liu G, et al. 2018. Global warming transforms coral reef assemblages. Nature 556(7702):492–496. [DOI] [PubMed] [Google Scholar]
- Inoue J, Satoh N.. 2019. ORTHOSCOPE: an automatic web tool for phylogenetically inferring bilaterian orthogroups with user-selected taxa. Mol Biol Evol. 36(3):621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IPCC. 2013. Summary for policymakers. In: Stocker TF, Qin D, Plattner GK, Tignor M, Allen SK, Boschung J, Nauels A, Xia Y, Bex V, Midgley PM, editors. Climate change 2013: the physical science basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge/New York: Cambridge University Press. [Google Scholar]
- Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30(9):1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouiaei M, Sunagar K, Federman Gross A, Scheib H, Alewood PF, Moran Y, Fry BG.. 2015. Evolution of an ancient venom: recognition of a novel family of cnidarian toxins and the common evolutionary origin of sodium and potassium neurotoxins in sea anemone. Mol Biol Evol. 32(6):1598–1610. [DOI] [PubMed] [Google Scholar]
- Kajitani R, Toshimoto K, Noguchi H, Toyoda A, Ogura Y, Okuno M, Yabana M, Harada M, Nagayasu E, Maruyama H, et al. 2014. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 24(8):1384–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalturin K, Shinzato C, Khalturina M, Hamada M, Fujie M, Koyanagi R, Kanda M, Goto H, Anton-Erxleben F, Toyokawa M, et al. 2019. Medusozoan genomes inform the evolution of the jellyfish body plan. Nat Ecol Evol. 3(5):811–822. [DOI] [PubMed] [Google Scholar]
- Kielbasa SM, Wan R, Sato K, Horton P, Frith MC.. 2011. Adaptive seeds tame genomic sequence comparison. Genome Res. 21(3):487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitahara MV, Cairns SD, Stolarski J, Blair D, Miller DJ.. 2010. A comprehensive phylogenetic analysis of the Scleractinia (Cnidaria, Anthozoa) based on mitochondrial CO1 sequence data. PLoS One 5(7):e11490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitahara MV, Fukami H, Benzoni F, Huang D.. 2016. The new systematics of Scleractinia: integrating molecular and morphological evidence. In: Goffredo S, Dubinsky Z, editors. The cnidaria, past, present and future: the world of Medusa and her sisters. Cham (Switzerland: ): Springer International Publishing. p. 41–59. [Google Scholar]
- Knowlton N, Brainard RE, Fisher R, Moews M, Plaisance L, Caley MJ.. 2010. Coral reef biodiversity. In: McIntyre AD, editor. Life in the world's oceans: diversity, distribution, and abundance. Chichester (United Kingdom: ): Wiley-Blackwell. p. 65–79. [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL.. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 305(3):567–580. [DOI] [PubMed] [Google Scholar]
- Kvitt H, Rosenfeld H, Zandbank K, Tchernov D.. 2011. Regulation of apoptotic pathways by Stylophora pistillata (Anthozoa, Pocilloporidae) to survive thermal stress and bleaching. PLoS One 6(12):e28665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Rodrigue N, Stubbs D, Richer J.. 2013. PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst Biol. 62(4):611–615. [DOI] [PubMed] [Google Scholar]
- Li W, Godzik A.. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22(13):1658–1659. [DOI] [PubMed] [Google Scholar]
- Liew YJ, Aranda M, Voolstra CR.. 2016. Reefgenomics.Org - a repository for marine genomics data. Database (Oxford). 2016:baw152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loya Y, Sakai K, Yamazato K, Nakano Y, Sambali H, van Woesik R.. 2001. Coral bleaching: the winners and the losers. Ecol Lett. 4(2):122–131. [Google Scholar]
- Luo YJ, Kanda M, Koyanagi R, Hisata K, Akiyama T, Sakamoto H, Sakamoto T, Satoh N.. 2018. Nemertean and phoronid genomes reveal lophotrochozoan evolution and the origin of bilaterian heads. Nat Ecol Evol. 2(1):141–151. [DOI] [PubMed] [Google Scholar]
- Luo YJ, Takeuchi T, Koyanagi R, Yamada L, Kanda M, Khalturina M, Fujie M, Yamasaki S, Endo K, Satoh N.. 2015. The Lingula genome provides insights into brachiopod evolution and the origin of phosphate biomineralization. Nat Commun. 6(1):8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Economo EP, Satoh N.. 2018. The roles of introgression and climate change in the rise to dominance of Acropora corals. Curr Biol. 28(21):3373–3382.e5. [DOI] [PubMed] [Google Scholar]
- Mao Y, Satoh N.. 2019. A likely ancient genome duplication in the speciose reef-building coral genus, Acropora. iScience 13:20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall PA, Baird AH.. 2000. Bleaching of corals on the Great Barrier Reef: differential susceptibilities among taxa. Coral Reefs 19(2):155–163. [Google Scholar]
- McInerney FA, Wing SL.. 2011. The Paleocene-Eocene thermal maximum: a perturbation of carbon cycle, climate, and biosphere with implications for the future. Annu Rev Earth Planet Sci. 39(1):489–516. [Google Scholar]
- Meher PK, Sahu TK, Saini V, Rao AR.. 2017. Predicting antimicrobial peptides with improved accuracy by incorporating the compositional, physico-chemical and structural features into Chou's general PseAAC. Sci Rep. 7(1):42362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya A, Sakamaki K, Mason BM, Huisman L, Foret S, Weiss Y, Bull TE, Tomii K, Imai K, Hayward DC, et al. 2016. Functional conservation of the apoptotic machinery from coral to man: the diverse and complex Bcl-2 and caspase repertoires of Acropora millepora. BMC Genomics 17(1):62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T. 2017. Mass coral bleaching event in Sekisei lagoon observed in the summer of 2016. J Jpn Coral Reef Soc. 19(1):29–40. [Google Scholar]
- Ohno S. 1970. Evolution by gene duplication. London: George Allen & Unwin.
- Olde K, Jarvis I, Uličný D, Pearce MA, Trabucho-Alexandre J, Čech S, Gröcke DR, Laurin J, Švábenická L, Tocher BA.. 2015. Geochemical and palynological sea-level proxies in hemipelagic sediments: a critical assessment from the Upper Cretaceous of the Czech Republic. Palaeogeogr Palaeoclimatol Palaeoecol. 435:222–243. [Google Scholar]
- Prada C, Hanna B, Budd AF, Woodley CM, Schmutz J, Grimwood J, Iglesias-Prieto R, Pandolfi JM, Levitan D, Johnson KG, et al. 2016. Empty niches after extinctions increase population sizes of modern corals. Curr Biol. 26(23):3190–3194. [DOI] [PubMed] [Google Scholar]
- Price AL, Jones NC, Pevzner PA.. 2005. De novo identification of repeat families in large genomes. Bioinformatics 21(Suppl 1):i351–i358. [DOI] [PubMed] [Google Scholar]
- Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov VV, et al. 2007. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317(5834):86–94. [DOI] [PubMed] [Google Scholar]
- Quinn PK, Bates TS.. 2011. The case against climate regulation via oceanic phytoplankton sulphur emissions. Nature 480(7375):51–56. [DOI] [PubMed] [Google Scholar]
- Raina JB, Dinsdale EA, Willis BL, Bourne DG.. 2010. Do the organic sulfur compounds DMSP and DMS drive coral microbial associations? Trends Microbiol. 18(3):101–108. [DOI] [PubMed] [Google Scholar]
- Raina JB, Tapiolas DM, Foret S, Lutz A, Abrego D, Ceh J, Seneca FO, Clode PL, Bourne DG, Willis BL, et al. 2013. DMSP biosynthesis by an animal and its role in coral thermal stress response. Nature 502(7473):677–680. [DOI] [PubMed] [Google Scholar]
- Ramos-Silva P, Kaandorp J, Huisman L, Marie B, Zanella-Cleon I, Guichard N, Miller DJ, Marin F.. 2013. The skeletal proteome of the coral Acropora millepora: the evolution of calcification by co-option and domain shuffling. Mol Biol Evol. 30(9):2099–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards ZT, Miller DJ, Wallace CC.. 2013. Molecular phylogenetics of geographically restricted Acropora species: implications for threatened species conservation. Mol Phylogenet Evol. 69(3):837–851. [DOI] [PubMed] [Google Scholar]
- Roach MJ, Schmidt SA, Borneman AR.. 2018. Purge Haplotigs: allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinformatics 19(1):460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano SL, Palumbi SR.. 1996. Evolution of scleractinian corals inferred from molecular systematics. Science 271(5249):640–642. [Google Scholar]
- Santodomingo N, Wallace CC, Johnson KG.. 2015. Fossils reveal a high diversity of the staghorn coral genera Acropora and Isopora (Scleractinia: Acroporidae) in the Neogene of Indonesia. Zool J Linn Soc. 175(4):677–763. [Google Scholar]
- Schuster F. 2000. Oligocene and Miocene examples of Acropora-dominated palaeoenvironments: Mesohellenic Basin (NW Greece) and northern Gulf of Suez (Egypt). Proceedings of the 9th International Coral Reef Symposium. Vol. 1. 2000 October 23–27: p. 23–27; Bali, Indonesia: Ministry of Environment, Indonesian Institute of Sciences, International Society for Reef Studies.
- Shinn EA. 1966. Coral growth-rate, an environmental indicator. J Paleontol. 40:233–240. [Google Scholar]
- Shinzato C, Inoue M, Kusakabe M.. 2014. A snapshot of a coral “holobiont”: a transcriptome assembly of the scleractinian coral, porites, captures a wide variety of genes from both the host and symbiotic zooxanthellae. PLoS One 9(1):e85182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinzato C, Shoguchi E, Kawashima T, Hamada M, Hisata K, Tanaka M, Fujie M, Fujiwara M, Koyanagi R, Ikuta T, et al. 2011. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476(7360):320–323. [DOI] [PubMed] [Google Scholar]
- Shumaker A, Putnam HM, Qiu H, Price DC, Zelzion E, Harel A, Wagner NE, Gates RD, Yoon HS, Bhattacharya D.. 2019. Genome analysis of the rice coral Montipora capitata. Sci Rep. 9(1):2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Smit A, Hubley R, Green P.. 1996. –2010. RepeatMasker Open-3.0. Available form: http://www.repeatmasker.org.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Keller O, Gunduz I, Hayes A, Waack S, Morgenstern B.. 2006. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 34(Web Server):W435–W439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagawa S, DeSalvo MK, Voolstra CR, Reyes-Bermudez A, Medina M.. 2009. Identification and gene expression analysis of a taxonomically restricted cysteine-rich protein family in reef-building corals. PLoS One 4(3):e4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunda W, Kieber DJ, Kiene RP, Huntsman S.. 2002. An antioxidant function for DMSP and DMS in marine algae. Nature 418(6895):317–320. [DOI] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P.. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34(Web Server):W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi T, Yamada L, Shinzato C, Sawada H, Satoh N.. 2016. Stepwise evolution of coral biomineralization revealed with genome-wide proteomics and transcriptomics. PLoS One 11(6):e0156424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchernov D, Kvitt H, Haramaty L, Bibby TS, Gorbunov MY, Rosenfeld H, Falkowski PG.. 2011. Apoptosis and the selective survival of host animals following thermal bleaching in zooxanthellate corals. Proc Natl Acad Sci U S A. 108(24):9905–9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium. 2018. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 46(5):2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallina SM, Simo R.. 2007. Strong relationship between DMS and the solar radiation dose over the global surface ocean. Science 315(5811):506–508. [DOI] [PubMed] [Google Scholar]
- Veron JEN. 2000. Corals of the world. Townsville (QLD): Australian Institute of Marine Science.
- Voolstra C, Miller D, Ragan M, Hoffmann A, Hoegh-Guldberg O, Bourne D, Ball E, Ying H, Foret S, Takahashi S, et al. 2015. The ReFuGe 2020 Consortium—using “omics” approaches to explore the adaptability and resilience of coral holobionts to environmental change. Front Mar Sci 2:17583.
- Voolstra CR, Li Y, Liew YJ, Baumgarten S, Zoccola D, Flot JF, Tambutte S, Allemand D, Aranda M.. 2017. Comparative analysis of the genomes of Stylophora pistillata and Acropora digitifera provides evidence for extensive differences between species of corals. Sci Rep. 7(1):17583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9(11):e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace C. 1999. Staghorn corals of the world: a revision of the genus Acropora. Collingwood (VIC: ): CSIRO Publishing. [Google Scholar]
- Wang D, Zhang Y, Zhang Z, Zhu J, Yu J.. 2010. KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinformatics. 8(1):77–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Liew YJ, Li Y, Zoccola D, Tambutte S, Aranda M.. 2017. Draft genomes of the corallimorpharians Amplexidiscus fenestrafer and Discosoma sp. Mol Ecol Resour. 17(6):e187–e195. [DOI] [PubMed] [Google Scholar]
- Waterhouse RM, Seppey M, Simao FA, Manni M, Ioannidis P, Klioutchnikov G, Kriventseva EV, Zdobnov EM.. 2018. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol Biol Evol. 35(3):543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson C. 2008. Status of coral reefs of the world. Townsville (QLD): Global Coral Reef Monitoring Network and Reef And Rainforest Research Center.
- Yellowlees D, Rees TA, Leggat W.. 2008. Metabolic interactions between algal symbionts and invertebrate hosts. Plant Cell Environ. 31(5):679–694. [DOI] [PubMed] [Google Scholar]
- Ying H, Cooke I, Sprungala S, Wang W, Hayward DC, Tang Y, Huttley G, Ball EE, Foret S, Miller DJ.. 2018. Comparative genomics reveals the distinct evolutionary trajectories of the robust and complex coral lineages. Genome Biol. 19(1):175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying H, Hayward DC, Cooke I, Wang W, Moya A, Siemering KR, Sprungala S, Ball EE, Foret S, Miller DJ.. 2019. The whole-genome sequence of the coral Acropora millepora. Genome Biol Evol. 11(5):1374–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachos J, Pagani M, Sloan L, Thomas E, Billups K.. 2001. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science 292(5517):686–693. [DOI] [PubMed] [Google Scholar]
- Zachos JC, Dickens GR, Zeebe RE.. 2008. An early Cenozoic perspective on greenhouse warming and carbon-cycle dynamics. Nature 451(7176):279–283. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article have been registered at GenBank under the BioProject accession PRJDB8519. Genome assemblies have been deposited at the DNA DataBank of Japan/European Nucleotide Archive/GenBank under accession numbers BLEZ01000000 (Acropora acuminata), BLFA01000000 (Acropora awi), BLFB01000000 (Acropora cytherea), BLFC01000000 (Acropora digitifera), BLFD01000000 (Acropora echinata), BLFE01000000 (Acropora florida), BLFF01000000 (Acropora gemmifera), BLFG01000000 (Acropora hyacinthus), BLFH01000000 (Acropora intermedia), BLFI01000000 (Acropora microphthalma), BLFJ01000000 (Acropora muricata), BLFL01000000 (Acropora nasta), BLFM01000000 (Acropora selago), BLAZ01000000 (Acropora tenuis), BLFN01000000 (Acropora yongei), BLFK01000000 (Astreopora myriophthalma), BLFO01000000 (Montipora cactus), and BLFP01000000 (Montipora efflorescens). Genome browsers for the 18 acroporid genomes are available at the Marine Genomics Unit web site (https://marinegenomics.oist.jp/gallery), and nucleotide and translated amino acid sequences of the acroporid gene models are available at ORTHOSCOPE (https://www.orthoscope.jp).