Nonnative species destabilize coastal ecosystems and microbial symbionts may facilitate their spread by enhancing host survival and fitness. However, we know little of the microorganisms that live inside invasive species and whether they change as the host spreads to new areas. In this study, we investigated the microbial communities of an introduced ascidian (Clavelina oblonga) and tracked symbiont changes across locations within the host’s native and invasive ranges. Ascidians in the invasive range had less-diverse microbiomes, as well as lower host haplotype diversity, suggesting that specific colonies reach new locations and carry select symbionts from native populations (i.e., founder effects). Further, ascidians in the invasive range hosted a different composition of symbionts, including microbes with the potential to aid in processes related to invasion success (e.g., nutrient cycling). We conclude that the putative functionality and observed flexibility of this introduced ascidian microbiome may represent an underappreciated factor in the successful establishment of nonnative species in new environments.
KEYWORDS: 16S rRNA, ascidian, invasive, microbiome, tunicate
ABSTRACT
Ascidians are prolific colonizers of new environments and possess a range of well-studied features that contribute to their successful spread, but the role of their symbiotic microbial communities in their long-term establishment is mostly unknown. In this study, we utilized next-generation amplicon sequencing to provide a comprehensive description of the microbiome in the colonial ascidian Clavelina oblonga and examined differences in the composition, diversity, and structure of symbiont communities in the host’s native and invasive ranges. To identify host haplotypes, we sequenced a fragment of the mitochondrial gene cytochrome c oxidase subunit I (COI). C. oblonga harbored a diverse microbiome spanning 42 bacterial and three archaeal phyla. Colonies in the invasive range hosted significantly less diverse symbiont communities and exhibited lower COI haplotype diversity than colonies in the native range. Differences in microbiome structure were also detected across colonies in the native and invasive range, driven largely by novel bacteria representing symbiont lineages with putative roles in nitrogen cycling. Variability in symbiont composition was also observed among sites within each range. Together, these data suggest that C. oblonga hosts a dynamic microbiome resulting from (i) reductions in symbiont diversity due to founder effects in host populations and (ii) environmental selection of symbiont taxa in response to new habitats within a range. Further investigation is required to document the mechanisms behind these changes and to determine how changes in microbiome structure relate to holobiont function and the successful establishment of C. oblonga worldwide.
IMPORTANCE Nonnative species destabilize coastal ecosystems and microbial symbionts may facilitate their spread by enhancing host survival and fitness. However, we know little of the microorganisms that live inside invasive species and whether they change as the host spreads to new areas. In this study, we investigated the microbial communities of an introduced ascidian (Clavelina oblonga) and tracked symbiont changes across locations within the host’s native and invasive ranges. Ascidians in the invasive range had less-diverse microbiomes, as well as lower host haplotype diversity, suggesting that specific colonies reach new locations and carry select symbionts from native populations (i.e., founder effects). Further, ascidians in the invasive range hosted a different composition of symbionts, including microbes with the potential to aid in processes related to invasion success (e.g., nutrient cycling). We conclude that the putative functionality and observed flexibility of this introduced ascidian microbiome may represent an underappreciated factor in the successful establishment of nonnative species in new environments.
INTRODUCTION
The growing problem of marine invasive species has caused severe ecological and economic impacts worldwide (1–3). There is evidence that invasions are increasing in frequency, reflecting an increase in both international shipping and aquaculture (4), which remain the two primary routes of introduction (2, 5, 6). International shipping activities facilitate the spread of nonnative species that are carried within ballast water or attached to ship hulls (2, 7). Aquaculture activities often involve the purposeful transportation of nonnative species outside their ranges in order to cultivate them elsewhere; however, accidental releases and unintentional introduction of associated “hitchhiking” species are common (8, 9).
Ascidians are sessile, benthic filter-feeding invertebrates (phylum Chordata) and are readily transported by maritime vessels, allowing some species to colonize artificial substrates outside their native range (10–12). The introduction of nonnative ascidians has received substantial attention due to the global distribution of many species, recurrent population outbreaks, and associated negative ecological impacts (11). Ascidians can also cause significant economic loss as common fouling organisms of shellfish aquaculture components, such as shells and cages, and often result in reduced shellfish growth (13) and displacement of shellfish (14). Aside from their detrimental effects on the aquaculture industry, nonnative ascidians can also disrupt natural ecosystems by outcompeting native species and eventually becoming dominant members of benthic communities (10, 15, 16).
Ascidians are aggressive competitors for space and resources via high reproduction outputs to quickly colonize available surfaces (17), rapid growth to facilitate overgrowth of other sessile species (15), and secondary metabolite production to deter predation, settling of competitor species, and fouling by epibionts (18–23). Recent evidence has suggested that at least some of these secondary metabolites are produced by symbiotic microorganisms residing within the ascidian tunic, suggesting that microbial symbionts play a critical role in host survival and invasive potential (24, 25).
Ascidians host diverse and highly specific assemblages of symbiotic microorganisms (25–30). At least some of these associations are hypothesized to be mutualistic, with the host providing protective habitat for the symbionts and, in return, the symbionts fulfilling a variety of roles that are beneficial to the host (31). Various symbiont guilds, such as photosynthetic taxa and nitrogen fixers, have been shown to provide photosynthate and fixed nitrogen to the host ascidian, therefore playing an important role in host nutrition (31, 32). Some symbiont metabolic pathways may provide advantageous services to the host; for example, ammonia-oxidizing and nitrite-oxidizing symbionts may remove and recycle waste ammonium (27, 30, 33). Moreover, some bacteria present in the ascidian microbiome are capable of heavy metal processing (30), which may confer a significant advantage to survive in the polluted harbor environments where some invasive ascidians thrive. Most recently, a global study of a worldwide invasive ascidian revealed correlations between microbiome structure and temperature range across sites (34), indicating a role for microbial symbionts in thermal adaptation of the host. Indeed, it has been hypothesized that the services that some symbiont associates carry out within the holobiont may contribute to the successful establishment of ascidians in nonnative areas (25, 30).
Clavelina oblonga (order Aplousobranchia, family Clavelinidae) is a colonial ascidian native to the East Coast of the United States (35) and invasive to Brazil, Panama, the Azores (36), Africa (37), and the Mediterranean Sea (38). Within its invasive range, C. oblonga is found on natural and artificial substrates (36) and was found to negatively impact aquaculture through fouling (38, 39). The recent spread of C. oblonga to natural habitats along the European Atlantic coast highlights its invasive potential (38), although the species’ effect on natural biota is currently unknown. Part of the successful introduction of C. oblonga in several regions of the world may be due to its ability to travel attached to ship hulls and thrive in both natural and polluted areas (36, 40). In this study, we utilized next-generation sequencing to characterize the symbiont community in C. oblonga. We examined the differences in the composition, diversity, and structure of C. oblonga’s symbiont community from the host’s native (Florida and North Carolina) and invasive (Brazil, Italy, and Spain) ranges and further investigated the potential contribution of microbial symbionts to ascidians’ successful establishment in new habitats.
RESULTS
Symbiotic microbial community composition and diversity.
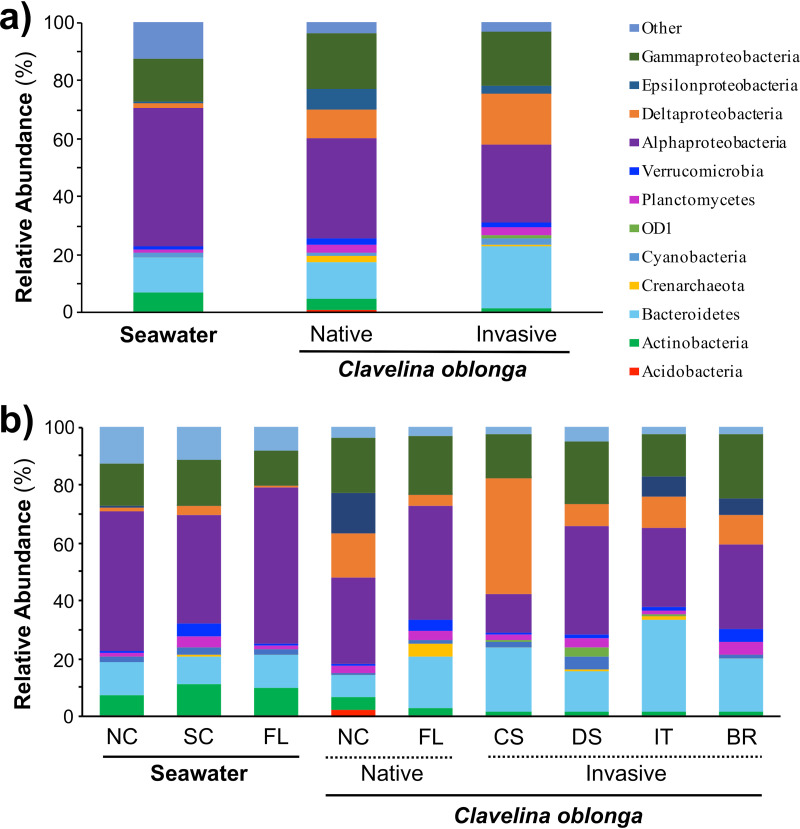
C. oblonga hosted a diverse microbial community consisting of 42 bacterial phyla and 3 archaeal phyla, as well as 163 operational taxonomic units (OTUs) not identified to the phylum level (bacteria, n = 150; archaea, n = 13). The microbial community was dominated by Alphaproteobacteria (average relative abundance = 29.5%), Gammaproteobacteria (18.8%), Bacteroidetes (18.7%), and Deltaproteobacteria (14.7%), together comprising 81.7% of the symbiont community (Fig. 1). All three archaeal phyla were present at each location, and Crenarchaeota was the most abundant archaeal phylum, comprising 1.1% of the average total abundance. Two-thirds of the OTUs identified were rare symbionts (relative abundance, <0.05%) and together comprised 3.3% of all sequence reads. In contrast, these taxa were more abundant and comprised 12.4% of the seawater community, which also exhibited clear phylum-level differences in microbiome composition compared to C. oblonga. Microbial communities in seawater exhibited significant increases in the relative abundances of Actinobacteria (P < 0.001), Alphaproteobacteria (P < 0.001), and Cyanobacteria (P = 0.011), and significant decreases in Bacteroidetes (P = 0.017), Deltaproteobacteria (P < 0.001), and OD1 (P < 0.001) compared to ascidian-associated communities (see Table S1 in the supplemental material).
FIG 1.
Composition of microbial symbionts in the colonial ascidian Clavelina oblonga and ambient seawater, showing the average relative abundances of microbial taxa by source (ambient seawater, C. oblonga) and range (native versus invasive) (a) and by sampling location: North Carolina (NC) and Florida (FL) from the native range and Cadis, Spain (CS), Ebro Delta, Spain (DS), Italy (IT), and Brazil (BR) from the invasive range (b). Symbionts are classified at the phylum level, except Proteobacteria, which are classified at the class level, and rare microbial taxa (contributing <1% to the average abundance across all sites) are classified as “Other.”
The dominant taxa identified in the microbiome of C. oblonga exhibited different relative abundances in hosts from native and invasive ranges. Specifically, Bacteroidetes had significantly elevated abundance in the invasive range (P = 0.037), while Acidobacteria and Actinobacteria had significantly elevated abundances in the native range (P = 0.002 and P = 0.006, respectively [Fig. 1a and Table S1]). Notably, there was high variability across locations within these ranges (Table S2). For example, Crenarchaeota differed significantly across the native range (P = 0.025), comprising 4.4% of the microbial community in C. oblonga from Florida but less than 0.1% of the community in North Carolina (Table S2). Similarly, Epsilonproteobacteria comprised 14.2% of the microbial community in North Carolina but less than 0.2% of the community in Florida (Fig. 1b and Table S2).
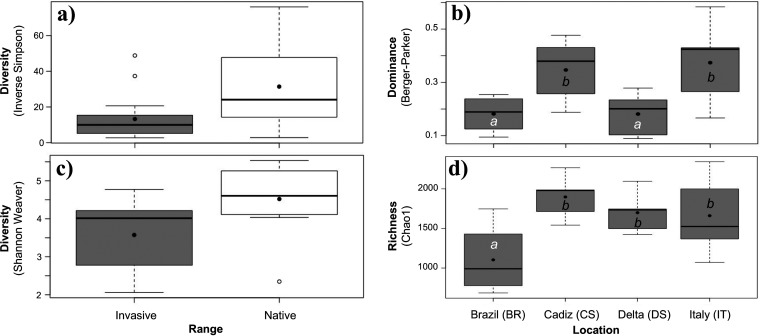
In addition to broad compositional differences, colonies of C. oblonga exhibited significantly lower symbiont community diversity in the invasive range than in the native range (inverse Simpson and Shannon-Weaver [Table 1 and Fig. 2a and c]). Comparisons among locations within each range revealed no significant differences between native locations, while locations within the invasive range exhibited significant differences in the expected richness and dominance indices (Table S3). Specifically, colonies from Brazil exhibited lower expected richness than those at other invasive sites, and colonies from Brazil and the Ebro Delta, Spain, exhibited lower average dominance than other invasive locations (Fig. 2b and d).
TABLE 1.
Analyses of variance results comparing diversity indices of symbiont communities in Clavelina oblonga from native and invasive rangesa
| Diversity index | Value for: |
df | F ratio | P value | |
|---|---|---|---|---|---|
| Native range | Invasive range | ||||
| Observed richness | 1,189 ± 122 | 966 ± 71 | 1, 27 | 2.824 | 0.104 |
| Expected richness, Chao | 1,894 ± 166 | 1,616 ± 105 | 1, 27 | 2.205 | 0.149 |
| Simpson evenness | 0.026 ± 0.004 | 0.017 ± 0.005 | 1, 27 | 1.250 | 0.273 |
| Berger-Parker | 0.186 ± 0.047 | 0.275 ± 0.032 | 1, 27 | 2.535 | 0.123 |
| Inverse Simpson | 31.4 ± 7.4 | 13.3 ± 2.7 | 1, 27 | 7.790 | 0.010* |
| Shannon-Weaver | 4.50 ± 0.30 | 3.57 ± 0.21 | 1, 27 | 6.968 | 0.014* |
Asterisks indicate significant differences between the native and invasive ranges.
FIG 2.
Diversity of symbiont communities in Clavelina oblonga from invasive (gray boxes) and native (white boxes) locations. Significant differences were detected between ranges based on the Inverse Simpson index (a) and the Shannon-Weaver index (c). Significant differences were detected among locations in the invasive range for evenness (b) and richness (d). Data are shown as boxplots depicting median values (central line), mean values (solid black dots), and first and third quartiles (box size). Error bars represent maximum and minimum values up to 2 standard deviations, with white dots representing outliers. Different letters indicate significantly different means among locations.
Symbiont microbial community structure.
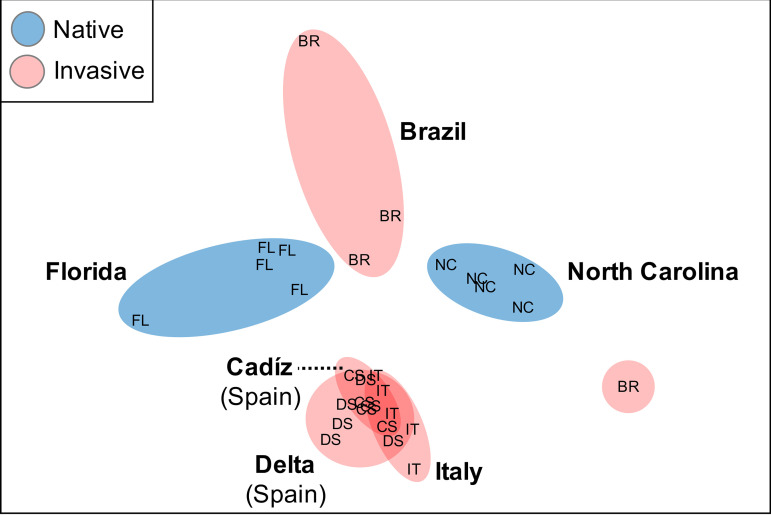
Symbiont community structure of C. oblonga colonies differed significantly from seawater communities (Fig. S1; PERMANOVA, F1,40 = 15.287 and P = 0.001) and averaged 74.1% (Bray-Curtis) dissimilarity between the native and invasive ranges, though these differences in structure across ranges were not significant (PERMANOVA, F1,23 = 1.6982 and P = 0.116). Further, significant differences in microbiome structure were observed across locations within each range (permutational multivariate analysis of variance [PERMANOVA], F4,23 = 2.9429 and P = 0.001 [Table 2]). Greater similarity of symbiont communities was observed across locations within the invasive range (average = 36.0%) than in the native range (32.3%), with fewer OTUs contributing to ∼70% of the similarity within the invasive range (n = 9) than in the native range (n = 56 OTUs). These data indicate reduced variability in community structure within the invasive range, and supporting this, the invasive range communities were more clustered in ordination plots than were the native range (Fig. 3). While no significant difference in dispersion were detected between ranges (permutational multivariate analyses of dispersion [PERMDISP], F1,27 = 0.4014 and P = 0.608), dispersion differed significantly across locations within each range (PERMDISP, F5,23 = 5.973 and P = 0.018). No significant difference in dispersion was detected between the two native locations, while 4 of 6 pairwise comparisons between invasive locations exhibited significant difference in dispersion (Table 2). Notably, 3 of the 4 significant pairwise tests involved colonies from Brazil, which exhibited higher variability in microbiome structure than other invasive locations (Table 2), consistent with ordination clustering (Fig. 3). While the Brazilian sites spanned a broad latitudinal range, no relationship between C. oblonga collection location and symbiont community similarity was observed in the Brazil replicates (Fig. 3).
TABLE 2.
PERMANOVA and PERMDISP results comparing symbiotic community structure in Clavelina oblonga across ranges and locationsa
| Range | Pairwise comparison | PERMANOVA |
PERMDISP |
||
|---|---|---|---|---|---|
| t | P | t | P | ||
| Invasive | Brazil-Spain (C) | 1.7792 | 0.009* | 7.5535 | 0.009* |
| Brazil-Spain (D) | 1.8216 | 0.009* | 5.1595 | 0.007* | |
| Brazil-Italy | 1.5205 | 0.013* | 5.6265 | 0.006* | |
| Spain (C)-Spain (D) | 1.4725 | 0.005* | 2.3698 | 0.099 | |
| Spain (C)-Italy | 1.4559 | 0.007* | 3.0743 | 0.044* | |
| Spain (D)-Italy | 1.2517 | 0.033* | 0.3774 | 0.744 | |
| Native | Florida-North Carolina | 2.1662 | 0.014* | 1.0981 | 0.534 |
Asterisks indicate significant differences between pairwise site comparisons within the invasive and native ranges. PERMANOVA, permutational multivariate analyses of variance; PERMDISP, permutational multivariate analyses of dispersion.
FIG 3.
Nonmetric multidimensional scaling (nMDS) plot visualizing the similarity of symbiotic microbial communities in Clavelina oblonga from invasive (red) and native (blue) locations. Letters indicate different collection locations: Florida (FL), North Carolina (NC), Brazil (BR), Ebro Delta, Spain (DS), Cadis, Spain (CS), and Italy (IT).
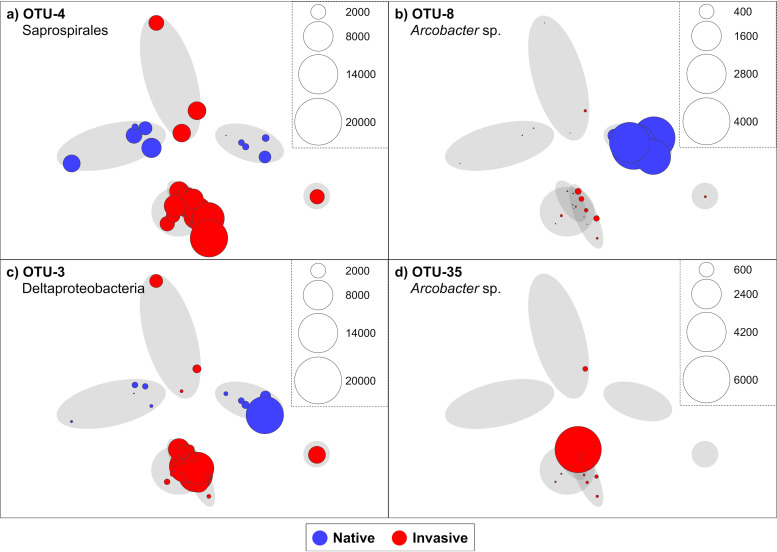
Differences in microbiome structure between colonies in native and invasive ranges were largely driven by 11 symbiont OTUs, which collectively contributed to ∼38% of the observed dissimilarity (Table 3). Notably, these OTUs were absent or exhibited low relative abundances (<0.4%) in seawater samples, with nearly all (10 of 11 OTUs) exhibiting significantly higher relative abundances in ascidian hosts (Table S4, Metastats, P < 0.010). The two largest contributors to community structure differences between ranges were OTU3 (class Deltaproteobacteria) and OTU4 (order Saprospirales), which contributed 10.6% and 8.3%, respectively, and had pairwise identities of 91.2% to the closest known organisms (Table 3). The average abundances of both these OTUs were higher in the invasive range than in the native range (Fig. 4a and c); however, only for OTU4 was this difference significant (Table 3, Metastats, P < 0.001). The relative abundance of OTU4 was relatively constant in C. oblonga colonies within each range (Fig. 4a) and was the largest contributor to microbiome similarity within both ranges, at 33.2% and 11.0% for the invasive and native ranges, respectively. In comparison, OTU3 contributed 3.3% and 17.2% to the similarity of microbial communities within the native and invasive ranges, respectively.
TABLE 3.
Symbiont OTUs that contributed to 38% of the dissimilarity between symbiotic communities in Clavelina oblonga from native and invasive ranges (SIMPER analysis)a
| OTU | Phylum (lowest taxonomy) | Characteristic traits | Avg relative abundance |
% Contribution | BLAST match |
|||
|---|---|---|---|---|---|---|---|---|
| Native | Invasive | P | Source (accession no.) | % ID | ||||
| 3 | Proteobacteria (c. Deltaproteobacteria) | 7.1 | 14.5 | 0.310 | 10.56 | Chiton gill tissue (HE663396) | 91.2 | |
| 4 | Bacteroidetes (o. Saprospirales) | 5.9 | 17.5 | 0.001* | 8.29 | Pu-erh tea (KT360474) | 91.2 | |
| 2 | Proteobacteria (f. Rhodobacteraceae) | Chemoheterotroph, hydrolyzes urea (49) | 7.8 | 10.4 | 0.545 | 6.98 | Culture (NR_114028) | 98.8 |
| 8 | Proteobacteria (g. Arcobacter) | Chemoheterotroph, reduces nitrate (52) | 5.7 | 0.1 | 0.003* | 3.61 | Marine sediment (JX391310) | 100.0 |
| 6 | Crenarchaeota (g. Nitrosopumilus) | NH4+ oxidation and vitamin B12 synthesis (51) | 2.1 | 0.5 | 0.315 | 1.44 | Seawater (KX950758)† | 100.0 |
| 11 | Proteobacteria (Vibrio shiloi) | Heavy metal resistance (50), coral pathogen (54) | 1.9 | 0.7 | 0.217 | 1.31 | Doclea sp. host (MG077075) | 100.0 |
| 15 | Proteobacteria (f. Endozoicimonaceae) | 0.3 | 2.1 | 0.049* | 1.27 | Styela clava host (KU648381) | 99.6 | |
| 51 | Proteobacteria (f. Rhodobacteraceae) | 1.9 | 0.3 | 0.638 | 1.25 | Seawater (NR_132291.1) | 100.0 | |
| 24 | Proteobacteria (c. Deltaproteobacteria) | 0.4 | 1.6 | 0.155 | 1.09 | Subseafloor sediment (JQ989870) | 90.8 | |
| 25 | Proteobacteria (c. Gammaproteobacteria) | 0.6 | 1.4 | 0.480 | 1.01 | Galaxea fascicularis host (KU353970) | 95.2 | |
| 35 | Proteobacteria (g. Arcobacter) | Chemoheterotroph, reduces nitrate (52) | 0.0 | 1.5 | 0.001* | 0.95 | Seawater (KX179260)† | 100.0 |
Shown are phylum- and lowest-level taxonomy (class [c.], order [o.], family [f.], and genus [g.]) and characteristic traits (gray boxes indicate OTUs with taxonomic resolution too low to assign functionality). Average relative abundance of each OTU in the native and invasive range is shown, with asterisks signifying significant differences between ranges (P < 0.05, Metastats). BLAST matches include the source and pairwise identity (ID) of the closest known relative for each OTU, with daggers (†) indicating that the BLAST result is an example chosen from a list of equally matched pairwise identity sequences.
FIG 4.
Relative abundances of symbiont OTUs in Clavelina oblonga from invasive (red circles) and native (blue circles) locations. Shaded circles indicate sample clusters by locations (from Fig. 3) and overlaid circles represent the read abundances of OTU4 Saprospirales (a), OTU8 (Arcobacter sp.) (b), OTU3 (Deltaproteobacteria) (c), and OTU35 (Arcobacter sp.) (d) at each location.
Variability between and within sampling locations also impacted microbiome comparisons between the two ranges of C. oblonga. For example, symbiont OTU8 (genus Arcobacter) contributed 3.6% to the dissimilarity between the ranges and exhibited significantly higher average abundance in the native range (Table 3, P = 0.003), due to elevated abundance in a single location (Fig. 4b). This bacterium exhibited elevated abundance at one native site (North Carolina [11.3%]) yet was present in very low abundance at the other (Florida [0.005%]). Furthermore, there was variability in the relative abundances of some symbiont OTUs among hosts at the same location. Symbiont OTU15 (family Endozoicimonaceae) contributed 1.3% to overall microbiome dissimilarity between ranges (Table 3) and exhibited significantly higher relative abundance in C. oblonga colonies from the invasive range (P < 0.05), yet it displayed elevated relative abundance in only one replicate from the Ebro Delta, Spain (27.9%), and was less abundant in other replicates from the same location (<3.8%). Similarly, symbiont OTU35 (genus Arcobacter) contributed to 1.0% of the dissimilarity and was present only in the invasive range (Table 2) yet at low relative abundances in all replicates from the invasive range (<0.29%) except from a single colony from Italy (26.5% [Fig. 4d]). OTU51 (family Rhodobacteraceae) displayed a similar trend within the native range, exhibiting high abundance (17.2%) in one Florida replicate compared to other colonies at that location (<0.77%).
Mantel tests revealed significant correlations between the similarity of microbial communities in C. oblonga and the geographic distance between all colonies (r = 0.682 and P < 0.001), colonies in the native range only (r = 0.826 and P < 0.001), and colonies in the invasive range only (r = 0.763 and P < 0.001). Partitioning data by range strengthened the explanatory power of regression lines, from 47% (all data) to 68% (native range) and 58% (invasive range) of the variation in ascidian microbiomes accounted for by geographic distance (Fig. S2). This trend was due to the rapid decay in symbiont communities with the native range (slope = −0.038) compared to the invasive range (slope = −0.003).
Host genetic diversity.
In total, we generated 14 cytochrome c oxidase subunit I (COI) sequences for C. oblonga from Beaufort, NC (n = 5), Fernandina Beach, FL (n = 5), Pier Enseada Suá in Vitória (Brazil, n = 2), Pier Baia Golfinhos (Brazil, n = 1), and Pier Florianópolis (Brazil, n = 1). The additional 15 COI sequences analyzed were from samples collected in Spain (Cádiz and Ebro Delta) and Italy (Taranto) obtained previously by Ordóñez et al. (38). Together, these 29 COI sequences grouped into three distinct haplotypes. All Mediterranean and Brazilian colonies were identical to haplotype 3 (GenBank accession number JN703739) described by Rocha et al. (36), along with colonies from Fernandina Beach, FL. Two additional haplotypes were detected in Beaufort, NC, and named in this study haplotype 4 and haplotype 5 to continue the descriptions used by Rocha et al. (36). Haplotype 4 was represented by 3 sequences obtained from Beaufort (CO.3Z, CO.5Z, and HS36.08), identical to a previously characterized sequence for the same species and site (GenBank accession number KY111417) in the work of Villalobos et al. (35), and differed from haplotype 3 by two mutations. Haplotype 5 was previously unreported, also found in Beaufort in two samples (HS36.09 and HS36.10), and differed from haplotype 3 by three mutations and from haplotype 4 by two mutations. Thus, haplotype diversity was higher in the native range of the species (n = 3) than in the invasive range (n = 1).
DISCUSSION
The composition, diversity, and structure of symbiont communities in the colonial ascidian C. oblonga were investigated across locations in the host’s native and invasive ranges. Overall, C. oblonga possessed a diverse symbiont community consisting of 7,702 prokaryotic OTUs representing 42 bacterial phyla and 3 archaeal phyla. Four bacterial taxa dominated the microbiome of C. oblonga (totaling 81.7% relative abundance): Alphaproteobacteria, Gammaproteobacteria, Deltaproteobacteria, and Bacteroidetes, consistent with previous investigations of invasive ascidian microbiomes (30, 41, 42). C. oblonga colonies exhibited significantly less diverse symbiont communities in the invasive range than in the native range, with differences in community structure between the ranges driven largely by the relative abundance of a few, novel bacterial lineages. Variability within sampling ranges and sampling locations also contributed to the observed dissimilarity, allowing for the identification of host-specific and site-specific members of the C. oblonga microbiome. The observed microbiome trends, in conjunction with the putative functionality associated with these symbiont taxa, indicate that colonization events of new habitats and subsequent adaptation shape the microbiome of the globally distributed invasive ascidian C. oblonga.
C. oblonga colonies exhibited less diverse symbiont communities in the invasive range than in the native range, mirroring patterns of host haplotype diversity. In addition to significant differences in two diversity metrics between ranges (inverse Simpson and Shannon-Weaver), all diversity metrics trended lower in the invasive range than in the native range, with the average observed richness (S), expected richness (Chao), and evenness (Simpson) reduced by 18.7%, 14.7%, and 35.0%, respectively. Similarly, the average dominance index (Berger-Parker) did not differ significantly between ranges but was 48.2% higher in the invasive range than in the native range. Founder effects, whereby the genetic diversity of a species decreases within their invasive range as a result of few initial colonizers (43, 44), have also been reported for ascidians (45). Accordingly, the reduction in symbiont diversity in the host’s invasive range observed in this study may result from founder effects on the symbiont taxa, concomitant with a reduction in the host gene pool. In other studies, the species compositions of invertebrate microbiomes have shown spatial variations that parallel host biogeography (for an example, see reference 46) and host haplotype (for an example, see reference 77). C. oblonga possessed lower COI haplotype diversity within the invasive range (1 unique haplotype) than in the native range (3 unique haplotypes). However, all of the Florida colonies (native) analyzed in this study did share a haplotype with the colonies within the invasive range. Lack of COI variation has been previously observed in other invasive sites: Brazil, Panama and the Azores (36), and in the Mediterranean Sea (38). Therefore, reduction in symbiont diversity appeared to be linked to a founder effect in the host.
In addition to differences in diversity, symbiont composition and community structure were also dissimilar between the invasive and native ranges of C. oblonga. However, this trend was not statistically significant, due to variability in symbiont structure within each range, in particular among colonies collected from Brazil. Indeed, significant differences in symbiont structure were detected among locations within the native and invasive ranges. Notably, these differences were greater among locations in the native range, despite closer geographic proximity. For example, colonies in the native range were separated by ca. 500 km and hosted distinct symbiont communities, while colonies in the Mediterranean Sea (invasive range) were separated by ca. 2,000 km yet hosted similar microbiomes. Accordingly, significant correlations between the similarity of microbial communities in C. oblonga and the geographic distance were detected, with greater distance-decay relationships across native populations. On average, symbiont communities decreased in similarity by 3.8% every 100 km within the native range, compared to 0.3% every 100 km within the invasive range. While additional sampling is needed to clarify the source and extent of symbiont variability in C. oblonga, including more sites in the native range and greater representation of the extensive Brazilian coastline, these results indicate a more homogenous microbiome in introduced populations of C. oblonga and greater biogeographic structure in native populations.
The two largest contributors to the observed dissimilarity in symbiont community structure between native and invasive colonies of C. oblonga were a bacterium in the order Saprospirales (OTU4) and a bacterium in the class Deltaproteobacteria (OTU3). Of the two, OTU4 was the only one that presented significantly higher relative abundances in colonies within the invasive range than in the native range, and it represented the largest contributor to the similarity between colonies in the invasive range. Notably, the type species of the order Saprospirales is Saprospira grandis, a coastal marine bacterium with the ability to capture and prey on other marine bacteria (47). Thus, the high abundance of this OTU in colonies within the invasive range may contribute to the reduction in diversity observed in invasive C. oblonga colonies. Indeed, significant negative correlations between OTU4 relative abundance and diversity metrics were detected across all C. oblonga colonies, with OTU4 relative abundance alone explaining 15% to 47% of the variation in microbiome diversity metrics (Fig. S3). Compared to that of OTU4, the average relative abundance of OTU3 was more variable within each range. For example, 3 of the C. oblonga colonies collected from the Ebro Delta, Spain (invasive range), exhibited reduced abundance of OTU3 (<1.4%) within the symbiont community, while 2 colonies exhibited elevated abundances (8.8 and 16.1%). Similarly, even though OTU3 was, on average, more abundant in invasive hosts, one native colony (North Carolina) exhibited an elevated abundance of OTU3 within the symbiont community (57.9% [Fig. 4c]). These data suggest that variability between and within sampling locations also impacts microbiome comparisons between the two ranges of C. oblonga. OTU3 and OTU4 both exhibited low pairwise identities (<92%) to the closest known bacteria, consistent with OTUs considered representative of new families (48), and provide further evidence for the novelty of ascidian-associated bacteria. Other large contributors to symbiont community differences between ranges exhibited closer matches to described bacteria and archaea, including symbionts with putative roles in hydrolyzing urea (49), resistance to heavy metals (50), ammonia oxidation, synthesis of B vitamins (51), and nitrate reduction (52).
OTU11 (Vibrio shiloi) and OTU6 (Nitrosopumilus sp.) both exhibited elevated abundance within native-range C. oblonga colonies compared to the invasive range, where the abundance of both OTUs was low (<1%). Vibrio shiloi is a known coral pathogen implicated in coral bleaching events (53, 54) that also forms associations with other ascidian species (30). The presence of a putative coral pathogen in apparently healthy ascidians further suggests that invasive ascidians may act as disease vectors (30), similar to other invasive taxa (55). OTU6 (Nitrosopumilus sp.) matched with 100% pairwise identity to Nitrosopumilus oxyclinae, an ammonia-oxidizing archaeon (51). Members of the genus Nitrosopumilus have often been described in association with ascidians (27, 30, 33, 41), and related members of the phylum Thaumarchaeota (formerly Crenarchaeota) have been reported in association with other invertebrate hosts (e.g., sponges [56]). The majority of the nitrogenous waste produced by ascidians occurs in the form of ammonium (57), and the resultant high levels of ammonium within the tunic may act as the substrate for ammonia oxidizer metabolism (27, 33, 58). The presence of a Nitrosopumilus sp. (OTU6) within the symbiotic community of C. oblonga further supports previous evidence that ascidian symbionts play a relatively unrecognized role in nitrogen cycling in the ocean (27, 33).
Two additional symbiont taxa with putative roles in nitrogen cycling exhibited elevated abundance in invasive C. oblonga colonies but occurred in moderate abundance (OTU2) or were absent (OTU35) in native colonies. OTU2 was the third greatest contributor to microbiome dissimilarity between the two ranges and matched closely (98.8% pairwise identity) to Maritimibacter akaliphilus, a chemoheterotroph which can use nitrate as a terminal electron acceptor (59) and hydrolyze urea (49). OTU35 matched to the genus Arcobacter, which contains species known to reduce nitrate (52). OTU35 was present only in the symbiont community of invasive colonies, although additional OTUs affiliated with the genus Arcobacter were detected in colonies from both ranges (e.g., OTU8). Potential denitrifying symbionts have been previously identified in ascidians (30), and in other invertebrate species, these symbionts confer evolutionary advantages in low-oxygen environments (60, 61) and supplement host nutrition (62). Harbor systems commonly exhibit elevated nitrate concentrations (for an example, see reference 63), and nitrate-reducing symbionts may aid in colonization and establishment in these regions by lowering nitrate concentrations within the ascidian tunic to tolerable levels.
By investigating C. oblonga from multiple locations in its native and invasive ranges, our study revealed a variety of associations with symbiotic microorganisms that are likely maintained by a “leaky” model of symbiont transmission (i.e., combination of vertical transmission from parent colonies and horizontal uptake from the local environment, reviewed in references 64 and 65). Similar symbiont membership across locations (e.g., OTU6 Nitrosopumilus occurred in every colony studied) suggests relatively stable host-symbiont relationships maintained by vertical transmission. Indeed, C. oblonga colonies reproduce by internal fertilization and produce well-developed brooded larvae (38), a reproductive strategy that allows for parent-to-offspring symbiont transmission in other brooding ascidians (28). Other symbiont taxa detected in C. oblonga were present only in some locations (e.g., OTU35) or matched to environmental bacteria (e.g., seawater and sediment), indicating some role of horizontal transmission in symbiont acquisition and community structure. This process may contribute to the variability observed within symbiotic communities at the location and replicate scale, resulting in dynamic fraction of C. oblonga symbiotic communities as observed in other ascidian species (34, 41, 42). Further investigation is required to understand the complex mechanisms behind loss of symbiont diversity and changes in symbiont community structure in invasive ascidians over time and to fully appreciate how this relates to species successful establishment in disparate habitats.
MATERIALS AND METHODS
Sample collection and DNA extraction.
Ascidian samples were collected from harbors and marinas in two sites within the native range of C. oblonga (Fernandina Beach, FL, and Beaufort, NC) and three countries within the invasive range (Table S5): Brazil (Pier Baia Golfinhos, Pier Enseada Suá in Vitória, and Pier Florianópolis), Italy (Taranto), and Spain (Cádiz and Alfacs Bay, Ebro Delta). Five biological replicates (i.e., samples from different colonies) were collected from all locations, except for Brazil (Pier Baia Golfinhos, n = 1; Pier Enseada Suá, n = 2; and Pier Florianópolis, n = 1), and stored at −20°C in 100% ethanol. Seawater samples (500 ml) were collected from four sites within the native range: Fernandina Beach (n = 3) and Smyrna Beach, FL (n = 3), Sunset Cay, SC (n = 3), and Wrightsville Beach, NC (n = 4), concentrated on 0.2-μm filters, and stored at –80°C. Colonial ascidians consist of individual animals (zooids) embedded in an extracellular cellulose (tunicine) matrix (tunic). Microbial associates commonly occur in the inner tunic (not exposed to the environment), while fouling and food microbes reside on the tunic surface and in the zooid gut, respectively. Accordingly, ascidian samples were dissected under a stereomicroscope to separate zooids (rich in host DNA) from the tunic (rich in symbiont DNA). Zooids were further dissected to separate the branchial sac for host genotyping. To reduce epibiont inclusion, tunic pieces that visually lacked epibionts were selected from each colony for symbiont characterization and washed several times in ethanol to remove loosely associated cells. DNA extractions were conducted separately on pieces of tunic, branchial sac, and seawater filters using the DNeasy blood and tissue kit (Qiagen). All samples were processed separately for host barcoding and symbiont characterization (see below), with resulting data grouped by the factors range (native versus invasive) and site (locations within each range).
Host ascidian barcoding.
Host genetic diversity was assessed by sequencing a fragment (ca. 678) of the mitochondrial gene cytochrome c oxidase subunit I (COI) using the primer set LCO1490 and HCO2198 (66) and PCR conditions described by Villalobos et al. (35). Sequences were obtained in an Applied Biosystems 3500 genetic analyzer available at University of North Carolina Wilmington (UNCW) Center for Marine Science and analyzed using the software Geneious v8 (Biomatters, Auckland, New Zealand). A maximum likelihood phylogeny was constructed using Mega v6.06 (67) with sequences obtained in this study and retrieved from GenBank, the general time-reversible (GTR) model, gamma distribution (G) with invariant sites (I), the nearest-neighbor-interchange heuristic method, and 100 bootstrap replicates. All COI sequences have been deposited in GenBank (accession numbers MK397817 to MK397830).
Microbiome sequencing and statistical analyses of microbial communities.
To characterize the symbiotic communities within C. oblonga, the V4 regions of 16S rRNA genes (ca. 300 bp) were amplified using primers 515f and 806r (68) and sequenced on an Illumina MiSeq platform at Molecular Research LP (Shallowater, TX).
Sequence processing.
Raw sequences were processed using the mothur software package (version 1.39.5 [69]) using a modified version (Table 4) of a previously described pipeline (70). Briefly, low-quality sequences were removed and the remaining sequences aligned to the SILVA database (release 128, nonredundant, mothur formatted). Poorly aligned sequences were removed and the remaining alignments trimmed to the V4 region. Chimeric and nontarget sequences (eukaryotic 18S rRNA, mitochondria, chloroplasts, and unknown) were removed. Operational taxonomic units (OTUs) were formed by creating a pairwise distance matrix and clustering sequences at 97% identity using the OptiClust algorithm (71). Rare OTUs, defined as those represented by fewer than 10 sequence reads across all samples, were removed. The data were then subsampled to standardize sequencing depth across host individuals (n = 22,065), and subsequent analyses were conducted on the subsampled data set.
TABLE 4.
Bioinformatics pipeline for raw sequence data processing in mothur (version 1.39.5), showing commands, input file types and settings for each step
| Command | Input files | Settings |
|---|---|---|
| unique.seqs | fasta | |
| align.seqs | fasta, reference | reference=silva.nr_v128.V4 |
| screen.seqs | fasta, group, name | start=1967, end=11549 |
| filter.seqs | fasta | vertical=T, trump=. |
| pre.culster | fasta, group, name | diffs=2 |
| chimera.uchime | fasta, group, name | dereplicate=t, reference=self |
| remove.seqs | fasta, group, name, accnos | |
| classify.seqs | fasta, group, name | reference=gg_13_5_99.fasta, taxonomy=gg_13_5_99.pds.tax, cutoff=60 |
| remove.lineage | fasta, group, name, taxonomy | taxon=Chloroplast-mitochondria-unknown-eukaryota |
| filter.seqs | fasta | vertical=T, trump=. |
| cluster | column, name | cutoff=0.03 |
| remove.rare | list, group | nseqs=10, label=0.03 |
| classify.otu | list, name, taxonomy | label=0.03 |
| get.oturep | fasta, group, name, list | method=abundance, label=0.03 |
| make.shared | list, group | label=0.03 |
| sub.sample | list, group | size=22065, persample=t |
| list.otulabels | list | |
| get.otulabels | accnoss, constaxonomy | |
| make.shared | list, group |
To investigate microbiome differences between host ranges and among geographic locations therein, statistical analyses included the factors range (invasive versus native) and location (among sites within each range). Statistical analyses of compositional differences (relative abundances of taxonomic groups) were conducted in SigmaPlot (version 11), including one-way analyses of variance (ANOVA) with Tukey pairwise post hoc testing. When distributions failed normality tests (Shapiro-Wilk, P < 0.05), ANOVA were performed on ranked data (Kruskal-Wallis) and pairwise post hoc testing was performed using Dunn’s method. Alpha diversity metrics for observed OTU richness (S), expected OTU (Chao), the Simpson evenness index, the Berger-Parker dominance index, the inverse Simpson index, and the Shannon-Weaver diversity index were calculated in mothur. Analyses of variance (ANOVA) were used to statistically compare the diversity indices between and within ranges and create boxplots using RStudio v3.2.1 (72). Based on OTU relative abundances, Bray-Curtis similarity matrices were created in PRIMER (version 7) after a square root transformation and visualized using nonmetric multidimensional scaling (nMDS) plots. Permutational multivariate analyses of variance (PERMANOVA) were performed in PRIMER for statistical comparisons of symbiont community structure between ranges (native versus invasive) and among sites, with multiple pairwise comparisons conducted for significant main test PERMANOVA results. Permutational multivariate analyses of dispersion (PERMDISP) were used for statistical comparisons of dispersion variability. To determine OTUs contributing to community-level differentiation, similarity percentage (SIMPER) analyses were conducted in PRIMER, to determine the percentage contribution of OTUs to the observed difference in symbiont community structure between the ranges. Representative sequences from OTUs that contributed >50% in total to the dissimilarity between the ranges were further categorized using BLASTn to identify closest-match sequences in GenBank (73). Sequence comparisons were based on highest percent identity matches, and when multiple matches of equal percentage were presented, only sequences described in published articles were reported. METASTATS analyses (74) were conducted in mothur to determine OTUs exhibiting significantly different relative abundances between the ranges (native versus invasive) and hosts (ascidian versus seawater). To test for isolation-by-distance effects (i.e., distance-decay relationships) in ascidian microbiomes, correlations between symbiont community similarity (Bray-Curtis) and geographic distance were assessed with Mantel tests conducted in R v3.3.3 using the package ade4 (75). Mantel tests were conducted on three data partitions: all locations, native locations only, and invasive locations only.
Data availability.
Raw sequence data were deposited in the Sequence Read Archive of the National Center for Biotechnology Information (SRA NCBI; accession no. SRP199333). Sequence data for seawater samples from Wrightsville Beach, NC, and C. oblonga samples from Beaufort, NC, were published previously (30, 76) and retrieved from the NCBI (accession no. SRP106072 and SRP125054). All COI sequences have been deposited in GenBank (accession no. MK397817 to MK397830).
Supplementary Material
ACKNOWLEDGMENTS
We thank X. Turon, V. Ordóñez, R. da Rocha, J. Evans, B. Counts, and H. Sihaloho for providing some of the samples.
This work was supported by a UNCW CMS Pilot grant to S.L.-L. and by grant CTM2017-88080 (AEI/EDER, UE).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Bax N, Williamson A, Aguero M, Gonzalez E, Geeves W. 2003. Marine invasive alien species: a threat to global biodiversity. Mar Policy 27:313–323. doi: 10.1016/S0308-597X(03)00041-1. [DOI] [Google Scholar]
- 2.Molnar JL, Gamboa RL, Revenga C, Spalding MD. 2008. Assessing the global threat of invasive species to marine biodiversity. Front Ecol Environ 6:485–492. doi: 10.1890/070064. [DOI] [Google Scholar]
- 3.Rivlov C, Crooks JA. 2009. Marine bioinvasions: conservation hazards and vehicles for ecological understanding, p 3–11. In Rivlov C, Crooks JA (ed), Biological invasions in marine ecosystems: ecological, management and geographic perspectives. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 4.Cohen AN, Carlton JT. 1998. Accelerating invasion rate in a highly invaded estuary. Science 279:555–558. doi: 10.1126/science.279.5350.555. [DOI] [PubMed] [Google Scholar]
- 5.Ruiz GM, Carlton JT, Grosholz ED, Hines AH. 1997. Global invasions of marine and estuarine habitats by non-indigenous species: mechanisms, extent, and consequences. Am Zool 37:621–632. doi: 10.1093/icb/37.6.621. [DOI] [Google Scholar]
- 6.Ruiz GM, Carlton JT. 2003. Invasion vectors: a conceptual framework for management, p 459–504. In Ruiz GM, Carlton JT (ed), Invasive species: vectors and management strategies. Island Press, Washington, DC. [Google Scholar]
- 7.Sylvester F, Kalaci O, Leung B, Lacoursière-Roussel A, Murray CC, Choi FM, Bravo MA, Therriault TW, MacIsaac HJ. 2011. Hull fouling as an invasion vector: can simple models explain a complex problem? Hull fouling invasion vector. J Appl Ecol 48:415–423. doi: 10.1111/j.1365-2664.2011.01957.x. [DOI] [Google Scholar]
- 8.Goldburg RJ, Elliott MS, Naylor RL. 2001. Marine aquaculture in the United States: environmental impacts and policy options. Pew Oceans Commission, Arlington, VA. [Google Scholar]
- 9.Naylor RL, Williams S, Strong D. 2001. Aquaculture—a gateway for exotic species. Science 294:1655–1656. doi: 10.1126/science.1064875. [DOI] [PubMed] [Google Scholar]
- 10.Lambert C, Lambert G. 2003. Persistence and differential distribution of nonindigenous ascidians in harbors of the Southern California Bight. Mar Ecol Prog Ser 259:145–161. doi: 10.3354/meps259145. [DOI] [Google Scholar]
- 11.Lambert G. 2007. Invasive sea squirts: a growing global problem. J Exp Mar Biol Ecol 342:3–4. doi: 10.1016/j.jembe.2006.10.009. [DOI] [Google Scholar]
- 12.Simkanin C, Davidson IC, Dower JF, Jamieson G, Therriault TW. 2012. Anthropogenic structures and the infiltration of natural benthos by invasive ascidians. Mar Ecol 33:499–511. doi: 10.1111/j.1439-0485.2012.00516.x. [DOI] [Google Scholar]
- 13.Carman MR, Morris JA, Karney RC, Grunden DW. 2010. An initial assessment of native and invasive tunicates in shellfish aquaculture of the North American east coast. J Appl Ichthyol 26:8–11. doi: 10.1111/j.1439-0426.2010.01495.x. [DOI] [Google Scholar]
- 14.Fletcher L, Forrest B, Bell J. 2013. Impacts of the invasive ascidian Didemnum vexillum on green-lipped mussel Perna canaliculus aquaculture in New Zealand. Aquacult Environ Interact 4:17–30. doi: 10.3354/aei00069. [DOI] [Google Scholar]
- 15.Bullard SG, Lambert G, Carman MR, Byrnes J, Whitlatch RB, Ruiz G, Miller RJ, Harris L, Valentine PC, Collie JS, Pederson J, McNaught DC, Cohen AN, Asch RG, Dijkstra J, Heinonen K. 2007. The colonial ascidian Didemnum sp. A: current distribution, basic biology and potential threat to marine communities of the northeast and west coasts of North America. J Exp Mar Biol Ecol 342:99–108. doi: 10.1016/j.jembe.2006.10.020. [DOI] [Google Scholar]
- 16.Rius M, Turon X, Marshall DJ. 2009. Non-lethal effects of an invasive species in the marine environment: the importance of early life-history stages. Oecologia 159:873–882. doi: 10.1007/s00442-008-1256-y. [DOI] [PubMed] [Google Scholar]
- 17.Bourque D, Davidson J, MacNair NG, Arsenault G, LeBlanc AR, Landry T, Miron G. 2007. Reproduction and early life history of an invasive ascidian Styela clava Herdman in Prince Edward Island, Canada J Exp Mar Biol Ecol 342:78–84. doi: 10.1016/j.jembe.2006.10.017. [DOI] [Google Scholar]
- 18.Paul V, Lindquist N, Fenical W. 1990. Chemical defenses of the tropical ascidian Atapozoa sp. and its nudibranch predators Nembrotha spp. Mar Ecol Prog Ser 59:109–118. doi: 10.3354/meps059109. [DOI] [Google Scholar]
- 19.Davis AR. 1991. Alkaloids and ascidian chemical defense: evidence for the ecological role of natural products from Eudistoma olivaceum. Mar Biol 111:375–379. doi: 10.1007/BF01319409. [DOI] [Google Scholar]
- 20.Wahl M, Jensen P, Fenical W. 1994. Chemical control of bacterial epibiosis on ascidians. Mar Ecol Prog Ser 110:45–57. doi: 10.3354/meps110045. [DOI] [Google Scholar]
- 21.Tarjuelo I, López-Legentil S, Codina M, Turon X. 2002. Defence mechanisms of adults and larvae of colonial ascidians: patterns of palatability and toxicity. Mar Ecol Prog Ser 235:103–115. doi: 10.3354/meps235103. [DOI] [Google Scholar]
- 22.López-Legentil S, Turon X, Schupp P. 2006. Chemical and physical defenses against predators in Cystodytes (Ascidiacea). J Exp Mar Biol Ecol 332:27–36. doi: 10.1016/j.jembe.2005.11.002. [DOI] [Google Scholar]
- 23.Morris J. 2009. Impact of the invasive colonial tunicate Didemnum vexillum on the recruitment of the bay scallop (Argopecten irradians irradians) and implications for recruitment of the sea scallop (Placopecten magellanicus) on Georges Bank. Aquat Invasions 4:207–211. doi: 10.3391/ai.2009.4.1.21. [DOI] [Google Scholar]
- 24.Schmidt EW, Nelson JT, Rasko DA, Sudek S, Eisen JA, Haygood MG, Ravel J. 2005. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc Natl Acad Sci U S A 102:7315–7320. doi: 10.1073/pnas.0501424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tianero MDB, Kwan JC, Wyche TP, Presson AP, Koch M, Barrows LR, Bugni TS, Schmidt EW. 2015. Species specificity of symbiosis and secondary metabolism in ascidians. ISME J 9:615–628. doi: 10.1038/ismej.2014.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Donia MS, Fricke WF, Partensky F, Cox J, Elshahawi SI, White JR, Phillippy AM, Schatz MC, Piel J, Haygood MG, Ravel J, Schmidt EW. 2011. Complex microbiome underlying secondary and primary metabolism in the tunicate-Prochloron symbiosis. Proc Natl Acad Sci U S A 108:E1423–E1432. doi: 10.1073/pnas.1111712108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Erwin PM, Pineda MC, Webster N, Turon X, López-Legentil S. 2014. Down under the tunic: bacterial biodiversity hotspots and widespread ammonia-oxidizing archaea in coral reef ascidians. ISME J 8:575–588. doi: 10.1038/ismej.2013.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.López-Legentil S, Turon X, Espluga R, Erwin PM. 2015. Temporal stability of bacterial symbionts in a temperate ascidian. Front Microbiol 6:1022. doi: 10.3389/fmicb.2015.01022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.López-Legentil S, Turon X, Erwin PM. 2016. Feeding cessation alters host morphology and bacterial communities in the ascidian Pseudodistoma crucigaster. Front Zool 13:2. doi: 10.1186/s12983-016-0134-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans JS, Erwin PM, Shenkar N, López-Legentil S. 2017. Introduced ascidians harbor highly diverse and host-specific symbiotic microbial assemblages. Sci Rep 7:11033. doi: 10.1038/s41598-017-11441-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirose E, Maruyama T. 2004. What are the benefits in the ascidian-Prochloron symbiosis? Endocytobiosis Cell Res 15:51–62. [Google Scholar]
- 32.Kühl M, Larkum AWD. 2004. The microenvironment and photosynthetic performance of Prochloron sp. in symbiosis with didemnid ascidians, p 273–290. In Seckbach J. (ed), Cellular origin and life in extreme habitats: symbioses, mechanisms and model systems. Kluwer Academic Publishers, Dordrecht, the Netherlands. [Google Scholar]
- 33.Martínez-García M, Stief P, Díaz-Valdés M, Wanner G, Ramos-Esplá A, Dubilier N, Antón J. 2008. Ammonia-oxidizing Crenarchaeota and nitrification inside the tissue of a colonial ascidian. Environ Microbiol 10:2991–3001. doi: 10.1111/j.1462-2920.2008.01761.x. [DOI] [PubMed] [Google Scholar]
- 34.Casso M, Turon M, Marco N, Pascual M, Turon X. 2020. The microbiome of the worldwide invasive ascidian Didemnum vexillum. Front Mar Sci 7:201. [Google Scholar]
- 35.Villalobos S, Lambert G, Shenkar N, López-Legentil S. 2017. Distribution and population dynamics of key ascidians in North Carolina harbors and marinas. Aquat Invasions 12:447–458. doi: 10.3391/ai.2017.12.4.03. [DOI] [Google Scholar]
- 36.Rocha R, Kremer L, Fehlauer-Ale K. 2012. Lack of COI variation for Clavelina oblonga (Tunicata, Ascidiacea) in Brazil: evidence for its human-mediated transportation? Aquat Invasions 7:419–424. doi: 10.3391/ai.2012.7.3.012. [DOI] [Google Scholar]
- 37.Pérès J. 1949. Contribution à l’étude des ascidies de la côte occidentale d’Afrique. Bull Inst Fondam Afr Noire 11:159–207. [Google Scholar]
- 38.Ordóñez V, Pascual M, Fernández-Tejedor M, Turon X. 2016. When invasion biology meets taxonomy: Clavelina oblonga (Ascidiacea) is an old invader in the Mediterranean Sea. Biol Invasions 18:1203–1215. doi: 10.1007/s10530-016-1062-0. [DOI] [Google Scholar]
- 39.Rocha R. 2009. Bivalve cultures provide habitat for exotic tunicates in southern Brazil. Aquat Invasions 4:195–205. doi: 10.3391/ai.2009.4.1.20. [DOI] [Google Scholar]
- 40.Monniot F. 1972. Ascidies aplousobranches des Bermudes. 1. Polyclinidae et Polycitorida. Bull Museum Natl Hist Nat 61:949–962. [Google Scholar]
- 41.Erwin PM, Carmen Pineda M, Webster N, Turon X, López-Legentil S. 2013. Small core communities and high variability in bacteria associated with the introduced ascidian Styela plicata. Symbiosis 59:35–46. doi: 10.1007/s13199-012-0204-0. [DOI] [Google Scholar]
- 42.Evans JS, Erwin PM, Shenkar N, López-Legentil S. 2018. A comparison of prokaryotic symbiont communities in nonnative and native ascidians from reef and harbor habitats. FEMS Microbiol Ecol 94:fiy139. doi: 10.1093/femsec/fiy139. [DOI] [PubMed] [Google Scholar]
- 43.Sakai AK, Allendorf FW, Holt JS, Lodge DM, Molofsky J, With KA, Baughman S, Cabin RJ, Cohen JE, Ellstrand NC, McCauley DE, O’Neil P, Parker IM, Thompson JN, Weller SG. 2001. The population biology of invasive species. Annu Rev Ecol Syst 32:305–332. doi: 10.1146/annurev.ecolsys.32.081501.114037. [DOI] [Google Scholar]
- 44.Dlugosch KM, Parker IM. 2008. Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol Ecol 17:431–449. doi: 10.1111/j.1365-294X.2007.03538.x. [DOI] [PubMed] [Google Scholar]
- 45.Pérez-Portela R, Turon X, Bishop J. 2012. Bottlenecks and loss of genetic diversity: spatio-temporal patterns of genetic structure in an ascidian recently introduced in Europe. Mar Ecol Prog Ser 451:93–105. doi: 10.3354/meps09560. [DOI] [Google Scholar]
- 46.Rubio-Portillo E, Kersting DK, Linares C, Ramos-Esplá AA, Antón J. 2018. Biogeographic differences in the microbiome and pathobiome of the coral Cladocora caespitosa in the western Mediterranean Sea. Front Microbiol 9:22. doi: 10.3389/fmicb.2018.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lewin RA. 1997. Saprospira grandis: a flexibacterium that can catch bacterial prey by “ixotrophy.” Microb Ecol 34:232–236. doi: 10.1007/s002489900052. [DOI] [PubMed] [Google Scholar]
- 48.Rosselló-Móra R, Amann R. 2015. Past and future species definitions for Bacteria and Archaea. Syst Appl Microbiol 38:209–216. doi: 10.1016/j.syapm.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 49.Lee K, Choo Y-J, Giovannoni SJ, Cho J-C. 2007. Maritimibacter alkaliphilus gen. nov., sp. nov., a genome-sequenced marine bacterium of the Roseobacter clade in the order Rhodobacterales. Int J Syst Evol Microbiol 57:1653–1658. doi: 10.1099/ijs.0.64960-0. [DOI] [PubMed] [Google Scholar]
- 50.Reshef L, Ron E, Rosenberg E. 2008. Genome analysis of the coral bleaching pathogen Vibrio shiloi. Arch Microbiol 190:185–194. doi: 10.1007/s00203-008-0388-0. [DOI] [PubMed] [Google Scholar]
- 51.Qin W, Heal KR, Ramdasi R, Kobelt JN, Martens-Habbena W, Bertagnolli AD, Amin SA, Walker CB, Urakawa H, Könneke M, Devol AH, Moffett JW, Armbrust EV, Jensen GJ, Ingalls AE, Stahl DA. 2017. Nitrosopumilus maritimus gen. nov., sp. nov., Nitrosopumilus cobalaminigenes sp. nov., Nitrosopumilus oxyclinae sp. nov., and Nitrosopumilus ureiphilus sp. nov., four marine ammonia-oxidizing archaea of the phylum Thaumarchaeota. Int J Syst Evol Microbiol 67:5067–5079. doi: 10.1099/ijsem.0.002416. [DOI] [PubMed] [Google Scholar]
- 52.Vandamme P, Falsen E, Rossau R, Hoste B, Segers P, Tytgat R, De Ley J. 1991. Revision of Campylobacter, Helicobacter, and Wolinella taxonomy: emendation of generic descriptions and proposal of Arcobacter gen. nov. Int J Syst Bacteriol 41:88–103. doi: 10.1099/00207713-41-1-88. [DOI] [PubMed] [Google Scholar]
- 53.Loya Y, Banin E, Rosenberg E, Stackebrandt E, Kushmaro A. 2001. Vibrio shiloi sp. nov., the causative agent of bleaching of the coral Oculina patagonica. Int J Syst Evol Microbiol 51:1383–1388. doi: 10.1099/00207713-51-4-1383. [DOI] [PubMed] [Google Scholar]
- 54.Kushmaro A, Loya Y, Fine M, Rosenberg E. 1996. Bacterial infection and coral bleaching. Nature 380:396. doi: 10.1038/380396a0. [DOI] [Google Scholar]
- 55.Pimentel D, Zuniga R, Morrison D. 2005. Update on the environmental and economic costs associated with alien-invasive species in the United States. Ecol Econ 52:273–288. doi: 10.1016/j.ecolecon.2004.10.002. [DOI] [Google Scholar]
- 56.López-Legentil S, Erwin PM, Pawlik JR, Song B. 2010. Effects of sponge bleaching on ammonia-oxidizing Archaea: distribution and relative expression of ammonia monooxygenase genes associated with the barrel sponge Xestospongia muta. Microb Ecol 60:561–571. doi: 10.1007/s00248-010-9662-1. [DOI] [PubMed] [Google Scholar]
- 57.Goodbody I. 1965. Nitrogen excretion in Ascidiacea: II. Storage excretion and the uricolytic enzyme system. J Exp Biol 42:299–305. [Google Scholar]
- 58.Parry DL. 1985. Nitrogen assimilation in the symbiotic marine algae Prochloron spp. Mar Biol 87:219–222. doi: 10.1007/BF00397797. [DOI] [Google Scholar]
- 59.Takai K, Moyer CL, Miyazaki M, Nogi Y, Hirayama H, Nealson KH, Horikoshi K. 2005. Marinobacter alkaliphilus sp. nov., a novel alkaliphilic bacterium isolated from subseafloor alkaline serpentine mud from Ocean Drilling Program Site 1200 at South Chamorro Seamount, Mariana Forearc. Extremophiles 9:17–27. doi: 10.1007/s00792-004-0416-1. [DOI] [PubMed] [Google Scholar]
- 60.Hentschel U, Felbeck H. 1995. Nitrate respiration in chemoautotrophic symbionts of the bivalve Lucinoma aequizonata is not regulated by oxygen. Appl Environ Microbiol 61:1630–1633. doi: 10.1128/AEM.61.4.1630-1633.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hentschel U, Felbeck H. 1993. Nitrate respiration in the hydrothermal vent tubeworm Riftia pachyptila. Nature 366:338–340. doi: 10.1038/366338a0. [DOI] [Google Scholar]
- 62.Hentschel U, Hand SC, Felbeck H. 1996. The contribution of nitrate respiration to the energy budget of the symbiont-containing clam Lucinoma aequizonata: a calorimetric study. J Exp Biol 199:427–433. [DOI] [PubMed] [Google Scholar]
- 63.Estacio FJ, García-Adiego EM, Fa DA, García-Gómez JC, Daza JL, Hortas F, Gómez-Ariza JL. 1997. Ecological analysis in a polluted area of Algeciras Bay (southern Spain): external ‘versus’ internal outfalls and environmental implications. Mar Pollut Bull 34:780–793. doi: 10.1016/S0025-326X(97)00046-5. [DOI] [Google Scholar]
- 64.Bright M, Bulgheresi S. 2010. A complex journey: transmission of microbial symbionts. Nat Rev Microbiol 8:218–230. doi: 10.1038/nrmicro2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kojima A, Hirose E. 2012. Transmission of cyanobacterial symbionts during embryogenesis in the coral reef ascidians Trididemnum nubilum and T. clinides (Didemnidae, Ascidiacea, Chordata). Biol Bull 222:63–73. doi: 10.1086/BBLv222n1p63. [DOI] [PubMed] [Google Scholar]
- 66.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3:294–299. [PubMed] [Google Scholar]
- 67.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weigel BL, Erwin PM. 2016. Intraspecific variation in microbial symbiont communities of the sun sponge, Hymeniacidon heliophila, from intertidal and subtidal habitats. Appl Environ Microbiol 82:650–658. doi: 10.1128/AEM.02980-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Westcott SL, Schloss PD. 2016. OptiClust: improved method for assigning amplicon-based sequence data to operational taxonomic units. bioRxiv doi: 10.1101/096537. [DOI] [PMC free article] [PubMed]
- 72.R Core Team. 2017. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 73.Altschul S, Gish W, Miller W, Myers E, Lipman D. 1990. Basic local alignment search tool. J Mol Biol 215:403–310. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 74.White JR, Nagarajan N, Pop M. 2009. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol 5:e1000352. doi: 10.1371/journal.pcbi.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dray S, Dufour A-B. 2007. The ade4 package: implementing the duality diagram for ecologists. J Stat Softw 22:1–20. [Google Scholar]
- 76.Evans JS, López-Legentil S, Erwin PM. 2018. Comparing two common DNA extraction kits for the characterization of symbiotic microbial communities from ascidian tissue. Microbes Environ 33:435–439. doi: 10.1264/jsme2.ME18031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Marino C, Pawlik J, López-Legentil S, Erwin P. 2017. Latitudinal variation in the microbiome of the sponge Ircinia campana correlates with host haplotype but not anti-predatory chemical defense. Mar Ecol Prog Ser 565:53–66. doi: 10.3354/meps12015. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequence data were deposited in the Sequence Read Archive of the National Center for Biotechnology Information (SRA NCBI; accession no. SRP199333). Sequence data for seawater samples from Wrightsville Beach, NC, and C. oblonga samples from Beaufort, NC, were published previously (30, 76) and retrieved from the NCBI (accession no. SRP106072 and SRP125054). All COI sequences have been deposited in GenBank (accession no. MK397817 to MK397830).