Serpentinization of ultramafic rock can generate conditions favorable for microbial methane (CH4) cycling, including the abiotic production of hydrogen (H2) and possibly CH4. Systems of low-temperature serpentinization are geobiological targets due to their potential to harbor microbial life and ubiquity throughout Earth’s history. Biomass in fracture waters collected from the Samail Ophiolite of Oman, a system undergoing modern serpentinization, yielded DNA and RNA signatures indicative of active microbial methanogenesis and methanotrophy. Intriguingly, transcripts for proteins involved in methanogenesis were most abundant in the most highly reacted waters that have hyperalkaline pH and elevated concentrations of H2 and CH4. These findings suggest active biological methane cycling in serpentinite-hosted aquifers, even under extreme conditions of high pH and carbon limitation. These observations underscore the potential for microbial activity to influence the isotopic composition of CH4 in these systems, which is information that could help in identifying biosignatures of microbial activity on other planets.
KEYWORDS: Ophiolite, serpentinization, environmental microbiology, geomicrobiology, methane, methanogens
ABSTRACT
Serpentinization can generate highly reduced fluids replete with hydrogen (H2) and methane (CH4), potent reductants capable of driving microbial methanogenesis and methanotrophy, respectively. However, CH4 in serpentinized waters is thought to be primarily abiogenic, raising key questions about the relative importance of methanogens and methanotrophs in the production and consumption of CH4 in these systems. Herein, we apply molecular approaches to examine the functional capability and activity of microbial CH4 cycling in serpentinization-impacted subsurface waters intersecting multiple rock and water types within the Samail Ophiolite of Oman. Abundant 16S rRNA genes and transcripts affiliated with the methanogenic genus Methanobacterium were recovered from the most alkaline (pH, >10), H2- and CH4-rich subsurface waters. Additionally, 16S rRNA genes and transcripts associated with the aerobic methanotrophic genus Methylococcus were detected in wells that spanned varied fluid geochemistry. Metagenomic sequencing yielded genes encoding homologs of proteins involved in the hydrogenotrophic pathway of microbial CH4 production and in microbial CH4 oxidation. Transcripts of several key genes encoding methanogenesis/methanotrophy enzymes were identified, predominantly in communities from the most hyperalkaline waters. These results indicate active methanogenic and methanotrophic populations in waters with hyperalkaline pH in the Samail Ophiolite, thereby supporting a role for biological CH4 cycling in aquifers that undergo low-temperature serpentinization.
IMPORTANCE Serpentinization of ultramafic rock can generate conditions favorable for microbial methane (CH4) cycling, including the abiotic production of hydrogen (H2) and possibly CH4. Systems of low-temperature serpentinization are geobiological targets due to their potential to harbor microbial life and ubiquity throughout Earth’s history. Biomass in fracture waters collected from the Samail Ophiolite of Oman, a system undergoing modern serpentinization, yielded DNA and RNA signatures indicative of active microbial methanogenesis and methanotrophy. Intriguingly, transcripts for proteins involved in methanogenesis were most abundant in the most highly reacted waters that have hyperalkaline pH and elevated concentrations of H2 and CH4. These findings suggest active biological methane cycling in serpentinite-hosted aquifers, even under extreme conditions of high pH and carbon limitation. These observations underscore the potential for microbial activity to influence the isotopic composition of CH4 in these systems, which is information that could help in identifying biosignatures of microbial activity on other planets.
INTRODUCTION
Life in deep subsurface environments is dependent on lithosphere-derived nutrients to drive metabolism and biosynthesis (i.e., chemosynthesis). Water-rock interactions are one potential source of nutrients that can be used by biological systems to generate chemical energy. During the hydration of olivine and/or pyroxene in ultramafic rocks, the oxidation of ferrous iron coupled with the reduction of water can generate molecular hydrogen (H2) through the geological process of serpentinization (1, 2). Elevated dissolved H2 concentrations can drive the reduction of inorganic carbon (CO2) to generate formate (HCOO−) and carbon monoxide (CO) (3), as well as methane (CH4) and additional light hydrocarbons, through abiotic reactions at low temperature (<100°C) (4, 5). Serpentinization-impacted waters often have very low oxidation-reduction potentials, have pH values of 8 to greater than 12, and can have nM to mM concentrations of H2 and CH4 that can serve as electron donors to fuel microbial metabolism (6–12). Zones of active, low-temperature serpentinization exist beneath the water table within ophiolites, portions of the oceanic crust and upper mantle that have been tectonically emplaced onto a continent. Ophiolites, such as the Samail Ophiolite in the Sultanate of Oman, provide an accessible venue to study the subsurface biosphere in bedrock environments undergoing serpentinization (7, 13).
Current data suggest CH4 in ophiolites is generated abiotically at low temperatures or is primarily relict from early high-temperature water/rock reactions that trapped fluids and gases in fluid inclusions, which are later released during weathering (5, 14). The abiotic sources of CH4 in these systems are inferred by studies of stable isotope compositions showing CH4 enriched in 13C (5). Alternatively, several types of microorganisms can produce CH4, including methanogenic Archaea that can generate CH4 from a variety of substrates, including H2/CO2, CO, formate, a variety of methylated substrates, and acetate (15), of which many have been detected in waters that have been subjected to serpentinization (7, 9, 16–18). Although methanogens differ in their substrate use, all require the methyl-coenzyme M reductase (MCR) enzyme complex (encoded by mcrABG) for the terminal step of methanogenesis. Some H2-dependent methanogens can also use formate as a source of electrons to reduce CO2 instead of H2 via the activity of formate dehydrogenase (encoded by fdhAB) (19). Conversely, anaerobic methanotrophs oxidize CH4 with a variety of terminal electron acceptors, including sulfate (SO42−), nitrate (NO3−), nitrite (NO2−), and metals, likely via the reverse methanogenesis pathway (20). In addition to anaerobes, aerobic methanotrophs catalyze the oxidation of CH4 (to methanol) using the particulate or soluble methane monooxygenase enzymes, encoded by the pmoABC and mmoXYZ genes, respectively (21, 22). In the second step of CH4 oxidation, methanol dehydrogenases (MDH) oxidize methanol to formaldehyde. The mxaF gene encodes the large subunit of the NAD-independent MDH known to proteobacterial methanotrophs (23). These genes, therefore, can serve as key putative markers of methanogenesis and methanotrophy in natural systems. The presence of genes that encode [NiFe]-hydrogenases and carbon cycling processes can provide further insight into the specific electron donors capable of fueling these cells in an environment.
Several environments impacted by the process of serpentinization show evidence of microbial methanogenesis and methanotrophy. For example, the detection of key genes required for methanogenesis and/or methanotrophy from the Voltri Massif (Italy), the Samail Ophiolite (Oman), and the Santa Elena Ophiolite (Costa Rica) suggests that these processes are active in these systems (9, 18, 24). The case for the presence of these organisms in ophiolites is bolstered by the detection of 16S rRNA genes affiliated with known CH4 cycling organisms (6, 7, 9, 25). Methanotrophic ANME-1 archaea have been detected via high-throughput sequencing methods in the Voltri Massif and Cabeço de Vide aquifers of Italy and Portugal, respectively (17, 18). Furthermore, the composition and 13C enrichment of archaeal lipids from the Chimaera ophiolite of Turkey provides evidence of archaeal methanogenesis under inorganic carbon limitation at that site (26). Organisms collected from the Samail Ophiolite and the Cedars (California) show CH4 production when amended with 14C- or 13C-labeled substrates in activity assays, respectively (11, 24), and the incubations conducted with organisms from the Samail Ophiolite also show labeled substrate assimilation into biomass (24). Additionally, an enrichment culture of a methanogen of the genus Methanobacterium grown from alkaline waters of the Samail Ophiolite was active over a pH range of 6.9 to 10.1 and showed an ability to use HCO3− and CaCO3 as a C source (27). Yet, other sites, including the Coast Range Ophiolite Microbial Observatory (CROMO) (California) and the Tablelands Ophiolite (Newfoundland, Canada), show no evidence of microbial methanogenesis, suggesting the presence of unknown factors that limit the distribution of CH4 metabolisms at these sites (28, 29). Therefore, while incubation and cultivation studies show microbial activity when amended with substrate, they may not be representative of activity in the modern subsurface of ophiolites. Consequently, the environmental conditions conducive to methanogenic activity require further investigation.
The Samail Ophiolite is the largest (approximately 15,000 km3) and best-exposed ophiolite on Earth, with zones in the mantle peridotite section currently undergoing serpentinization largely below 60°C (7, 30–34). In 1983 to 1985 and 2004 to 2005, the Sultanate of Oman drilled several wells into the ophiolite, making it an accessible location to sample subsurface fracture waters and to investigate the microbial contribution to CH4 cycling in a low-temperature continental serpentinizing environment. Previous work has detected μM to mM concentrations of dissolved H2 and CH4 in aquifer waters, with the CH4 in hyperalkaline waters displaying unusually high δ13C values (up to +3‰ VPDB [Vienna Pee Dee Belemnite]) that do not fall within typical ranges of microbial CH4 (7, 27, 32, 35). This suggests either an abiotic origin for CH4 or extensive biological production and/or consumption of CH4 that is already enriched in δ13C. Here, we apply genomic and transcriptomic sequencing approaches to biomass collected from these same sites to better define the distribution and putative activity of microbial methanogens and methanotrophs within the Samail Ophiolite.
RESULTS
Geochemical characterization of subsurface fracture waters.
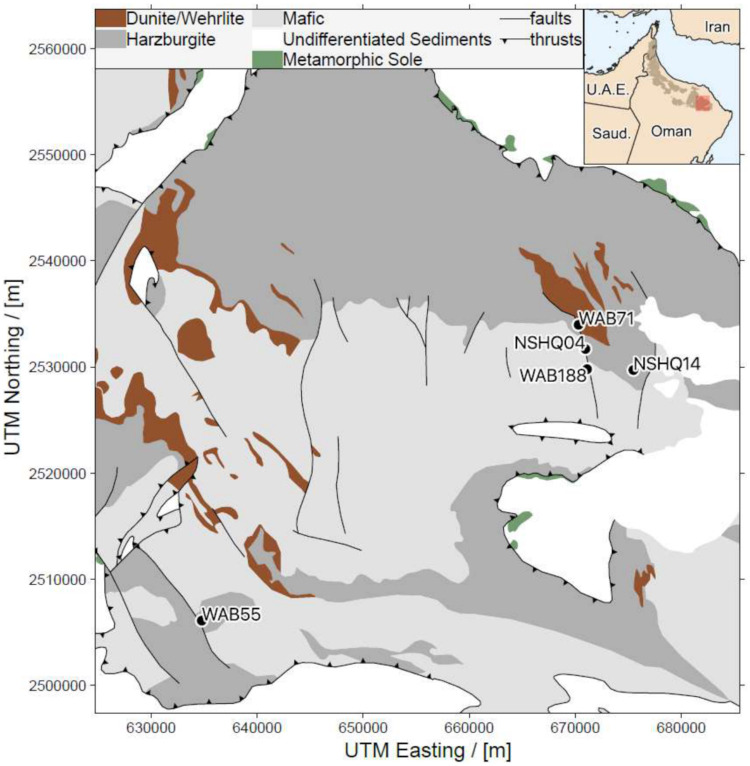
Fracture waters from five preexisting wells in the Samail Ophiolite, Oman (Fig. 1), were sampled in February of 2017 for planktonic biomass for use in geochemical (Table 1) and DNA- and RNA-based-analyses. Well NSHQ14 was sampled at a depth of 50 m (NSHQ14B) and 85 m (NSHQ14C). Waters recovered from wells drilled in peridotite bedrock (NSHQ14 and WAB71) had hyperalkaline pH (pH, >10), with waters from NSHQ14C at a 85-meter depth exhibiting the highest measured pH (11.3) and H2 concentration (253 μM) of any of the sampled wells. The waters recovered from wells drilled near the “contact” or subsurface faulted boundary between gabbro and peridotite bedrock (NSHQ04 and WAB55) had alkaline pH, with values of 10 for NSHQ04 and 9.2 for WAB55. The pH of gabbro-hosted well WAB188 was not measured in 2017, but previous observations recorded values of 8.7 and 7.6 in 2015 and 2016, respectively (6).
FIG 1.
Geological map of a portion of the Samail Ophiolite (created in the program qGIS using data from Nicolas et al. [30]) showing sampling locations in the Wadi Tayin massif (adapted from Nothaft et al. [91] with permission).
TABLE 1.
Geochemical composition of waters sampled from wells that intersect peridotites or that lie at the boundary of peridotites and gabbros in the Samail Ophiolite in 2017a
| Parameter | Data for well/condition (pump depth [m]) |
LOQ | ||||||
|---|---|---|---|---|---|---|---|---|
| NSHQ14 |
WAB71 (70) | NSHQ04 (5.8) | WAB55 (30) | WAB188 (78) | Rain | |||
| 50 | 85 | |||||||
| Well type | Peridotite | Peridotite | Peridotite | Contact | Contact | Contact | ||
| pH | 11.05 | 11.28 | 10.59 | 10 | 9.22 | NA | - | |
| Temp (°C) | 34.4 | 36.3 | - | - | - | - | - | |
| Eh (mV) | −415 | −253 | - | - | - | 214 | - | |
| H2 (μM) | 32.5 | 253 | 0.59 | BLOQ | BLOQ | 0.99 | - | 0.05 |
| CH4 (μM) | 53.5 | 106 | 14.8 | 483 | 0.106 | 1.83 | - | 0.015 |
| DIC (mM) | 0.05 | 0.13 | 0.12 | 0.04 | 2.90 | 3.00 | - | 0.02 |
| CO (μM) | BLOQ | BLOQ | BLOQ | BLOQ | BLOQ | BLOQ | - | 0.28 |
| ∑ Na (mM) | 8.35 | 10.21 | 4.95 | 10.40 | 4.12 | 3.49 | 0.25 | 5.85 × 10−3 |
| ∑ Ca (mM) | 3.66 | 4.34 | 4.07 | 7.79 | 0.05 | 1.33 | 0.52 | 1.6 × 10−4 |
| ∑ Mg (mM) | 0.01 | 0.02 | BLOQ | 0.02 | 2.75 | 1.44 | 0.15 | 5.9 × 10−5 |
| ∑ Mn (μM) | 7.83 × 10−3 | 3.3 × 10−2 | BLOQ | 0.03 | 0.02 | 0.14 | 1.0 × 10−4 | 7.38 × 10−4 |
| ∑ Al (mM) | BLOQ | 2.0 × 10−3 | 1.8 × 10−3 | 2.0 × 10−3 | BLOQ | BLOQ | 1.0 × 10−3 | 7.6 × 10−4 |
| ∑ Fe (mM) | 1.5 × 10−4 | 1.8 × 10−3 | 1.6 × 10−4 | 8.2 × 10−4 | 2.5 × 10−3 | 3.8 × 10−4 | 3.6 × 10−4 | 6 × 10−6 |
| ∑ Si (mM) | 8 × 10−3 | 6 × 10−3 | 2.1 × 10−2 | 3.6 × 10−2 | 3 × 10−3 | 0.37 | 0.08 | 4 × 10−4 |
| ∑ K (mM) | 0.20 | 0.25 | 0.25 | 0.29 | 0.21 | 3.9 × 10−2 | 7.1 × 10−2 | 8.3 × 10−4 |
| NH4+ (μM) | 14.2 | 13.0 | 100.0 | 55.5 | BLOQ | BLOQ | - | 1.0 |
| SO42− (mM) | 0.13 | 2 × 10−3 | 0.04 | 0.68 | 0.88 | 1.13 | 0.17 | 1.04 × 10−3 |
| NO3− (mM) | BLOQ | BLOQ | BLOQ | BLOQ | 0.14 | 0.12 | 0.26 | 1.61 × 10−3 |
| Cl− (mM) | 14.28 | 16.20 | 11.59 | BLOQ | 7.24 | 5.04 | 0.40 | 2.82 × 10−3 |
| Br− (mM) | 0.02 | 2.5 × 10−2 | 1.2 × 10−2 | 2.7 × 10−2 | 5 × 10−3 | 2 × 10−3 | BLOQ | 1.79 × 10−4 |
For comparison, data are presented on the geochemical composition of a single sample of rainwater collected from the Samail Ophiolite in 2017. ∑ indicates the value is the sum of all species of an element. Values below the limit of quantification (BLOQ) are indicated, and dashes (-) indicate that measurements were not taken. Concentrations of PO42− were below the limit of quantification in all samples. The pH of NSHQ04 was taken with a colorimetric indicator strip. Measurement of the pH of WAB188 was not possible in 2017 and previous measurements recorded in 2015 and 2016 were 8.7 and 7.6, respectively (7).
CH4 was detected in every well, with the highest concentration (483 μM) measured in contact well NSHQ04. Dissolved inorganic carbon (DIC) was detected in low (<0.2 mM) concentrations in the hyperalkaline wells and in greater concentrations (up to 3 mM) in WAB188 and WAB55. Peridotite wells had lower concentrations of potential electron acceptors (e.g., SO42−, NO3−, and NO2−) than the contact wells (Table 1). Trace metal and nonmetal elemental concentrations for all wells are in Table S1 in the supplemental material.
Previous studies grouped the waters in each of the wells sampled herein as either type I, type II, or crust/mantle contact waters based on water geochemistry, specifically water pH and concentrations of Ca and Mg (31, 33, 36–39). In agreement with prior classifications, NSHQ14, WAB71, and NSHQ04 were dominated by Ca/OH− waters typical of closed-system serpentinization (termed type II waters), WAB55 intersects Mg/HCO3− waters more typical of open-system serpentinization (termed type I waters), and gabbro-hosted WAB188 contains waters typical of a contact well near the crust/mantle boundary.
Diversity of 16S rRNA genes and transcripts in subsurface fracture waters.
Biomass was concentrated from subsurface type I and II waters by filtration (0.22 μm) and processed for DNA and RNA; the RNA was converted to cDNA. The V4/5 hypervariable regions of both 16S rRNA genes and their transcripts (cDNA) were amplified, sequenced, and clustered into amplicon sequence variants (ASVs). An overview of the most abundant ASVs from each well (see Fig. S1 in the supplemental material), sequencing metrics (see Table S2 in the supplemental material), and rarefaction curves of observed species richness (see Fig. S2 in the supplemental material) are in the supplemental information. DNA/RNA extraction, PCR, and the reverse transcriptase (RT)-negative controls produced low numbers of sequence reads and did not resemble sampled well community 16S rRNA gene compositions (see Fig. S3 in the supplemental material). Eukaryotic 18S rRNA sequence counts were low (0.11% of all sequences from all wells), and these sequences were deemed contaminants in this investigation due to the low read counts and compositional similarity to laboratory controls.
Numerous sequences were detected with close affiliation to known CH4-producing and -consuming organisms. Based on homology to cultivars, the most abundant sequences corresponding to methanogenic taxa were those affiliated with the genus Methanobacterium, a methanogen within the Euryarchaeota phylum (40). This Methanobacterium 16S rRNA gene ASV was one of the most abundant sequences detected in both type I and type II well waters (Fig. S1). Other lesser abundant ASVs showed homology to other characterized methanogenic archaea. The most abundant sequences related to methanotrophs were those showing homology to Methylococcus, an aerobic methanotroph within Gammaproteobacteria (41). Less abundant ASVs affiliated with putative anaerobic CH4-oxidizing taxa were also detected in well water communities, including ANME-1 (20) and “Candidatus Methylomirabilis” of the NC10 group (42).
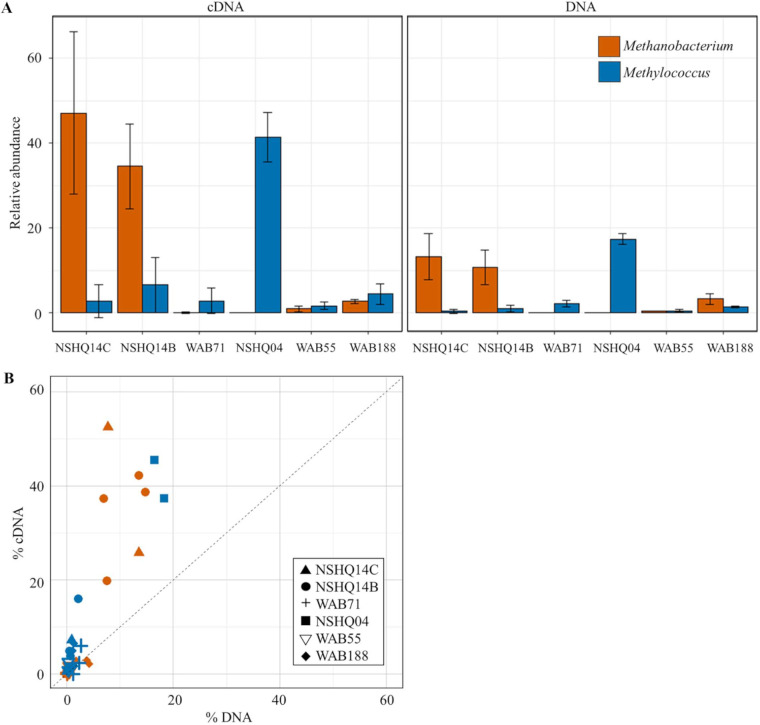
Stark differences in ASV abundance were apparent between the DNA and cDNA fractions within samples. The mean abundance of 16S rRNA genes (DNA) and transcripts (cDNA) of two taxa putatively involved in CH4 cycling is shown in Fig. 2. The mean percentage of sequences of ASVs affiliated with the most abundant methanogen Methanobacterium was greater in the cDNA fraction for NSHQ14 (Fig. 2A). At the 85-m depth for NSHQ14, 47.1% of the cDNA reads were attributed to Methanobacterium compared with 13.3% of the DNA reads. Similarly, Methanobacterium constituted 34.5% of the cDNA and 10.7% of the DNA reads at the 50-m depth. The proportion of Methanobacterium reads in NSHQ14 varied more widely in the cDNA (19.9% to 62.9%) than in the DNA (6.9% to 18.6%) (Fig. 2B). In contrast to NSHQ14, the Methanobacterium ASVs represented less than 5% of the reads in any sample from other wells. Similarly, the Methylococcus-affiliated ASV comprised a greater mean percentage of the cDNA (6.6%, 2.8%) compared with the DNA (1.0%, 0.4%) in NSHQ14 at 50 m and 85 m. In hyperalkaline contact well NSHQ04, Methylococcus was 41.4% of the cDNA and 17.4% of the DNA. The anaerobic methanotroph-affiliated ASVs (ANME-1, “Ca. Methylomirabilis”) had low read abundances (<0.5%) in both the DNA fractions in hyperalkaline peridotite water samples.
FIG 2.
(A) Mean relative abundance of the most abundant taxa putatively involved in CH4 production and consumption in the cDNA and DNA of biomass collected from well waters of the Samail Ophiolite. (B) Percentage of the top putative methanogenic and methanotrophic organisms inferred by SSU rRNA sequence abundance of ASVs with homology to Methanobacterium (red/orange) and Methylococcus (blue). Each point represents a biological replicate.
Metagenomic characterization of subsurface fracture water communities.
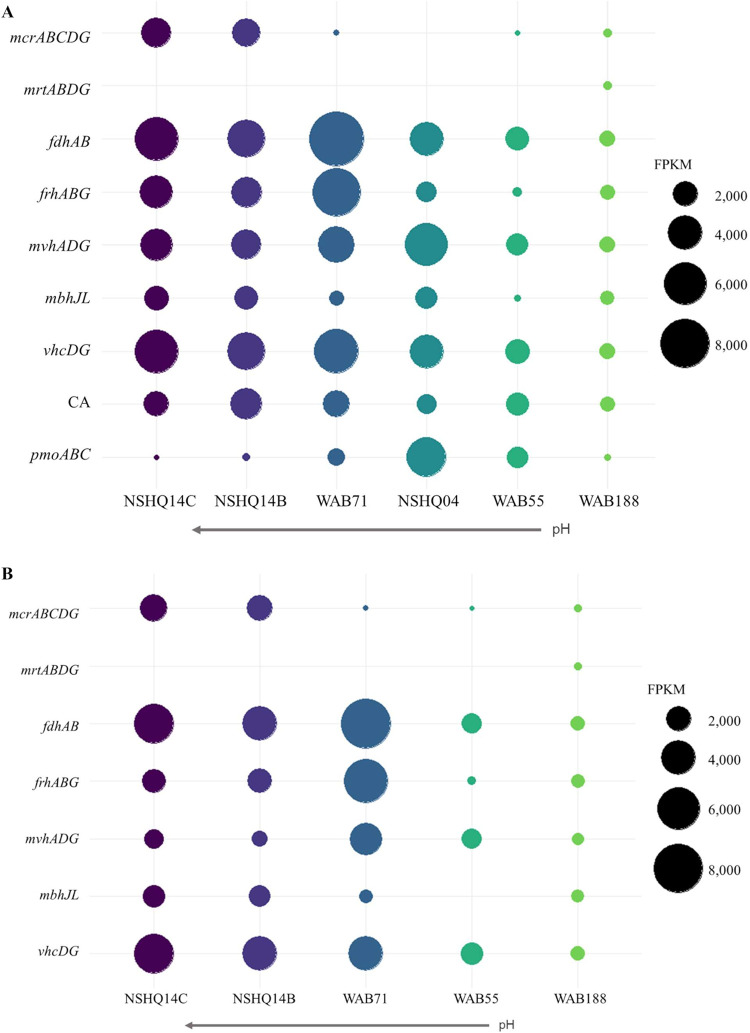
Key genes encoding proteins involved in methanogenesis were detected in assembled metagenomic sequences from the type II hyperalkaline waters of the peridotite-hosted wells NSHQ14 and WAB71 (Fig. 3A). Metagenomic assemblies from both depth intervals at NSHQ14 harbored genes coding for MCR (mcrABCDG) and tetrahydromethanopterin S-methyltransferase (MTR; mtrABCDEFGH) operons. The MCR and MTR homologs were colocalized on the same contigs recovered from both 50-m and 85-m depth intervals in NSHQ14, and the McrA sequences were most closely affiliated with McrA from a cultivated Methanobacterium sp. (GenBank accession TMS43336.1). Genes for various [NiFe]-hydrogenases involved in supplying reductant or balancing osmotic potential in methanogens (15) were also identified. These include active site subunits of membrane-bound, ion translocating [NiFe]-hydrogenases (mbhJL), putative bifurcating hydrogenases (mvhADG), and F420- or cytochrome-reducing hydrogenases (frhABG and vhcD, respectively) (43). Like MCR and MTR, these homologs were detected in assemblies from both depths of NSHQ14 and showed close homology to various cultivated Methanobacterium sp. CH4 cycling genes identified in metagenomes with homology to proteins from Methanobacterium sp. (with an E value of <1 × 10−6, >30% amino acid identity over >50% of the length) are shown in Fig. 3B.
FIG 3.
(A) Fragments per kilobase of exon per million reads (FPKM) of key functional genes of interest for CH4 cycling metabolisms. The methyl-coenzyme M I (mcrABGCD), methyl-coenzyme M II (mrtABGD), formate dehydrogenase (fdhAB), carbonic anhydrase (CA), particulate methane monooxygenase (pmoABC), and methanogenic [NiFe]-hydrogenase (frh, mvh, mbh, and vhc) enzymes from assembled metagenomes are shown. Notably, FPKM values are comparable within each sample (shown by color) but not across samples. Wells are ordered by decreasing fluid pH. (B) FPKM of CH4 cycling genes that are homologous to proteins from Methanobacterium sp. (E value of < 1 × 10−6; >30% amino acid identity over >50% of the length).
In metagenomic assemblies from the peridotite-hosted well WAB71, only an amino acid sequence for mcrG was detected, and it exhibited homology to mcrG of a Methanophagales sp. of the ANME-1 group (GenBank accession RZN33282.1). Among the communities in type I well waters, those from WAB188 showed the greatest capability for hydrogenotrophic methanogenesis (Fig. 3A). The operons encoding MCR (mcrABCDG) and MTR (mtrABCDEFGH) were colocalized on a single contig, while the MCR II (mrtBDGA) operon was found on a separate contig, and homologs of [NiFe]-hydrogenases (mvhDG, frhABG, vhcG, and mbhJL) were detected. Homologs of each protein were closely related to cultivated Methanobacterium sp. (mcrA, GenBank accession WP_048081846.1; mrtA, GenBank accession AXV36901.1). In comparison, metagenomic assemblies from WAB55 (type I) were found to contain only homologs of mcrCG (related to Methanobacterium; GenBank accession WP_048081846.1), and assemblies from NSHQ04 encoded only mtbB homologs most closely related to halophilic methanogenic taxa (Methanonatronarchaeum thermophilum, GenBank accession OUJ19070; and Methanohalophilus sp., GenBank accession OBZ35607.1). These findings point to methanogens being less abundant in WAB55 and NSHQ04 than in WAB188 and NSHQ14, thereby leading to incomplete representation of pathways involved in their energy metabolism in metagenomic assemblies.
Homologs of fdhAB encoding the subunits of the formate dehydrogenase enzyme were detected in all well metagenomes, with some sequences homologous to Methanobacterium strains (Fig. 3). Homologs of carbonic anhydrase (CA) genes (can, cynT, and cah) were detected in all metagenomes. Most CA sequences were identified most closely to nonmethanogenic/methanotrophic organisms, but CA sequences from NSHQ14 and WAB188 were found to be most closely related to methanogens and methanotrophs, including Methanobacterium and Methylococcus strains.
Homologs of protein-coding genes associated with methanotrophy were detected in metagenomic sequences, consistent with 16S rRNA gene and transcript data suggesting the potential importance of methanotrophy in the Samail Ophiolite. Homologs of pmoABC were detected in every well (Fig. 3A), often with one or more slightly divergent copies. These homologs were most closely related to those identified in previously characterized aerobic Methylococcus sp. Homologs of protein-coding genes affiliated with soluble methane monooxygenases (mmoXYZ) or of the large subunit of methanol dehydrogenase (mxaF) were not detected in any metagenome from the subsurface water communities.
Metatranscriptomic characterization of subsurface fracture water communities.
Transcripts of genes involved in various CH4-cycling processes were detected in subsurface waters from the Samail Ophiolite. Transcript abundances were normalized to counts per million reads (CPM) and lowly expressed transcripts (<1 CPM in all samples) were removed as possible contaminants. The results for all transcripts investigated are in Table 2.
TABLE 2.
Genes targeted to investigate CH4-cycling metabolisms in the Samail Ophiolite
| Gene target | Gene product(s) | Enzymatic function/pathway in CH4 cycling | Reference | Transcripts (CPM)a
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NSHQ14C | NSHQ14B | WAB71 | NSHQ04 | WAB55 | WAB188 | No RT | ||||
| mcrABG, mrtABG | Methyl-coenzyme M reductase I and II subunits | Catalyzes the final step in methanogenesis | Reeve et al. (88) | 236.3 | 510.6 | 0.2 | 0 | 0 | 9.4 | 0 |
| mvhADG | F420-nonreducing hydrogenase subunits | Cytoplasmic [NiFe]-hydrogenase | Thauer et al. (15) | 0.5 | 0 | 0 | 4.1 | 0 | 0 | 0 |
| frhABG | F420-reducing hydrogenase subunits | 10.1 | 6.3 | 0.1 | 0 | 0 | 0.3 | 0.2 | ||
| vhcADG | F420-nonreducing hydrogenase Vhc subunits | 5.2 | 1.5 | 0.8 | 4.4 | 0 | 3.4 | 0 | ||
| vhuADGU | F420-nonreducing hydrogenase Vhu subunits | 0 | 5.1 | 0 | 0 | 0 | 2.7 | 0 | ||
| mbhJL | Energy-converting [NiFe]-hydrogenase Mbh subunits | Membrane-associated energy-converting [NiFe]-hydrogenase | 6.0 | 3.2 | 0 | 0 | 0 | 0 | 0 | |
| ehaNO | Energy-converting [NiFe]-hydrogenase Eha subunits | NA | NA | NA | NA | NA | NA | NA | ||
| echCE | Energy-converting [NiFe]-hydrogenase Ech subunits | NA | NA | NA | NA | NA | NA | NA | ||
| ebhMN | Energy-converting [NiFe]-hydrogenase Ebh subunits | NA | NA | NA | NA | NA | NA | NA | ||
| fdhAB | Formate dehydrogenase subunits | Catalyzes the oxidation of formate to reduce CO2 to CH4 | Shuber et al. (19) | 55.0 | 41.7 | 6.9 | 103.7 | 3.3 | 24.7 | 4.5 |
| ackA | Acetate kinase | Acetoclastic methanogenesis | Fournier et al. (89) | 3.1 | 1.5 | 3.0 | 305.2 | 7.1 | 4.4 | 4.2 |
| pta | Phosphate acetyltransferase | 2.7 | 8.7 | 14.3 | 15.8 | 17.1 | 18.8 | 12.7 | ||
| mtaA | Methylcobamide:CoM methyltransferase | Methylotrophic methanogenesis | Crespo-Medina et al. (9) | 0 | 0 | 0 | 0 | 0 | 5.4 | 0 |
| mtbB | Dimethylamine methyltransferase | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| mtmB | Monomethylamine methyltransferase | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| mttB | Trimethylamine methyltransferase | Paul et al. (90) | 0 | 0 | 0 | 14.6 | 0 | 1.7 | 1.8 | |
| mtsA | Methylated-thiol-coenzyme M methyltransferase | NA | NA | NA | NA | NA | NA | NA | ||
| pmoABC | Particulate CH4 monoxygenase subunits | Catalyzes the oxidation of CH4 to methanol | Stainthorpe et al. (21) | 72.2 | 37.2 | 1.8 | 1650.9 | 14.2 | 38.4 | 0 |
| mmoXYZ | Soluble CH4 monooxygenase subunits | Csáki et al. (22) | NA | NA | NA | NA | NA | NA | NA | |
| mxaF | Methanol dehydrogenase alpha subunit | Catalyzes the oxidation of methanol to formaldehyde | Lau et al. (23) | 18.3 | 5.3 | 5.0 | 513.1 | 8.4 | 6.6 | 3.3 |
| cynT, cah, can | Carbonic anhydrase | Catalyzes the interconversion of HCO3− + H+ to CO2 + H2O | Smith et al. (54) | 757.3 | 363.1 | 16.0 | 8.1 | 0.8 | 12.5 | 10.6 |
| cdhAB | Ni-containing CO dehydrogenase subunits | NiCODH, catalyzes the reversible oxidation of CO to CO2 | Fones et al. (24) | 32.8 | 9.6 | 0.9 | 25.2 | 0 | 3.0 | 0 |
| cooS | Ni-containing CO dehydrogenase | |||||||||
| cutSML | Mo-containing CO dehydrogenase subunits | MoCODH, catalyzes the reversible oxidation of CO to CO2 | 29.7 | 16.7 | 13.1 | 16.6 | 7.6 | 26.8 | 4.5 | |
| coxSML | Mo-containing CO dehydrogenase subunits | |||||||||
Transcript counts per million reads (CPM) from metatranscriptomes are given for each gene and well. NA indicates that transcripts were not detected in annotations from any sample. No RT, a negative control wherein no reverse transcriptase enzyme was added to PCR.
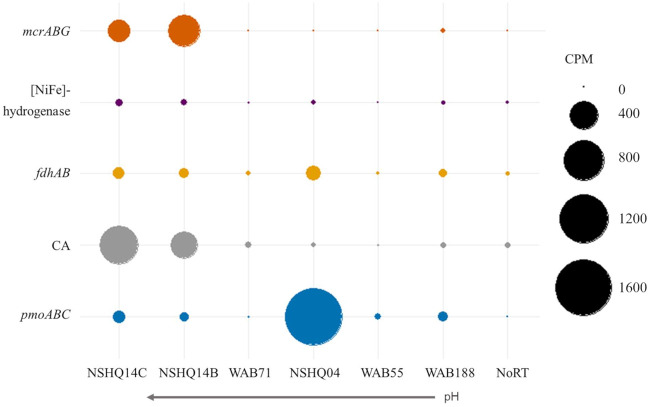
The abundances of transcripts in CPM for MCR (mcrABG), [NiFe]-hydrogenases (mvhADG, frhABG, vhcADG, vhuADGU, and mbhJL), pmoABC, CA, and fdhAB are shown in Fig. 4. MCR was expressed in NSHQ14, WAB188, and WAB71. NSHQ14 extracts contained the greatest expression of MCR transcripts, with 510.6 CPM at the 50-m depth interval. To further evaluate the energy metabolism of putative methanogens in the Samail Ophiolite, the abundance of transcripts affiliated with [NiFe]-hydrogenases was examined. After filtering out transcripts with low normalized expression (<1 CPM), homologs of [NiFe]-hydrogenases common to characterized methanogens were detected primarily in NSHQ14C and NSHQ14B with CPM of 21.7 and 16.1, respectively (Table 2; Fig. 4). No transcription of energy-converting [NiFe]-hydrogenases was detected. Transcripts for formate dehydrogenases (fdhAB) were most abundant in NSHQ04 (103.7 CPM) and NSHQ14C (55 CPM). Transcripts for fdhAB and [NiFe]-hydrogenases were not highly expressed relative to MCR in NSHQ14.
FIG 4.
Transcript counts per million reads (CPM) for genes of interest in CH4 cycling normalized by the TMM method. Reads with homology to transcripts for methyl-coenzyme M reductase (mcrABGCD) and the colocalized tetrahydromethanopterin S-methyltransferase subunits (mtrACDEH), particulate CH4 monooxygenase (pmoABC), formate dehydrogenase alpha subunit (fdhA), and carbonic anhydrase (CA) are shown. A negative control wherein no reverse transcriptase was added to the PCR (NoRT) had no transcripts for any gene within this subset.
The carbon monoxide dehydrogenase (CODH) and carbonic anhydrase (CA) enzymes could generate inorganic carbon from CO or carbonates as another source of CO2 for H2-dependent methanogens and autotrophs under carbon limitation in serpentinizing environments. Transcripts affiliated with Ni-containing CODH and Mo-containing CODH homologs were detected throughout the ophiolite waters. NSHQ14C contained the largest CPM of both Ni-CODH and Mo-CODH with 32.8 and 29.7 CPM, respectively. CA transcripts were similarly observed across all well extracts, with the greatest expression at the 85-m depth interval (757 CPM) and 50-m depth interval (363 CPM) of NSHQ14.
Aligned with the presence of genes encoding the three subunits of particulate methane monooxygenases (pmoABC) in metagenomic assemblies from all wells examined, transcripts for pmoABC were detected in extracts from all wells, with the greatest expression in NSHQ04 (1,650.9 CPM) and NSHQ14C (72.2 CPM). Transcripts for mxaF were similarly expressed with 513.1 CPM in NSHQ04 and 18.3 CPM in NSHQ14C. No transcripts of soluble CH4 monooxygenase subunit genes (mmoXYZ) were detected in transcriptomes from any well.
DISCUSSION
Active microbial methanogenesis in subsurface waters of the Samail Ophiolite.
The enrichment of Methanobacterium in the cDNA fraction relative to the DNA fraction of 16S rRNA in extracts from NSHQ14 suggests this organism is active. This finding agrees with previous studies of planktonic communities from this well showing 14CH4 production from 14C-labeled bicarbonate (24) and the detection of Methanobacterium-affiliated 16S rRNA genes (6, 7). However, cDNA and DNA comparisons can be impacted by differences in sequencing library sizes, affecting the ratio of Methanobacterium-affiliated small subunit (SSU) rRNA genes. In addition to SSU rRNA, genes for the biosynthesis of MCR were detected on single contigs assembled from NSHQ14 and WAB188 metagenomes that share close homology to Methanobacterium sp. The homology of this MCR to Methanobacterium sp. was previously reported by Fones et al. (24), where the authors found low homolog counts per Mbp assembled for MCR. In this work, these single contigs of Methanobacterium MCR have >2,000 fragments per kilobase of exon per million reads (FPKM), indicating adequate sequencing coverage and Methanobacterium as the primary methanogenic strain. Metagenomic sequences can offer evidence of functional capability but not cellular activity, as these genes may be from dormant or dead organisms. Microbial methanogenic activity was evidenced by the detection of MCR transcripts in NSHQ14. Transcription of mcr genes has been correlated with methanogenic activity in other environments and incubation experiments containing Methanobacterium (44–46). Collectively, these molecular observations suggest Methanobacterium to be an active organism in the hyperalkaline (pH 11.3) waters of NSHQ14 and thus represent an important extension of the pH spectrum (4.5 to 10.2) (47) where methanogenesis is commonly observed.
SSU rRNA gene sequencing from other deep subsurface ecosystems (e.g., references 6, 18, and 48–50) that have geochemical similarity to NSHQ14 and WAB188 suggests Methanobacterium is a cosmopolitan organism in other highly reduced, high pH environments. For example, in the pH 9.1, H2- and CH4-containing waters of a deep fault within the Driefontein Mine of South Africa, Methanobacterium codominate the microbial community with a sulfate-reducing bacterium (50). SSU rRNA sequences affiliated with members of the family Methanobacteriaceae (of which Methanobacterium belongs to) have also been identified in pH 11.5 springs of the Santa Elena ophiolite; yet, ANME-1 organisms dominate the archaeal communities in these waters (9). Likewise, 16S rRNA gene and metagenomic data suggest the presence of Methanobacterium in hyperalkaline waters (pH 11.8 to 12.3) in the Voltri Massif of Italy; however, subsequent incubation experiments did not confirm methanogenic activity (18). The transcriptional evidence presented herein confirms methanogenic metabolic activity at pH 11.3, a unit above the pH of 10.2 at which methanogenesis is typically observed (47). In the context of previous observations of a biological reduction of HCO3− to CH4 in incubations from NSHQ14C (24), these data strongly point to a microbial contribution to CH4 in waters impacted by serpentinization in the Samail Ophiolite, in particular in hyperalkaline, highly reacted waters with dissolved inorganic carbon levels below detection.
Genes and transcripts of the energy-converting [NiFe]-hydrogenases Eha/Ehb typical of Methnobacterium sp. were not detected. However, genes for another energy-converting [NiFe]-hydrogenase (mbhJL) and a subtype of the hydrogenase Mvh (vhcD) displayed homology to Methanobacterium sp., although these genes are not found in this cytochrome-lacking methanogen (15). This finding may indicate an annotation mischaracterization for Mbh and Vhc in our study. Interestingly, while [NiFe]-hydrogenase genes were identified in metagenomic assemblies in Samail Ophiolite waters, transcripts corresponding to these genes were not in high abundance. [NiFe]-hydrogenases are requisite for hydrogenotrophic methanogenesis (15), and thus, the evidence for their low transcription CPM is surprising. It is possible that this observation is attributable to limitations and/or sequencing biases imposed by the low biomass associated with these samples or that [NiFe]-hydrogenase transcripts are inherently less stable and thus degrade rapidly.
A subset of methanogens can use the formate dehydrogenase enzyme (encoded by fdhAB) to use formate as a methanogenic substrate as an alternative to using hydrogenases to activate H2 as a source of electrons for methanogenesis (19). Furthermore, Fones et al. (24) found that life in alkaline Samail Ophiolite waters is likely carbon limited rather than energy limited, and formate may be a favored carbon source in alkaline waters. The hydrogenotrophic Methanobacterium sp. may therefore use formate instead of H2/CO2 as a methanogenic substrate under carbon limitation and abundant H2 in this environment. Transcripts of fdhAB had higher CPM than [NiFe]-hydrogenases in hyperalkaline NSHQ14 but were not highly expressed relative to MCR transcripts in this well. This finding suggests formate marginally augments the energy metabolism of methanogens in NSHQ14 and may point to a different and yet-to-be-defined mechanism of generating reductant in methanogens (51).
Other compounds, such as CO or mineral carbonates, might be a source of inorganic carbon for autotrophs in serpentinizing environments. Homologs of CODH were detected in NSHQ14 metagenomes previously (24), but low abundances of transcripts for these genes in the communities examined indicate that CO is not a predominant source of carbon or energy for the microbial populations. Alternatively, mineral carbonates may provide a source of carbon for autotrophs. Mineral carbonate dissolution in seawater has been correlated with activity of microbial carbonic anhydrase (CA) (52), an enzyme that catalyzes the interconversion of carbonic acid with HCO3− and H+ (53). CA may therefore play a role in liberating inorganic carbon from mineral carbonates to make it bioavailable by shifting the equilibrium dissolution of carbonate toward HCO3−. Of the three (α, β, and γ) classes of CA, genes for the β and γ types have been identified in strains of the hydrogenotrophic methanogen Methanobacterium thermoautotrophicum and acetoclastic methanogen Methanosarcina thermophila, respectively (54). In M. thermoautotrophicum, where CO2 is in demand for hydrogenotrophic methanogenesis, the β CA could be used to interconvert HCO3− and CO2 and/or concentrate CO2 near the formylmethanofuran dehydrogenase for the first step of methanogenesis, analogous to β CA and the CO2-fixing enzyme in the photosynthetic organism (55).
In NSHQ14, the CPM of CA transcripts increased with increasing depth and pH, indicating CA may be generating the carbon needed by autotrophic methanogens as the water becomes more alkaline. Consistent with a potential for CA in mitigating inorganic carbon limitation for autotrophs in serpentinizing systems, homologs of this gene were detected in genomes of Serpentinomonas strains isolated from a hyperalkaline serpentinization site in The Cedars, California (56). Indeed, this strain was shown to be capable of autotrophic growth on solid calcium carbonate (CaCO3), H2, and O2 (56). Furthermore, Miller et al. (27) observed a Methanobacterium strain cultured from the Samail Ophiolite to be capable of growth on H2 and solid CaCO3 as the sole carbon source. Collectively, these observations and CA transcription suggest that the dissolution of carbonates may be a source of inorganic carbon for methanogens in high pH, carbon-limited environments such as NSHQ14.
Alternatively, carbonic anhydrase activity may be driven by microorganisms scavenging bicarbonate introduced into NSHQ14 via limited mixing of type I and II waters within the aquifer or small amounts of atmospheric CO2 introduced during sample collection. Zwicker et al. (26) describes this scenario where a mixture of type I and II fluids with minimal CO2 is the carbon source for methanogens in the Chimaera ophiolite of Turkey. Furthermore, the isotopic composition of archaeal lipid biomarkers from the C-limited conditions of the Chimaera ophiolite show enrichment in 13C, and the authors conclude that microbially produced CH4 may contribute to the abiotic CH4 pool in that system (26). Similarly, the high H2 concentrations in NSHQ14 likely make scavenging any available bicarbonate via CA favorable for autotrophs and methanogens.
Methanotrophy in subsurface waters of the Samail Ophiolite.
All well waters contained SSU rRNA gene sequences affiliated with the aerobic bacterial methanotroph Methylococcus, with the greatest relative abundance observed in the CH4-rich groundwaters of NSHQ04. This finding is consistent with the detection of pmo genes and their transcripts in NSHQ04 and, to a lesser extent, NSHQ14. Notably, NSHQ04 was sampled shallowly at 6 m due to blockage within the well. Waters sampled from this depth might have been infused with atmospheric oxygen to a greater extent than waters from other wells that were sampled much deeper, and this may have led to the increased relative proportions of aerobic methanotrophs relative to methanogens in samples collected in NSHQ04. Consistent with this hypothesis, previous sequencing work on 16S rRNA genes recovered from deeper within this well, prior to the blockage, detected sequences affiliated with Methanobacterium (7, 27). Thus, aerobic methanotrophs are components of communities in subsurface waters of Oman, most notably in more oxidizing waters near zones of mixing between aquifer waters and/or the atmosphere.
In addition to aerobic methanotrophy, DNA sequencing suggests the possibility of anaerobic CH4-oxidizing organisms in several of the communities sampled from the Samail Ophiolite. Several sequences with homology to the ANME-1 group of Archaea, organisms putatively involved in anaerobic oxidation of CH4 (AOM) (20), were detected in the hyperalkaline wells WAB71 and NSHQ14. Likewise, mcrG sequences with homology to ANME-1 were also detected in WAB71. The lack of a full complement of MCR homologs for putative ANME-1 organisms in these wells is likely attributable to their low abundance, as gauged by 16S rRNA gene and transcript sequencing, which likely limited their representation in our metagenomic sequences. Nonetheless, the presence of ANME in serpentinization-impacted waters is consistent with previous reports of the evidence of these guilds in Chimaera, Santa Elena, and Cabeço de Vide serpentinizing environments (9, 17, 26). However, the lack of detected transcripts affiliated with these organisms and their genes and their limited representation in metagenomic sequences, together, suggest that AOM is of minimal importance in the waters in the Samail Ophiolite.
A subsurface CH4 cycle impacted by microbial activity.
Dissolved CH4 in ophiolite waters is often enriched in 13C, and this evidence has been used to suggest that CH4 is primarily abiogenic (57, 58). However, methanogens can produce CH4 enriched in 13C under inorganic C-limited conditions. For example, a Methanobacterium strain isolated from NSHQ04 (pH 10) was shown to generate CH4 that was markedly enriched in 13C (−28‰ VPDB) when using calcium carbonate mineral (−0.1‰ VPDB) as the sole C source, in particular when cultivated in medium with pH values greater than 9 (27). Previous CH4 isotopic measurements of NSHQ14 waters reported a δ13Cof up to +3‰ VPDB (7), and our study found transcription of carbonic anhydrase specific to this well. The dominance of Methanobacterium in our analysis of SSU rRNA genes and MCR transcripts in NSHQ14 (pH 11.3) indicates that methanogenesis is occurring at high environmental pH values in this system, and it is possible that these cells are using C liberated from carbonate minerals as a carbon source. The use of carbon liberated from carbonate minerals by methanogens under hyperalkaline, DIC-limited conditions may thus be contributing to the environmental CH4 pool; yet, their contribution may be obscured by the unusual isotopic signatures associated with this environment.
The opposing process of microbial methanotrophy can impact the C isotopic composition of CH4 by preferentially using 12CH4 leading to 13C enrichment (59). Like the Samail Ophiolite waters, those of the Santa Elena Ophiolite host CH4 that is unusually enriched in 13C and contain methanogenic and methanotrophic microorganisms, including ANME-1 and Methanobacterium sp. (9). The detection of CH4 that is enriched in 13C combined with evidence for potential methanogens/methanotrophs in geographically distinct ophiolites warrants further studies focused on the interplay between organisms involved in CH4 cycling and their effect on the δ13C of CH4 in environments influenced by serpentinization.
Additional work is also needed to better understand potential electron donors that fuel methanogenesis in environments that are impacted by the process of serpentinization. The high concentration of H2 in some serpentinizing environments has been used to suggest that these systems not only are conducive to hosting robust communities of hydrogenotrophic methanogens but also may have been prime environments for the origin of this process (60). This argument was based on the extremely low reduction potentials associated with waters in active serpentinizing systems, a feature that should allow for the facile reduction of low potential ferredoxin (Fd) with H2. Reduced, low-potential Fd is required during the reduction of CO2 to formylmethanofuran during the first step of autotrophic methanogenesis (61). However, transcripts for [NiFe]-hydrogenases that can catalyze the reduction of Fd with H2 in Methanobacterium (group 4 Eha/Ehb or group 3c Mvh) (43) were detected in low abundance in our analysis of the NSHQ14 RNA pool, pointing toward the potential importance of other electron donors (e.g., formate and CO) capable of fueling methanogenesis in environments impacted by serpentinization.
The process of serpentinization creates additional challenges for autotrophs, including methanogens. Serpentinization generates waters with high pH and Ca, which leads to low-aqueous DIC in systems closed to atmospheric CO2. Nonetheless, the data presented here indicate that autotrophic Methanobacterium members are active under such conditions, which indirectly shows that they are meeting demands for cytoplasmic CO2 in a yet-to-be-defined mechanism. Carbonic anhydrase transcripts within NSHQ14 indicate that cells may be capable of interconverting bicarbonate introduced from fluid mixing or liberated from dissolution of carbonate minerals to meet CO2 demands. However, the source of bicarbonate remains unknown, and it is unclear whether carbonate dissolution rates are sufficient to meet this CO2 demand through equilibration or if cells actively promote dissolution. These possibilities are likely to have an influence on the isotopic composition of CH4 produced during methanogenesis. Additional physiological studies of these organisms and their mechanisms of acquiring cytoplasmic CO2 need to be conducted.
MATERIALS AND METHODS
Site description and geochemical characterization of subsurface waters.
Five preexisting water wells drilled in the mantle section of the Samail Ophiolite by the Oman Ministry of Regional Municipalities and Water Resources were sampled in February of 2017. Well waters were collected for geochemistry and cellular biomass from borehole NSHQ14 at 50 m (NSHQ14B) and 85 m (NSHQ14C); and from boreholes WAB188, WAB71, and WAB55 using a Grundfos SQ2-85 submersible pump (Grundfos Pumps Corp., Denmark, Netherlands) and a splitting manifold with field-washed Tygon tubing. Borehole NSHQ04 was sampled with a small Typhoon pump (Proactive Env. Products, Bradenton, FL).
At each well, the pump, manifold, tubing, and filter housing were field washed by running the pump for 20 to 30 minutes (approximately ≥100-liter throughput). Well waters were then passed through a 0.22-μm polycarbonate filter (MilliporeSigma, Burlington, MA) and collected in 15-ml Falcon tubes (Corning Inc., Corning, NY) for analyses of anion and cation concentrations, with the latter acidified with nitric acid (for a solution with pH of <2) in the field at the time of collection. Cations and anions were quantified using inductively coupled plasma atomic emission spectroscopy (ICP-AES; Optima 5300, Perkin-Elmer, Fremont, CA) and ion chromatography (IC; ICS-90; Dionex, Sunnyvale, CA), respectively, at the Colorado School of Mines. For DIC analyses, 6-ml aliquots of water were transferred from the sample collection vials (blue butyl-stoppered borosilicate glass) to 12.0-ml helium-purged Labco Exetainer tubes. To convert DIC species to CO2 for analysis, 0.5 ml of boiled 85% H3PO4 was added to the samples while still hot. Standards were made by weighing out CaCO3 in various amounts to Exetainer tubes, which were subsequently flushed with He and injected with 6 ml of boiled Milli-Q water while still hot. Acidification was performed at the same time and using the same methods for standards and samples. Standards and samples were centrifuged and then mixed on a shaker table for 12 to 18 hours to homogenize and equilibrate CO2. Headspace CO2 was then introduced via a Thermo Fisher GasBench II system to a Thermo Delta V Plus isotope ratio mass spectrometer for analysis.
Gas sampling was conducted using the bubble strip method (modified from reference 62). Details on bubble strip gas sampling are available online at https://doi.org/10.17504/protocols.io.2x5gfq6. Gas concentrations were measured using an SRI 8610C gas chromatograph (GC) with N2 as the carrier gas. H2, CO, CH4, and CO2 were separated with a 2-m by 1-mm ID micropacked ShinCarbon ST column. Peak intensities were measured concurrently using a thermal conductivity detector (TCD) and a flame ionization detector (FID) and calibrated with standard gas mixes (accuracy, ±2%; Supelco Analytical, Bellefonte, PA, USA). Measurement repeatability expressed as relative standard deviation is 5% over most of the calibrated range. We define the limit of quantitation as the signal at which the relative standard deviation increases to 20%.
Biomass collection, DNA/RNA extraction, quantification, and SSU rRNA gene and transcript sequencing.
Biomass was collected onto 0.22-μm polycarbonate filters. Once filters began to clog or appeared to hold particulates, they were removed from the housing and suspended in bead tubes with DNA/RNA shield lysis/stabilization solution (Zymo Research, Inc., Irvine, CA), which stabilizes nucleic acids at room temperature over several weeks. Samples were shipped to the Colorado School of Mines where cells were lysed by bead beating for a total of 5 minutes with rests (in intervals of 1 min for lysis and 1 min for rest) to cool the sample tubes to prevent RNA degradation. DNA and RNA were extracted in parallel using the microbiomics soil/fecal DNA miniprep extraction kit (Zymo Research, Inc.) following the manufacturer’s protocol. DNA was quantified postextraction by the Qubit double-stranded DNA (dsDNA) high-sensitivity (HS) assay (ThermoFisher Scientific, Waltham, MA). Recovered DNA and RNA were stored at −80°C.
A portion of the extracted RNA from each sample was converted to cDNA via reverse transcription-PCR (RT-PCR) with the qScript XLT one-step RT-PCR kit (Quanta Biosciences, Beverly, MA). Each 25-μl PCR mix contained one-step HiFi PCR ToughMix (1× concentration), one-step RT master mix (1×), 200 nM of the forward primer and 200 nM of the reverse primer (described below), nuclease-free water, and 10 μl of RNA sample template. A reaction in which no one-step RT master mix was added served as a negative control for the activity of the reverse transcriptase to ensure no extraneous DNA was being amplified. Reactions were run in a thermocycler with the lid preheated to 105°C; two initial steps of 48°C for 20 minutes and 94°C for 3 minutes; followed by 30 cycles of 94°C for 45 seconds, 50°C for 45 seconds, and 68°C for 90 seconds; and ending with a final extension of 68°C for 5 minutes and a hold at 4°C until removal from the thermocycler.
SSU rRNA genes were amplified from each DNA and cDNA sample via PCR with primers that span the V4 and V5 hypervariable regions of the 16S rRNA to produce gene fragments of ∼400 bp and ∼600 bp for Bacteria/Archaea and Eukarya, respectively. The 515-Y M13 and 926R primer set (modified from reference 63) most evenly amplifies this region of SSU rRNA from all three domains of life. The primers and PCR conditions used in this study are described previously (64). Technical replicate reactions for each sample, five extraction negative controls, and three negative PCR controls (no sample added) were amplified as well. Technical replicates were pooled and purified using Pure beads (Kapa Biosystems, Wilmington, MA) at a 1.0× ratio of beads to sample volume to retain any fragments of ≥250 bp long. Barcoding of sequences was carried out on the purified PCR products using a limited 6-cycle PCR (64). Replicate barcode reactions were pooled and purified with Kapa beads before quantification with the Qubit dsDNA HS assay. Final products were pooled in equimolar amounts before being concentrated to a final volume of 80 μl on an Ultracel-30K membrane (Millipore Sigma, Billerica, MA) within an Amicon Ultra 0.5-ml centrifugal filter (Millipore Sigma). Extraction blanks (no sample added) and negative PCR control reactions (no template added) were included in this sequenced pool. The prepared DNA/cDNA library was sequenced on a MiSeq instrument (Illumina Inc., San Diego, CA) at the Duke Center for Genomic and Computational Biology (https://www.genome.duke.edu) using V2 PE250 chemistry. Sequences produced from this effort are available on the Sequence Read Archive (NCBI) database under accession PRJNA560313.
Resultant FASTQ sequence files were demultiplexed and trimmed with Cutadapt (65). Reads were filtered by error rates, amplicon sequence variants (ASVs) were identified, and read pairs were merged to construct a sequence table with DADA2 in R (66, 67). Chimeric sequences were removed before taxonomic assignment against the SILVA r138 database (68). The protocol and resultant files for this effort are available online at https://github.com/danote/Samail_16S_compilation as “OM17.” The phyloseq and ggplot2 software packages were used for analysis and visualization of the sequence table (69, 70).
Metagenomic and metatranscriptomic library preparation and sequencing.
Metagenomic and metatranscriptomic libraries were prepared from six DNA samples from the five wells examined, namely, WAB55, WAB188, WAB71, NSHQ04, and NSHQ14, with separate libraries made for two different depths (50 m and 85 m) of NSHQ14. Metagenomic library preparation was conducted using the NexteraXT library preparation kit (Illumina Inc.) according to manufacturer’s instructions with 1-ng template DNA as the input. Metatranscriptomic libraries were generated by first incubating 10 μl of RNA templates in a reaction mix of 12.5 μl qScript XLT one-step RT-quantitative PCR (RT-qPCR) ToughMix (1× final concentration) (Qiagen, Beverly MA), 200 nM random hexamer primers, 1 μl of 25× qScript XLT one-step reverse transcriptase (RT), and 1.25 μl of nuclease-free water. An RT-negative control was run with the same components, but an additional 1 μl of nuclease-free water was added instead of the RT enzyme. Technical replicate reactions of 25 μl were produced for each sample and RT-negative control and were pooled after reverse transcription and amplification in separate thermocyclers with preheated bonnets. The reaction steps were 20 minutes at 48°C; 3 minutes at 94°C; followed by 30 cycles of 94°C for 45 seconds, 50°C for 45 seconds, and 68°C for 90 seconds; and ending with a final extension of 5 minutes at 68°C and a short 4°C hold. Library amplification and fragment size distribution were confirmed on a 2100 bioanalyzer with the DNA 7500 assay (Agilent Technologies, Santa Clara CA) for all libraries. Libraries were then pooled at an equimolar ratio and sequenced on an Illumina HiSeq 2500 instrument using V2 PE250 Rapid Run chemistry at the Duke Center for Genomic and Computational Biology. Metagenomic sequences are available in the MG-RAST database under accession numbers mgm4795805.3 to mgm4795809.3 and mgm4795811.3.
Raw metagenomic sequence read adapters were removed using PEAT (71), and individual samples were assembled using MEGAHIT (72) with a maximum kmer of 141. This work used the Extreme Science and Engineering Discovery Environment (XSEDE) (73) resource Comet at the San Diego Supercomputer Center through allocation TG-BIO180010. Protein-coding regions were identified and annotated with Prokka v1.12 and Prodigal v.2.6.3 with an E value threshold of 1 × 10−6 (74, 75), and the output translated protein files were also annotated with GHOSTX (76) to search for homologs involved in various methanogenesis and methanotrophic pathways (described in detail below). Protein sequences with homology to those involved in methanogenesis and methanotrophic pathways were subjected to reciprocal BLASTp analysis (77) to check annotation accuracy and homology to known methanogens/methanotrophs. Homology was determined if the query amino acid sequence was >30% identical over >50% of its length, with an E value below 1 × 10−6. Homolog counts were normalized by exon length and sequencing depth to fragments per kilobase of exon per million reads (FPKM) to ensure adequate coverage indicating a noncontaminant.
Metatranscriptomic sequences were trimmed and de novo coassembled into transcripts with Trinity v2.8.6 (78–80). Sequence reads from each sample were aligned to the assembled transcripts with RSEM and counted to generate an expression matrix (81). Coding regions of transcripts were identified with Transdecoder and annotated with Trinotate against the NCBI and Swiss-Prot databases and against the Pfam database with HMMER v3.3 (hmmer.org) using default parameters and an E value threshold of 1 × 10−6 (80, 82–84). Top hits were used as transcript identities. Exploratory analyses of transcript expression used R with the “edgeR” and “limma” packages (66, 85, 86). Transcript counts were normalized by the trimmed mean of M-values method (TMM). Transcripts with less than or equal to 10 counts per million (CPM) were removed, and transcript counts were renormalized by the TMM method for a comparison of counts across samples (87). Metatranscriptomic sequences are available at the Sequence Read Archive (NCBI) database under accession PRJNA560313 (SRR12816294 to SRR12816299).
Metagenome and metatranscriptomes were queried for key genes associated with methanogenesis and methanotrophic pathways, including genes for methyl coenzyme M reductases (mcrABGCD and mcrIIABGCD) (88), the subunits of [NiFe]-hydrogenases (15), formate dehydrogenase (fdhAB) (19), acetate kinase and phosphate acetyltransferase (ackA and pta) for acetoclastic methanogenesis (89), methylotrophic methanogenesis genes (mtaA, mtbB, mtmB, mttB, and mtsA) (9, 90), particulate and soluble methane monooxygenases (pmoABC and mmoXYZ, respectively) (21, 22), and the large subunit of methanol dehydrogenase (mxaF) (23). Transcripts of genes potentially involved in producing alternative inorganic carbon sources for methanogens, including carbonic anhydrases (can, cynT, and cah) (54) and carbon monoxide dehydrogenases (cooS, cdhAB, coxL, and cutL) (24) were also examined.
Data availability.
Unprocessed demultiplexed sequences produced for the SSU rRNA and metatranscriptomic analyses are available at the Sequence Read Archive (NCBI) database under BioProject accession PRJNA560313. SSU rRNA sequences are accessions SRR12495563 to SRR12495576, and metatranscriptomic sequences are accessions SRR12816294 to SRR12816299 in the SRA. Metagenomic sequences are available in the MG-RAST database under accession numbers mgm4795805.3 to mgm4795809.3 and mgm4795811.3 (https://www.mg-rast.org/linkin.cgi?project=mgp85625).
Supplementary Material
ACKNOWLEDGMENTS
We thank the Ministry of Regional Municipalities and Water Resources in the Sultanate of Oman (particularly engineer Said Al-Habsi, Rashid Al-Abri, engineer Salim Al-Khanbashi, Abdullah Al-Kasbi, and Said Al Mangji) and the Oman 2017 Bio Team for insightful discussion, collaboration, and sample collection.
This work was supported by the NASA Astrobiology Institute “Rock-Powered Life” NAI (NNA15BB02A). The funding agency had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We declare that the research herein was conducted in the absence of any commercial or financial relationships that could act as a potential conflict of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Sleep NH, Meibom A, Fridriksson T, Coleman RG, Bird DK. 2004. H2-rich fluids from serpentinization: geochemical and biotic implications. Proc Natl Acad Sci U S A 101:12818–12823. doi: 10.1073/pnas.0405289101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schulte M, Blake D, Hoehler T, McCollom T. 2006. Serpentinization and its implications for life on the early Earth and Mars. Astrobiology 6:364–376. doi: 10.1089/ast.2006.6.364. [DOI] [PubMed] [Google Scholar]
- 3.Seewald JS, Zolotov MY, McCollom T. 2006. Experimental investigation of single carbon compounds under hydrothermal conditions. Geochim Cosmochim Acta 70:446–460. doi: 10.1016/j.gca.2005.09.002. [DOI] [Google Scholar]
- 4.Proskurowski G, Lilley MD, Seewald JS, Früh-Green GL, Olson EJ, Lupton JE, Sylva SP, Kelley DS. 2008. Abiogenic hydrocarbon production at lost city hydrothermal field. Science 319:604–607. doi: 10.1126/science.1151194. [DOI] [PubMed] [Google Scholar]
- 5.Etiope G, Whiticar MJ. 2019. Abiotic methane in continental ultramafic rock systems: towards a genetic model. J Appl Geochem 102:139–152. doi: 10.1016/j.apgeochem.2019.01.012. [DOI] [Google Scholar]
- 6.Rempfert KR, Miller HM, Bompard N, Nothaft D, Matter JM, Kelemen P, Fierer N, Templeton AS. 2017. Geological and geochemical controls on subsurface microbial life in the Samail Ophiolite, Oman. Front Microbiol 8:56. doi: 10.3389/fmicb.2017.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller HM, Matter JM, Kelemen P, Ellison ET, Conrad ME, Fierer N, Ruchala T, Tominaga M, Templeton AS. 2016. Modern water/rock reactions in Oman hyperalkaline peridotite aquifers and implications for microbial habitability. Geochim Cosmochim Acta 179:217–241. doi: 10.1016/j.gca.2016.01.033. [DOI] [Google Scholar]
- 8.Suzuki S, Ishii S, Hoshino T, Rietze A, Tenney A, Morrill PL, Inagaki F, Kuenen JG, Nealson KH. 2017. Unusual metabolic diversity of hyperalkaliphilic microbial communities associated with subterranean serpentinization at the Cedars. ISME J 11:2584–2598. doi: 10.1038/ismej.2017.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crespo-Medina M, Twing KI, Sánchez-Murillo R, Brazelton WJ, McCollom TM, Schrenk MO. 2017. Methane dynamics in a tropical serpentinizing environment: the Santa Elena Ophiolite, Costa Rica. Front Microbiol 8:916. doi: 10.3389/fmicb.2017.00916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brazelton WJ, Nelson B, Schrenk MO. 2012. Metagenomic evidence for H2 oxidation and H2 production by serpentinite-hosted subsurface microbial communities. Front Microbiol 2:268. doi: 10.3389/fmicb.2011.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kohl L, Cumming E, Cox A, Rietze A, Morrissey L, Lang SQ, Richter A, Suzuki S, Nealson KH, Morrill PL. 2016. Exploring the metabolic potential of microbial communities in ultra-basic, reducing springs at the Cedars, CA, USA: experimental evidence of microbial methanogenesis and heterotrophic acetogenesis. J Geophys Res Biogeosci 121:1203–1220. doi: 10.1002/2015JG003233. [DOI] [Google Scholar]
- 12.Canovas PA, Hoehler T, Shock EL. 2017. Geochemical bioenergetics during low-temperature serpentinization: an example from the Samail ophiolite, Sultanate of Oman. J Geophys Res Biogeosci 122:1821–1847. doi: 10.1002/2017JG003825. [DOI] [Google Scholar]
- 13.Mayhew LE, Ellison ET, McCollom TM, Trainor TP, Templeton AS. 2013. Hydrogen generation from low-temperature water-rock reactions. Nature Geosci 6:478–484. doi: 10.1038/ngeo1825. [DOI] [Google Scholar]
- 14.Grozeva NG, Klein F, Seewald JS, Sylva SP. 2020. Chemical and isotopic analyses of hydrocarbon-bearing fluid inclusions in olivine-rich rocks. Philos Trans R Soc A Math Phys Eng Sci 378. doi: 10.1098/rsta.2018.0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thauer RK, Kaster A-K, Goenrich M, Schick M, Hiromoto T, Shima S. 2010. Hydrogenases from methanogenic Archaea, nickel, a novel cofactor, and H2 storage. Annu Rev Biochem 79:507–536. doi: 10.1146/annurev.biochem.030508.152103. [DOI] [PubMed] [Google Scholar]
- 16.Brazelton WJ, Schrenk MO, Kelley DS, Baross JA. 2006. Methane- and sulfur-metabolizing microbial communities dominate the Lost City hydrothermal field ecosystem. Appl Environ Microbiol 72:6257–6270. doi: 10.1128/AEM.00574-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tiago I, Veríssimo A. 2013. Microbial and functional diversity of a subterrestrial high pH groundwater associated to serpentinization. Environ Microbiol 15:1687–1706. doi: 10.1111/1462-2920.12034. [DOI] [PubMed] [Google Scholar]
- 18.Brazelton WJ, Thornton CN, Hyer A, Twing KI, Longino AA, Lang SQ, Lilley MD, Früh-Green GL, Schrenk MO. 2017. Metagenomic identification of active methanogens and methanotrophs in serpentinite springs of the Voltri Massif, Italy. PeerJ 5:e2945. doi: 10.7717/peerj.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shuber AP, Orr EC, Recny MA, Schendel PF, May HD, Schauer NL, Ferry JG. 1986. Cloning, expression, and nucleotide sequence of the formate dehydrogenase genes from Methanobacterium formicicum. J Biol Chem 261:12942–12947. [PubMed] [Google Scholar]
- 20.Cui M, Ma A, Qi H, Zhuang X, Zhuang G. 2015. Anaerobic oxidation of methane: an “active” microbial process. Microbiologyopen 4:1–11. doi: 10.1002/mbo3.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stainthorpe AC, Lees V, Salmond GPC, Dalton H, Murrell JC. 1990. The methane monooxygenase gene cluster of Methylococcus capsulatus (Bath). Gene 91:27–34. doi: 10.1016/0378-1119(90)90158-N. [DOI] [PubMed] [Google Scholar]
- 22.Csáki R, Bodrossy L, Klem J, Murrell JC, Kovács KL. 2003. Genes involved in the copper-dependent regulation of soluble methane monooxygenase of Methylococcus capsulatus (Bath): cloning, sequencing and mutational analysis. Microbiology 149:1785–1795. doi: 10.1099/mic.0.26061-0. [DOI] [PubMed] [Google Scholar]
- 23.Lau E, Fisher MC, Steudler PA, Cavanaugh CM. 2013. The methanol dehydrogenase gene, mxaF, as a functional and phylogenetic marker for proteobacterial methanotrophs in natural environments. PLoS One 8:e56993. doi: 10.1371/journal.pone.0056993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fones EM, Colman DR, Kraus EA, Nothaft DB, Poudel S, Rempfert KR, Spear JR, Templeton AS, Boyd ES. 2019. Physiological adaptations to serpentinization in the Samail Ophiolite, Oman. ISME J 13:1750–1762. doi: 10.1038/s41396-019-0391-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sánchez-Murillo R, Gazel E, Schwarzenbach EM, Crespo-Medina M, Schrenk MO, Boll J, Gill BC. 2014. Geochemical evidence for active tropical serpentinization in the Santa Elena Ophiolite, Costa Rica: an analog of a humid early Earth? Geochem Geophys Geosyst 15:1783–1800. doi: 10.1002/2013GC005213. [DOI] [Google Scholar]
- 26.Zwicker J, Birgel D, Bach W, Richoz S, Smrzka D, Grasemann B, Gier S, Schleper C, Rittmann SKMR, Koşun E, Peckmann J. 2018. Evidence for archaeal methanogenesis within veins at the onshore serpentinite-hosted Chimaera seeps, Turkey. Chem Geol 483:567–580. doi: 10.1016/j.chemgeo.2018.03.027. [DOI] [Google Scholar]
- 27.Miller HM, Chaudhry N, Conrad ME, Bill M, Kopf SH, Templeton AS. 2018. Large carbon isotope variability during methanogenesis under alkaline conditions. Geochim Cosmochim Acta 237:18–31. doi: 10.1016/j.gca.2018.06.007. [DOI] [Google Scholar]
- 28.Twing KI, Brazelton WJ, Kubo MDY, Hyer AJ, Cardace D, Hoehler TM, McCollom TM, Schrenk MO. 2017. Serpentinization-influenced groundwater harbors extremely low diversity microbial communities adapted to high pH. Front Microbiol 8:308. doi: 10.3389/fmicb.2017.00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morrill PL, Brazelton WJ, Kohl L, Rietze A, Miles SM, Kavanagh H, Schrenk MO, Ziegler SE, Lang SQ. 2014. Investigations of potential microbial methanogenic and carbon monoxide utilization pathways in ultra-basic reducing springs associated with present-day continental serpentinization: the Tablelands, NL, CAN. Front Microbiol 5:613. doi: 10.3389/fmicb.2014.00613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicolas A, Bouldier F, Ildefonse B, Ball E. 2000. Accretion of Oman and United Arab Emirates ophiolite—discussion of a new structural map. Mar Geophys Res 21:147–180. doi: 10.1023/A:1026769727917. [DOI] [Google Scholar]
- 31.Kelemen PB, Matter J. 2008. In situ carbonation of peridotite for CO2 storage. Proc Natl Acad Sci U S A 105:17295–17300. doi: 10.1073/pnas.0805794105. [DOI] [Google Scholar]
- 32.Neal C, Stanger G. 1983. Hydrogen generation from mantle source rocks in Oman. Earth Planet Sci Lett 66:315–320. doi: 10.1016/0012-821X(83)90144-9. [DOI] [Google Scholar]
- 33.Paukert AN, Matter JM, Kelemen PB, Shock EL, Havig JR. 2012. Reaction path modeling of enhanced in situ CO2 mineralization for carbon sequestration in the peridotite of the Samail Ophiolite, Sultanate of Oman. Chem Geol 330–331:86–100. doi: 10.1016/j.chemgeo.2012.08.013. [DOI] [Google Scholar]
- 34.Kelemen PB, Matter J, Streit EE, Rudge JF, Curry WB, Blusztajn J. 2011. Rates and mechanisms of mineral carbonation in peridotite: natural processes and recipes for enhanced, in situ CO2 capture and storage. Annu Rev Earth Planet Sci 39:545–576. doi: 10.1146/annurev-earth-092010-152509. [DOI] [Google Scholar]
- 35.Etiope G. 2017. Methane origin in the Samail ophiolite: comment on “Modern water/rock reactions in Oman hyperalkaline peridotite aquifers and implications for microbial habitability.” Geochim Cosmochim Acta 197:467–470. doi: 10.1016/j.gca.2016.08.001. [DOI] [Google Scholar]
- 36.Neal C, Stanger G. 1985. Past and present serpentinisation of ultramafic rocks; an example from the Semail ophiolite nappe of northern Oman, p 249–275. In The chemistry of weathering. Springer Netherlands, Dordrecht, Netherlands. [Google Scholar]
- 37.Drever JI. 1985. The chemistry of weathering. Springer Netherlands, Dordrecht, Netherlands. [Google Scholar]
- 38.Chavagnac V, Monnin C, Ceuleneer G, Boulart C, Hoareau G. 2013. Characterization of hyperalkaline fluids produced by low-temperature serpentinization of mantle peridotites in the Oman and Ligurian ophiolites. Geochem Geophys Geosyst 14:2496–2522. doi: 10.1002/ggge.20147. [DOI] [Google Scholar]
- 39.Barnes I, O'Neil JR. 1969. The relationship between fluids in some fresh alpine-type ultramafics and possible modern serpentinization, western United States. Geol Soc America Bull 80:1947–1960. doi: 10.1130/0016-7606(1969)80[1947:TRBFIS]2.0.CO;2. [DOI] [Google Scholar]
- 40.Boone DR. 2015. Methanobacterium In Bergey’s Manual of Systematics of Archaea and Bacteria. John Wiley & Sons, Ltd, Chichester, UK. doi: 10.1002/9781118960608.gbm00495. [DOI] [Google Scholar]
- 41.Bowman JP. 2015. Methylococcus In Bergey’s Manual of Systematics of Archaea and Bacteria. John Wiley & Sons, Ltd, Chichester, UK: https://onlinelibrary.wiley.com/doi/abs/10.1002/9781118960608.gbm01181. [Google Scholar]
- 42.Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MMM, Schreiber F, Dutilh BE, Zedelius J, de Beer D, Gloerich J, Wessels HJCT, van Alen T, Luesken F, Wu ML, van de Pas-Schoonen KT, Op den Camp HJM, Janssen-Megens EM, Francoijs K-J, Stunnenberg H, Weissenbach J, Jetten MSM, Strous M. 2010. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464:543–548. doi: 10.1038/nature08883. [DOI] [PubMed] [Google Scholar]
- 43.Peters JW, Schut GJ, Boyd ES, Mulder DW, Shepard EM, Broderick JB, King PW, Adams MWW. 2015. [FeFe]- and [NiFe]-hydrogenase diversity, mechanism, and maturation. Biochim Biophys Acta 1853:1350–1369. doi: 10.1016/j.bbamcr.2014.11.021. [DOI] [PubMed] [Google Scholar]
- 44.Munk B, Bauer C, Gronauer A, Lebuhn M. 2012. A metabolic quotient for methanogenic Archaea. Water Sci Technol 66:2311–2317. doi: 10.2166/wst.2012.436. [DOI] [PubMed] [Google Scholar]
- 45.Morgan RM, Pihl TD, Nölling J, Reeve JN. 1997. Hydrogen regulation of growth, growth yields, and methane gene transcription in Methanobacterium thermoautotrophicum δH. J Bacteriol 179:889–898. doi: 10.1128/jb.179.3.889-898.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Freitag TE, Prosser JI. 2009. Correlation of methane production and functional gene transcriptional activity in a peat soil. Appl Environ Microbiol 75:6679–6687. doi: 10.1128/AEM.01021-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taubner RS, Schleper C, Firneis MG, Rittmann SKMR. 2015. Assessing the ecophysiology of methanogens in the context of recent astrobiological and planetological studies. Life 5:1652–1686. doi: 10.3390/life5041652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woycheese KM, Meyer-Dombard DR, Cardace D, Argayosa AM, Arcilla CA. 2015. Out of the dark: transitional subsurface-to-surface microbial diversity in a terrestrial serpentinizing seep (Manleluag, Pangasinan, the Philippines). Front Microbiol 6:44. doi: 10.3389/fmicb.2015.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blank JG, Green SJ, Blake D, Valley JW, Kita NT, Treiman A, Dobson PF. 2009. An alkaline spring system within the Del Puerto Ophiolite (California, USA): a mars analog site. Planet Space Sci 57:533–540. doi: 10.1016/j.pss.2008.11.018. [DOI] [Google Scholar]
- 50.Moser DP, Gihring TM, Brockman FJ, Fredrickson JK, Balkwill DL, Dollhopf ME, Lollar BS, Pratt LM, Boice E, Southam G, Wanger G, Baker BJ, Pfiffner SM, Lin LH, Onstott TC. 2005. Desulfotomaculum and Methanobacterium spp. dominate a 4- to 5-kilometer-deep fault. Appl Environ Microbiol 71:8773–8783. doi: 10.1128/AEM.71.12.8773-8783.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boone DR, Castenholz RW. 2001. The Archaea and the deeply branching phototrophic bacteria, 2nd ed Springer-Verlag, New York, NY. [Google Scholar]
- 52.Subhas AV, Adkins JF, Rollins NE, Naviaux J, Erez J, Berelson WM. 2017. Catalysis and chemical mechanisms of calcite dissolution in seawater. Proc Natl Acad Sci U S A 114:8175–8180. doi: 10.1073/pnas.1703604114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Capasso C, Supuran CT. 2015. An overview of the alpha-, beta- and gamma-carbonic anhydrases from Bacteria: can bacterial carbonic anhydrases shed new light on evolution of bacteria? J Enzyme Inhib Med Chem 30:325–332. doi: 10.3109/14756366.2014.910202. [DOI] [PubMed] [Google Scholar]
- 54.Smith KS, Jakubzick C, Whittam TS, Ferry JG. 1999. Carbonic anhydrase is an ancient enzyme widespread in prokaryotes. Proc Natl Acad Sci U S A 96:15184–15189. doi: 10.1073/pnas.96.26.15184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith KS, Ferry JG. 1999. A plant-type (β-class) carbonic anhydrase in the thermophilic methanoarchaeon Methanobacterium thermoautotrophicum. J Bacteriol 181:6247–6253. doi: 10.1128/JB.181.20.6247-6253.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suzuki S, Kuenen JG, Schipper K, Van Der Velde S, Ishii S, Wu A, Sorokin DY, Tenney A, Meng X, Morrill PL, Kamagata Y, Muyzer G, Nealson KH. 2014. Physiological and genomic features of highly alkaliphilic hydrogen-utilizing Betaproteobacteria from a continental serpentinizing site. Nat Commun 5:3900. doi: 10.1038/ncomms4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Etiope G, Ehlmann BL, Schoell M. 2013. Low temperature production and exhalation of methane from serpentinized rocks on Earth: a potential analog for methane production on Mars. Icarus 224:276–285. doi: 10.1016/j.icarus.2012.05.009. [DOI] [Google Scholar]
- 58.Milkov AV, Etiope G. 2018. Revised genetic diagrams for natural gases based on a global dataset of >20,000 samples. Org Geochem 125:109–120. doi: 10.1016/j.orggeochem.2018.09.002. [DOI] [Google Scholar]
- 59.Whiticar MJ. 1999. Carbon and hydrogen isotope systematics of bacterial formation and oxidation of methane. Chem Geol 161:291–314. doi: 10.1016/S0009-2541(99)00092-3. [DOI] [Google Scholar]
- 60.Boyd ES, Amenabar MJ, Poudel S, Templeton AS. 2020. Bioenergetic constraints on the origin of autotrophic metabolism. Philos Trans A Math Phys Eng Sci 378:20190151. doi: 10.1098/rsta.2019.0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thauer RK, Kaster AK, Seedorf H, Buckel W, Hedderich R. 2008. Methanogenic archaea: ecologically relevant differences in energy conservation. Nat Rev Microbiol 6:579–591. doi: 10.1038/nrmicro1931. [DOI] [PubMed] [Google Scholar]
- 62.Kampbell DH, Wilson JT, McInnes DM. 1998. Determining dissolved hydrogen, methane, and vinyl chloride concentrations in aqueous solution on a nanomolar scale with the bubble strip method, p 176–190. In Proceedings of the 1998 Conference on Hazardous Waste Research, Snowbird, UT, 18 to 21 May 1998. [Google Scholar]
- 63.Parada AE, Needham DM, Fuhrman JA. 2016. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol 18:1403–1414. doi: 10.1111/1462-2920.13023. [DOI] [PubMed] [Google Scholar]
- 64.Kraus EA, Beeler SR, Mors RA, Floyd JG, Stamps BW, Nunn HS, Stevenson BS, Johnson HA, Shapiro RS, Loyd SJ, Spear JR, Corsetti FA. 2018. Microscale biosignatures and abiotic mineral authigenesis in Little Hot Creek, California. Front Microbiol 9:997. doi: 10.3389/fmicb.2018.00997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 66.R Core Team. 2013. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 67.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yilmaz P, Parfrey LW, Yarza P, Gerken J, Pruesse E, Quast C, Schweer T, Peplies J, Ludwig W, Glöckner FO. 2014. The SILVA and “all-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res 42:D643–D648. doi: 10.1093/nar/gkt1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McMurdie PJ, Holmes S. 2013. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wickham H. 2016. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York, NY. [Google Scholar]
- 71.Li YL, Weng JC, Hsiao CC, Chou M, Te Tseng CW, Hung JH. 2015. PEAT: an intelligent and efficient paired-end sequencing adapter trimming algorithm. BMC Bioinformatics 16:S2. doi: 10.1186/1471-2105-16-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li D, Liu CM, Luo R, Sadakane K, Lam TW. 2015. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 73.Towns J, Cockerill T, Dahan M, Foster I, Gaither K, Grimshaw A, Hazlewood V, Lathrop S, Lifka D, Peterson GD, Roskies R, Scott JR, Wilkins-Diehr N. 2014. XSEDE: accelerating scientific discovery. Comput Sci Eng 16:62–74. doi: 10.1109/MCSE.2014.80. [DOI] [Google Scholar]
- 74.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 75.Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Suzuki S, Kakuta M, Ishida T, Akiyama Y. 2014. GHOSTX: an improved sequence homology search algorithm using a query suffix array and a database suffix array. PLoS One 9:e103833. doi: 10.1371/journal.pone.0103833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, Di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, Macmanes MD, Ott M, Orvis J, Pochet N, Strozzi F, Weeks N, Westerman R, William T, Dewey CN, Henschel R, Leduc RD, Friedman N, Regev A. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bryant DM, Johnson K, DiTommaso T, Tickle T, Couger MB, Payzin-Dogru D, Lee TJ, Leigh ND, Kuo TH, Davis FG, Bateman J, Bryant S, Guzikowski AR, Tsai SL, Coyne S, Ye WW, Freeman RM, Peshkin L, Tabin CJ, Regev A, Haas BJ, Whited JL. 2017. A tissue-mapped axolotl de novo transcriptome enables identification of limb regeneration factors. Cell Rep 18:762–776. doi: 10.1016/j.celrep.2016.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.The UniProt Consortium. 2019. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res 47:D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.NCBI Resource Coordinators. 2018. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 46:D8–D13. doi: 10.1093/nar/gkx1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Robinson M, McCarthy D, Smyth G. 2010. edgeR: a Bioconductor pacakge for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ritchie M, Phipson B, Wu D, Hu Y, Law C, Shi W, Smyth G. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Robinson MD, Oshlack A. 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Reeve JN, Nölling J, Morgan RM, Smith DR. 1997. Methanogenesis: genes, genomes, and who’s on first? J Bacteriol 179:5975–5986. doi: 10.1128/JB.179.19.5975-5986.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fournier GP, Gogarten JP. 2008. Evolution of acetoclastic methanogenesis in Methanosarcina via horizontal gene transfer from cellulolytic Clostridia. J Bacteriol 190:1124–1127. doi: 10.1128/JB.01382-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Paul L, Ferguson DJ, Krzycki JA. 2000. The trimethylamine methyltransferase gene and multiple dimethylamine methyltransferase genes of Methanosarcina barkeri contain in-frame and read- through amber codons. J Bacteriol 182:2520–2529. doi: 10.1128/jb.182.9.2520-2529.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nothaft DB, Templeton AS, Rhim JH, Wang DT, Labidi J, Miller HM, Boyd ES, Matter JM, Ono S, Young ED, Kopf SH, Kelemen PB, Conrad ME. 2020. Geochemical, biological and clumped isotopologue evidence for substantial microbial methane production under carbon limitation in serpentinites of the Samail Ophiolite, Oman. ESSoAr. doi: 10.1002/ESSOAR.10504124.1. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Unprocessed demultiplexed sequences produced for the SSU rRNA and metatranscriptomic analyses are available at the Sequence Read Archive (NCBI) database under BioProject accession PRJNA560313. SSU rRNA sequences are accessions SRR12495563 to SRR12495576, and metatranscriptomic sequences are accessions SRR12816294 to SRR12816299 in the SRA. Metagenomic sequences are available in the MG-RAST database under accession numbers mgm4795805.3 to mgm4795809.3 and mgm4795811.3 (https://www.mg-rast.org/linkin.cgi?project=mgp85625).